Abstract
Expression of the trypanosomal mitochondrial genome requires the insertion and deletion of uridylyl residues at specific sites in pre-mRNAs. RET2 terminal uridylyl transferase (TUTase) is an integral component of the RNA editing core complex (RECC) and is responsible for the guide RNA-dependent U-insertion reaction. By analyzing RNAi-based knock-in Trypanosoma brucei cell lines, purified editing complex and individual protein, we have investigated RET2’s association with the RECC. In addition, the U-insertion activity exhibited by RET2 as RECC subunit was compared with characteristics of the monomeric protein. We show that RET2 interaction with RECC is accomplished via a protein-protein contact between its middle domain and a structural subunit MP81. The recombinant RET2 catalyzes a faithful editing on gapped (pre-cleaved) double-stranded RNA substrates and this reaction requires an internal monophosphate group at the 5′-end of the mRNA 3′-cleavage fragment. However, RET2 processivity is limited to insertion of three Us. Incorporation into the RECC voids the internal phosphate requirement and allows filling of longer gaps similar to those observed in vivo. Remarkably, monomeric and RECC-embedded enzymes display a similar bimodal activity: the distributive insertion of a single uracil is followed with a processive extension limited by the number of guiding nucleotides. Based on the RNA substrate specificity of RET2 and purine-rich nature of U-insertion sites, we propose that the distributive +1 insertion creates a substrate for the processive gap-filling reaction. Upon base-pairing of the +1-extended 5′-cleavage fragment with a guiding nucleotide, this substrate is recognized by RET2 in a different mode as compared to the product of the initial nucleolytic cleavage. Therefore, RET2 distinguishes base-pairs in gapped RNA substrates which may constitute an additional checkpoint contributing to overall fidelity of the editing process.
Keywords: Trypanosoma, mitochondria, RNA editing, TUTase, RNAi
Introduction
The mitochondrial genome of Kinetoplastids (kinetoplast, kDNA) represents one of the most complex DNA structures found in nature. The kinetoplast is a dense catenated network of ~50 maxicircles and ~10,000 minicircles. Maxicircles, ranging from 20 to 40 kb, encode conventional mitochondrial genes such as rRNAs and subunits of respiratory complexes.1 Twelve of the eighteen protein-coding transcripts require post-transcriptional uridine insertion/deletion editing to generate translation-competent mRNAs.2-4 These changes correct frame-shifts, create start and stop codons, and, for some transcripts, generate large portions of their open reading frames.5-7 Editing is directed by short (~60 nucleotides, nt) trans-acting guide RNAs (gRNAs) encoded primarily in the minicircles8 or, in the case of subunit 2 of cytochrome oxidase, by a cis-interacting element located in the 3′-untranslated region (UTR).9
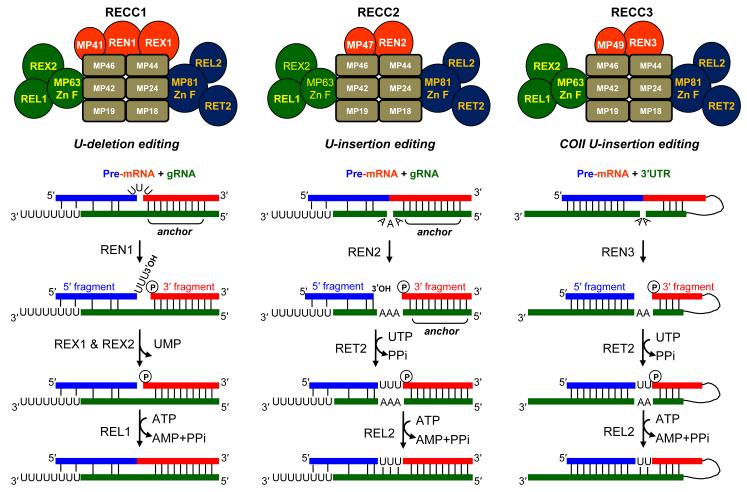
The endonucleolytic, exonucleolytic, nucleoside transfer, and ligation activities are catalyzed by the ~1.2 MDa RNA editing core complex (RECC), also referred to as the 20S editosome or L-complex, which consists of ~20 polypeptides (reviewed in 10-12). The nomenclature of editing complexes and proteins proposed in Simpson et al.13 has been adopted throughout this paper. In Trypanosoma brucei, three forms of RECC are distinguished by association with distinct RNase III-type endonucleases, REN1, REN2 and REN3, which are proposed to cleave at U-deletion,14 U-insertion,15 and cis-guided editing16 sites, respectively (Fig. 1). Additional compositional differences among the RECC’s include structural proteins (MP41, MP47,and MP49) and the exclusive presence of REX1 in the U-deletion RECC1.16, 17 Although only RECC1 is presumed to be active in the U-deletion editing, each RECC variant contains the components of both U-insertion and U-deletion pathways. These enzymatic cascades are mediated by spatially separate trimeric subcomplexes consisting of 1) a TUTase or 3′-5′ U-specific exonuclease, 2) a C2H2-zinc finger scaffolding protein, and 3) an RNA ligase.18 Editing is initiated by mRNA cleavage immediately upstream of the “anchor” duplex formed between gRNA’s 5′-region and the pre-edited mRNA. Resultant 5′- and 3′- mRNA cleavage fragments are thought to be bridged by gRNA. For U-deletion, unpaired Us are removed by either REX1 or REX2 exonucleases.19, 20 The post-cleavage U-insertion site represents a gapped double stranded RNA (dsRNA) in which the 5′-mRNA cleavage fragment terminates with a hydroxyl group while the 3′ cleavage fragment is 5′-phosphorylated (Fig. 1). Uracils are inserted by RNA Editing Terminal Uridylyl Transferase 2 (RET2) according to the number of guiding purine nucleotides. Finally, the cleavage fragments are re-ligated by editing ligases REL1 and REL2 producing an mRNA that is complementary to the gRNA.
Fig. 1.
Schematic representation of the U-deletion (left), U-insertion (middle), and cis-editing (right) enzymatic cascades catalyzed by RECC1, RECC2 and RECC3, respectively. RECC – RNA editing core complex. U-insertion and U-deletion subcomplexes are depicted in blue and green, respectively. MP: mitochondrial protein; REX: RNA editing exonuclease; REN: RNA editing endonuclease; REL: RNA editing ligase; RET: RNA editing TUTase; gRNA: guide RNA; UTR: untranslated region; anchor: 5-15-nt long double-stranded region formed by the 5′-portion of the gRNA and pre-edited mRNA. Initial endonucleolytic mRNA cleavage occurs at the first unpaired nucleotide upstream of the anchor.
In addition to RET2, two other mitochondrial TUTases are known in trypanosomes: RET1, which interacts with multiple complexes (reviewed in 11) to uridylylate guide RNAs,21 ribosomal RNAs22, 23 and mRNAs,23-25 and MEAT1, which interacts with an RECC-like complex, but also exists as an unassociated protein.26 In contrast, RET2 is maintained only as a subunit of the U-insertion subcomplex and is the sole nucleotidyl transferase of the RECC (Fig. 1). RET2 binds to the core complex via direct contact with MP81 zinc finger protein27, 28 and its RNAi knockdown in insect form parasites29 or gene knockout in bloodstream form parasites30 results in a severe growth inhibition. Loss of RET2 abolishes U-insertion activity and decreases REL1 and MP81 protein levels but has no effect on the U-deletion activity or the overall RECC integrity.29
U-insertion editing is a unique method of transferring genetic information which does not rely on template-dependent nucleic acid polymerization, often allowing guanosine residue-guided U-insertion.28, 29 The uridine specificity is determined by the RET2’s intrinsic selectivity for UTP30 and not by the nature of the guiding nucleotides. Recombinant RET2 is exclusively UTP-specific and predominantly adds one uracil to a single-stranded RNA (ssRNA) terminating with adenine or guanine at the terminal base, but is virtually inactive on ssRNAs with Us at the 3′ end.28, 31 However, faithful insertion editing of a synthetic pre-cleaved editing substrate 32 has been shown using editing complexes purified to various degrees32-34 and recombinant RET230 (reviewed in 11, 35). Therefore, specificity for a gapped dsRNA appears to be a key factor contributing to the overall fidelity of U-insertion editing.
To understand the contributions of RET2’s intrinsic properties and those conferred by its association with RECC, we investigated structural elements responsible for complex recruitment and enzymatic properties exhibited by RET2 as a RECC subunit and as an individual protein. By creating RNAi-based knock-in (iCODA26) cell lines of insect (procyclic) Trypanosoma brucei and analyzing the activities of the purified RECC and the recombinant RET2 (rRET2) on various RNA substrates, we show that RET2 interaction with RECC is accomplished via a protein-protein contact between its middle domain and MP81 protein. Reaction catalyzed by rRET2 on gapped (pre-cleaved) double-stranded RNA substrates requires an internal monophosphate and is limited to insertion of three Us. Purified RECC does not require an internal phosphate and is capable of filling gaps similar to the longest ones observed in vivo. Despite moderate differences in catalytic efficiency, individual and RECC-embedded enzymes display a similar bimodal activity: the distributive insertion of a single uracil is followed by a processive gap-filling reaction. We propose that the +1 insertion, which creates a substrate for the processive reaction, is likely followed by dissociation of RECC from RNA substrate. Upon base-pairing of the +1U-extended 5′-cleavage fragment with a guiding nucleotide, this substrate is recognized by RET2 in a different mode as compared to the product of the initial nucleolytic cleavage. Therefore, prior to processive U-insertion, RET2 distinguishes base-pairs in gapped RNA substrates which may be an additional checkpoint contributing to overall fidelity of the editing process. It is possible that the lack of +1U base-pairing would create a substrate for the 3′-5′ U-specific exonuclease REX2 acting as a proofreading enzyme within the U-insertion site. This hypothesis rationalizes an evolutionary pressure to maintain U-deletion activity within RECC2 and RECC3, which are dedicated to U-insertion sites (Fig. 1).
Results
Functional RNAi complementation and isolation of the active RET2 complex
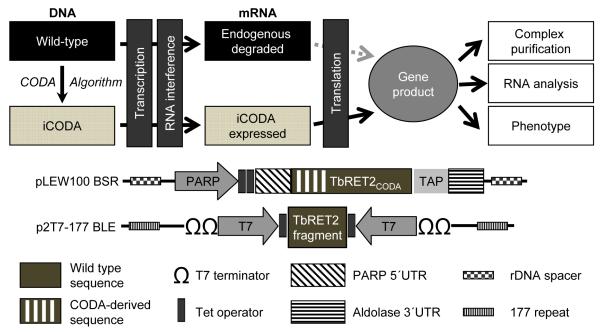
To determine the protein module responsible for docking into the RECC and to assess the effects of RECC-incorporation on RET2’s catalytic parameters, we have further developed the iCODA (RNAi resistant genes via computationally-optimized DNA assembly) technology26 to enable affinity purification of wild-type and mutated proteins from a genetic background lacking the endogenous RET2 (Fig. 2). This methodology was previously employed to address potential RNAi off-targeting in knockdown studies of the mitochondrial editosome-like complex associated TUTase 1 (MEAT1)26. As diagrammed in Fig. 2, RET2 knockdown was performed by cloning a fragment corresponding to positions 41-545 of the RET2 gene between opposing T7 RNA polymerase promoters and tet operators within the p2T7-177 vector.36 The RET2 knock-in was accomplished by inducible co-expression of the RNAi-resistant gene which contained at least one silent mutation per 12 base-pairs within the RNAi-targeted region. A C-terminal TAP tag37 was also incorporated to allow affinity purification of RET2-iCODA protein from cells depleted of the endogenous protein.
Fig. 2.
Schematic representation of iCODA, an RNAi-based inducible knock-in strategy in procyclic form of T. brucei. Silent mutations (at least 1 per 12 base-pairs) were introduced into the RET2 gene region targeted by RNAi to minimize potential effects on translation49 and to prevent transcript targeting by the RNAi machinery. The co-expression of both RET2-iCODA protein and RET2 RNAi cassette is controlled by tet operators positioned downstream of a procyclic acidic repetitive protein promoter (PARP), which is recognized by RNA polymerase I, and T7 RNA polymerase promoter, respectively. Co-expression is performed in T. brucei strain 29-13 which constitutively expresses T7 RNA polymerase and tet repressor.40 BSR: blasticidin resistance gene; BLE: phleomycin resistance gene; 177 repeat: transcriptionally-silent 177-bp satellite repeat sequence;50 rDNA spacer: transcriptionally-silent spacers between ribosomal RNA genes.
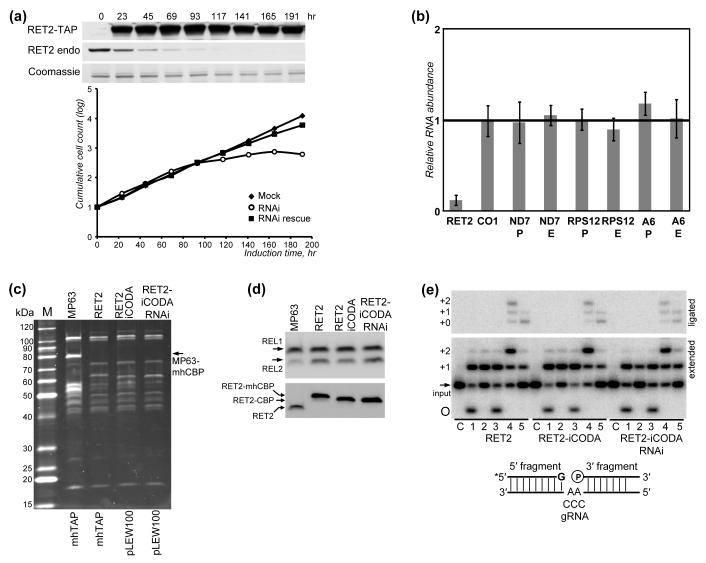
While RET2 RNAi knockdown caused growth inhibition after ~80 hrs of induction, no significant changes in division time were observed for cells co-expressing the RNAi cassette and RET2-iCODA protein. Western blotting analysis demonstrated a depletion of the endogenous RET2 by RNAi and inducible expression of RET2-iCODA, which indicates a functional RNAi rescue (Fig. 3a). To verify unaltered levels of mitochondrial RNAs in RET2-iCODA/RNAi cells, quantitative RT-PCR (qRT-PCR) was used to measure the relative abundances of select never-edited, three pre-edited, and corresponding edited mRNAs (Fig. 3b). While the endogenous RET2 mRNA was degraded by more than 90%, all mitochondrial mRNAs tested remained virtually unaffected, confirming intact U-insertion editing activity in RET2 iCODA/RNAi cells.
Fig. 3.
Functional complementation of RET2 RNAi knockdown by co-expression of the RNAi-resistant transcript. (a) Growth kinetics of RET2-RNAi and RET2-iCODA/RNAi cell lines. Mock: uninduced RNAi cell; RNAi: tet-induced RNAi cells; RNAi rescue: tet-induced RET2-iCODA RNAi cells. Immunoblotting of endogenous and iCODA-derived RET2 in RNAi rescue cells is shown above the graph panel. (b) Quantitative RT-PCR analysis of mitochondrial mRNAs and endogenous RNAi-targeted RET2 transcript in RET2-iCODA/RNAi cells. RNA levels were normalized to α-tubulin mRNA. P: pre-edited mRNA; E: edited mRNA. Error bars: standard deviation of three replicates. Thick line at 1: no change in mRNA relative abundance; bars above and below represent an increase or decrease, respectively. (c) TAP-purified RNA editing core complexes were separated on an 8-16% gradient SDS-PAGE gel and stained with Sypro Ruby. Cell lines and genetic constructs used for expression of TAP-tagged proteins are listed above and below the gel, respectively. The pLEW79-based mhTAP vector51 was used for overexpression of MP63 structural protein and WT RET2. The pLEW100-BSR-based TAP vector52 was used for expression iCODA-RET2. mhCBP: 6His tag plus calmodulin binding peptide which remain on the tagged protein upon TEV protease cleavage. (d) TAP-purified complexes from (c) were analyzed by self-adenylation of RNA editing ligases in the presence of [α-32P]ATP (top panel) and by Western blotting with anti-RET2 antibodies (bottom panel). (e) U-insertion editing activity of TAP-purified complexes on pre-cleaved editing substrates (diagrammed). Asterisk: radiolabeled 5′-fragment. Top panel: ligated products; bottom panel: products of U-addition to the 5′-fragment. C: control, input RNA; 1: 5′-fragment; 2: 5′- fragment +gRNA; 3: 5′-fragment +3′-fragment; 4: full assembly (5′-fragment + gRNA + 3′-fragment) with AA as guiding nucleotides; 5: full assembly with CCC as guiding nucleotides; circle: circularized 5′-fragment.
The presence of both endogenous and TAP-tagged RNA editing ligases in RECC purified from L. tarentolae29 and T. brucei18 suggested that at least some components may be present in more than one copy per complex. To confirm RET2-iCODA incorporation into RECC and to assess its stoichiometry within RECC, we have purified complexes from cells overexpressing TAP-tagged structural subunit MP63, wild type (WT) RET2 and RET2-iCODA, and cells co-expressing RET2-iCODA and RET2 RNAi. The resultant protein profiles (Fig. 3c) and self-adenylation signals of RNA editing ligases (Fig. 3d, upper panel) were nearly identical among all three RET2 preparations, demonstrating equal complex integration of RET2-iCODA. Immunoblotting also showed similar levels of tagged RET2 polypeptides, which are slightly larger because of residual tags as compared to the endogenous RET2 in MP63-purified complex (Fig 3d, bottom panel). Most importantly, the lack of the endogenous RET2 in complexes purified from either high- or moderate-level expression systems (mhTAP and pLEW100, respectively) demonstrated that there is a single RET2 molecule per RECC.
Finally, to compare the in vitro U-insertion activities of all three RET2 complexes, affinity-purified fractions were tested in a pre-cleaved RNA editing assay.32 Synthetic RNAs (5′-fragment, 3′-fragment and gRNA) were annealed to form a gapped double-stranded RNA mimicking an editing substrate in which the mRNA-gRNA hybrid had already undergone endonucleolytic cleavage. No apparent differences in U-addition to ssRNA substrate and U-insertion into fully-assembled pre-cleaved editing substrates programmed for +2U insertion were found among respective complexes (Fig. 3e). To conclude, complex purified from RET2-iCODA/RNAi cells has similar activity with those purified from RET2-overexpressing cell lines.
Middle domain is required for RET2 incorporation into the RECC
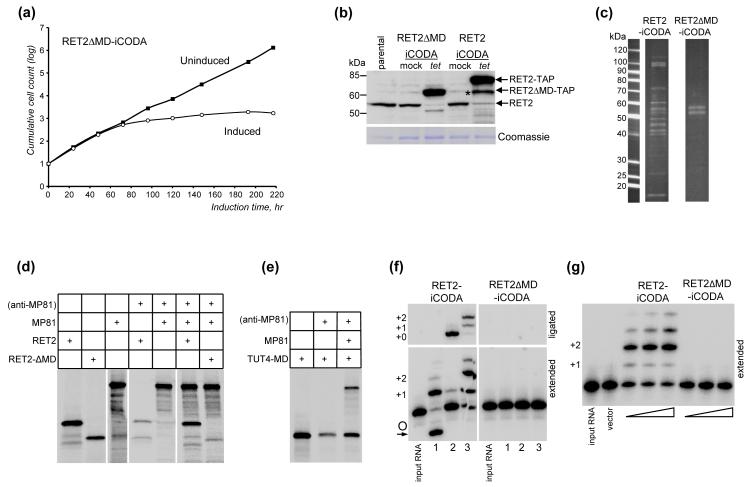
Superpositioning of RET230 and the smallest known TUTase, TUT4,38 crystal structures revealed a compact middle domain (MD, ~110 amino-acids) inserted within the RET2’s N-terminal catalytic domain (NTD). The site of MD insertion is highly-conserved among trypanosomal uridylyl transferases and non-canonical poly(A) polymerases, but the primary structures of middle domains are highly divergent.11 The deleterious effect of MD deletion on ssRNA-specific RET1 TUTase activity suggested that middle domains may be functionally important.39 Indeed, the presence of a divergent module within an otherwise conserved NTD suggests that the MD may act as a function-specific adaptor for the catalytic “bi-domain” formed by NTD and CTD.30 We hypothesized that the middle domain is required for binding to the zinc-finger containing structural subunit MP8118, 28 and, therefore, RET2 docking into U-insertion sub-complex. To investigate the role of MD in RET2-MP81 binding in vivo, we have replaced residues 151-264 with a Gly-Ser-Gly-Ser linker to accommodate a ~2.5Å distance between the points of exit and return within the NTD. Co-expression of RET2ΔMD-iCODA with RET2 RNAi induced a severe growth inhibition similar to that of RET2 RNAi, demonstrating the deletion mutant was unable to compensate for knockdown of the endogenous protein (Fig. 4a, b).
Fig. 4.
Deletion of the middle domain disrupts RET2 incorporation into RECC and RET2-mediated U-insertion. (a) Growth kinetics of RET2ΔMD-iCODA/RNAi cells. (b) Western blotting analysis of RET2 knockdown and RET2ΔMD-TAP expression after 70 hr of induction. RET2-iCODA/RNAi cells were analyzed alongside to demonstrate similar expression levels of RET2ΔMD and full-length RET2. The proteolysis product sporadically appearing in RET2-iCODA/RNAi cells is indicated by asterisk. (c) TAP-purified complexes from RET2-iCODA/RNAi and RET2ΔMD-iCODA/RNAi cells were separated on an 8-16% gradient SDS-PAGE gel and stained with Sypro Ruby. (d) Co-immunoprecipitation of RET2 and RET2ΔMD with MP81 protein. All polypeptides were synthesized in the reticulocyte transcription-translation system in the presence of [35S] methionine. (e) Co-immunoprecipitation of in vitro-synthesized [35S]-labeled MP81 and TbTUT4 TUTase with grafted RET2’s middle domain. (f) U-insertion and RNA ligase activities of TAP-purified fractions shown in (c). Protein amounts were normalized by Sypro stained gel bands. Assay was carried out with 2 μl of purified fraction, 50 nM RNA, 100 μM UTP for 1 hr followed by 30 minute incubation in the presence of 100 μM ATP. 1: 5′-fragment; 2: +0 insertion substrate; 3: +2 insertion substrate. (g) U-insertion activity of in vitro synthesized RET2 and RET2ΔMD proteins. Assay was carried out with 0.5, 1.0 and 3.0 μl of rabbit reticulocyte lysate (TnT Quick Coupled Transcription/ Translation System, Promega) in the presence of 25 nM of RNA substrate and 100 μM of UTP for 1 hr. Vector: expression plasmid pET15b (Novagen).
Immunoblotting of cell lysates from parental 29-13,40 RET2ΔMD-iCODA and RET2-iCODA cell lines demonstrated that mutated and full-length proteins were expressed at similar levels while the endogenous RET2 was effectively depleted (Fig. 4b). Tandem affinity purification of complexes from RET2-iCODA and RET2ΔMD-iCODA cells produced a typical RECC SDS gel profile for the former and two protein bands for the latter (Fig. 4c). To determine proteins associated with RET2 and RET2ΔMD, both affinity-purified fractions were subjected to sequential Lys-C and trypsin proteolysis and LC-MS/MS analysis. All nineteen proteins that are currently viewed as subunits of the T. brucei core editing complex16, 17, 41 have been identified in RET2-associated complex with high confidence (Table 1), including RET2 (42 peptides, 68% coverage) and MP81 (56 peptides, 66% coverage). The shorter RET2ΔMD bait protein produced 9 peptides (28% coverage) while only four peptides were detected for MP81 (7% coverage). The decreased MP81:RET2 peptide yield ratio demonstrates a significant loss of RET2-MP81 interaction in vivo and implicates the middle domain as RECC docking interface.
Table 1.
Mass spectrometry analysis of affinity-purified RET2 and RET2ΔMD complexes. Established components of the core editing complex (20S editosome) are listed.17, 41 Number of unique peptides identified by LC MS/MS is indicated for each polypeptide.
| RET2 | RET2ΔMD | ||||||
|---|---|---|---|---|---|---|---|
| Protein | Peptides | Protein | Peptides | Protein | Peptides | Protein | Peptides |
| MP81 | 56 | MP41 | 31 | RET2 | 42 | MP81 | 4 |
| MP63 | 40 | MP24 | 17 | REX1 | 38 | RET2 | 9 |
| MP49 | 10 | MP19 | 12 | REX2 | 53 | ||
| MP47 | 35 | MP18 | 10 | REL1 | 32 | ||
| MP46 | 26 | REN1 | 39 | REL2 | 38 | ||
| MP44 | 30 | REN2 | 38 | ||||
| MP42 | 28 | REN3 | 28 | ||||
We next examined the role of the middle domain in RET2:MP81 interaction in vitro. In agreement with affinity purification data, the stoichiometric co-immunoprecipitation of in vitro-synthesized [35S]-labeled MP81 was observed with RET2 but not with RET2ΔMD protein (Fig. 4d). To address the possibility of conformational changes or folding problems caused by the middle domain deletion, we inquired whether grafting of the middle domain would confer MP81 binding properties to a different protein. We chose a non-mitochondrial TUTase, TUT4, as a recipient of the RET2’s middle domain. Positions 130-132 of TbTUT4, which represent a short loop between the two enclosing β-strands that are nearly identical in RET2 and TUT4,38 have been replaced with RET2’s middle domain (positions 153-263, RET2 numbering). Although incubation of chimeric [35S]-labeled TbTUT4-MD protein with anti-MP81 antibody produced unusually high background, addition of MP81 into binding reaction increased the efficiency of TbTUT4-MD immunoprecipitation by approximately two-fold (Fig. 4e). Combined with negative co-immunoprecipitation of the full-length TbTUT4 and MP81 with anti-TUT4 or anti-MP81 antibody (not shown and 26), these data indicate a gain of MP81-binding function for the TbTUT4-MD protein. Collectively, our findings demonstrate that the middle domain is required for RET2 docking into the RECC via a direct contact with structural zinc finger protein MP81.
Middle domain is essential for RET2 enzymatic activity
To analyze the effects of MD deletion on catalysis, the ssRNA U-addition and dsRNA U-insertion activities of the full-length RET2 and RET2ΔMD affinity purified fractions were tested. The mRNA 5′-cleavage fragment was used as generic ssRNA substrate while the pre-cleaved editing substrate programmed for +2 insertion served as a model dsRNA (Fig. 4f). Taking into account the mass spectrometry analysis of RET2 and RET2ΔMD affinity purified fractions (Table 1), the complete lack of U-insertion editing activity in the RET2ΔMD fraction was most likely caused by MD deletion whereas the loss of RNA ligase activity is consistent with missing RNA ligases and most other editosome components. The detrimental effect of MD deletion on RET2 enzymatic activity was further confirmed using in vitro-synthesized RET2 and RET2ΔMD proteins and pre-cleaved RNA substrates (Fig. 4g).
To conclude, our data indicate that MD is required for both RECC docking and enzymatic activity of RET2. Although the MD deletion may have caused structural changes detrimental for MP81 binding and TUTase activity, available evidence suggests MD’s direct involvement in these functions. The overexpression of RET2ΔMD in T. brucei produced no discernable phenotype while a single-amino acid mutation in the catalytic site (D97A) led to a dramatic growth inhibition (not shown). Apparently, the dominant negative effect occurs only if the inactive protein associates with RECC. Finally, single-amino acid changes along the putative RNA binding path extending across the middle domain lead to inhibition of RET2 activity, but not RECC incorporation (Ringpis et al, submitted).
RECC and rRET2 show distinct RNA substrate requirements and processivity in vitro
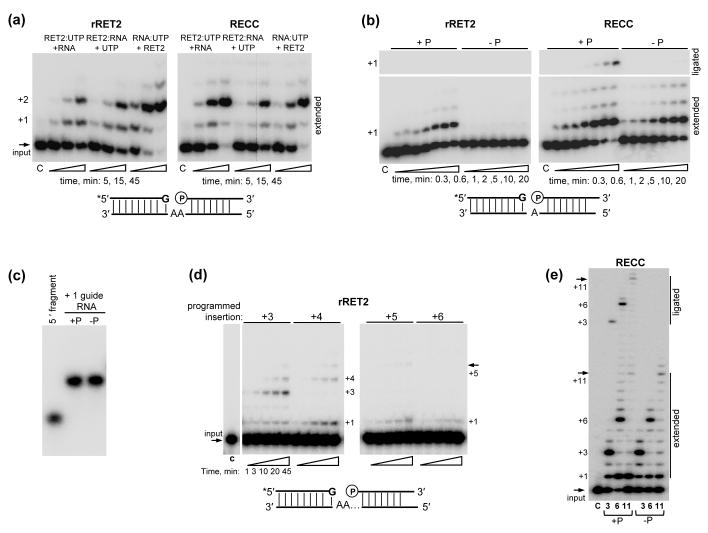
The order of RNA and UTP substrate addition has been shown to dramatically affect processivity of LtRET1 TUTase39, which is capable of UTP polymerization without RNA, and the efficiency of the TbTUT4-catalyzed reaction31. Combined with crystallographic analysis of TbTUT4-UTP,38 and TbTUT4-UTP-UMP31 complexes, these findings demonstrate that UTP can partially occupy the RNA binding site and compete with a polynucleotide substrate. The opposite situation of RNA substrate partially occupying UTP binding site also seems possible based on TbTUT4-UpU structure31 and close positioning of RET2’s UTP binding site and UTP/UMP binding site (site B), which is likely involved in RNA substrate positioning.30 To assess the order of substrates addition effect on rRET2- and RECC-catalyzed U-insertions, the reactions were started by adding RNA, UTP or protein to pre-incubated protein plus UTP, protein plus RNA and RNA plus UTP mixtures, respectively. Time-course analysis showed less efficient U-insertion when rRET2 was pre-incubated with either substrate as compared to the reaction initiated by enzyme addition (Fig. 5a, left panel) which is consistent with competition between the two substrates for the RET2 catalytic site. However, the efficiency of RECC-catalyzed reaction was unaffected by the order of substrate addition. Although we have not been able to accurately measure an apparent Kd for RET2 and RECC for technical reasons, these observations indirectly suggest that RECC’s higher affinity for RNA overcame competition with UTP for the RNA binding site (Fig. 5a, right panel).
Fig. 5.
RECC incorporation increases RET2’s RNA affinity and processivity. (a) Effect of the substrate addition order. Pre-cleaved editing assays were carried out with 20 nM of rRET2 (left panel) and 2.8 nM of RECC-embedded RET2 (concentration estimated by Western blotting with anti-RET2 antibody in reference to rRET2, right panel). Pre-cleaved RNA substrate programmed for +2-insertion and UTP were present at 100 nM and 100 μM, respectively. C: control, input RNA. (b) Effect of the phosphate group at the 5′-end of the 3′-cleavage fragment. Reactions were performed as in (a) with substrates programmed for +1 insertion for indicated periods of time. Top panels: ligated products. Bottom panels: U-insertion products. +P: 5′-phosphorylated 3′-cleavage product; −P: 3′-cleavage fragment with 5′-hydoxyl group. (c) Assembly of the pre-cleaved editing substrate. Annealed pre-cleaved RNA substrates from (b) were separated alongside 5′-cleavage fragment on a 10% acrylamide native Tris-HEPES gel. (d) Processivity of rRET2. U-insertion assay was performed with 20 nM of protein, 100 μM UTP and RNA substrates programmed for 3, 4, 5 and 6 U-insertions for indicated periods of time. (e) Processivity of RECC-catalyzed U-insertion reactions. Assays were performed for 45 min as in (d). RECC-embedded RET2 (2.8 nM) was incubated with radiolabeled 5′-cleavage fragment, phosphorylated (+P) or non-phosphorylated (−P) 3′-cleavage fragment and gRNAs with indicated gap sizes in the presence of 100 μM of UTP.
RET2, a member of the DNA Polymerase β (Pol β) superfamily of nucleotidyl transferases42, recognizes a dsRNA which resembles DNA Pol β’s gapped DNA substrate during nucleotide excision repair43. The 5′-monophosphate at the 3′-fragment is critical for DNA binding by the purified Pol β whose gap-filling capacity in vitro is limited to 3-4 nucleotides.44 This constraint is imposed by a proximity requirement between the 5′-fragment’s hydroxyl group and the internal monophosphate. The monophosphate moiety, which participates in the post-U-insertion ligation of 5′- and 3′-fragments,32 is also critical for rRET2’s activity on dsRNA with a two-nucleotide gap.26 To minimize proximity effects, we have compared rRET2 and RECC activities on RNA substrates bearing a single-nucleotide gap. As expected, rRET2 efficiently inserted one uracil residue into editing substrates possessing a 5′-phosphorylated 3′-cleavage fragment, but was inactive in the absence of an internal monophosphate (Fig. 5b, left panel). In contrast, RECC-catalyzed +1 U-insertion equally efficient on substrates possessing or lacking the 5′-monophosphate group. As expected, the ligation reaction was completely blocked for the latter substrate (Fig. 5b, right panel).
To ensure that the +1 U-insertion occurs within a double-stranded RNA and not with unassembled 5′-fragment, the annealed tripartite substrates with 5′-phosphorylated or dephosphorylated 3′-fragments were separated on a native gel alongside with the 5′-fragment (Fig. 5c). In conclusion, recognition of a monophosphate appears to be a conserved feature of Pol-β family members acting on double-stranded gapped substrates. In RECC, lack of this positive contribution is likely compensated by its higher-affinity for RNA, which is consistent with order-of-addition experiments (Fig. 5a).
Previous studies using partially-purified mitochondrial extracts,34 purified editing complexes,28, 33 and recombinant RET230 have demonstrated guided U-insertions of up to three uracil residues. However, U-insertions in vivo range from a single nucleotide to 12 uridines.45 Therefore, we next determined whether rRET2 and RECC can act on longer gaps. Recombinant RET2 efficiently filled gaps of up to three nucleotides, but its activity declined sharply on editing substrates programmed for more insertions (Fig. 5d). This observation is consistent with a monophosphate requirement suggesting that, similar to DNA Pol β, the 3′-hydroxyl must be tethered in close proximity (1-3-nt gap) to the monophosphate group to be recognized by the enzyme. In contrast, the RECC-embedded RET2 achieved up to 11 guided U-insertions irrespective of the phosphate presence (Fig. 5e). Thus, association with RECC stimulates RET2’s processivity and enables filling of longer gaps in double-stranded RNA substrates.
Distributive insertion of the first U generates a substrate for a processive gap filling
We have noticed that a single uracil is not only inserted with similar efficiency into a +1-programmed substrate by both rRET2 and RECC (Fig. 5b, Table 2), but the +1U is also a prominent product on dsRNAs with longer gaps. It has also been shown that the partially-purified editing complex adds a single U to a 5′-cleavage product regardless of the base-pairing capacity with corresponding guiding nucleotide.34 We hypothesized that a distributive +1 insertion acts as an additional specificity mechanism by which the presence of a correct base-pairing with gRNA is monitored: if +1-addition hybridizes with a guiding purine, the editing complex re-binds and catalyzes a processing gap filling.
Table 2.
Steady-state kinetic parameters of UTP incorporation by rRET2 and RECC.
| enzyme | Substrate | Km, UTP (μM) |
± | kcat (min−1) |
± | kcat / Km (min−1 M−1) |
|---|---|---|---|---|---|---|
| rRET2 | 0U | 2.5 | 1.9 | 0.02 | 0.002 | 8.0×103 |
| RECC | 0U | 0.4 | 0.05 | 0.04 | 0.0004 | 1.0×105 |
| rRET2 | 1U | 12.6 | 3.2 | 0.20 | 0.0007 | 1.6×104 |
| RECC | 1U | 20.8 | 5.9 | 0.09 | 0.005 | 4.0×103 |
It has been shown that TUT431 and MEAT126 TUTases prefer uracils at the terminal base of the single-stranded RNA substrate while RET2 prefers terminal purines.28, 31 This is expected since endonucleolytic cleavage produces As or Gs as the terminal base in the 5′-cleavage fragment in ~99% of all U-insertion editing sites.45 We first asked whether RET2’s catalytic parameters in +1U-insertion reaction are affected by its incorporation into RECC and the nature of the 3′-nucleotide in the 5′-fragment. The apparent Km and rate constants of UTP incorporation were determined for dsRNA substrates programmed for a +1-insertion and G or U as terminal bases of the 5′-cleavage fragment (Table 2). Moderate differences in catalytic efficiencies of rRET2 and RECC on both substrates suggest that neither the chemical nature of the terminal base nor complex association exert significant effects on +1 U-insertion activity.
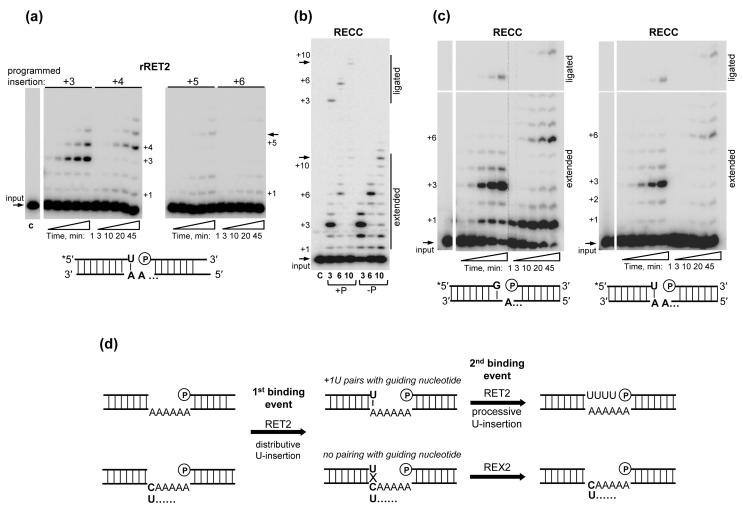
We next inquired whether the capacity to fill longer gaps would be altered by the presence of a terminal U-A base-pair in 5′-fragment-gRNA hybrid, which imitates a pre-cleaved substrate following a single-round of U-insertion. The activity of the recombinant RET2 was indeed stimulated by the presence of a terminal U-residue not only yielding prominent +3 and +4 signals but also reaching +5-insertion (Fig. 6a). The RECC-catalyzed reaction was similar on substrates ending with G (Fig. 5d) or U (Fig. 6b) in terms of processivity and yield of guided products and was not affected by the lack of phosphate group. The prominent +1U insertion, however, was not observed on RNA substrates in which the 5′-fragment already contained a uracil at the 3′-end. To further strengthen this conclusion, the RECC-catalyzed U-insertion was tested on both RNA substrates as a function of the reaction time (Fig. 6c). Apparently, the +1 is a predominant insertion to the purine-terminated 5′-fragment and is more prominent on the longer 6-nt gap (Fig. 6c, left panel) likely because the shorter 3-nt gap is filled more efficiently. In the case of 5′-fragment terminating with U, virtually no accumulation of +1 product was detected (Fig. 6c, right panel). These findings suggest that the RECC-embedded RET2 catalyzes a bimodal reaction: distributive addition of a single uracil to the 5′-fragment and, upon hybridization of this residue to a guiding purine on the opposite strand, a processive filling of the gap according to the number of guiding nucleotides.
Fig. 6.
Structure of the RNA substrate modulates RET2 activity. (a) Processivity of rRET2 on the gapped dsRNA assembled with 5′-fragment that already contained a single uracil at the 3′-end. U-insertion assay was performed with 20 nM of protein, and 100 μM UTP for indicated periods of time on RNA substrates programmed for +3, 4, 5 and 6 insertions. (b) Processivity of RECC on the RNA substrates, similar to those in (a), programmed for +3, +6, and +10 insertions. Assay was performed as in (a) with 2.8 nM of RECC-embedded RET2 for 45 min. RNAs with indicated gap sizes were assembled with phosphorylated (+P) and non-phosphorylated (−P) 3′-fragments. (c) Time course of RECC-catalyzed U-insertion on RNA substrates that imitate post-cleavage (left panel) and post-1U-insertion (right panel) intermediates programmed for +3 and +6 insertions. (d) A model of the U-insertion editing reaction catalyzed by RECC2.
Discussion
In this work, we have determined a specific domain required for RNA Editing TUTase 2 incorporation into RECC and compared enzymatic properties of monomeric and RECC-embedded RET2. We have utilized RNAi knockdown/knock-in cell lines of procyclic T. brucei, bacterial expression and in vitro protein synthesis systems to obtain highly-purified complexes and individual proteins. Activities of unassociated and RECC-bound RET2 were tested on synthetic RNA substrates which resemble intermediates of the U-insertion reaction and allow incorporation of up to 11 uridine residues. While endonuclease, exonuclease and RNA ligase activities are executed by two or more homologous enzymes, RNA Editing TUTase 2 represents the only nucleotidyl transferase found in all variants of the core complex. Hence, a combination of a novel genetic system (iCODA) and an available X-ray crystal structure30 presents a unique opportunity to determine whether association of editing enzymes into a stable complex alters their intrinsic substrate specificities and catalytic efficiencies. The iCODA methodology provided not only a means to study RET2 within the RECC context, but also established that a single RET2 molecule is present per complex. In contrast to dominant negative approaches, the iCODA system proved to be of value for in vivo analysis of mutant proteins lacking domains required for complex association. In addition, the successful knock-in confirmed exquisite specificity of RNAi knockdown in T. brucei – introduction of a single mismatch per twelve base-pairs into a synthetic gene was sufficient to confer an RNAi resistance to its transcript.
A pronounced sequence similarity between the N-terminal catalytic and C-terminal base-recognition domains of trypanosomal TUTases indicates conservation of these modules but provides few clues for the major differences in their enzymatic properties and capacities for specialized functions. Interactions with binding partners are likely to be carried out by function-specific auxiliary domains, such as RET2’s middle domain. Crystallographic studies of RET2 revealed that NTD and CTD share a large interface essentially creating a spherically-shaped catalytic “bi-domain.”30 The middle domain is inserted between two β-strands at the C-terminus of the NTD and folds out into the solvent while maintaining extensive interactions with the CTD. Positioning of the MD in respect to the catalytic cavity makes its contribution to UTP binding unlikely, but points to a potential role in RNA binding and/or protein-protein interactions. Indeed, MD deletion led to a complete enzyme inactivation and loss of association with the editing complex. The fact that RET2ΔMD was expressed at a level comparable to that of the full-length gene suggests that the folding was not dramatically affected by the MD deletion. Grafting the RET2’s middle domain into a topologically-conserved site within TUT4 TUTase38 led to a partial gain of interaction between the TUT4-MD chimeric protein and MP81, an established RET2 binding partner.18, 41 On the other hand, lack of TUTase activity in RET2ΔMD indicates that the middle domain may be involved in both complex association and RNA binding.
The accuracy of U-insertion into dsRNA is crucial for the overall fidelity of the editing process. Specificity for U is dictated by RET2’s intrinsic selectivity for the uracil base30 rather than by Watson-Crick base-pairing of the incoming UTP with guiding nucleotides.32, 34 The RET2’s substrate is generated by the endonucleolytic mRNA cleavage which leaves a phosphate group at the 5′ end of the 3′ fragment (Fig. 1). Consistent with previous study by Igo et al.32 performed with mitochondrial extract enriched for editing activity, the phosphate is not required for U-insertion by the RECC, but is essential for U-insertion by monomeric RET2. This may reflect mechanistic similarities between RNA editing and base excision repair enzymes, but not their respective complexes. Indeed, gapped DNA substrates are generated by the AP endo/exonuclease, which is homologous to the REX1/REX2 editing exonucleases, and then targeted by the DNA polymerase β.20, 46 The DNA repair polymerase activity depends on 5′ phosphate recognition44 to processively extend the 5′ fragment through the lesion but only if the gap does not exceed 4-5 nucleotides, which matches the intrinsic processivity limit of monomeric RET2.
The phosphate group requirement for RET2-catalyzed reaction indicates that the double-stranded “anchor” with an internal 5′-monophosphate constitutes the RET2 binding site (Fig. 1). In case of RECC, the lack of phosphate dependence and capacity to fill longer gaps emphasizes the contribution of other subunit(s) to RNA binding. If RET2, as core complex subunit, remains bound to the “anchor” duplex upon mRNA cleavage, the 3′-OH group of 5′-mRNA cleavage fragment must be somehow positioned in the vicinity of the active center. For short gaps, the 3′-hydoxyl group may be held in sufficiently close proximity by 1-3 guiding nucleotides; hence efficient U-insertion may not require additional RNA contacts outside of RET2. This hypothesis is consistent with very similar catalytic parameters for +1U-insertion reactions catalyzed by RET2 and RECC (Table 2). For longer gaps, a greater entropy cost of bringing the 3′-OH group into the RET2’s active site is likely to be compensated by interactions with other RECC subunits. Structural protein MP81, which binds both RET2 and REL2 ligase18 and has been shown to enhance RET2 activity in vitro,28 is a primary candidate for providing these additional RNA binding contacts. Collectively, our data and earlier findings from the Stuart laboratory28, 32, 34 suggest that complex association enhances RET2-catalyzed U-insertion editing by optimal scaffolding of RNA substrates with gaps longer than three nucleotides.
The endonucleolytic cleavage of typically purine-rich pre-edited mRNAs leaves As or Gs at the 3′-end of the 5′-mRNA cleavage fragment. In mitochondrial extract, a single U-insertion into a gapped dsRNA can be accomplished even if the opposite nucleotide in the gRNA (U or C) does not base-pair with the inserted uracil. The sequential +2 insertion, however, is inhibited.32 Thus, although in the majority of U-insertion editing sites first U is added to the mRNA fragment ending with purine, all sequential Us are added to RNA ending with uracil. We propose that the distributive +1-insertion acts as a “sampling mechanism” to ensure that a purine nucleotide occupies a base-pairing position in the opposite guiding strand. When the +1-extended product hybridizes to the guiding purine, RET2 recognizes the base-pair of the opposite geometry in a different mode which triggers processive U-insertion. It remains to be elucidated whether editing complex fully dissociates after +1U insertion and then rebinds in a different mode. The thermodynamic contribution of +1U insertion to stabilizing the gRNA — 5′ cleavage fragment binding is apparently neglligable.34 However, TUT4 structure and RET2 homology modeling imply that the RNA’s terminal base pair must enter the stacking interaction with the uracil base of the bound UTP.31 Such interaction would favor UTP—3′-U stacking, which may stimulate RET2’s processivity, while UTP—3′-purine stacking would be sub-optimal for processive activity because of base translation. To conclude, the +1 insertion likely serves a dual function of verifying purine nucleotide occupancy in complementary gRNA and stimulating processive gap filling. In this scenario, an unpaired uracil may be removed by the U-deletion activity upon RECC re-binding, which would explain an evolutionary pressure to keep components of the U-deletion pathway in a core complex dedicated to U-insertion (Fig. 1). It is plausible that REX2 acts on U-insertion editing sites as a proofreading enzyme.
Materials and Methods
Cell cultures, RNAi and RNA analysis
The RET2 RNAi expression plasmid was generated by cloning positions 41-545 of the RET2 gene into a p2T7-177 vector.36 Clonal tetracycline-inducible RNAi cell lines were obtained by transfection of this construct into procyclic T. brucei strain 29-1340 followed by selection by limiting dilution. RNAi was induced by the addition of 1 μg/ml tetracycline and cells were diluted every 24 hours to ~1×106/ml. Isolation of total RNA and quantitative real-time PCR analysis of mitochondrial RNAs was performed as previously described.25, 26, 47
iCODA
To generate the RNAi-resistant fragment of RET2coda, the following sequence was designed, assembled from oligonucleotides, cloned into pTOPO vector and verified by sequencing: 5′-ATGTTGATGCACACAGCACCCTGGTTGCACATGAGGCTTAGTAGGTTGTT TAGGCAGAGTCCACTTAGTTTGCCGAGTACTAAACTTAACCCTAGTCCTG ATCATTACGCAGTTTGGGGTAAGGCAATTATGGCCGAAAATAATCGTAGG GTAGGACCTGAGCACATGTTTCGTACAGCAATTAGGGCACAGCAGCAACT TCAGGGTTTAGCTGATAAATGGACGCCTGACGCAAAGGTGTACTGTTGTG GTAGTATGGTGACGTACGGTCAGATGGAGTGGGGTAGTGATCTTGATTTA GCATGTATGTTTGATGATCCATACCCTAGTCATGAGGTTCAGGCTAAGCG TACCGATAAGTTGTGGACAGTGATTAAGCGCTACGTCCCACACTACTTGA GGAACAATCTTTTAGGTCTGACTGAAGCGCGTACACCAGTAGTAAAGTTG AGGTTTGCTAACGATGAAAAGGTAGCTAGGGCTAGGTACACACCATTGAG CGAAGAAGAGGACCGTAAGGCACGTACAGCATTGCTTGATGTTAGG-3′.
To generate pLEW100-RET2-iCODA-TAP-BSR, the iCODA fragment was PCR-amplified with A222 and A223 primers. The recipient vector was amplified by inverted PCR from pLEW100-RET2-TAP-BSR with A220 and A221 primers. Both fragments were digested with HindIII, gel-purified, and ligated to replace the first 546 bps of the RET2 gene.
Clonal cell lines for RET2-iCODA/RET2 RNAi were generated by co-transfecting pLEW100-RET2-iCODA-TAP-BSR and p2T7-177-RET2 into procyclic T. brucei strain 29-1340 and subsequent selection of cells resistant to blasticidin, phleomycin, hygromycin, and neomycin by limiting dilution. Concurrent RET2-iCODA-TAP expression and RET2 RNAi were induced by the addition of tetracycline at 1 μg/ml.
Protein purification and in vitro synthesis
A RET2-iCODA gene lacking 21 codons at the 5′-end (putative mitochondrial importation signal) was PCR-amplified and cloned into pET15b E. coli expression vector. Recombinant RET2-iCODA was purified as previously described.35 For expression in T. brucei, the RET2 gene was cloned into pLEW79-BLE35 and pLEW100-BSR vectors. Tandem affinity purification of RECC-embedded RET2 from whole cell lysates (~2×1010 cells) was performed using a modified protocol.35 Briefly, transgenic cell lines were grown in 2 liters of SDM-79 medium supplemented with 10% heat-inactivated fetal bovine serum, 50 μg/ml of Geneticin (G418), 50 μg/ml of hygromycin, 10 μg/ml of hemin, and 2.5 μg/ml phleomycin and/or 10 μg/ml blasticidin. Cells were induced with 1 μg/ml tetracycline, harvested after 48-72 hours of induction, and centrifuged in a fixed-angle rotor at 5,000 g for 10 minutes. Subsequent purification was performed at 4°C. Cell pellet was resuspended in 6 ml of extraction buffer (50 mM Tris-HCl, pH 7.6, 150 mM KCl, 2 mM EDTA). Detergent NP-40 was added to 0.3% plus 0.3 tablet of Complete™ (Roche) protease inhibitors. Extract was sonicated three times at 9W for 10 sec and then centrifuged at 100,000 g for 15 min at 4°C in a TLA 100 rotor (Beckman). Supernatant was harvested and pellet was resuspended with 6 ml of extraction without NP-40 followed by another round of sonication. Pooled supernatants were filtered through a 0.45-μm membrane and incubated with 0.3 mL of IgG Sepharose resin (GE Healthcare) for 1 hour. Material was transferred to a gravity-flow column and washed with 6 volumes of IgG Binding Buffer (25 mM Tris-HCl, pH 7.6, 150 mM KCl, 1 mM EDTA, and 0.1% NP-40) followed by 2 column volumes of TEV Cleavage Buffer (IgG Binding Buffer plus 1mM DTT). Column was sealed and incubated overnight with gentle agitation with 1.5 ml TEV Cleavage Buffer supplemented with 150 units of tobacco etch virus (TEV) protease and 0.02 tablet of Complete™ protease inhibitor. The IgG column was drained directly onto 300 μl of calmodulin resin. The IgG column was washed with an additional 3 ml of Calmodulin Binding Buffer (25 mM Tris-HCl, pH 7.6, 150 mM KCl, 0.1% NP-40, 10 mM β-mercaptoethanol, 1 mM magnesium acetate, 1 mM imidazole, and 2 mM CaCl2) and the washes were also loaded on the calmodulin resin. The suspension was supplemented with 6 μl of 1 M CaCl2 and then incubated with gentle agitation for 1 hour. Suspension was transferred to a gravity-flow column and washed with 6 column volumes of Calmodulin Binding Buffer. Complexes were then eluted with 25 mM Tris-HCl, pH 7.6, 150 mM KCl, 3 mM EGTA, 1mM EDTA, and 10% glycerol.
pET15b-based vectors were used to clone genes of interest under control of T7 RNA polymerase promoter. 35S-labelled proteins were synthesized with TNT Quick Coupled Transcription/Translation System (Promega). Briefly, 1 μg of plasmid DNA, 2 μl of [35S]methionine, and 40 μl of TnT Quick Master Mix are combined in a 50 μl reaction volume, and incubated for 90 minutes at 30°C.
RNA substrates, enzymatic assays and native gels
Editing ligase self-adenylation reactions were performed as described.35, 48 Pre-cleaved editing assays were carried out with 5-20 nM recombinant RET2, or 2 μl TAP-purified complexes or 0.5-2 μl of TNT lysate in 20μl of 50 mM HEPES, pH 8.0, 10 mM MgCl2, 50 mM KCl, 1 mM DTT, 100 μM UTP. RNA substrates35 were assembled from chemically synthesized radiolabeled 5′-fragment, in vitro transcribed guide RNA and chemically synthesized 3′-fragement. Respective RNAs were mixed at 1:1.5:3 molar ratios in 20 mM HEPES-KOH buffer, pH 7.5 and 50 mM of KCl, heated to 65°C and cooled to 4°C over 30 minutes period of time. The assembly of dsRNAs was verified by 10% native gel electrophoresis in 50 mM Tris — 50 mM HEPES buffer (pH 7.8, without adjustment). RNA substrates were used in U-insertion reactions at final concentration of 0.05-0.1 μM. For double-stranded RNAs, concentration of the 5′-fragment was assumed. Reactions were terminated by addition of two volumes of 0.5 M sodium acetate (pH 5.0), 10 mM EDTA, 0.1% SDS, and 2 μg of glycogen (Ambion). RNA was extracted with phenol-chloroform, precipitated with ethanol, and resuspended in 95% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol FF, and 10 mM EDTA. The sample was denatured by heating at 80°C for 2 min and analyzed on a 15% acrylamide/urea gel. Steady-state kinetic parameters of UMP incorporation were obtained with reaction times and UTP concentrations varying from 0.33 to 5 minutes and 0.5-150 μM, respectively.
Western blotting and immunoprecipitation
Rabbit polyclonal antibodies against full-length recombinant RET2 were affinity-purified on antigen column. For Western blotting, ~3×106 cells were dissolved in SDS loading buffer, separated on an 8-16% gradient SDS-PAGE gel, and transferred to a nitrocellulose membrane. For co- immunoprecipitation assays, equal volumes of TnT lysates expressing 35S-labeled proteins or containing empty vectors were combined for 15 minutes at 30°C. Magnetic Protein G Dynabeads (Invitrogen) were coated with purified anti-MP81 monoclonal antibodies in IP buffer (20 mM Tris-HCl, pH 7.8, 150 mM NaCl, and 0.1% Triton X-100) and then saturated with 10 mg/ml of BSA in IP buffer. For a single IP, 5 μl of beads suspension and 1 μg of total immunoglobulin were used. Beads were re-suspended in 100 μl of IP buffer plus 10 mg/ml of BSA and incubated with the 100 μl TnT reaction for 1 h at 4 °C with constant mixing. Beads were pelleted, washed three times with 1 ml of IP buffer for 15 min, resuspended in 1x SDS loading buffer, separated on an SDS-PAGE gel, and then transferred to a nitrocellulose membrane for visualization on a phosphor storage screen.
Mass-spectrometric analysis by LC MS/MS
LC MS/MS was carried out by nanoflow reverse phase liquid chromatography (RPLC) (Eksigent, CA) coupled on-line to a Linear Ion Trap (LTQ)-Orbitrap mass spectrometer (Thermo-Electron Corp). The LC analysis was performed using a capillary column (100 μm ID × 150 mm long) packed with Polaris C18-A resin (Varian Inc., CA) and the peptides were eluted using a linear gradient of 2% to 35% B in 85 min at a flow of 300 nL/min (solvent A: 100% H2O/0.1% formic acid; solvent B: 100 % acetonitrile/0.1% formic acid). A cycle of one full FT scan mass spectrum (350-2000 m/z, resolution of 60,000 at m/z 400) followed by ten data-dependent MS/MS acquired in the linear ion trap with normalized collision energy (setting of 35%). Target ions already selected for MS/MS were dynamically excluded for 30 s.
Monoisotopic masses of parent ions and corresponding fragment ions, parent ion charge states and ion intensities from the tandem mass spectra (MS/MS) were obtained by using in-house software with Raw_Extract script from Xcalibur v2.4. Following automated data extraction, resultant peak lists for each LC MS/MS experiment were submitted to the development version of Protein Prospector (UCSF) for database searching similarly as described (Wang and Huang, 2008 ). The T. brucei database v4 (www.genedb.org, 09/01/2006, 8303 sequence entries) was used for database searching. Trypsin was set as the enzyme with a maximum of two missed cleavage sites. The mass tolerance for parent ion was set as ±20 ppm, whereas ±0.5 Da tolerance was chosen for the fragment ions. Chemical modifications such as protein N-terminal acetylation, methionine oxidation, N-terminal pyroglutamine, and deamidation of asparagine were selected as variable modifications during database search. The Search Compare program in Protein Prospector was used for summarization, validation and comparison of results. To determine the expectation value cutoff that corresponds to a percent false positive (% FP) rate, each project was searched against a normal database concatenated with the reversed form of the database. An algorithm in Search Compare automatically plots the expectation values versus % FP rate for each search result. Based on these results, we chose an expectation value cutoff for all peptides corresponding to ≤0.025% FP. At this false positive rate, false protein hits from decoy database was not observed. Positive protein identification was based on at least three peptides.
Supplementary Material
Acknowledgements
We thank Ronald Etheridge and James Weng for help with qRT-PCR and complex purification and members of the Aphasizhev lab for stimulating discussions. We thank George Cross and Elisabetta Ullu for sharing trypanosomal strains and plasmids. Monoclonal antibody against MP81 was a kind gift of Ken Stuart. This work was supported by the NIH grant AI064653 to RA. GR was supported by the UCI Biomedical Informatics Training Program. All materials described in this article are available in accordance with NIH grant policy on sharing of Biomedical Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This manuscript has been approved by all authors. Authors declare no conflicts of interest or competing financial interests.
References
- 1.Lukes J, Guilbride DL, Votypka J, Zikova A, Benne R, Englund PT. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell. 2002;1:495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stuart K, Allen TE, Heidmann S, Seiwert SD. RNA editing in kinetoplastid protozoa. Microbiol. Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stuart K, Panigrahi AK, Schnaufer A, Drozdz M, Clayton C, Salavati R. Composition of the editing complex of Trypanosoma brucei. Philos. Trans. R. Soc. Lond B Biol Sci. 2002;357:71–79. doi: 10.1098/rstb.2001.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson L, Sbicego S, Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benne R, Van den Burg J, Brakenhoff J, Sloof P, Van Boom J, Tromp M. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 6.Bhat GJ, Koslowsky DJ, Feagin JE, Smiley BL, Stuart K. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell. 1990;61:885–894. doi: 10.1016/0092-8674(90)90199-o. [DOI] [PubMed] [Google Scholar]
- 7.Koslowsky DJ, Bhat GJ, Perrollaz AL, Feagin JE, Stuart K. The MURF3 gene of T. brucei contains multiple domains of extensive editing and is homologous to a subunit of NADH dehydrogenase. Cell. 1990;62:901–911. doi: 10.1016/0092-8674(90)90265-g. [DOI] [PubMed] [Google Scholar]
- 8.Sturm NR, Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990;61:879–884. doi: 10.1016/0092-8674(90)90198-n. [DOI] [PubMed] [Google Scholar]
- 9.Golden DE, Hajduk SL. The 3′-untranslated region of cytochrome oxidase II mRNA functions in RNA editing of African trypanosomes exclusively as a cis guide RNA. RNA. 2005;11:29–37. doi: 10.1261/rna.7170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Aphasizhev R, Aphasizheva I. Terminal RNA uridylyltransferases of trypanosomes. Biochim. Biophys. Acta. 2008;1779:270–280. doi: 10.1016/j.bbagrm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 15.Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA Editing in Trypanosoma brucei requires three different editosomes. Mol. Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnaufer A, Ernst NL, Palazzo SS, O’Rear J, Salavati R, Stuart K. Separate Insertion and Deletion Subcomplexes of the Trypanosoma brucei RNA Editing Complex. Mol Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 19.Kang X, Gao G, Rogers K, Falick AM, Zhou S, Simpson L. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13944–13949. doi: 10.1073/pnas.0604476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers K, Gao G, Simpson L. Uridylate-specific 3′ 5′-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J. Biol. Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- 21.Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome Mitochondrial 3′ Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-insertion/deletion RNA Editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 22.Adler BK, Harris ME, Bertrand KI, Hajduk SL. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol. Cell. Biol. 1991;11:5878–5884. doi: 10.1128/mcb.11.12.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aphasizheva I, Aphasizhev R. RET1-catalyzed Uridylylation Shapes the Mitochondrial Transcriptome in Trypanosoma brucei. Mol. Cell. Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Decker CJ, Sollner-Webb B. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell. 1990;61:1001–1011. doi: 10.1016/0092-8674(90)90065-m. [DOI] [PubMed] [Google Scholar]
- 25.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aphasizheva I, Ringpis GE, Weng J, Gershon PD, Lathrop RH, Aphasizhev R. Novel TUTase associates with an editosome-like complex in mitochondria of Trypanosoma brucei. RNA. 2009;15:1322–1337. doi: 10.1261/rna.1538809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drozdz M, Palazzo SS, Salavati R, O’Rear J, Clayton C, Stuart K. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 2002;21:1791–1799. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst NL, Panicucci B, Igo RP, Jr., Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 29.Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual Role of the RNA Substrate in Selectivity and Catalysis by Terminal Uridylyl Transferases. Proc Natl Acad Sci U S A. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igo RP, Palazzo SS, Burgess ML, Panigrahi AK, Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Igo RP, Jr., Lawson SD, Stuart K. RNA sequence and base pairing effects on insertion editing in Trypanosoma brucei. Mol Cell Biol. 2002;22:1567–1576. doi: 10.1128/mcb.22.5.1567-1576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aphasizhev R, Aphasizheva I. RNA Editing Uridylyltransferases of Trypanosomatids. Methods Enzymol. 2007;424:51–67. doi: 10.1016/S0076-6879(07)24003-0. [DOI] [PubMed] [Google Scholar]
- 36.Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 37.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 38.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J. Mol. Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aphasizheva I, Aphasizhev R, Simpson L. RNA-editing terminal uridylyl transferase 1: identification of functional domains by mutational analysis. J. Biol. Chem. 2004;279:24123–24130. doi: 10.1074/jbc.M401234200. [DOI] [PubMed] [Google Scholar]
- 40.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 41.Schnaufer A, Wu M, Park YJ, Nakai T, Deng J, Proff R, Hol WG, Stuart KD. A protein-protein interaction map of trypanosome ~20S editosomes. J. Biol. Chem. 2010;285:5282–5295. doi: 10.1074/jbc.M109.059378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holm L, Sander C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 43.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase Beta. Chem. Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 44.Prasad R, Beard WA, Wilson SH. Studies of gapped DNA substrate binding by mammalian DNA polymerase beta. Dependence on 5′-phosphate group. J. Biol. Chem. 1994;269:18096–18101. [PubMed] [Google Scholar]
- 45.Ochsenreiter T, Cipriano M, Hajduk SL. KISS: the kinetoplastid RNA editing sequence search tool. RNA. 2007;13:1–4. doi: 10.1261/rna.232907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang X, Rogers K, Gao G, Falick AM, Zhou S, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-Binding Complex from Mitochondria of Trypanosomatids. Mol. Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelletier M, Read LK, Aphasizhev R. Isolation of RNA Binding Proteins Involved in Insertion/deletion Editing. Methods Enzymol. 2007;424:69–96. doi: 10.1016/S0076-6879(07)24004-2. [DOI] [PubMed] [Google Scholar]
- 49.Larsen LS, Wassman CD, Hatfield GW, Lathrop RH. Computationally Optimised DNA Assembly of synthetic genes. Int. J. Bioinform. Res. Appl. 2008;4:324–336. doi: 10.1504/IJBRA.2008.019578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sloof P, Bos JL, Konings AF, Menke HH, Borst P, Gutteridge WE, Leon W. Characterization of satellite DNA in Trypanosoma brucei and Trypanosoma cruzi. J. Mol. Biol. 1983;167:1–21. doi: 10.1016/s0022-2836(83)80031-x. [DOI] [PubMed] [Google Scholar]
- 51.Jensen BC, Kifer CT, Brekken DL, Randall AC, Wang Q, Drees BL, Parsons M. Characterization of protein kinase CK2 from Trypanosoma brucei. Mol Biochem Parasitol. 2007;151:28–40. doi: 10.1016/j.molbiopara.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelly S, Reed J, Kramer S, Ellis L, Webb H, Sunter J, Salje J, Marinsek N, Gull K, Wickstead B, Carrington M. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol. 2007;154:103–109. doi: 10.1016/j.molbiopara.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.