Abstract
Alzheimer disease (AD) is a neurodegenerative disorder characterized clinically by progressive memory loss and subsequent dementia and neuropathologically by senile plaques, neurofibrillary tangles, and synapse loss. Interestingly, a small percentage of individuals with normal antemortem psychometric scores meet the neuropathological criteria for AD (termed `preclinical' AD (PCAD)). In this study, inferior parietal lobule (IPL) from PCAD and control subjects were compared for oxidative stress markers by immunochemistry, amyloid beta-peptide by ELISA, and identification of protein expression differences by proteomics. We observed a significant increase in highly insoluble monomeric Aβ42, but no significant differences in oligomeric Aβ nor in oxidative stress measurements between controls and PCAD subjects. Expression proteomics identified proteins whose trends in PCAD are indicative of cellular protection, possibly correlating with previous studies showing no cell loss in PCAD. Our analyses may reveal processes involved in a period of protection from neurodegeneration that mimic the clinical phenotype of PCAD.
Keywords: Preclinical Alzheimer, oxidative stress, amyloid beta, proteomics, brain, inferior parietal lobule, IPL
Introduction
Alzheimer disease (AD) is one of the leading causes of death among the elderly. About 50 percent of persons aged 85 or older are at risk for developing this neurodegenerative disease for which currently there is no cure or prevention. Patients with AD progress from stages of mild memory impairment to complete dementia. A definitive diagnosis of AD is made only after clinically observable symptoms of dementia are accompanied by the postmortem identification of two neuropathological hallmarks: senile plaques (SPs) and neurofibrillary tangles (NFTs), among other indices.
Senile plaques are mainly composed of amyloid-beta peptide (Aβ), the latter produced via sequential cleavages of amyloid precursor protein (APP) by beta- and gamma-secretases (Haass et al. 1992, Shoji et al. 1992, Seubert et al. 1992). Although several peptides of varying length can be formed from these cleavages, research shows that the 42-amino acid Aβ is most toxic, possibly upon self-association into oligomers (Pike et al. 1991, Geula et al. 1998, Li et al. 2009a, Shankar et al. 2008). Oligomers of Aβ have been heavily implicated in the initiation and pathogenesis of AD, while monomeric forms of Aβ have been suggested to be less harmful, and even neuroprotective. Aβ-mediated oxidative stress, possibly through the Met at position 35, has also been suggested to underlie AD (Butterfield et al. 2010, Butterfield et al. 2001, Markesbery 1999).
Neurofibrillary tangles consist of hyperphosphorylated tau protein, a microtubule-associated structural protein. Other conditions, such as frontotemporal degeneration and Pick's disease, among others, also contain tau-related deposits (Lee et al. 2001). Balanced kinase-phosphatase reactions regulate the biological function of this protein in neurons; disruption of these events can cause over-phosphorylation of tau, leading to protein breakdown and subsequent development of tangles and dysfunction of anterograde and retrograde transport. Both tau and phosphorylated tau in cerebrospinal fluid (CSF) have been proposed as biomarkers for the prediction and diagnosis of AD in living patients (for tau as a possible biomarker review, see (Aluise et al. 2008)).
As mentioned previously, SPs, NFTs, and dementia are all requirements for diagnosis of AD. Adding to the complexity of this disease, brains from a number of cognitively intact individuals at autopsy reveal an extensive SP and NFT load, indicating the possibility of a preclinical or presymptomatic disease state. We have tentatively chosen to define preclinical AD (PCAD) as those individuals with sufficient AD pathological alterations to meet National Institute on Aging-Reagan Institute (NIA-RI) intermediate or, rarely, high likelihood criteria (Braak stage III or higher and moderate or frequent neuritic plaque scores) who had normal cognitive function as shown by antemortem psychometric test scores within the normal range after adjustment for age and education; the classification of these individuals as PCAD is consistent with a consensus report in 2009 from seven independent hospitals and aging centers, including the University of Kentucky, detailing this condition in cognitively normal aged individuals (Price et al. 2009). Data on these individuals are rather scarce due to the rarity of sample availability; however, in addition to high levels of pathological hallmarks and no observable behavioral/memory problems, other anatomical/biochemical features of PCAD include neuronal hypertrophy (Iacono et al. 2009), no hippocampal cell loss (West et al. 2004), increased synaptic plasticity (O'Brien et al. 2009), and alterations in zinc transporters (Lyubartseva et al. 2009). In the current study, we examined the inferior parietal lobule (IPL) of PCAD subjects and controls for changes in oxidative damage to proteins, levels of Aβ42 monomers and Aβ oligomers, and proteomics-determined differences in protein levels that may shed light on biochemical events in PCAD brain.
Participants
PCAD and control brains
Frozen IPL samples were obtained from 12 subjects with PCAD and 12 age-matched subjects who were cognitively intact without postmortem neuropathologic changes of AD (controls). The Rapid Autopsy Program at the University of Kentucky Alzheimer's Disease Clinical Center (UK ADC) obtained samples with extremely short post mortem intervals (PMIs) (Table 1). A short PMI is advantageous in proteomics studies using human tissue since PMI related artifacts are minimized and results more likely reflect the intrinsic condition in PCAD brain.
Table 1.
Patient demographicsa
| Group | Age | Gender | Brain weight (g) | PMI (h) | Braak (median) | MMSE (median) |
|---|---|---|---|---|---|---|
| Control | 85.7 (1.96) | 8F/ 4M | 1218.33 (44.16) | 2.99 (0.75) | I | 29 |
| PCAD | 84.3 (1.37) | 9F/ 3M | 1201.30 (36.14) | 2.76 (0.61) | IV* | 29 |
| Group | Age | Sex | Brain Weight (g) | PMI (h) | Braak | MMSE |
|---|---|---|---|---|---|---|
| Control | 75 | Female | 1080 | 3.50 | 1 | 29 |
| Control | 86 | Female | 1310 | 2.25 | 2 | 30 |
| Control | 86 | Female | 1300 | 3.75 | 1 | 29 |
| Control | 81 | Male | 1410 | 2.00 | 2 | 30 |
| Control | 87 | Male | 1230 | 2.40 | 2 | 29 |
| Control | 74 | Male | 1440 | 4.00 | 1 | 26 |
| Control | 91 | Female | 1230 | 1.75 | 2 | 29 |
| Control | 95 | Female | 1130 | 3.50 | 0 | 26 |
| Control | 90 | Male | 1310 | 3.50 | 0 | 30 |
| Control | 93 | Female | 1080 | 2.75 | 2 | 29 |
| Control | 86 | Female | 1200 | 3.50 | 0 | 28 |
| Control | 84 | Female | 900 | 3.00 | 1 | 26 |
| PCAD | 88 | Female | 1240 | 2.75 | 6 | 29 |
| PCAD | 80 | Male | 1430 | 3.00 | 4 | 27 |
| PCAD | 84 | Female | 1020 | 2.75 | 4 | 29 |
| PCAD | 75 | Male | 1240 | 4.00 | 5 | 28 |
| PCAD | 89 | Female | 1210 | 1.75 | 4 | 30 |
| PCAD | 86 | Female | 1240 | 2.41 | 4 | 28 |
| PCAD | 87 | Female | 1180 | 2.25 | 4 | 30 |
| PCAD | 84 | Female | 1300 | 3.25 | 4 | 29 |
| PCAD | 90 | Female | 1110 | 3.50 | 3 | 29 |
| PCAD | 88 | Female | 1070 | 2.25 | 3 | 29 |
| PCAD | 77 | Male | 1340 | 2.75 | 4 | 30 |
| PCAD | 84 | Female | 1035 | 2.50 | 4 | 30 |
p<0.05
mean unless otherwise noted (+/− SEM)
All participants underwent annual mental status testing and annual physical and neurological exams, as a part of the UK ADC normal volunteer longitudinal aging study and did not have a history of dementia or other neurologic disorders. All participants had test scores in the normal range. Neuropathologic evaluation of the brain samples used as controls revealed only age associated gross and histopathologic alterations. Hematoxylin-eosin and modified-Bielschowsky staining, amyloid β-peptide (Aβ) antibody (10D5), and alpha-synuclein immunohistochemistry were used on multiple neocortical, hippocampal, entorhinal, amygdala, brainstem and cerebellum sections for diagnosis. All procedures to obtain human brain samples were approved by the University of Kentucky Institutional Review Board and experiments were approved by the Executive Committee of the University of Kentucky ADC.
Treatment of samples
Brain samples for oxidative stress analyses and proteomics were homogenized and suspended in Media I buffer containing protease inhibitors: leupeptin (0.5 mg/mL), pepstatin (0.7 μg/mL), and aprotinin (0.5 μg/mL). Homogenates were centrifuged at 2,000 ×g for 5 min to remove debris. Protein concentration in the supernatant was determined by the BCA protein assay (Pierce, Rockford, IL, USA).
Amyloid beta solubility
The amount of Aβ in tissue samples was determined using a three-step serial extraction procedure. This approach takes advantage of progressively more denaturing conditions to serially extract Aβ that is progressively more insoluble, and is followed by the quantitative measurement of Aβ by ELISA. This is a standard procedure in our laboratory, and details of the methodology and antibodies used are available (Das et al. 2003, McGowan et al. 2005, Murphy et al. 2007). Briefly, tissue was homogenized using a PowerMax AHS200 homogenizer in standard PBS buffer (pH = 7.4, 1.0 ml/150 mg of tissue) with a complete protease inhibitor cocktail included (Amresco). The supernatant was collected following centrifugation at 20,000×g for 30 min at 4 °C, to sediment fibrillar material. The pellet was re-extracted by brief sonication (10 × 0.5 s microtip pulses at 100 W; Fisher Sonic Dismembrator, Model 500) in 2% SDS, centrifuged (as above, at 10 °C), and the supernata nt again collected. The remaining pellet was finally extracted by sonication in 70% formic acid (FA), and centrifuged at 20,000×g for 1 h at 4 °C. Sample ext racts were stored frozen at − 80°C until time of assay.
Prior to assay, FA-extracted material was neutralized by 1:20 dilution in TP buffer (1 M Tris base, 0.5 M Na2HPO4), followed by further dilution (at least 1:1) in AC buffer [0.02 M sodium phosphate buffer (pH 7), 0.4 M NaCl, 2 mM EDTA, 0.4% Block Ace (Serotec), 0.2% BSA, 0.05% CHAPS, and 0.05% NaN3]. PBS and SDS-soluble fractions were diluted in AC buffer alone. Final dilutions were determined empirically based on signals obtained in pilot studies. Standard curves of synthetic Aβ42 peptide were prepared in dilution buffer. All standards and samples were run at least in duplicate. Sandwich ELISA for monomeric Aβ42 was conducted using antibodies Ab9 (human sequence Aβ1–16, used for capture) and 2.1.3 (end specific for Aβ42, used for capture). The selectivity of the Ab9 antibody for Aβ monomers was demonstrated in our previous report (Murphy et al. 2007). Biotinylated-4G8 antibody was used for detection. Single site assays for SDS-soluble, oligomeric Aβ were performed in a similar manner, except using antibody 4G8 (against Aβ17–42) for capture and biotinylated-4G8 for detection, and comparing signals against synthetic oligomeric Aβ, as described (LeVine 2004). Plates (Immulon 4HBX) were coated with antibody in PBS (0.5 μg/well used for all capture antibodies), and blocked with a solution of Synblock (Pierce, as per the manufacturer's instructions). Detection antibodies were used at 1 μg / ml in buffer DB [0.02 M sodium phosphate buffer (pH 7), 0.4 M NaCl, 2 mM EDTA, 1% BSA, and 0.00002% thimerosal]. Neutravidin-HRP (Pierce) was used at 0.1 μg / ml. Following development with TMB reagent (Kirkegaard and Perry Laboratories, Gaithersburg, MD), plates were stopped with 6% o-phosphoric acid and read at 450 nm using a BioTek multiwell plate reader.
Oxidative stress assays
Protein carbonyl assay
Samples (5 μl), 12% SDS (5 μl), and 10 μl of 10 times-diluted 2,4-dinitrophenylhydrazine (DNPH) from a 200 mM stock solution were incubated at room temperature for 20 min, followed by neutralization with 7.5 μl neutralization solution (2 M Tris in 30% glycerol). This neutralized solution (250 ng protein) was loaded in each well on a nitrocellulose membrane under vacuum using a slot–blot apparatus. The bicinchoninic acid (BCA, Pierce) assay was used for protein estimation. The membrane was blocked in blocking buffer (3% bovine serum albumin) in PBS 0.01% (w/v) sodium azide and 0.2% (v/v) Tween 20 for 3 h and incubated with a 1:100 dilution of anti-DNP polyclonal antibody in PBS containing 0.01% (w/v) sodium azide and 0.2% (v/v) Tween 20 for 2 h. The membrane was washed three times in PBS following primary antibody incubation at intervals of 5 min each. The membrane was incubated following washing with an anti-rabbit IgG alkaline phosphatase-linked secondary antibody diluted in PBS in a 1:8000 ratio for 1 h. The membrane was washed for three times in PBS for 5 min and developed in Sigmafast tablets, [5-bromo-4-chloro-3-indolyl phosphate/Nitro blue tetrazolium substrate (BCIP/NBT substrate)]. Blots were dried, scanned with Adobe Photoshop, and quantified with Scion Image (PC version of Macintosh compatible NIH image). Controls included samples treated with NaBH4 to reduce carbonyls to alcohol moieties prior to adding DNPH, in which case no immunochemical signal was observed (data not shown).
3-nitrotyrosine (3-NT) and protein-bound HNE assays
In addition to protein carbonyls, other indices of oxidative stress investigated in this study were 3-nitrotyrosine (3-NT) and protein-bound 4-hydroxynonenal (HNE). Lipid peroxidation can lead to HNE, which can covalently modify arginine, lysine, or histidine residues via Michael addition (Butterfield et al. 2001). Samples for 3NT and protein-bound HNE determination were solubilized with Laemmli buffer (5uL), and loaded (250ng) to apparatus under vacuum. Membrane development was similar to protein carbonyls except a primary antibody for 3-nitrotyrosine (Sigma Aldrich) or protein-bound HNE (Chemicon, Rosemont, IL) was used. Controls with only secondary antibody showed no or minimal background signal (data not shown).
Two-dimensional gel electrophoresis
Protein samples (250 μg) were precipitated by adding ice-cold 100% trichloroacetic acid (TCA) to a final concentration of 15% for 10 min on ice. Precipitates were centrifuged for 2 min at 14,000 ×g at 4 °C. The pellet was retained and washed three times with 1 ml of 1:1 (v/v) ethyl acetate/ethanol. The final pellet was dissolved in rehydration buffer (8 M urea, 2 M thiourea, 2% CHAPS, 0.2% (v/v) biolytes, 50 mM dithiothreitol (DTT), and bromophenol blue). Samples were sonicated in 15s intervals three times each.
Two-dimensional polyacrylamide gel electrophoresis was performed with a Bio-Rad IEF Cell system using 110-mm pH 3–10 immobilized pH gradients (IPG) strips and Criterion 8–16% resolving gels. IPG strips were actively rehydrated at 50 V 20 °C followed by isoelectric focusing: 300 V for 2 h linear gradient, 1200 V for 4 h slow gradient, 8000 V for 8 h linear gradient, and 8000 for 10 h rapid gradient. Gel strips were equilibrated for 10 min prior to second-dimension separation in solution A [0.375 M Tris–HCl (pH 8.8), 6 M urea (Bio-Rad, Hercules, CA), 2% (w/v) sodium dodecyl sulfate (SDS), 20% (v/v) glycerol, and 0.5% dithiothreitol (Bio-Rad, Hercules, CA)], and then re-equilibrated for 10 min in solution A containing 4.5% iodoacetamide (IA) instead of dithiothreitol. Control and EAD strips were placed on the 8–16% Criterion gels, unstained molecular standards were applied, and electrophoresis was performed at 200 V for 65 min.
SYPRO Ruby staining
Gels were fixed in a solution containing 10% (v/v) methanol, 7% (v/v) acetic acid for 20 min and stained overnight at room temperature with agitation in 50 ml of SYPRO Ruby gel stain (Bio-Rad, Hercules, CA, USA). Gels were then destained with 50 ml deionized water overnight.
In gel digestion
Samples were prepared according to the method described by Thongboonkerd et al. 2002 (Thongboonkerd et al. 2002). Briefly, the protein spots were cut and removed from the gel with a clean razor blade. The gel pieces were placed into individual, clean 1.5 ml microcentrifuge tubes and kept overnight at − 20 °C. The gel pieces were thawed an d washed with 0.1 M ammonium bicarbonate (NH4HCO3) (Sigma) for 15 min at room temperature. Acetonitrile (Sigma) was added to the gel pieces and incubated for an additional 15 min. The liquid was removed and the gel pieces were allowed to dry. The gel pieces were rehydrated with 20 mM DTT (Bio-Rad, Hercules, CA, USA) in 0.1 M NH4HCO3 (Sigma) and incubated for 45 min at 56 °C. The DTT was removed and replaced with 55 mM IA (Bio-Rad) in 0.1 M NH4HCO3 for 30 min in the dark at room temperature. The liquid was drawn off and the gel pieces were incubated with 50 mM NH4HCO3 at room temperature for 15 min. Acetonitrile was added to the gel pieces for 15 min at room temperature. All solvents were removed and the gel pieces were allowed to dry for 30 min. The gel pieces were rehydrated with addition of a minimal volume of 20 ng/μl modified trypsin (Promega, Madison, WI, USA) in 50 mM NH4HCO3. The gel pieces were chopped and incubated with shaking overnight (~18 h) at 37 °C
Analysis of gel images
Images from SYPRO Ruby stained gels, used to measure protein content, were obtained using a phosphorimager (excitation = 470 nm, emission = 618 nm, Molecular Dynamics, Sunnyvale, CA, USA). PDQuest 2-D Analysis Software (Bio-Rad) was used to match and analyze visualized protein spots among differential gels. The average mode of background subtraction was used to normalize intensity values, which represent the amount of protein per spot. After completion of spot matching, the normalized intensity of each protein spot from individual gels was compared between groups using statistical analysis. Statistical significance was assessed using a two-tailed Student's t-test. P-values less than 0.05 were considered significant.
Mass spectrometry
Tryptic peptides were analyzed with an automated nanospray Nanomate (Advion Biosystems) Orbitrap XL MS (ThermoFinnigan) platform. The Orbitrap MS was operated in a data-dependent mode whereby the 8 most intense parent ions measured in the FT at 60,000 resolution were selected for ion trap fragmentation with the following conditions: injection time 50 ms, 35% collision energy, MS/MS spectra were measured in the FT at 7500 resolution, and dynamic exclusion was set for 120 seconds. Each sample was acquired for a total of ~2.5 minutes. MS/MS spectra were searched against the International Protein Index (IPI) _Mouse Database using SEQUEST and search results were filtered with the following criteria: Xcorr > 1.5, 2.0, 2.5, 3.0 for +1, +2, +3, and +4 charge states,respectively, ΔCn >0.1, and P-value (protein and peptide) < 0.01. IPI accession numbers were cross-correlated with SwissProt accession numbers for final protein identification.
Results
Monomeric, but not oligomeric, amyloid β-peptide levels are elevated in PCAD IPL relative to control
Using the system of insolubility staging of Aβ described in Methods above, we quantified levels of monomeric Aβ42 as soluble (PBS), moderately soluble (SDS), and completely insoluble (FA) fractions. Although we observed a significant increase in the total level of monomeric Aβ42 (FA + PBS + SDS), we observed significant differences only in the FA fraction, while SDS and PBS fractions showed no significant differences (Table 2). Furthermore, mean levels of oligomeric Aβ were decreased in PCAD IPL relative to control IPL (Table 2); although not significantly.
Table 2.
Amyloid beta-peptide levels in control and PCAD cases
| Group | Aβ42 PBS& | Aβ42 SDS& | Aβ42 FA& | Total Aβ42& | Oligomer% |
|---|---|---|---|---|---|
| Control | 0.2 | 2.7 (1.3) | 250.7 (72.5) | 253.6 (72.5) | 7.0 (2.0) |
| PCAD | 0 | 28.2 (13.6) | 1962 (610.0)* | 1982 (617.1)* | 3.3 (2.6) |
p<0.05, mean (+/− SEM)
units in pg/mL
arbitrary units
Analysis of oxidative and nitrosative modifications to brain proteins in PCAD IPL
Oxidative stress has been observed in several regions of the AD brain in many independent reports. Studies have shown oxidative damage to proteins, lipids, DNA, RNA, and carbohydrates in AD brain (Markesbery 1999, Markesbery & Lovell 2007, Butterfield et al. 2001). Oxidative damage to biomolecules and altered levels of brain-resident antioxidants have also been observed in MCI (Butterfield et al. 2007, Sultana et al. 2008, Sultana et al. 2007b, Lovell & Markesbery 2007, Markesbery et al. 2005, Markesbery & Lovell 2007, Butterfield et al. 2001, Sultana et al. 2006, Markesbery 1999, Smith & Perry 1996, Sultana et al. 2010). Because oxidative damage to cellular components is capable of causing mitochondrial impairment, structural damage, and cell death, it is believed that the neuronal death in the AD brain is a possible consequence of oxidative stress (Sultana et al. 2009).
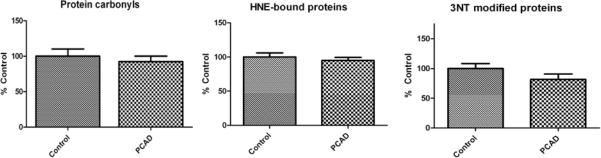
Protein carbonyls can result from direct oxidative damage to the protein backbone, by oxidation of key amino acid side chains, or attack by a moiety containing a protein carbonyl (Butterfield & Stadtman 1997). Carbonyls alter the 3D conformational structure of proteins, generally compromising activity (Butterfield & Stadtman 1997). 3NT and HNE modification of proteins also can contort protein conformation, resulting in loss of activity (Reed et al. 2008a, Reed et al. 2008b, Reed et al. 2009). Immunochemical analyses allow for the global measure of oxidative modifications in a given tissue or fluid. Using this method, we observed no significant differences in the global levels of protein carbonyls, 3NT, or protein-bound HNE between PCAD and control samples (Fig 1).
Figure 1.
Global oxidative stress measurements in control and PCAD IPL. Markers of oxidative and nitrosative damage to proteins were assessed immunochemically. No significant differences were observed between PCAD IPL and control IPL for total protein carbonyls, protein-bound HNE, or 3-nitrotyrosine (n=12 per group).
Proteomics determined differences in expression of brain proteins in PCAD IPL relative to healthy controls
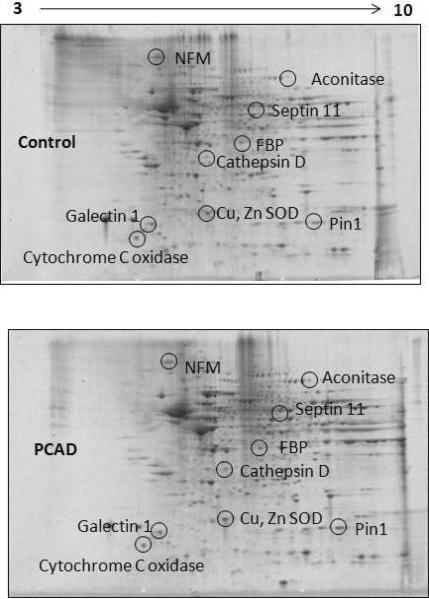
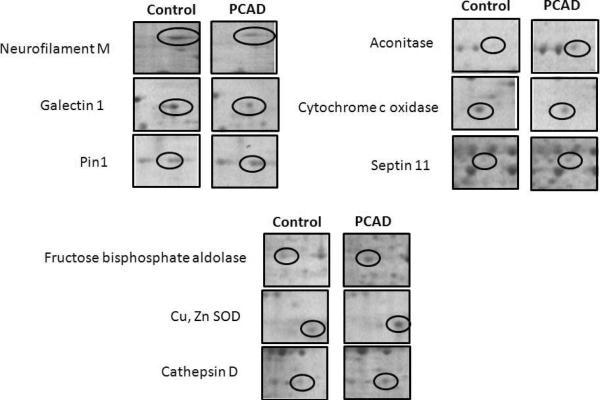
Proteomics has been used to determine differential levels of specific brain proteins in AD and MCI (Butterfield & Sultana 2008, Butterfield & Sultana 2007). Two dimensional gel electrophoresis is one of the most commonly used methods to separate proteins from a biological tissue or fluid to investigate differences in protein levels to shed light on a specific pathology. Here, we report findings of differential levels of proteins in PCAD IPL relative to healthy controls. We observed significant differences in the levels of 9 protein spots in PCAD IPL relative to controls. Six proteins were identified with increased levels in PCAD IPL relative to controls, while 3 proteins were identified as having lower levels (Table 3, Fig 2). Figure 3 displays expanded versions of each altered gel spot in PCAD and control IPL to more clearly show differences in spot intensity. MS/MS analysis of these spots and subsequent interrogation of protein databases identified the proteins with increased levels as cathepsin D, peptidyl prolyl cis-trans isomerase A1 (Pin1), fructose biphosphate aldolase (FBP), aconitase, Cu,Zn superoxide dismutase (SOD), and septin 11, while galectin 1, neurofilament protein-medium subunit, and cytochrome c oxidase had lower levels in PCAD IPL. A 2D gel map of spots and corresponding identities is shown in Figure 2 to convey correspondence between geographical spot location and approximate pI and molecular weight of these proteins in Table 3. A brief overview of these proteins, their respective functions, and expression trends in PCAD IPL relative to controls is given below.
Table 3.
Proteomics-determined differences in protein levels in PCAD IPL
| Protein | SwissProt Accession | Peptide Hits | p of id# | p of change% | Fold change | Regulation in PCAD | MW&/pl |
|---|---|---|---|---|---|---|---|
| Galectin-1 | P09832 | 13 | 1 e -11 | 0.019 | 0.22 | Down | 14/5.18 |
| Neurofilament medium subunit (NFM) | P07197 | 36 | 1 e -30 | 0.006 | 0.35 | Down | 102/4.75 |
| Aconitase (precursor) | Q99798 | 6 | 4 e -05 | 0.002 | 1.46 | Up | 85/7.32 |
| Fructose bisphosphate aldolase | P00441 | 6 | 3e -08 | 0.033 | 1.66 | Up | 48/7.98 |
| Cytochrome c oxidase (precursor) | P20674 | 4 | 4 e -09 | 0.033 | 0.23 | Down | 16/6.36 |
| Cathepsin D (precursor) | P07339 | 12 | 7 e -08 | 0.019 | 1.33 | Up | 45/6.10 |
| Cu, Zn SOD | P09772 | 3 | 1e -04 | 0.033 | 1.3 | Up | 16/5.68 |
| Peptidyl prolyl isomerase 1 (Pin1) | P62937 | 3 | 5 e -05 | 0.046 | 1.31 | Up | 18/7.82 |
| Septin 11 | Q9NVA2 | 4 | 3 e -09 | 0.047 | 1.64 | Up | 49/6.37 |
refers to probability associated with SEQUEST score
refers to p value of change in spot density between groups
approximate molecular weight, in kDa
Figure 2.
Representative two-dimensional gel map of proteins that show differential levels in PCAD IPL compared to control IPL. MS-identified spots showing significant alterations in PCAD relative to control are displayed in representative gels with corresponding identities.
Figure 3.
Enhanced images of identified proteins with altered levels in PCAD IPL relative to controls, to more clearly show reported changes in spot density.
Discussion
More than 5 million Americans are affected by AD, a number that is expected to more than triple by the year 2030 with the aging Baby Boomer generation. Patients with AD exhibit memory loss, with later progression to dementia and an extremely compromised quality of life. Biochemically, patients with AD have severe synaptic and neuronal loss (corresponding to decreased brain volumes relative to brains of subjects without dementia), as well as increased levels of two pathological hallmarks of the disease, SPs and NFTs. Oxidative stress has also been demonstrated in brains of patients with AD, MCI, and AD mouse models (Markesbery 1999, Butterfield et al. 2001). Our laboratory showed that a component of SPs, Aβ42, is capable of causing oxidative stress through free radical reactions, establishing a possible link between AD pathology and oxidative stress. A large body of evidence shows the toxicity of this species is predicated on its association with itself, as oligomers have been deemed toxic (Shankar et al. 2008), while monomers appear to be less toxic, and even in some studies, protective (Giuffrida et al. 2009).
There is a subclass of individuals who exhibit no memory alterations demonstrable on psychometric examination but at autopsy have demonstrative SP and NFT pathology. Several explanations have been proposed to possibly explain this anomaly in the scope of AD. One hypothesis is cognitive reserve, that some individuals with more synapses are capable of biochemical adjustment to pathology that protects the brain and, therefore, quality of life, from AD pathogenesis. Others opine that PCAD is a normal stage of AD progression, where, had the patients lived long enough, they would eventually have developed the MCI/AD diagnosis. While this latter scenario is a possibility, we posit that the former hypothesis can better explain PCAD based on our data and prevailing literature on this subject.
Subjects with PCAD have no significant brain cell loss or neuronal atrophy relative to healthy controls (Iacono et al. 2009, West et al. 2004). In our study, the mean brain weight of the PCAD group is almost identical to that of the control group, thus supporting this observation (Table I). Despite the fact that the 12 PCAD subjects exhibited significantly elevated levels of SPs and NFTs (Braak staging, Table I), Mini Mental State Examinaton scores were not different between controls and PCAD, and all PCAD patients scores were within the normal range (>25). It has been proposed that the preservation of brain cells in PCAD corresponds to better memory, while cell loss likely contributes to cognitive decline in MCI and AD. The connection between AD pathological hallmarks and brain cell death/dementia is still debatable, despite intense research effort. Because a growing body of evidence suggests that oxidative stress mediated by Aβ, possibly involving oligomerization of Aβ (Shankar et al. 2007, Walsh & Selkoe 2007, Haass & Selkoe 2007), is the propagator of cell death in AD (Butterfield et al. 2001, Markesbery 1999), we sought to investigate this phenomenon in the scope of PCAD.
In this study, we observed no significant differences between PCAD and control groups in any of the oxidative stress measurements (Fig 1). We suggest that the preservation of the oxidative status of proteins may contribute to the preservation of cellular vitality. Furthermore, as oxidative stress in AD has been viewed as a consequence of Aβ oligomerization, we observed significant increases in insoluble monomeric Aβ42 and total monomeric Aβ42, but not oligomeric Aβ; this is consistent with the notion that, although Aβ is increasingly produced in PCAD, excess oligomers are not formed, and therefore, do not cause toxicity associated with extensive oxidative modification.
Another aim of this study was to use proteomics to investigate which IPL proteins, if any, exhibit differences in levels in PCAD relative to controls without AD pathology. Here, our proteomics analysis revealed 9 brain proteins whose levels change in PCAD IPL relative to control. Many of these proteins and their trends in PCAD imply increased protection of intracellular components; however, these trends may also signify the earliest alterations of brain proteins in AD, before the onset of clinically observable symptoms. These proteins are involved in cytoskeletal maintenance, antioxidant defense, apoptosis, synaptic integrity, and energy metabolism. A brief overview of each protein and its contribution to a description of PCAD is given below.
Pin1 is a peptidyl-prolyl cis/trans isomerase that catalyzes the cis-trans isomerization of pThr-Pro and pSer-Pro residues of target proteins, including tau and APP (Schutkowski et al. 1998, Pastorino et al. 2006). This enzyme has also been shown to be necessary for cellular entry into mitosis (Lu et al. 1996, Shen et al. 1998). Pin1 is down-regulated and also oxidized in AD and MCI brain, resulting in decreased activity (Sultana et al. 2006, Butterfield et al. 2006). Overexpression of Pin1 can suppress tauopathy in transgenic animals (Lim et al. 2008) and reduce Aβ secretion in cultures (Pastorino et al. 2006). Here, we observed an increase in Pin1 levels in PCAD IPL relative to control; we suggest that the augmented levels of this protein in PCAD IPL signal a compensatory response to attempt to suppress increased levels of amyloid β-peptide, SP, and NFT.
Neurofilament proteins are a structural family of proteins found specifically in neurons. Three types of neurofilaments exist: light (~68 kDa), medium (~150kDa), and heavy (~210 kDa). Previously, a 94% decrease in expression of the gene coding for the medium neurofilament (NFM) subunit was detected in AD (Kittur et al. 1994). Because NFM is found in amyloid plaques in AD (Schmidt et al. 1991), we propose the decreased level of this protein observed in PCAD in the current study is the result of plaque localization and, therefore, resistant to solubility under SDS-PAGE conditions; it would be interesting to know if the gene level of NFM is lower in PCAD as in AD. Another possibility for our result is that the decreased level of this protein is an extremely early event in AD. If PCAD is a predisposition to eventual dementia, our data may reflect differences observed in CSF, as neurofilament proteins can be increased in AD patients (Elder et al. 1998, Rosengren et al. 1999).
Fructose bisphosphate aldolase (FBPA) and aconitase are enzymes involved in energy metabolism. FBPA converts fructose 1,6-biphosphate to glyceraldehyde 3-phosphate and dihydroxyacetone phosphate in glycolysis, while aconitase converts citrate to isocitrate in the tricarboxylic acid cycle. We previously observed significantly increased levels of FBPA in AD hippocampus relative to controls (Sultana et al. 2007a). A common feature of AD is reduced glucose metabolism, possibly due to the oxidation/down regulation of glycolytic or TCA enzymes; previous studies from our lab report significant increases in the oxidation of proteins involved with glycolysis and TCA cycle, and significant decreases in TCA enzyme levels in AD brain (Sultana et al. 2007a, Butterfield et al. 2007). In PCAD IPL, we observed significant increases in the levels of both FBPA and aconitase. Fluorodeoxyglucose positron emission tomography scanning in healthy individuals showed differences in glucose metabolism that correlated with AD biomarkers in CSF, suggesting that glucose changes occur before onset of clinical symptoms (Petrie et al. 2009), possibly reflecting changes in glucose metabolizing enzymes observed here in PCAD brain.
Galectins are carbohydrate-binding proteins with particular binding affinity for beta-galactosides. Galectins are thought to be synthesized on ribosomes in the cytosol (Liu 2002). Although multifunctional, a major action of galectin proteins is regulation of apoptosis. In this study, we observed significantly decreased levels of galectin-1 in PCAD IPL relative to controls. Other studies reported this protein to have a pro-apoptotic role, possibly through activation of extracellular-related kinase 2 (ERK-2) (Vespa et al. 1999), induction of AP-1 (Rabinovich et al. 2000), downregulation of anti-apoptotic Bcl-2 (Rabinovich et al. 2000), or activation of caspases (Rabinovich et al. 2002, Hsu et al. 2006). Based on this role, our result of decreased levels of pro-apoptotic galectin-1 appears to be consistent with the lack of cell loss of PCAD neurons relative to controls.
Septins are a family of GTP binding proteins that have roles in cytokinesis, membrane/synaptic remodeling, and cellular compartmentalization. Septin 11 plays a major role in dendritic arborization and GABAergic synaptic connectivity (Li et al. 2009b), and knockdown of septin 11 in hippocampal cell culture reduces dendritic arborization and decreases GABAergic synapse contacts these neurons receive (Li et al. 2009b). In AD, septin proteins are reportedly co-localized with NFTs (Kinoshita et al. 1998). In our study, we observed significantly increased levels of septin 11 in PCAD IPL relative to controls, which may be related to the neuronal hypertrophy observed in PCAD (Iacono et al. 2009), or increased synaptic plasticity of these neurons (O'Brien et al. 2009).
Cathepsin D has been previously implicated in AD, both at the protein and genetic levels. Cathepsin D is an aspartyl protease, highly concentrated in lysosomes for protein degradation. Cathepsin D was initially observed to have APP processing activity (Dreyer et al. 1994, Evin et al. 1995, Higaki et al. 1996); however, it has also been reported that murine neurons lacking cathepsin D had no effect on amyloidogenic processing (Saftig et al. 1996). Cathepsin D has been observed in SPs (Cataldo & Nixon 1990), and its mRNA has been observed to be increased in AD (Cataldo et al. 1995), although another study found no differences between AD and control brain levels of cathepsin D (Kohnken et al. 1995). In APP/PS1 transgenic mice, cathepsin D load increased with Aβ deposition, an observation that may contribute to the explanation of why levels of this protein were increased in our study (Howlett et al. 2008). Furthermore, our result may reflect decreased levels of this protein found in CSF in AD (Castano et al. 2006); this possibility remains to be seen, however, and more studies are needed to decisively address this point.
Cu, Zn SOD is one of the more highly expressed endogenous antioxidant proteins. This enzyme catalyzes the dismutation of superoxide (O2.−) to H2O2 and O2. Levels of Cu, Zn SOD mRNA were significantly increased in AD IPL relative to controls (Aksenov et al. 1998). In the current study, Cu, Zn SOD protein level was also increased in PCAD IPL relative to controls (Fig 2). We previously showed that the expression level of this protein was increased in canines with behavioral enrichment, as well as behavioral enrichment combined with enriched antioxidant diet (Opii et al. 2008). In the same study, black/white reversal behavior and spatial learning correlated directly with Cu, Zn SOD levels (Opii et al. 2008). Therefore, if results from the canine study (Opii et al. 2008) are translatable to humans, the levels of this enzyme and subsequent scavenging of O2.− radicals may be strongly correlated with memory preservation in PCAD.
Cytochrome c oxidase (COX), or complex IV, a protein involved in the electron transport chain on the inner membrane of the mitochondria, transfers electrons to oxygen in the mitochondrial matrix for reduction to water to power the proton gradient for ATP production. Studies in AD reported decreased COX efficiency in vulnerable AD brain regions (Kish et al. 1992, Mutisya et al. 1994). Here, we observed decreased levels of this protein in PCAD IPL compared to controls. Interestingly, COX deficiency can decrease oxidative damage, amyloid plaques, beta secretase activity, and Aβ42 levels in neurons (Fukui et al. 2007), possibly contributing to memory and cellular conservation in PCAD.
In summary, our data support the notion that in PCAD, resistance to AD-related dementia exists. No difference in oxidative stress, but differences in the levels of brain proteins in PCAD compared to control subjects, suggest PCAD individuals may compensate for increased AD-like pathology to preserve cognitive function. Bradley et al. (2010) also showed no significant differences in protein carbonyls and protein-bound acrolein in hippocampal/parahippocampal gyrus (HPG), superior and medial temporal gyrus (SMTG), or cerebellum from PCAD patients, consistent with our data presented here from PCAD IPL (Bradley et al. 2010). However, the same report showed significantly increased protein-bound HNE only in HPG, but not the other regions examined (Bradley et al. 2010). No differences in protein-bound HNE were observed here in PCAD IPL, possibly suggesting (and supported by the previous report) a regional disparity in either formation of or susceptibility to HNE and HNE-related protein modifications.
Unfortunately, a large caveat to working with PCAD autopsy-obtained samples is the lack of knowledge as to future cognitive status, i.e., whether these people would have eventually developed MCI/AD. Continued studies of PCAD patients (antemortem) using imaging and biomarker techniques may be able to address this issue. Nevertheless, our data may reveal a period of protection from neurodegeneration consistent with the clinical phenotype of PCAD.
Acknowledgements
This work was supported in part by NIH grants to DAB [AG-05119]. We are grateful to the Clinical and Neuropathology Cores of the University of Kentucky Alzheimer's Disease Clinical Center for providing well characterized tissue samples from volunteers in the longitudinal normal aging study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aksenov MY, Tucker HM, Nair P, Aksenova MV, Butterfield DA, Estus S, Markesbery WR. The expression of key oxidative stress-handling genes in different brain regions in Alzheimer's disease. J Mol Neurosci. 1998;11:151–164. doi: 10.1385/JMN:11:2:151. [DOI] [PubMed] [Google Scholar]
- Aluise CD, Sowell RA, Butterfield DA. Peptides and proteins in plasma and cerebrospinal fluid as biomarkers for the prediction, diagnosis, and monitoring of therapeutic efficacy of Alzheimer's disease. Biochim Biophys Acta. 2008;1782:549–558. doi: 10.1016/j.bbadis.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley MA, Markesbery WR, Lovell MA. Increased levels of 4-hydroxynonenal and acrolein in the brain in preclinical Alzheimer's disease (PCAD) Free Radic Biol Med. 2010 doi: 10.1016/j.freeradbiomed.2010.02.016. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Drake J, Pocernich C, Castegna A. Evidence of oxidative damage in Alzheimer's disease brain: central role for amyloid beta-peptide. Trends Mol Med. 2001;7:548–554. doi: 10.1016/s1471-4914(01)02173-6. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Galvan V, Lange MB, et al. In vivo oxidative stress in brain of Alzheimer disease transgenic mice: Requirement for methionine 35 in amyloid beta-peptide of APP. Free Radic Biol Med. 2010;48:136–144. doi: 10.1016/j.freeradbiomed.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Poon HF, St Clair D, Keller JN, Pierce WM, Klein JB, Markesbery WR. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer's disease. Neurobiol Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Reed TT, Perluigi M, De Marco C, Coccia R, Keller JN, Markesbery WR, Sultana R. Elevated levels of 3-nitrotyrosine in brain from subjects with amnestic mild cognitive impairment: implications for the role of nitration in the progression of Alzheimer's disease. Brain Res. 2007;1148:243–248. doi: 10.1016/j.brainres.2007.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Stadtman ER. Protein oxidation processes in aging brain. Adv. Cell Aging Gerontol. 1997;2:161–191. [Google Scholar]
- Butterfield DA, Sultana R. Redox proteomics identification of oxidatively modified brain proteins in Alzheimer's disease and mild cognitive impairment: insights into the progression of this dementing disorder. J Alzheimers Dis. 2007;12:61–72. doi: 10.3233/jad-2007-12107. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Sultana R. Redox proteomics: understanding oxidative stress in the progression of age-related neurodegenerative disorders. Expert Rev Proteomics. 2008;5:157–160. doi: 10.1586/14789450.5.2.157. [DOI] [PubMed] [Google Scholar]
- Castano EM, Roher AE, Esh CL, Kokjohn TA, Beach T. Comparative proteomics of cerebrospinal fluid in neuropathologically-confirmed Alzheimer's disease and non-demented elderly subjects. Neurol Res. 2006;28:155–163. doi: 10.1179/016164106X98035. [DOI] [PubMed] [Google Scholar]
- Cataldo AM, Barnett JL, Berman SA, Li J, Quarless S, Bursztajn S, Lippa C, Nixon RA. Gene expression and cellular content of cathepsin D in Alzheimer's disease brain: evidence for early up-regulation of the endosomal-lysosomal system. Neuron. 1995;14:671–680. doi: 10.1016/0896-6273(95)90324-0. [DOI] [PubMed] [Google Scholar]
- Cataldo AM, Nixon RA. Enzymatically active lysosomal proteases are associated with amyloid deposits in Alzheimer brain. Proc Natl Acad Sci U S A. 1990;87:3861–3865. doi: 10.1073/pnas.87.10.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P, Howard V, Loosbrock N, Dickson D, Murphy MP, Golde TE. Amyloid-beta immunization effectively reduces amyloid deposition in FcRgamma−/− knock-out mice. J Neurosci. 2003;23:8532–8538. doi: 10.1523/JNEUROSCI.23-24-08532.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer RN, Bausch KM, Fracasso P, et al. Processing of the pre-beta-amyloid protein by cathepsin D is enhanced by a familial Alzheimer's disease mutation. Eur J Biochem. 1994;224:265–271. doi: 10.1111/j.1432-1033.1994.00265.x. [DOI] [PubMed] [Google Scholar]
- Elder GA, Friedrich VL, Jr., Bosco P, Kang C, Gourov A, Tu PH, Lee VM, Lazzarini RA. Absence of the mid-sized neurofilament subunit decreases axonal calibers, levels of light neurofilament (NF-L), and neurofilament content. J Cell Biol. 1998;141:727–739. doi: 10.1083/jcb.141.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evin G, Cappai R, Li QX, Culvenor JG, Small DH, Beyreuther K, Masters CL. Candidate gamma-secretases in the generation of the carboxyl terminus of the Alzheimer's disease beta A4 amyloid: possible involvement of cathepsin D. Biochemistry. 1995;34:14185–14192. doi: 10.1021/bi00043a024. [DOI] [PubMed] [Google Scholar]
- Fukui H, Diaz F, Garcia S, Moraes CT. Cytochrome c oxidase deficiency in neurons decreases both oxidative stress and amyloid formation in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:14163–14168. doi: 10.1073/pnas.0705738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula C, Wu CK, Saroff D, Lorenzo A, Yuan M, Yankner BA. Aging renders the brain vulnerable to amyloid beta-protein neurotoxicity. Nat Med. 1998;4:827–831. doi: 10.1038/nm0798-827. [DOI] [PubMed] [Google Scholar]
- Giuffrida ML, Caraci F, Pignataro B, et al. Beta-amyloid monomers are neuroprotective. J Neurosci. 2009;29:10582–10587. doi: 10.1523/JNEUROSCI.1736-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Schlossmacher MG, Hung AY, et al. Amyloid beta-peptide is produced by cultured cells during normal metabolism. Nature. 1992;359:322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Higaki J, Catalano R, Guzzetta AW, Quon D, Nave JF, Tarnus C, D'Orchymont H, Cordell B. Processing of beta-amyloid precursor protein by cathepsin D. J Biol Chem. 1996;271:31885–31893. doi: 10.1074/jbc.271.50.31885. [DOI] [PubMed] [Google Scholar]
- Howlett DR, Bowler K, Soden PE, et al. Abeta deposition and related pathology in an APP × PS1 transgenic mouse model of Alzheimer's disease. Histol Histopathol. 2008;23:67–76. doi: 10.14670/HH-23.67. [DOI] [PubMed] [Google Scholar]
- Hsu DK, Yang RY, Liu FT. Galectins in apoptosis. Methods Enzymol. 2006;417:256–273. doi: 10.1016/S0076-6879(06)17018-4. [DOI] [PubMed] [Google Scholar]
- Iacono D, Markesbery WR, Gross M, Pletnikova O, Rudow G, Zandi P, Troncoso JC. The Nun study: clinically silent AD, neuronal hypertrophy, and linguistic skills in early life. Neurology. 2009;73:665–673. doi: 10.1212/WNL.0b013e3181b01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Kinoshita M, Akiyama H, Tomimoto H, Akiguchi I, Kumar S, Noda M, Kimura J. Identification of septins in neurofibrillary tangles in Alzheimer's disease. Am J Pathol. 1998;153:1551–1560. doi: 10.1016/S0002-9440(10)65743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kish SJ, Bergeron C, Rajput A, Dozic S, Mastrogiacomo F, Chang LJ, Wilson JM, DiStefano LM, Nobrega JN. Brain cytochrome oxidase in Alzheimer's disease. J Neurochem. 1992;59:776–779. doi: 10.1111/j.1471-4159.1992.tb09439.x. [DOI] [PubMed] [Google Scholar]
- Kittur S, Hoh J, Endo H, Tourtellotte W, Weeks BS, Markesbery W, Adler W. Cytoskeletal neurofilament gene expression in brain tissue from Alzheimer's disease patients. I. Decrease in NF-L and NF-M message. J Geriatr Psychiatry Neurol. 1994;7:153–158. doi: 10.1177/089198879400700305. [DOI] [PubMed] [Google Scholar]
- Kohnken RE, Ladror US, Wang GT, Holzman TF, Miller BE, Krafft GA. Cathepsin D from Alzheimer's-diseased and normal brains. Exp Neurol. 1995;133:105–112. doi: 10.1006/exnr.1995.1013. [DOI] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- LeVine H., 3rd Alzheimer's beta-peptide oligomer formation at physiologic concentrations. Anal Biochem. 2004;335:81–90. doi: 10.1016/j.ab.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Li S, Hong S, Shepardson NE, Walsh DM, Shankar GM, Selkoe D. Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron. 2009a;62:788–801. doi: 10.1016/j.neuron.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Serwanski DR, Miralles CP, Nagata K, De Blas AL. Septin 11 is present in GABAergic synapses and plays a functional role in the cytoarchitecture of neurons and GABAergic synaptic connectivity. J Biol Chem. 2009b;284:17253–17265. doi: 10.1074/jbc.M109.008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Balastik M, Lee TH, et al. Pin1 has opposite effects on wild-type and P301L tau stability and tauopathy. J Clin Invest. 2008;118:1877–1889. doi: 10.1172/JCI34308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FT. Galectins: novel anti-inflammatory drug targets. Expert Opin Ther Targets. 2002;6:461–468. doi: 10.1517/14728222.6.4.461. [DOI] [PubMed] [Google Scholar]
- Lovell MA, Markesbery WR. Oxidative damage in mild cognitive impairment and early Alzheimer's disease. J Neurosci Res. 2007;85:3036–3040. doi: 10.1002/jnr.21346. [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- Lyubartseva G, Smith JL, Markesbery WR, Lovell MA. Alterations of Zinc Transporter Proteins ZnT-1, ZnT-4 and ZnT-6 in Preclinical Alzheimer's Disease Brain. Brain Pathol. 2009 doi: 10.1111/j.1750-3639.2009.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markesbery WR. The role of oxidative stress in Alzheimer disease. Arch Neurol. 1999;56:1449–1452. doi: 10.1001/archneur.56.12.1449. [DOI] [PubMed] [Google Scholar]
- Markesbery WR, Kryscio RJ, Lovell MA, Morrow JD. Lipid peroxidation is an early event in the brain in amnestic mild cognitive impairment. Ann Neurol. 2005;58:730–735. doi: 10.1002/ana.20629. [DOI] [PubMed] [Google Scholar]
- Markesbery WR, Lovell MA. Damage to lipids, proteins, DNA, and RNA in mild cognitive impairment. Arch Neurol. 2007;64:954–956. doi: 10.1001/archneur.64.7.954. [DOI] [PubMed] [Google Scholar]
- McGowan E, Pickford F, Kim J, et al. Abeta42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 2005;47:191–199. doi: 10.1016/j.neuron.2005.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP, Beckett TL, Ding Q, Patel E, Markesbery WR, St Clair DK, LeVine H, 3rd, Keller JN. Abeta solubility and deposition during AD progression and in APPxPS-1 knock-in mice. Neurobiol Dis. 2007;27:301–311. doi: 10.1016/j.nbd.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Mutisya EM, Bowling AC, Beal MF. Cortical cytochrome oxidase activity is reduced in Alzheimer's disease. J Neurochem. 1994;63:2179–2184. doi: 10.1046/j.1471-4159.1994.63062179.x. [DOI] [PubMed] [Google Scholar]
- O'Brien RJ, Resnick SM, Zonderman AB, et al. Neuropathologic Studies of the Baltimore Longitudinal Study of Aging (BLSA) J Alzheimers Dis. 2009 doi: 10.3233/JAD-2009-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opii WO, Joshi G, Head E, Milgram NW, Muggenburg BA, Klein JB, Pierce WM, Cotman CW, Butterfield DA. Proteomic identification of brain proteins in the canine model of human aging following a long-term treatment with antioxidants and a program of behavioral enrichment: relevance to Alzheimer's disease. Neurobiol Aging. 2008;29:51–70. doi: 10.1016/j.neurobiolaging.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L, Sun A, Lu PJ, et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528–534. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- Petrie EC, Cross DJ, Galasko D, Schellenberg GD, Raskind MA, Peskind ER, Minoshima S. Preclinical evidence of Alzheimer changes: convergent cerebrospinal fluid biomarker and fluorodeoxyglucose positron emission tomography findings. Arch Neurol. 2009;66:632–637. doi: 10.1001/archneurol.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike CJ, Walencewicz AJ, Glabe CG, Cotman CW. In vitro aging of beta-amyloid protein causes peptide aggregation and neurotoxicity. Brain Res. 1991;563:311–314. doi: 10.1016/0006-8993(91)91553-d. [DOI] [PubMed] [Google Scholar]
- Price JL, McKeel DW, Jr., Buckles VD, et al. Neuropathology of nondemented aging: presumptive evidence for preclinical Alzheimer disease. Neurobiol Aging. 2009;30:1026–1036. doi: 10.1016/j.neurobiolaging.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich GA, Alonso CR, Sotomayor CE, Durand S, Bocco JL, Riera CM. Molecular mechanisms implicated in galectin-1-induced apoptosis: activation of the AP-1 transcription factor and downregulation of Bcl-2. Cell Death Differ. 2000;7:747–753. doi: 10.1038/sj.cdd.4400708. [DOI] [PubMed] [Google Scholar]
- Rabinovich GA, Ramhorst RE, Rubinstein N, Corigliano A, Daroqui MC, Kier-Joffe EB, Fainboim L. Induction of allogenic T-cell hyporesponsiveness by galectin-1-mediated apoptotic and non-apoptotic mechanisms. Cell Death Differ. 2002;9:661–670. doi: 10.1038/sj.cdd.4401009. [DOI] [PubMed] [Google Scholar]
- Reed T, Perluigi M, Sultana R, Pierce WM, Klein JB, Turner DM, Coccia R, Markesbery WR, Butterfield DA. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer's disease. Neurobiol Dis. 2008a;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Reed TT, Owen J, Pierce WM, Sebastian A, Sullivan PG, Butterfield DA. Proteomic identification of nitrated brain proteins in traumatic brain-injured rats treated postinjury with gamma-glutamylcysteine ethyl ester: insights into the role of elevation of glutathione as a potential therapeutic strategy for traumatic brain injury. J Neurosci Res. 2009;87:408–417. doi: 10.1002/jnr.21872. [DOI] [PubMed] [Google Scholar]
- Reed TT, Pierce WM, Jr., Turner DM, Markesbery WR, Butterfield DA. Proteomic identification of nitrated brain proteins in early Alzheimer's disease inferior parietal lobule. J Cell Mol Med. 2008b doi: 10.1111/j.1582-4934.2008.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren LE, Karlsson JE, Sjogren M, Blennow K, Wallin A. Neurofilament protein levels in CSF are increased in dementia. Neurology. 1999;52:1090–1093. doi: 10.1212/wnl.52.5.1090. [DOI] [PubMed] [Google Scholar]
- Saftig P, Peters C, von Figura K, Craessaerts K, Van Leuven F, De Strooper B. Amyloidogenic processing of human amyloid precursor protein in hippocampal neurons devoid of cathepsin D. J Biol Chem. 1996;271:27241–27244. doi: 10.1074/jbc.271.44.27241. [DOI] [PubMed] [Google Scholar]
- Schmidt ML, Lee VM, Trojanowski JQ. Comparative epitope analysis of neuronal cytoskeletal proteins in Alzheimer's disease senile plaque neurites and neuropil threads. Lab Invest. 1991;64:352–357. [PubMed] [Google Scholar]
- Schutkowski M, Bernhardt A, Zhou XZ, Shen M, Reimer U, Rahfeld JU, Lu KP, Fischer G. Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry. 1998;37:5566–5575. doi: 10.1021/bi973060z. [DOI] [PubMed] [Google Scholar]
- Seubert P, Vigo-Pelfrey C, Esch F, et al. Isolation and quantification of soluble Alzheimer's beta-peptide from biological fluids. Nature. 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- Shankar GM, Bloodgood BL, Townsend M, Walsh DM, Selkoe DJ, Sabatini BL. Natural oligomers of the Alzheimer amyloid-beta protein induce reversible synapse loss by modulating an NMDA-type glutamate receptor-dependent signaling pathway. J Neurosci. 2007;27:2866–2875. doi: 10.1523/JNEUROSCI.4970-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar GM, Li S, Mehta TH, et al. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Stukenberg PT, Kirschner MW, Lu KP. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 1998;12:706–720. doi: 10.1101/gad.12.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Golde TE, Ghiso J, et al. Production of the Alzheimer amyloid beta protein by normal proteolytic processing. Science. 1992;258:126–129. doi: 10.1126/science.1439760. [DOI] [PubMed] [Google Scholar]
- Smith MA, Perry G. Alzheimer disease: protein-protein interaction and oxidative stress. Bol Estud Med Biol. 1996;44:5–10. [PubMed] [Google Scholar]
- Sultana R, Boyd-Kimball D, Cai J, Pierce WM, Klein JB, Merchant M, Butterfield DA. Proteomics analysis of the Alzheimer's disease hippocampal proteome. J Alzheimers Dis. 2007a;11:153–164. doi: 10.3233/jad-2007-11203. [DOI] [PubMed] [Google Scholar]
- Sultana R, Boyd-Kimball D, Poon HF, et al. Oxidative modification and down-regulation of Pin1 in Alzheimer's disease hippocampus: A redox proteomics analysis. Neurobiol Aging. 2006;27:918–925. doi: 10.1016/j.neurobiolaging.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Sultana R, Perluigi M, Butterfield DA. Oxidatively modified proteins in Alzheimer's disease (AD), mild cognitive impairment and animal models of AD: role of Abeta in pathogenesis. Acta Neuropathol. 2009;118:131–150. doi: 10.1007/s00401-009-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, Perluigi M, Newman SF, Pierce WM, Cini C, Coccia R, Butterfield A. Redox Proteomic Analysis of Carbonylated Brain Proteins in Mild Cognitive Impairment and Early Alzheimer's Disease. Antioxid Redox Signal. 2010;12:327–336. doi: 10.1089/ars.2009.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, Piroddi M, Galli F, Butterfield DA. Protein levels and activity of some antioxidant enzymes in hippocampus of subjects with amnestic mild cognitive impairment. Neurochem Res. 2008;33:2540–2546. doi: 10.1007/s11064-008-9593-0. [DOI] [PubMed] [Google Scholar]
- Sultana R, Reed T, Perluigi M, Coccia R, Pierce WM, Butterfield DA. Proteomic identification of nitrated brain proteins in amnestic mild cognitive impairment: a regional study. J Cell Mol Med. 2007b;11:839–851. doi: 10.1111/j.1582-4934.2007.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongboonkerd V, Luengpailin J, Cao J, Pierce WM, Cai J, Klein JB, Doyle RJ. Fluoride exposure attenuates expression of Streptococcus pyogenes virulence factors. J Biol Chem. 2002;277:16599–16605. doi: 10.1074/jbc.M200746200. [DOI] [PubMed] [Google Scholar]
- Vespa GN, Lewis LA, Kozak KR, Moran M, Nguyen JT, Baum LG, Miceli MC. Galectin-1 specifically modulates TCR signals to enhance TCR apoptosis but inhibit IL-2 production and proliferation. J Immunol. 1999;162:799–806. [PubMed] [Google Scholar]
- Walsh DM, Selkoe DJ. A beta oligomers - a decade of discovery. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- West MJ, Kawas CH, Stewart WF, Rudow GL, Troncoso JC. Hippocampal neurons in pre-clinical Alzheimer's disease. Neurobiol Aging. 2004;25:1205–1212. doi: 10.1016/j.neurobiolaging.2003.12.005. [DOI] [PubMed] [Google Scholar]