Abstract
It is commonly believed that neurons remain in G0 phase of the cell cycle indefinitely. Cell cycle reentry, however, is known to contribute to neuronal apoptosis. Moreover, recent evidence demonstrates the expression of cell cycle proteins in differentiated neurons under physiological conditions. Functional roles of such expression remain unclear. Since DNA repair is generally attenuated by differentiation in most cell types, cell cycle-associated events in postmitotic cells may reflect the need to reenter the cell cycle to activate DNA repair. We show that cyclin C-directed pRb-dependent G0 exit activates the non-homologous end joining pathway of DNA repair (NHEJ) in postmitotic neurons. Using RNA interference, we found that abrogation of cyclin C-mediated exit from G0 compromised DNA repair but did not initiate apoptosis. Forced G1 entry combined with prevention of G1→S progression triggered NHEJ activation even in the absence of DNA lesions, but did not induce apoptosis in contrast to unrestricted progression through G1→S. We conclude that G0→G1 transition is functionally significant for NHEJ repair in postmitotic neurons. These findings reveal the importance of cell cycle activation for controlling both DNA repair and apoptosis in postmitotic neurons and underline the particular role of G1→S progression in apoptotic signaling, providing new insights into the mechanisms of DNA damage response in postmitotic neurons.
Keywords: neuron, DNA repair, cell cycle, DNA damage, apoptosis
Introduction
Due to continuous exposure to genotoxic stress resulting from exogenous and endogenous sources, protection of genomic integrity is a major challenge for living cells. Failure to repair DNA lesions often leads to cell death, genomic instability, and tumorigenesis [1, 2]. Terminally differentiated neurons are highly susceptible to oxidative DNA damage due to their high rate of oxidative metabolism [3]. For this reason, DNA repair is highly important for these cells [2, 4].
The mechanisms of DNA repair has been investigated mainly in proliferating cells. In these cells, the cell cycle machinery is a part of the DNA damage response (DDR), involved in both DNA repair and apoptosis [5]. DNA repair in terminally differentiated cells is not expected to be linked to the cell cycle. However, increasing evidence indicates that the cell cycle machinery plays a key role in different processes in terminally differentiated cells [6–11]; for example, cell cycle activation is essential for apoptotic signaling in neurons [3, 12, 13]. Expression of cell cycle proteins has been observed at physiological conditions in terminally differentiated neurons [11], but the functional relevance of this expression remains unknown. Since DNA repair is generally attenuated by differentiation in most types of cells [14–16], the expression of cell cycle-related proteins may reflect the need of resting cells to reenter the cell cycle in order to activate DNA repair. Our recent data demonstrating that cell cycle-related proteins are expressed in neurons exposed to repairable DNA damage also support the existence of a link between the cell cycle machinery and DNA repair in postmitotic cells [17].
Although cells in most mammalian tissues enter a terminally arrested (G0) phase at some point during their life, the mechanisms regulating the maintenance of G0 and G0→G1 transition are not fully understood. The retinoblastoma protein (pRb) plays a crucial role in cell cycle regulation [18]. Recently, the cyclin C/CDK3 complex was shown to facilitate exit from G0 into the cell cycle by pRb phosphorylation at serines (S) 807 and 811. This cyclin C-associated kinase activity peaked shortly after mitogenic stimulation of quiescent cells in early G1 [19]. During exit from G0, cyclin C/CDK3 complex directed pRb phosphorylation at S807/811 in a temporal pattern that preceded pRb phosphorylation by cyclin D/CDK4, cyclin D/CDK6, and cyclin E/CDK2 [19]. The levels of cyclin E protein and associated kinase activity rise in late G1 phase [20].
Because DSBs are the most lethal form of DNA damage [21], maintenance of genomic integrity depends on an efficient and accurate DSB repair. Mammalian cells repair DSBs by two pathways: homologous recombination and non-homologous end-joining (NHEJ). NHEJ is predominant in mammalian cells [22]. NHEJ starts with the binding of Ku70/Ku80 (Ku) heterodimer to the broken DNA ends. Ku facilitates recruitment of Artemis-DNA–protein kinase catalytic subunit (PKcs) complex, which processes the ends to prepare them for ligation [22].
In this study, we found that the activation of NHEJ in postmitotic neurons was accompanied by phosphorylation of pRb at S807/811 directed by cyclin C-associated kinase activity, which was previously shown to be sufficient for G0→G1 transition [19]. The abrogation of cell cycle entry compromised NHEJ repair, while forcing G1 entry caused NHEJ activation even in the absence of DNA lesions. Together, these resultssuggest the need for resti ng cells to re-enter the cell cycle to activate the NHEJ repair machinery.
Results
5 μM H2O2 generates repairable DSBs in postmitotic neurons
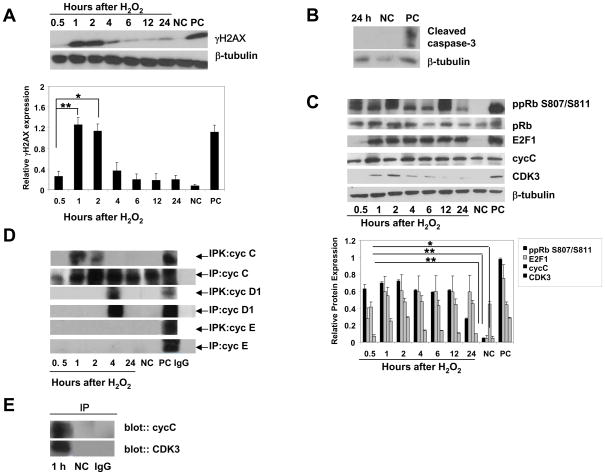
To investigate the effect of non-lethal DSBs on neurons, we exposed postmitotic rat cortical neurons to repairable DNA damage generated by hydrogen peroxide (H2O2). H2O2, a product of normal oxygen metabolism, extensively used to induce oxidative stress in cell culture cell models and is known to induce DSBs [17, 21]. Cultures of cortical neurons according to our previous results yielded ~ 99% pure neuronal populations. The purity of neuronal populations was controlled by the expression of specific neuronal marker, NeuN (data not shown). Previously, we demonstrated that 5 μM H2O2 induces repairable DSBs and does not induce apoptosis in postmitotic cortical neurons [17]. We used phosphorylated H2AX (γH2AX) as a marker of DSB formation [17]. The kinetics of γH2AX accumulation demonstrates the repairable character of DSBs induced by 5 μM H2O2, as illustrated by a significant increase in γH2AX expression compared with untreated neurons, followed by its significant reduction (Fig. 1A). The analysis of apoptotic markers, including cleavage of caspase 3 (Fig. 1B) and examination of apoptotic neuron nuclei (data not shown), revealed that by 24 h after exposure to 5 μM H2O2, the neuronal cultures did not undergo apoptosis, consistent with our data on γH2AX expression. Previously, we demonstrated the repairable character of generated by 5 μM H2O2 DSBs using comet assay and by testing apoptotic markers, cleavage of nuclear (Mcm3) and cytoplasmic (β-actin) caspase 3 substrates [17].
Figure 1. DSB induction is associated with cell cycle entry of postmitotic cortical neurons.
(A, B) Immunoblot analysis of γH2AX and apoptotic caspase-3 cleavage in neurons treated with 5 μM H2O2. Negative control (NC), untreated cultures, positive control (PC), extract from staurosporine-treated Jurkat cells. The values are the means and SD (n = 5); *p < 0.002; **p < 0.001.
(C) Immunoblot analysis of the expression of cell cycle-related proteins in neurons exposed to 5 μM H2O2. Negative control (NC), untreated neurons, positive control (PC), extracts from proliferating HeLa cells. The values are the means and SD (n = 4); *p < 0.002; **p < 0.001 compared with corresponding NC.
(D) Lysates from cortical neurons were prepared at the indicated times and subjected to immune precipitation (IP) using anticyclin C, anticyclin D1, and anticyclin E. Precipitates were tested for in vitro kinase activity using Rb 769 as substrate (IPK). Negative control (NC), untreated neurons; positive control (PC), proliferating HeLa cells. Isospecific control (IgG), normal rabbit or mouse IgG.
(E) Lysates from cortical neurons treated with 5 μM H2O2 were subjected to immune precipitation followed by immune blot analysis. Anti-cyclin C precipitates were analyzed by anti-CDK3 antibody and anti-CDK3 precipitates were analyzed by anticyclin C antibody. Normal rabbit IgG (IgG) was used as a control for both anti-CDK3 and cyclin C antibodies.
G0→G1 transition in neurons exposed to DSBs
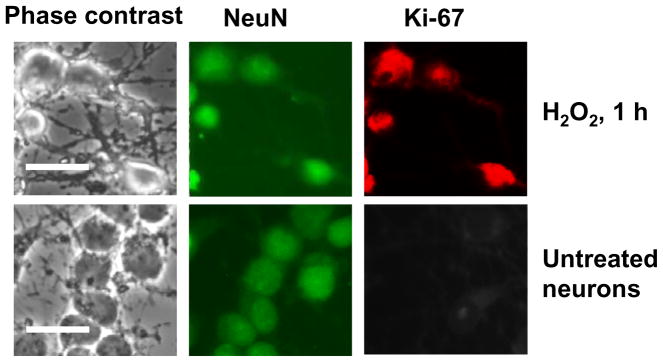
DSB induction was accompanied by early activation of cell cycle regulatory proteins, including phosphorylation of pRb at S807/811, required for G0 exit [19] and expression of transcription factor E2F1 (Fig. 1C). E2F1 gene expression is known to rise as cells exit from G0 and undergo G0→G1 transition [18]. Cyclin C levels were high in postmitotic neurons, consistent with those seen in G0-arrested cells of non-neuronal origin [19] and were not changed significantly after H2O2 treatment, although cyclin C levels were typically highest by 1 h after treatment. Cyclin C immune-precipitates from lysates of cortical neurons revealed pRb kinase activity at 1 and 2 h after H2O2 treatment (Fig. 1D). In contrast to cyclin C, its partner CDK3 was not expressed in G0 neurons but CDK3 levels significantly increased after H2O2 exposure (Fig. 1C). Coimmune precipitation experiments demonstrate that the anti-CDK3 precipitate contains cyclin C (Fig. 1E). pRb kinase activity associated with cyclin D1 was not seen until at least 4 h of treatment (Fig. 1D). No cyclin E-associated pRb kinase activity was found (Fig. 1D). Co-expression of the neuronal marker NeuN, and the marker of proliferating cells Ki-67, [17], was observed in neurons by 1 h of H2O2 exposure but not in untreated neurons (Fig. 2), confirming the entrance of postmitotic neurons into the cell cycle upon non-lethal DSB DNA damage. This suggests that the response to DSB formation in postmitotic neurons is associated with cyclin C-directed pRb phosphorylation that precedes pRb phosphorylation directed by cyclin D1, but is not associated with cyclin E-dependent pRb kinase activity (late G1). Such early pRb phosphorylation at S807/811 is consistent with previously observed pRb phosphorylation at S807/811 in T lymphocytes and T98G cells during exit from G0 [19, 23].
Figure 2. The co-expression of cell cycle and neuronal markers in neurons at 1 h of H2O2 exposure.
Co-expression of neuronal marker NeuN and a marker of active cell cycle, Ki-67 were detected by immunocytochemistry. Scale bars are 50 μm.
Postmitotic neurons activate NHEJ upon DNA DSB damage
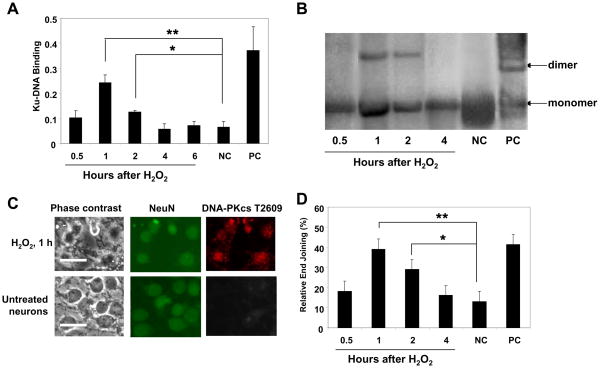
Next, we examined NHEJ activation in postmitotic neurons exposed to 5 μM H2O2. Since Ku-DNA binding is crucial for NHEJ-mediated repair [22], we first assessed the DNA-binding activity of Ku proteins in neuronal extracts by immune recognition of an epitope on Ku70 or Ku86 that is accessible upon DNA binding [24]. The analysis of NHEJ activity was performed based on in vitro end-joining of linearized plasmid DNA using cell-free extracts [25]. This DNA-end ligation assay, which results in the formation of DNA concatemers, requires a functional NHEJ apparatus, as seen in the absence of DNA oligomers when using extract from untreated (undamaged) neurons (Fig. 3B). Both Ku-DNA binding and end joining kinetics revealed an early NHEJ activation (at 1–2 h after H2O2 exposure), followed by a decrease of both parameters to the levels seen in untreated cells (Fig. 3A, B). To relate NHEJ activation to neurons, we assessed the co-expression of neuronal marker NeuN and DNA-PKcs phosphorylated at Threonine (T) 2609 as a marker of activated NHEJ. As previously described, DNA-PKcs phosphorylation at this site is required for NHEJ activation, and mutations at the T2609 cluster severely compromised the ability of DNA-PKcs to restore DSB repair defects in DNA-PKcs-deficient cells [26, 27]. Results of immunofluorescence analysis revealed the co-localization of both neuronal and NHEJ activation markers in cells at 1 h of H2O2 exposure but not in untreated cells (Fig. 3C). These data show that early and brief NHEJ activity is sufficient for DSB repair in postmitotic neurons exposed to a non-lethal DNA-damaging insult. Importantly, this timing of NHEJ activation coincided tightly with the timing of cyclin C-associated pRb kinase activity (Fig. 1D), essential for G0→G1transition in postmitotic cells. Interestingly, pRb kinase activity associated with cyclin D1 was observed outside of the time frame of NHEJ activity (at 4 h after H2O2 exposure), although this activity was indeed induced in postmitotic neurons after DSBs, in contrast to cyclin E-dependent pRb kinase activity, which was not induced (Fig. 1D).
Figure 3. NHEJ activation in postmitotic neurons exposed to 5 μM H2O2.
(A) Ku-DNA binding analysis of cortical neurons exposed to 5 μM H2O2. Negative control (NC), untreated cultures; positive control (PC), the Raji nuclear extract. The values are the means and SD (n = 4); *p < 0.02; **p < 0.005.
(B) The NHEJ analysis of cortical neurons exposed to 5 μM H2O2. DNA substrate was prepared by linearizing pGL3 basic was added to the NHEJ reaction. Products were subjected to electrophoresis (0.8% agarose gel) and Southern blot, hybridized with the 32P-labeled plasmid probe and visualized by autoradiography. Negative control (NC), untreated cultures; positive control (PC), T4 ligase treated linearized plasmid DNA (pGL3 basic/XhoI). The values are the means and SD (n = 4); *p < 0.05; **p < 0.002.
(C). Co-expression of neuronal marker NeuN and phosphorylated at Thr 2609 DNA-PKcs as a marker of activated NHEJ repair was detected by immunofluorescence. Scale bars are 50 μm.
The suppression of G0→G1 transition attenuates NHEJ activation
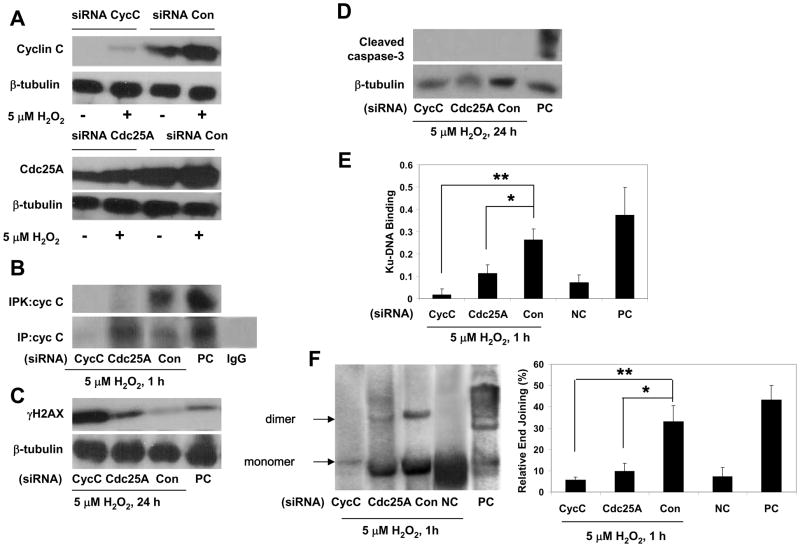
To determine the functional role of cell cycle activation and particular roles for cyclin C- and cyclin D-associated phosphorylation of pRb in DSB repair in postmitotic neurons, we prevented G0→G1 transition by silencing cyclin C or Cdc25A. Cyclin C suppression was previously shown to affect G0 exit [19]. Cdc25A is a member of the cell division cycle 25 (Cdc25) family of proteins that function by dephosphorylating CDK subunits, and thereby activating CDK/cyclin complex. Cdc25A is involved in the activation of cyclin E- and cyclin D1-associated kinase activities and pRb phosphorylation [28]. This suggests that Cdc25A suppression should not affect cyclin C-associated pRb phosphorylation which precedes pRb phosphorylation by cyclin D/CDK4/6 and cyclin E/CDK2.
The suppression of cyclin C in cortical neurons by cyclin C siRNA resulted in decreased levels of cyclin C protein and an almost complete blockade of cyclin C-associated pRb kinase activity (Fig. 4A, B), as well as cyclin D1-associated pRb kinase activity (data not shown). Cyclin C silencing compromised DSB repair in neurons exposed to 5 μM H2O2, in contrast to neurons transfected with control siRNA, as evidenced by the attenuation of both Ku-DNA binding and end-joining activities (Fig. 4E, F). The lack of decrease in γH2AX levels in neurons transfected with cyclin C siRNA that were treated with 5 μM H2O2 (Fig. 4C) suggests that these neurons failed to repair DSBs and supports the hypothesis that NHEJ is impaired by silencing cyclin C. Interestingly, compromised DSB repair in these neurons did not lead to apoptosis, as evidenced by the absence of caspase 3 cleavage. Basically the same pattern of change in NHEJ activity was observed in postmitotic neurons after reducing Cdc25A levels, although the reduction of NHEJ activity tended to be lower than in cells with suppressed cyclin C (Fig. 4E, F). This may reflect a smaller suppression of Cdc25A protein expression. Surprisingly, Cdc25A suppression affected cyclin C-associated pRb kinase activity (Fig. 4B), suggesting that Cdc25A is involved not only in cyclin E- and cyclin D1-associated kinase activities [28], but is also involved in cyclin C-associated kinase activity.
Figure 4. Blockade of cell cycle entry attenuates NHEJ activation in postmitotic neurons.
(A, B) Cortical neurons transfected either with cyclin C (cycC), Cdc25A or control (Con) siRNA were exposed to 5 μM H2O2 for the indicated durations and analyzed by immunoblotting for the expression of cyclin C and Cdc25A or by immune precipitation (IP) with anticyclin C antibody and following in vitro kinase activity assay using Rb679 as substrate (IPK). Positive control (PC), lysates from proliferating HeLa cells. Isospefic control for anticyclin C (IgG), normal rabbit IgG.
(C, D) Immunoblot analysis of DSBs (γH2AX) and apoptotic caspase-3 cleavage in neurons transfected either with cyclin C (cycC), Cdc25A or control (Con) siRNA and exposed to 5 μM H2O2 for indicated durations. Positive control (PC), extracts from staurosporine-treated apoptotic Jurkat cells.
(E) Ku-DNA binding of cortical neurons transfected with either cyclin C (cycC), Cdc25A or control (Con) siRNA and exposed to 5 μM H2O2 (1 h). Negative control (NC), untreated untransfected cultures; positive control (PC), the nuclear extract of Raji cells. The values are the means and SD (n = 4); *p < 0.01; **p < 0.001.
(F) The NHEJ analysis of cortical neurons transfected with either cyclin C (cycC), Cdc25A or control (Con) siRNA and exposed to 5 μM H2O2. Negative control (NC), untreated cultures; positive control (PC), T4 ligase treated linearized plasmid DNA (pGL3 basic/XhoI). The values are the means and SD (n = 4); *p < 0.005; **p < 0.001.
Forced G1 entry causes NHEJ activation in the absence of DSB lesions
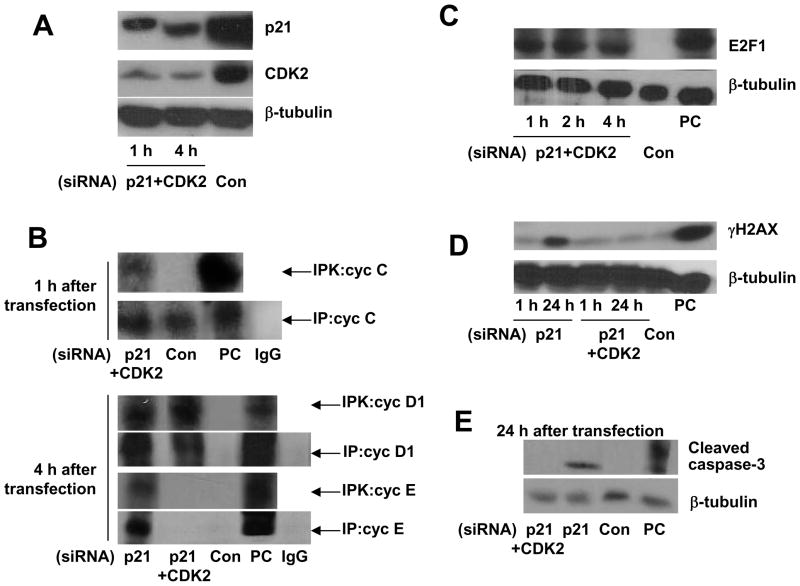
To further define the requirement of cell cycle reentry for NHEJ activation in postmitotic neurons, we examined the effects of forced cell cycle entry on NHEJ activation. To force cell cycle reactivation, we suppressed p21 expression by siRNA. p21 is a cyclin-dependent kinase inhibitor (CKI) which is also involved in DNA damage signaling and whose suppression by siRNA was previously shown to efficiently trigger DNA synthesis in primary, terminally differentiated skeletal muscle cells and quiescent fibroblasts [29, 30]. Reactivation of the cell cycle observed in quiescent cells upon over-expression of active cell cycle factors, E2F1 and Cdc25A, or upon suppression of p21 was accompanied by DNA replication and apoptosis [28, 29, 31, 32]. In our experiments, p21 siRNA, unlike a control siRNA, reduced p21 protein levels and led to entry into S phase, as demonstrated by the appearance of cyclin E-associated pRb kinase activity (Fig. 5A, B). Consistent with previous findings [29], postmitotic cortical neurons transfected with p21 siRNA underwent apoptosis by 24 h after transfection (Fig. 5E). To avoid the S phase entry and ensuing apoptosis and to determine the direct impact of G0→G1 transition on the DNA repair machinery in postmitotic neurons, we combinedp21 suppression with the targeting CDK2, critical for G1→S transition [18]. Since we observed cyclin E-associated pRb activity by 4 h and apoptosis by 24 h after p21 siRNA transfection (Fig. 5B, D, E), we decided to examine the expression of p21 and CDK2, as well as the expression of cell cycle markers (E2F1, cyclin C-, cyclin D1, and cyclin E-associated pRb kinase activities) in cortical neurons co-transfected with p21 and CDK2 as early as 1 h after transfection.
Figure 5. p21 silencing leads to S phase entry, while co-targeting p21 and CDK2 reactivates only G1.
(A) Cortical neurons were transfected with either cyclin p21 or control (Con) siRNA or co-transfected with p21 and CDK2 (p21/CDK2) siRNA. Expression of p21 and CDK3 was analyzed 1 and 4h after transfection.
(B) Cortical neurons were transfected either with p21, p21/CDK2 or control (Con) siRNA. In 1 and 4 h after transfection, lysates were subjected to immune precipitation (IP) using anti-cyclin C, anti-cyclin D1 or anti-cyclin E. Precipitates were tested for in vitro kinase activity using Rb679 as substrate (IPK). Positive control (PC), lysates from proliferating HeLa cells. Isospefic controls for anti-cyclin C, anti-cyclin D1 and anti-cyclin E (IgG), normal rabbit and mouse IgG.
(C) Immunoblot analysis of the expression of E2F1 in neurons transfection with p21/CDK2 or control (Con) siRNA. Positive control (PC), proliferating HeLa cells.
(D, E) Immunoblot analysis of γH2AX- or cleaved caspase-3 expression in neurons transfected with p21, p21/CDK2 or control (Con) siRNA. Positive control (PC), extract from staurosporine-treated apoptotic Jurkat cells. Note the appearance of γH2AX and the expression of cleaved caspase-3 only in cells transfected with p21, in contrast to neurons transfected with p21/CDK2 and control siRNA.
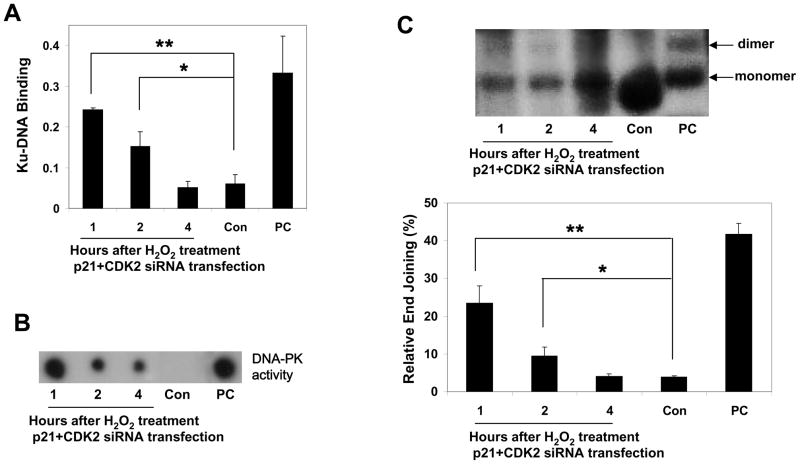
The specificity of the transfection was ascertained by the ability of siRNAs to lower p21- and CDK2 protein levels and to induce a massive early (by 1 h after transfection) re-entry of neurons into the cell cycle, but not S phase, as measured by E2F1 expression, cyclin C-, cyclin D1 but not cyclin E-associated pRb kinase activities, in contrast to the silencing of p21 alone (Fig. 5). The specificity of transfection was also supported by the absence of apoptosis usually associated with S phase entry (Fig. 5E), and DSBs (the lack of γH2AX expression which labels apoptotic cells) (Fig. 5D). To determine whether G0→G1 transition in postmitotic neurons is sufficient for initiation of NHEJ repair machinery, we determined the parameters of NHEJ activation in p21/CDK2 siRNA-transfected neurons. p21/CDK2 silencing initiated a significant increase in Ku-DNA binding and DNA-PK activity (Fig. 6A, B). DNA-PK activity assay is based on kinase activity analysis of whole DNA complex composed of two subunits, Ku and DNA-PKcs. We next determined whether G0→G1 transition induces end joining activation. NHEJ activity in p21/CDK2 siRNA-transfected neurons was significantly higher than in neurons transfected with control siRNA (Fig. 6C). Importantly, NHEJ activation was short (1 and 2 h after transfection) and coincided with the timing of increased cyclin C-associated pRb kinase activity (Fig. 5B), as it was observed in neurons exposed to 5 μM H2O2 (Fig. 1, 3).
Figure 6. Forcing neurons into the cell cycle activates NHEJ in postmitotic neurons.
(A) Ku-DNA binding analysis of cortical neurons transfected with p21/CDK2 or control (Con) siRNA. Positive control (PC), the nuclear extract of Raji cells. The values are the means and SD (n = 4); *p < 0.01; **p < 0.001.
(B) Cortical neurons were transfected with p21/CDK2 or control (Con) siRNA and after indicated time after transfection, DNA-PK activity was measured by kinase activity assay of whole DNA-PK complex. Positive control (PC), HeLa cells exposed to etoposide.
(C) The NHEJ analysis of cortical neurons transfected with either p21/CDK2 or control (Con) siRNA. Positive control (PC), T4 ligase treated linearized plasmid DNA (pGL3 basic/XhoI). The values are the means and SD (n = 4); *p <0.002; **p <0.001.
Discussion
The expression of various canonical cell cycle regulators in differentiated neurons of the adult brain observed under normal conditions suggests that they have a physiological function [6–11], but this function remains unknown. Given the attenuation of the DNA repair capacity following differentiation in most cell types [14–16], the expression of cell cycle-related proteins may reflect the need for resting cells to reenter the cell cycle to activate DNA repair. In mitotic cells, the cell cycle machinery is an important component of the cellular response to DNA damage, involved in the DNA repair and apoptosis [5]. We and others have demonstrated that cell cycle activation is essential for DNA damage-induced apoptosis [3, 12, 13]. Here, we show that kinase activity involved in the cell’s exit from G0, only recently established to be associated with cyclin C [19], is required for activation of NHEJ repair in postmitotic neurons. Both the increase in cyclin C-associated pRb kinase activity and NHEJ activation occurred shortly after cell exposure to H2O2 and showed close kinetics. The fact that these cells are undergoing G0→G1 transition is supported by our findings that E2F1 levels are elevated (this occurs as cells undergo G0→G1 transition [18]), and that pRb is phosphorylated at S807/811. Cyclin D1- (but not cyclin E) associated pRb kinase activity was also induced as a result of DSB DNA damage, although cyclin D1-associated pRb kinase activity occurred outside of the time frame of NHEJ activity, suggesting that cyclin D1-associated pRb kinase activity may not be implicated in NHEJ repair. Thus, activation of NHEJ repair in postmitotic neurons was associated with entering early G1, driven by cyclin C-associated pRb kinase activity, but it was not associated with late G1 progression directed by cyclin E-dependent pRb kinase activity.
The suppression of cyclin C by siRNA was shown to delay exit from G0 [19]. We found that ablation of cyclin C by siRNA significantly reduced cyclin C-associated pRb kinase activity induced by DNA damage, and attenuated NHEJ in neurons, as revealed by the increased levels of γH2AX. These changes, however, did not lead to apoptosis, suggesting that cell cycle activation is important for both DNA repair and apoptosis in postmitotic cells. Indeed, numerous studies have demonstrated that the suppression of cell cycle signaling leads to an attenuation of DNA damage-initiated neuronal apoptosis [13, 28, 33, 34]. Thus, preventing cell cycle activation disables both DNA repair and apoptosis, which may result in the accumulation of DNA lesions and cell death by a non-apoptotic mechanism. The effect of Cdc25A suppression on DSB DNA repair was not significantly different from the effect of cyclin C ablation, although the reduction of NHEJ activity was typically lower than in cells with reduced cyclin C levels. We found that decreasing Cdc25A levels by siRNA attenuated cyclin C-associated pRb kinase activity, providing evidence for the involvement of Cdc25A in this activity, in addition to previously established cyclin E- and cyclin D-associated pRb kinase activities [28]. Thus, impeding cyclin C-associated G0 exit attenuates NHEJ DNA repair. This finding underscores the functional significance of G0→G1 transition for NHEJ activation in postmitotic neurons.
Additional validation of the importance of early G1 for NHEJ repair in postmitotic neurons comes from our observation that forcing G1 entry of postmitotic neurons induces NHEJ activation in the absence of DSB lesions. Previously, cells in quiescent states, including differentiation were shown to be able to reenter the cell cycle simply by removing appropriate CKIs. Interference with p21 was sufficient to reactivate the cell cycle and DNA synthesis in terminally differentiated skeletal muscle cells and quiescent fibroblasts [29]. Reactivation of the cell cycle and DNA replication has also been documented in quiescent cells overexpressing E2F1 and Cdc25A [28, 29, 31, 32]. Such reactivation of the cell cycle and DNA replication were sufficient to promote neuronal death even in the absence of DNA damage [35, 36]. Given the possibility that DNA replication might play a decisive role in triggering apoptosis in differentiated cells forced into the cell cycle, we prevented DNA replication by silencing CDK2, an essential kinase for G1→S transition [18]. Indeed, joint silencing of p21 and CDK2 prevented both DNA replication and apoptosis but did permit the initiation of cyclin C-dependent pRb kinase activity, exit from G0 and NHEJ induction.
Thus, our results demonstrate that cyclin C-associated pRb kinase activity and associated progression through early G1 are crucial for NHEJ repair. This is evidenced by (1) the coincidence of the initiation of cyclin C-associated pRb kinase activity with NHEJ activation, (2) NHEJ activation in response to forced G0→G1 transition. While cyclin D1-associated pRb kinase activity was induced in postmitotic neurons in response to DSBs or p21/CDK2 silencing, it occurred after cyclin C-directed pRb kinase activity and after NHEJ was activated; thus, the role of cyclin D1 in NHEJ activation remains to be established, as are the mechanisms preventing cells from initiation of cyclin E-associated pRb kinase activity and G1→S progression.
In contrast with repairable DNA damage, which only requires progression through early G1, DNA damage leading to neuronal death requires the cell cycle machinery for entry into the S phase and DNA replication in addition to G0→G1 transition (cells cannot enter S phase without G0→G1 transition) [13, 29, 35–40]. Preventing DNA replication or S phase entry attenuated apoptosis, while unrestricted progressing through G1→S led to apoptosis. The fatal effect of DNA replication can be explained by the fact that DNA replication itself is a potential source of DNA damage [41], and unrepaired DNA lesions when they interfere with DNA replication, may lead to critical secondary lesions which in turn may activate apoptotic signaling [42]. Moreover, DNA replication in postmitotic neurons undergoing apoptosis involves a highly error prone polymerase beta (pol β), normally engaged in DNA repair [43] instead of DNA polymerases, essential for canonical DNA synthesis. This may amplify DNA damage and generate apoptotic signaling in neurons if the DNA damage is not repaired.
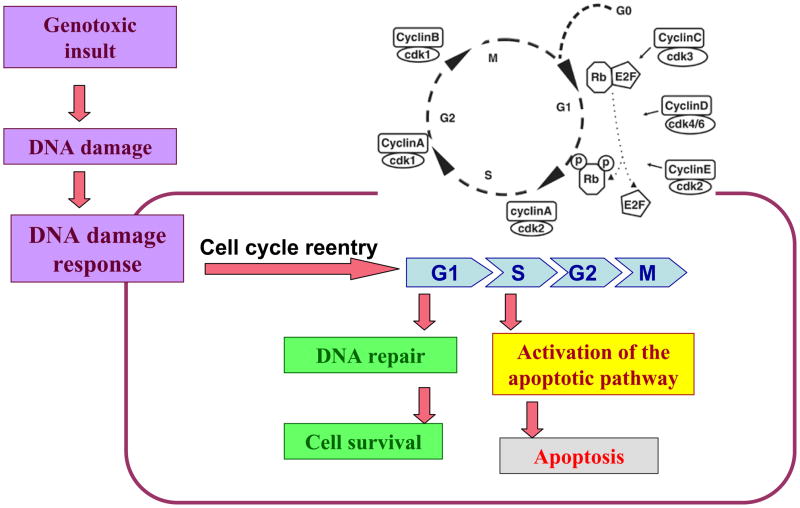
Depending on multiple factors, including the source and extent of an insult, DNA damage results in either repair of damaged DNA or in destruction of the cell by apoptosis. The mechanisms responsible for this decision are far from being elucidated. The two processes are not mutually exclusive and are orchestrated by components implicated in both processes. According to our results, cell cycle activation occurs in response to DNA damage and is involved in both DNA repair and apoptosis in postmitotic neurons. Figure 7 represents our model for the involvement of the cell cycle machinery in the DDR of postmitotic neurons. The cell cycle machinery may play a role in mechanisms that are part of the DDR orchestrated by the ataxia telangiectasia mutated (ATM) protein, the key regulator of the DSB signaling [44]. It may also involve elements of the DNA repair machinery such as Ku70/Ku86 proteins which, besides their vital role in DNA repair, have been implicated in apoptosis [45–47] and cell cycle regulation [48, 49].
Figure 7. Hypothesized role of cell cycle activation in response to DNA damage in postmitotic neurons.
DNA damage generated by a genotoxic insult results in activation of the DNA damage response (DDR). Cell cycle reentry which is an essential element of the DDR, plays a role in both DNA repair and apoptosis. G1→S transition is critical for apoptotic signaling.
Our studies were focused on postmitotic neurons. Whether the role for early G1 activation in DNA repair is limited to neurons and NHEJ or may function in other types of terminally differentiated cells and other types of DNA repair remains to be addressed.
The integrity of DNA is constantly being challenged by genotoxic stress resulting from exogenous sources and most importantly, by oxiradicals produced by normal metabolic processes [1, 2]. Due to a high rate of oxygen metabolism, the DNA of postmitotic neurons is under increased risk of damage from free radicals. For humans, given that the life span of postmitotic neurons is many decades, competent DNA repair could be critical for neuronal survival [3]. Since DNA damaging events occur continuously in living cells, the protective DNA repair mechanism should be available. NHEJ activity and the expression of cell cycle markers were indeed observed in neurons of normal brain [10, 11, 50, 51]. Here, we demonstrate that this is not a coincidence. In response to DNA damage, neurons reenter the cell cycle and activate NHEJ. This is a permanent process in normal brain and this explains why both the expression of cell cycle markers and NHEJ activity were observed in brain at physiological conditions. Results of microdialysis applied to direct measurement of H202 in the rat striatum demonstrate that the 5 μM H202 used in our study is close to concentrations of H202 in normal brain [52] underscoring the physiological implications of our results.
Materials and Methods
Antibody and reagents
Primary antibodies for immunoblotting and IP recognized cyclin C (polyclonal, 1:1000; Abcam), Rb (polyclonal; 1:1000; Pharmigen), phospho-Rb at S807/S811 (polyclonal; 1:1000; Abcam), CDK3 (polyclonal; 1:1000; Abcam), p21 (polyclonal; 1:1000; Abcam), CDK2 (polyclonal; 1:1000; Abcam), γH2AX (polyclonal; 1;1000; Abcam), cleaved caspase-3 (polyclonal; 1:1000; Millipore), cyclin E (monoclonal; 1:1000; Santa Cruz), cyclin D1 (polyclonal; 1:1000; Santa Cruz), Cdc25A (polyclonal; 1:1000; Santa Cruz), E2F1 (monoclonal; 1:1000; Sigma) or tubulin (polyclonal; 1:4000; Abcam). HRP-conjugated sheep anti-mouse IgG and donkey anti- rabbit IgG (1:5000; Amersham) were used as secondary antibodies. Primary antibody for immunofluorescence recognized Ki-67 (polyclonal; 1:50; Abcam), phosphorylated at Thr 2609 DNA-PKcs (polyclonal; 1:50; Genway) or NeuN (monoclonal, 1:500; Millipore). Texas Red-conjugated goat anti-rabbit (1:200; Santa Cruz), Alexa 488-conjugated goat anti-mouse (1:200; Invitrogen), and Alexa350-conjugated goat anti-mouse (1:200; Invitrogen) antibodies were used as secondary antibodies. Rb769, a substrate for the in vitro kinase assay, normal rabbit and normal mouse IgG were purchased from Santa Cruz.
Cell culture
All experiments involving the use of animals were approved by the IACUC at the TTUHSC. Primary cortical cell cultures were established from E18 Sprague-Dawley rats. The cells were plated according to procedures described earlier [36]. Following dissociation by mild trypsinization and trituration, cells were seeded onto plastic dishes or chamber slides precoated with 0.025 mg/ml poly-L-lysine, at a density of 1.3 × 103 neurons/mm2 in Neurobasal medium containing B-27 supplement, 1 mM HEPES, 2 mM glutamate and 0.001% gentamycin sulfate. All of the experiments were performed with five-day-old cultures. A fresh stock of 1 mM H2O2 (Sigma) was prepared in Neurobasal medium for each experiment and added at the indicated concentration.
Survival assays
Neuronal viability was assessed by quantifying apoptotic nuclei following the treatments. Cells were fixed and stained with DNA-binding dye Hoechst 33258 (1 μg/ml; Sigma), and the percentage of cells with apoptotic nuclei was calculated as described previously [36]. Nuclear staining was viewed and photographed using a Nikon Eclipse E800 fluorescence microscope equipped with a Spot digital camera and software. Apoptosis was also determined by immunoblot analysis for activated (cleaved) caspase-3 in cellular extracts from corresponding neuronal cultures. Extract from staurosporine-treated apoptotic Jurkat cells (Sigma) was used as a positive control.
Preparation of cell lysates
Cells were lysed after washing with cold PBS by incubation in ice-cold RIPA buffer containing a protease inhibitor cocktail (Calbiochem), 1 mM NaF and 0.5 mM Na3VO4before collection by scraping and sonicat ion and centrifugated at 14,000 rpm for 15 min at 4°C. The resulting supernatant was collected as the total cell extracts used for immunoblotting, IP, NHEJ assay and Ku-binding assays. For the preparation of nuclear extract used for immunoblotting of γH2AX, cortical neurons were lysed in ice-cold buffer containing protease inhibitor cocktail (Calbiochem) and incubated with hydrochloric acid (0.2 M) on ice for 30 min. After centrifugation, the acid-insoluble pellet was discarded, and the supernatant was dialyzed against 200 ml 0.1 M acetic acid twice (1–2 h each time) and then dialyzed against water.
Immunoblotting and immunoprecipitation
The protein concentrations of the lysates were measured with the Bio-Rad protein assay reagent. Total-cell or nuclear lysates (20–100 μg protein) or immune precipitates were separated by SDS-PAGE and electrophoretically transferred to PVDF membranes (Millipore) and probed with the appropriate antibodies. Blots were then incubated with HRP-conjugated secondary antibodies and developed using Enhanced Chemiluminescence Reagent Pico (Pierce). For immunoprecipitation, lysates (200–800 μg) were incubated with the appropriate antibody overnight at 4°C followed by incubation for 1 h with Protein A/G-Sepharose beads (Santa Cruz). Immune complexes were washed 2 times with RIPA buffer, boiled and separated by SDS-PAGE or used for the in vitro kinase assay. The immunoblot band intensities were normalized to β-tubulin or pRb (for ppRb S807/S811). Quantification was performed using Image software (Quantity one-4.6, Biorad).
Preparation of Substrate DNA and Analysis of NHEJ Activity
We designed the substrate DNA for the end-joining assay that generated a linear dimer as a reaction product without the formation of circular monomers by linearizing pGL3 basic (a kind gift from Dr. P. Makhov, Fox Chase Cancer Center) by XhoI enzyme. DNA fragments was purified on Qiagen spin columns, precipitated with 95% ethanol and resuspended to 1 μg/μl in Tris buffer (TE). End-joining assays in the total cell extracts dialized against 50 mM TRIS (pH 8,0), 20 mM potassium acetate, 20% glycerol, 1 mM EDTA, 5 mM cysteine were carried out as previously described [26, 27]. The dialyzed extracts were normalized for their respective total protein levels using the Bio-Rad protein assay. Reactions (40–60 μg protein) were carried out in 50 mM TRIS (pH 7.5), 0.5 mM Mg(OAc)2, 60 mM potassium acetate, 2 mM ATP, 1 mM DTT, 0.1 mg/ml BSA, and 0.5 μg of the linearized plasmid DNA. The samples were incubated at 37°C for 2 h and then deproteinized by phenol:chlorophorm:isoamyl alcohol solution (USB), and DNA was precipitated by ethanol and dissolved in TE buffer. The purified DNA substrate was separated by electrophoresis on 0.8% agarose gels. For Southern analysis, the gel was depurinated and blotted onto a nylon membrane (Hybond N+, Amersham) in denaturating buffer (alkaline transfer). The membrane was hybridized with the 32P-labeled plasmid (pGL3 basic) probe prepared by Random primer labeling kit (Stratagene). As a positive control, we used T4 ligase (New England Biolabs) treated linearized plasmid DNA (pGL3 basic/XhoI). The end joined products were visualized by autoradiography. Quantification of the Southern blot band intensity was performed with Image software (Quantity one-4.6, Biorad). The percentage of rejoining was calculated by the sum of dimer and multimer divided by the sum of monomer, dimer and multimer.
DNA-PK assay
The kinase activity of DNA-PK was determined using the Signa TECT™ DNA-dependent Protein Kinase Assay System (Promega). In brief, 10 μg of nuclear extract was incubated with an activator DNA, a biotinylated p53-derived peptide substrate, and [γ-32P] ATP at 30°C for 5 min. The reaction was terminated by adding termination buffer. Each termination reaction sample was spotted onto SAM2™ Biotin Capture Membrane and washed with 2M NaCl and 2M NaCl in 1% H3PO4. The SAM2TM Membrane squares were analyzed using Molecular Imager System (Bio-Rad).
siRNA transfection
siRNA oligonucleotides targeting cyclin C, Cdc25A, p21 and CDK2 (SMARTpool reagents, Dharmacon), each representing a cocktail of four siRNAs directed against different regions of corresponding genes, were designed and used according to the manufacturer’s guidelines. siRNA oligonucleotides targeting MAPK1 (Qiagen) was used as a control siRNA. Double stranded siRNAs were generated by adding corresponding mixture of siRNA nucleotides to siRNA buffer (Dharmacon) to obtain a 50 mM solution. The reaction mixture was heated to 90°C for 1 min and stored at −20°C. Transfection of RNA oligonucleotides was performed using the RNAi Starter Kit (Qiagen) according to the manufacturer’s recommendations, with a final oligonucleotide concentration of 50 nM or 100 nM (for cotransfection of p21- and CDK2- RNAi, 50 nM of each RNAi was used). After 24 h (for cyclin C and Cdc25A siRNA), the viability and protein expression were assessed in lysates collected in 1h after treatment of transfected cells with 5 μM H2O2 by Western blotting using primary antibodies recognizing either cyclin C or Cdc25A. The viability and protein expression in neurons transfected with p21 and p21/CDK2 siRNA were assessed after 1 and 4 h after transfection.
Measurement of Ku70/Ku86 (Ku)-DNA binding activity
Ku70/Ku86 DNA binding activity was analyzed by using a Ku70/Ku86 DNA Repair Kit (Active Motif). Briefly, cells were washed and resuspended in hypotonic buffer. The nuclear extract was isolated for Ku activity analysis. 5 μg of cell extract protein was loaded into the oligonucleotide-coated 96-well plate and incubated for 60 min at room temperature. Ku70 or Ku86 antibody was added and incubated for another 60 min. After washing, HRP-conjugated secondary antibody was added and incubated for 60 min. After washing, developing solution and stop solution were added. Optical density was read on a spectrophotometer at 450 nm. Each experiment was repeated three times, and data represent the mean ± SD of three separate determinations.
In vitro kinase assays
Cyclin C-, D1- and cyclin E-associated kinase activities were assayed as described [19]. Briefly, 200 μg protein from total cell lysate was incubated with either 2.5 μg anti-cyclin C, anti-cyclin D1 or anti-cyclin E overnight at 4°C. Then 20 μl of 50% slurry protein A/G agarose beads were added and incubated for 1–2 h at 4°C. The beads were washed twice with RIPA buffer, twice with kinase buffer (50 mM Tris HCl, pH 7.5, 150mM NaCl, 10 mM MgCl2, 1 mM DTT) and resuspended in 20 μl of kinase buffer containing 20 μM ATP, 1 μl [γ-32P]ATP (3000 Ci/mmol, PerkinElmer Life Sciences), 0.45 μg Rb 769 (Santa Cruz). The reaction was incubated for 30 min at 30°C. The reaction was stopped by adding 3 μl of 6 × SDS-PAGE sample buffer with following boiling. Samples were resolved by SDS-PAGE and electrophoretically transferred to PVDF membranes. The products of the kinase reaction were visualized by autoradiography.
Immunofluorescence
Neurons grown on glass cover slips were fixed in freshly prepared 4% formaldehyde for 30 min at 4°C and then permeabilized for 10 min in 0.1% Triton X-100 in 1% BSA prepared in PBS, blocked in 1% BSA for 1 h at room temperature, and double-stained with NeuN and Ki-67 or NeuN and Thr 2609 DNA-PKcs primary antibodies for 45min at 37°C followed by PBS/0.1% Triton washing and staining with appropriate secondary antibodies. Cover slips were mounted with Vectashield mounting medium (Vector Laboratories) and examined with a Nikon Eclipse E800 fluorescence microscope equipped with a Spot digital camera and software.
Statistical analyses
Statistical analyses were performed with Microsoft Excel and p values were obtained using ANOVA and Fisher’s post-hoc test. A p value < 0.05 was considered significant.
Acknowledgments
We thank Raul Perez-Olle for helpful discussions and P. Makhov, for pGL3 basic vector. This work was supported by the Garrison Institute on Aging, TTUHSC. MG was funded by the NIA-IRP, NIH.
Abbreviations
- CDK
cyclin-dependent kinase
- CKI
cyclin-dependent kinase inhibitor; DNA-PKcs
- DSB
double-strand break; DNA–protein kinase catalytic subunit
- NHEJ
non-homologous end joining
- pRb
retinoblastoma protein
- siRNA
small interfering RNA
References
- 1.Bassing CH, Alt FW. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Lavin MF, Kozlov S. DNA damage-induced signalling in ataxia-telangiectasia and related syndromes. Radiother Oncol. 2007;83:231–237. doi: 10.1016/j.radonc.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 3.Fishel ML, Vasko MR, Kelley MR. DNA repair in neurons: so if they don’t divide what’s to repair? Mutat Res. 2007;614:24–36. doi: 10.1016/j.mrfmmm.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Biton S, Barzilai A, Shiloh Y. The neurological phenotype of ataxia-telangiectasia: solving a persistent puzzle. DNA Repair (Amst) 2008;7:1028–1038. doi: 10.1016/j.dnarep.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Rhind N, Russell P. Checkpoints: it takes more than time to heal some wounds. Curr Biol 2000. 2000;10:R908–R911. doi: 10.1016/s0960-9822(00)00849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter JD. Think globally, translate locally: what mitotic spindles and neuronal synapses have in common. Proc Natl Acad Sci USA. 2001;98:7069–7071. doi: 10.1073/pnas.111146498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hlaing M, Shen X, Dazin P, Bernstein HS. The hypertrophic response in C2C12 myoblasts recruits the G1 cell cycle machinery. J Biol Chem. 2002;277:23794–23799. doi: 10.1074/jbc.M201980200. [DOI] [PubMed] [Google Scholar]
- 8.Stegmüller J, Bonni A. Moving past proliferation: new roles for Cdh1-APC in postmitotic neurons. Trends Neurosci 2005. 2005;28:596–601. doi: 10.1016/j.tins.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Ahuja P, Perriard E, Pedrazzini T, Satoh S, Perriard JC, Ehler E. Re-expression of proteins involved in cytokinesis during cardiac hypertrophy. Exp Cell Res. 2007;313:1270–8310. doi: 10.1016/j.yexcr.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Schmetsdorf S, Gärtner U, Arendt T. Constitutive expression of functionally active cyclin-dependent kinases and their binding partners suggests noncanonical functions of cell cycle regulators in differentiated neurons. Cereb Cortex. 2007;17:1821–1829. doi: 10.1093/cercor/bhl091. [DOI] [PubMed] [Google Scholar]
- 11.Schmetsdorf S, Arnold E, Holzer M, Arendt T, Gärtner U. A putative role for cell cycle-related proteins in microtubule-based neuroplasticity. Eur J Neurosci. 2009;29:1096–10107. doi: 10.1111/j.1460-9568.2009.06661.x. [DOI] [PubMed] [Google Scholar]
- 12.Becker EB, Bonni A. Cell cycle regulation of neuronal apoptosis in development and disease. Prog Neurobiol. 2004;72:1–25. doi: 10.1016/j.pneurobio.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Kruman II, Wersto RP, Cardozo-Pelaez F, Smilenov L, Chan SL, Chrest FJ, et al. Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron. 2004;41:549–561. doi: 10.1016/s0896-6273(04)00017-0. [DOI] [PubMed] [Google Scholar]
- 14.Muller C, Monferran S, Gamp AC, Calsou P, Salles B. Inhibition of Ku heterodimer DNA end binding activity during granulocytic differentiation of human promyelocytic cell lines. Oncogene. 2001;20:4373–4382. doi: 10.1038/sj.onc.1204571. [DOI] [PubMed] [Google Scholar]
- 15.McMurray CT. To die or not to die: DNA repair in neurons. Mutat Res. 2005;577:260–274. doi: 10.1016/j.mrfmmm.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Narciso L, Fortini P, Pajalunga D, Franchitto A, Liu P, Degan P, et al. Terminally differentiated muscle cells are defective in base excision DNA repair and hypersensitive to oxygen injury. Proc Natl Acad Sci U S A. 2007;104:17010–17015. doi: 10.1073/pnas.0701743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz EI, Smilenov LB, Price MA, Osredkar T, Baker RA, Ghosh S, et al. Cell cycle activation in postmitotic neurons is essential for DNA repair. Cell Cycle. 2007;6:318–329. doi: 10.4161/cc.6.3.3752. [DOI] [PubMed] [Google Scholar]
- 18.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 19.Ren S, Rollins BJ. Cyclin C/Cdk3 promotes Rb-dependent G0 exit. Cell. 2004;117:239–251. doi: 10.1016/s0092-8674(04)00300-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Ghio AJ, Gao M, Wei K, Rosen GD, Upadhyay D. Ambient particulate matter induces alveolar epithelial cell cycle arrest: role of G1 cyclins. FEBS Lett. 2007;581:5315–5320. doi: 10.1016/j.febslet.2007.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 22.Yano K, Morotomi-Yano K, Adachi N, Akiyama H. Molecular mechanism of protein assembly on DNA double-strand breaks in the non-homologous end-joining pathway. J Radiat Res (Tokyo) 2009;50:97–108. doi: 10.1269/jrr.08119. [DOI] [PubMed] [Google Scholar]
- 23.Lea NC, Orr SJ, Stoeber K, Williams GH, Lam EW, Ibrahim MA, et al. Commitment point during G0-->G1 that controls entry into the cell cycle. Mol Cell Biol. 2003;23:2351–2361. doi: 10.1128/MCB.23.7.2351-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grawunder U, Finnie N, Jackson SP, Riwar B, Jessberger R. Expression of DNA-dependent protein kinase holoenzyme upon induction of lymphocyte differentiation and V(D)J recombination. Eur J Biochem. 1996;241:931–940. doi: 10.1111/j.1432-1033.1996.00931.x. [DOI] [PubMed] [Google Scholar]
- 25.Diggle CP, Bentley J, Kiltie AE. Development of a rapid, small-scale DNA repair assay for use on clinical samples. Nucleic Acids Res. 2003;31:e83. doi: 10.1093/nar/gng083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, et al. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Qu D, Morris EJ, O’Hare MJ, Callaghan SM, Slack RS, et al. TheChk1/Cdc25A pathway as activators of the cell cycle in neuronal death induced by camptothecin. J Neurosci. 2006;26:8819–8828. doi: 10.1523/JNEUROSCI.2593-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pajalunga D, Mazzola A, Salzano AM, Biferi MG, De Luca G, Crescenzi M. Critical requirement for cell cycle inhibitors in sustaining nonproliferative states. J Cell Biol. 2007;176:807–818. doi: 10.1083/jcb.200608109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 31.Smith DS, Leone G, DeGregori J, Ahmed MN, Qumsiyeh MB, Nevins JR. Induction of DNA replication in adult rat neurons by deregulation of the retinoblastoma/E2F G1 cell cycle pathway. Cell Growth Differ. 2000;11:625–633. [PubMed] [Google Scholar]
- 32.Rogoff HA, Kowalik TF. Life, death and E2F: linking proliferation control and DNA damage signaling via E2F1. Cell Cycle. 2004;3:845–846. doi: 10.4161/cc.3.7.976. [DOI] [PubMed] [Google Scholar]
- 33.Di Giovanni S, Movsesyan V, Ahmed F, Cernak I, Schinelli S, Stoica B, et al. Cell cycle inhibition provides neuroprotection and reduces glial proliferation and scar formation after traumatic brain injury. Proc Natl Acad Sci U S A. 2005;102:8333–8338. doi: 10.1073/pnas.0500989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvira D, Yeste-Velasco M, Folch J, Casadesús G, Smith MA, Pallàs M, et al. Neuroprotective effects of caffeine against complex I inhibition-induced apoptosis are mediated by inhibition of the Atm/p53/E2F-1 path in cerebellar granule neurons. J Neurosci Res. 2007;85:3079–3088. doi: 10.1002/jnr.21427. [DOI] [PubMed] [Google Scholar]
- 35.Hou ST, Callaghan D, Fournier MC, Hill I, Kang L, Massie B, et al. The transcription factor E2F1 modulates apoptosis of neurons. J Neurochem 2000. 2000;75:91–100. doi: 10.1046/j.1471-4159.2000.0750091.x. [DOI] [PubMed] [Google Scholar]
- 36.O’Hare MJ, Hou ST, Morris EJ, Cregan SP, Xu Q, Slack RS, et al. Induction and modulation of cerebellar granule neuron death by E2F-1. J Biol Chem. 2000;275:25358–25364. doi: 10.1074/jbc.M001725200. [DOI] [PubMed] [Google Scholar]
- 37.Neve RL, McPhie DL. Dysfunction of amyloid precursor protein signaling in neurons leads to DNA synthesis and apoptosis. Biochim Biophys Acta 2007. 2007;1772:430–437. doi: 10.1016/j.bbadis.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein JA, Longo-Guess CM, Rossmann MP, Seburn KL, Hurd RE, Frankel WN, et al. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419:367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 39.Kuan CY, Schloemer AJ, Lu A, Burns KA, Weng WL, Williams MT, et al. Hypoxia-ischemia induces DNA synthesis without cell proliferation in dying neurons in adult rodent brain. J Neurosci 2004. 2004;24:10763–10772. doi: 10.1523/JNEUROSCI.3883-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Höglinger GU, Breunig JJ, Depboylu C, Rouaux C, Michel PP, Alvarez-Fischer D, et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc Natl Acad Sci USA. 2007;104:3585–3590. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruzankina Y, Asare A, Brown EJ. Replicative stress, stem cells and aging. Mech Ageing Dev. 2008;129:460–466. doi: 10.1016/j.mad.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Copani A, Sortino MA, Caricasole A, Chiechio S, Chisari M, Battaglia G, et al. Erratic expression of DNA polymerases by beta-amyloid causes neuronal death. FASEB J. 2002;16:2006–2008. doi: 10.1096/fj.02-0422fje. [DOI] [PubMed] [Google Scholar]
- 44.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Bio. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Yokota T, Gama V, Yoshida T, Gomez JA, Ishikawa K, et al. Bax-inhibiting peptide protects cells from polyglutamine toxicity caused by Ku70 acetylation. Cell Death Differ. 2007;14:2058–2067. doi: 10.1038/sj.cdd.4402219. [DOI] [PubMed] [Google Scholar]
- 47.Amsel AD, Rathaus M, Kronman N, Cohen HY. Regulation of the proapoptotic factor Bax by Ku70-dependent deubiquitylation. Proc Natl Acad Sci U S A. 2008;105:5117–5122. doi: 10.1073/pnas.0706700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bürckstümmer T, Bennett KL, Preradovic A, Schütze G, Hantschel O, Superti-Furga G, et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 49.Rampakakis E, Di Paola D, Zannis-Hadjopoulos M. Ku is involved in cell growth, DNA replication and G1→S transition. J Cell Sci. 2008;121:590–600. doi: 10.1242/jcs.021352. [DOI] [PubMed] [Google Scholar]
- 50.Ren K, Peña de Ortiz S. Non-homologous DNA end joining in the mature rat brain. J Neurochem. 2002;80:949–959. doi: 10.1046/j.0022-3042.2002.00776.x. [DOI] [PubMed] [Google Scholar]
- 51.Sharma S. Age-related nonhomologous end joining activity in rat neurons. Brain Res Bull. 2007;73:48–54. doi: 10.1016/j.brainresbull.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 52.Hyslop PA, Zhang Z, Pearson DV, Phebus LA. Measurement of striatal H2O2 by microdialysis following global forebrain ischemia and reperfusion in the rat: correlation with the cytotoxic potential of H2O2 in vitro. Brain Res. 1995;671:181–186. doi: 10.1016/0006-8993(94)01291-o. [DOI] [PubMed] [Google Scholar]