Abstract
Using an in solution based approach with a sub-proteomic fraction enriched in cardiac sarcomeric proteins; we identified protein abundance in ischemic and non-ischemic regions of rat hearts stressed by acute myocardial ischemia by ligating the left-anterior descending coronary artery in vivo for 1-hour without reperfusion. Sub-cellular fractionation permitted more in depth analysis of the proteome by reducing the sample complexity. A series of differential centrifugations produced nuclear, mitochondrial, cytoplasmic, microsomal, and sarcomeric enriched fractions of ischemic and non-ischemic tissue. The sarcomeric enriched fractions were labeled with isobaric tags for relative quantitation (iTRAQ), and then fractionated with an Agilent 3100 OFFGEL fractionator. The OFFGEL fractions were run on a Dionex U-3000 nano LC coupled to a ThermoFinnigan LTQ running in PQD (pulsed Q dissociation) mode. The peptides were analyzed using two search engines MASCOT (MatrixScience), and MassMatrix with false discovery rate of <5%. Compared to no fractionation prior to LC-MS/MS, fractionation with OFFGEL improved the identification of proteins approximately four fold. We found approximately 22 unique proteins in the sarcomeric enriched fraction had changed at least 20%. Our workflow provides an approach for discovery of unique biomarkers or changes in the protein profile of tissue in disorders of the heart.
Keywords: Ischemia, myocardium, differential centrifugation, sarcomere, MassMatrix
1. Introduction
In experiments reported here, we describe an approach for sub-proteomic analysis of rat myocardium stressed by an acute ligation of the coronary artery to simulate myocardial infarction (MI). Ischemic events are known to alter the myocardial protein profile and to induce contractile dysfunction after reperfusion [1,2]. There is substantial evidence that a major mechanism for the reduction in contractility following MI involves altered response of the myofilaments to Ca2+ [3-5]. These mechanisms include alterations in the chemical environment such as altered pH and inorganic phosphate concentration, but also are likely to involve altered protein profiles. We have therefore focused on the sarcomeric sub-proteome, which contains these myofilament proteins responsible for cardiac muscle contraction. Our studies compared protein profiles in the infarcted region of the myocardium with profiles in the adjacent non-ischemic regions.
To amplify our ability to detect modifications in the complex set of myocardial proteins, we reduced the complexity of the analysis by sub-cellular fractionation[6]. Sub-cellular fractionation (in our case the sarcomeric enriched fraction) selects for biologically associated proteins and reduces the large dynamic range of proteins found in whole homogenates of cardiac tissue [7,8]. Moreover, we employed a gel free approach for in solution proteomics, which provides the advantage of a workflow without gel based technology. Avoidance of gel based approaches overcomes typical problems of reduced enzyme accessibility to the protein, inefficient extraction of large peptides from the gel leading to reduced protein coverage and avoids the need to identify potentially hundreds of individual spots. Even though methods have been developed to overcome these drawbacks of gel based approaches, they are tedious in nature and involve transferring proteins to a membrane which is inefficient for large molecular weight proteins[9]. This problem has been addressed by the development of OFFGEL electrophoresis [10] in which proteins or peptides can be separated based on their isoelectric point in a liquid phase. OFFGEL electrophoresis has recently been compared to classic MudPIT experiments with comparable results[11], and when coupled with iTRAQ labeling may even improve proteome coverage[12]. In the current, study we combined rat cardiac sub-proteomic fractionation, iTRAQ, and OFFGEL as an approach to quantify protein abundance differences between ischemic and non-ischemic cardiac tissue. We found 22 proteins involved in regulating intracellular Ca2+, thin filament length, metabolism, cytoskeleton structure, and transport that changed significantly in abundance. Our data provide novel insights into mechanisms of ischemia induced cardiac disorders, and indicate potential biomarkers for acute MI.
2. Materials and methods
2.1. Coronary ligation and demarcation of ischemic and non-ischemic tissue
Rats were initially anesthetized with isoflurane (1.5%) and supplemented with etomidate (10 mg/kg BW; i.p.) for intubation. Surgical anesthesia was maintained using 1.5% isoflurane delivered through a vaporizer with 100% oxygen (compressed gas) connected in series to a rodent ventilator with the stroke volume set at 2.5 to 3.0 ml/min (based on body weight) and a respiration rate of 90 cycles per minute. A left thoracotomy was performed to expose the heart, and the pericardium was ruptured. The left coronary artery was permanently ligated 3 mm from the aortic ostium with an 8-0 monofilament polypropylene suture to produce ischemia for one hour. Rats were kept warm and remained anesthetized under isoflurane for the entire procedure. At the end of the ischemic period, the rats were heparanized (300 units; I.P). The hearts were harvested, placed in an ice-cold bath of PBS and the ascending aorta was cannulated and flushed with ice-cold PBS. Then 0.2 ml of 5% Evans Blue dye was infused retrograde into the ascending aorta to demarcate non-ischemic (Evans Blue) and ischemic (absence of Evans Blue) area [13]. The ischemic area of the left ventricle was dissected from the non-ischemic area, snap frozen in liquid nitrogen and analyzed separately.
2.2. Sub-cellular fractionation
Sub-cellular fractions were prepared from liquid nitrogen frozen Sprague Dawley rat cardiac tissue. All steps were performed on ice or at 4°C unless otherwise specified. The RCDC assay (BioRad) was used to determine protein concentration during fractionation following manufacturer's recommendations. Rat ventricular tissue (100mg wet weight) was finely diced and homogenized in 8 vol of 20mM Na 4O7P2·10 H2O, 20mM NaH2PO4, 1mM MgCl2, 0.303M sucrose, 0.5mM EDTA, pH 7.0, mammalian protease inhibitor cocktail 1:100 (Sigma) and phosphatase inhibitor cocktail 1:100 (Calbiochem) [14]. The tissue was homogenized (15 strokes) with a 2ml Dounce using both loose and tight pestles (A and B). After the initial homogenization, a sample was collected as fraction 1 or whole homogenate. The whole homogenate was centrifuged in a microfuge at 1000 g for 5 min and the supernatant was saved. The pellet was re-homogenized in 500μL of buffer with pestle A (ten strokes). The sample was then centrifuged at 1000 g for 5 min and the supernatant was pooled with the previous supernatant. The pellet was resuspended with the aid of a vortex in 500μL of buffer, and then centrifuged in a microfuge at 1000 g for 5 min, repeated once and all four supernatant fractions were pooled [15]. The pellet was kept on ice until enrichment of the sarcomeric, nuclear, and cytoplasmic proteins.
The pooled supernatant fractions were enriched for mitochondrial, microsomal, and some cytoplasmic proteins by centrifugation at 7000 g for 15 min. The pellet was resuspended in half the original volume and centrifuged again at 7000 g for 15 min [15], and the supernatant was saved for microsomal and cytoplasmic fractions. The pellet contained the crude enriched mitochondrial fraction 3. The pooled volume of the two supernatant fractions were determined, and KCl was added from solid powder to 0.6M final concentration, mixed carefully by inversion, incubated for 30 min on ice followed by centrifugation at 142,000 g for 30 min[14]. This step was repeated once. The pellet from the high speed spins was enriched microsomal fraction 4. The two supernatant fractions from the high speed spins were pooled and labeled as fraction 5, which contained mitochondrial, microsomal, and cytoplasmic fractions.
The pellet reserved for enrichment of the sarcomeric, nuclear, and cytoplasmic proteins was re-suspended in 60mM KCl, 20mM MOPS pH 7.0, 2mM MgCl2, 5mM EDTA, 1% (v/v) Triton X-100, mammalian protease inhibitor cocktail 1:100 (Sigma) and phophatase inhibitor cocktail 1:100 (Calbiochem) with 4 volumes relative to the original starting material [16]. The pellet was homogenized using the dounce homogenizer pestle B with at least 15 strokes then centrifuged at 2500 g for 5 min with three repeats. The supernatant fractions were pooled and saved as cytoplasmic containing fraction 6. The final pellet was resuspended and washed twice with 500 μl of 0.1M HEPES pH 8.0. The supernatants were discarded and the final pellet was solubilized in 200 μL of 0.1M HEPES pH 8.0, 8M urea and homogenized with a sonic dismembrator model 100 set at power level 1 (Fisher Scientific). The solubilized fraction was clarified by centrifugation at 20,000 g for 5 min and the supernatant fraction was kept as the enriched sarcomeric and nuclear fraction 2. Fraction 2 was the only fraction used for all subsequent experiments since the focus of this study was sarcomeric proteins.
2.3. SDS-PAGE and Western blot analysis
We employed SDS-PAGE and Western blot analysis to determine the enrichment of sub-cellular components. Samples were load equalized based on total protein content determined by the RCDC assay (BioRad) following manufacturer's recommendations. The SDS-PAGE resolving gel was 12% total acrylamide, 0.5% bis-acrylamide crosslinked, 10% (v/v) glycerol, pH 8.8 [17,18]. The stacking gel was 2.95% acrylamide, 15% N,N’ diallyltartardiamide (DATD), 10% (v/v) glycerol, pH 6.8 and 0.01% bromophenol blue [17,18]. After the gels finished running, they were either stained with Coomassie G-250 (Bio-Safe from BioRad) or immediately placed in transfer buffer (10mM CAPS pH 11.0) [19], transferred and analyzed as previously described [20]. The primary antibodies used were: tropomyosin CH1 (Iowa Hybridoma), RNA polymerase II (Active Motif), succinate dehydrogenase (SDH) (Molecular Probes), SERCA IIa (Affinity BioReagents), GAPDH (Santa Cruz Biotechnology), vimentin (Thermo Scientific), heat shock protein 70 (Cell Signaling), elongation factor TU (Abcam), myosin binding protein –C (kind gift from Dr. Richard Moss). The secondary antibodies used were goat anti-mouse IgG conjugated to peroxidase (Sigma) and goat anti-rabbit IgG conjugated to peroxidase (GE Heathcare). Western blots were developed with ECL-Plus detection reagents and Hyperfilm both from GE Healthcare. Hyperfilm was imaged with an Imagescanner III from GE Healthcare, and subsequently analyzed with ImageQuant TL (GE Healthcare) software to determine optical density values of bands for relative comparisons.
2.4. Trypsin digestion and iTRAQ labeling
The total protein concentration of the clarified sarcomeric enriched fraction was determined with an RC-DC assay (BioRad) following manufacturer's recommendations. Once the concentration was determined, 125μg of protein was digested with Trypsin Gold (TPCK treated from Promega). The protein sample was first reduced with 10mM DTT for 45 min at room temperature then treated with 40mM iodoacetamide for 45 min at room temperature. The sample was diluted to 1M urea finalconcentration with 100mM triethylammonium bicarbonate pH 8.5. Trypsin was added at 1:20 trypsin: protein ratio and allowed to digest overnight for approximately 16 hrs at 37°C.
Once the digestion was completed, the sample was concentrated in a centrifugal vacuum concentrator to less than 50μL but not to dryness. The remaining sample was labeled with iTRAQ reagents, using either 114 or 117 following the manufacturer's recommendations (Applied Biosystems). The ischemic and non-ischemic samples were labeled with alternating 114 and 117 reagents to correct for labeling bias. We used four biologic replicates analyzed in duplicate; two ischemic samples were labeled with reagent 114 and the other two with 117. The same with done for the non-ischemic samples. After one hour of incubation with the iTRAQ reagent, the ischemic sample and remote non-ischemic sample were equally mixed (reagent 114 and 117). The mixed sample was concentrated in a centrifugal vacuum concentrator to less than 100μL but not to dryness. To clean up the unreacted iTRAQ reagents and other contaminants, the sample (<100μL) was either diluted in 900μL of mobile phase (5% (v/v) ACN, 0.1% TFA) for a Vydac C18 4.6×150mm reverse phase column or 10mM KH2PO4, 25% ACN, pH3.0 for a strong cation exchange (SCX) cartridge ( Applied Biosystems) as recommended by manufacturer. The SCX cartridge was syringe driven, and the peptides were eluted with 10mM KH2PO4, 350mM KCL, 25% ACN, and pH 3.0. Peptides that were cleaned up with the strong cation cartridge also had to be loaded onto the Vydac column to remove contaminants prior to OFFGEL fractionation. The HPLC used for the Vydac column was a Dionex U-3000 analytical system equipped with a UV detector (214nm) run at 1 ml/min with separation solution B 95% ACN, 0.5% TFA run in a step wise gradient from 0.0% B for 10 min, 80% B for 10 min, 80%-100% in 2 min, held at 100% B for 5 min, 0.0% B for 7 min. All peptides eluted in one clean peak, which was verified with an immediate subsequent blank run using a traditional gradient to confirm all the peptides were eluted from the column. The collected and cleaned up peptide fraction was then concentrated in a centrifugal vacuum concentrator to dryness.
2.5. OFFGEL
The dried peptides were fractionated in-solution with an Agilent 3100 OFFGEL fractionator (Agilent Technologies). To focus the peptides based on their isoelectric point, 13 cm IPG strips, pH 3-10 were used with IPG buffer, pH 3-10 both from GE Healthcare [21], and a 12 well frame set from Agilent Technologies. The IPG strips and paper wicks were rehydrated with 4.8% glycerol (v/v), 2% IPG buffer (1:50) [21] at 40uL per well for 15 min. While the strips were rehydrating, the sample was solubilized in 1.8ml of the same rehydration buffer. After rehydration was complete, 150 μL of sample was added to each well, the wells were sealed, and mineral oil was added to each end of the strip. The strips were focused until 20kV·h was reached with a max voltage of 8000V, 50μA, 200mW, and a hold setting of 500V. After 24 hours of run time the paper wicks were changed with new wicks wetted with water. The runs took approximately 35-40 hours.
2.6. LC-MS/MS
After the samples had completed in the OFFGEL fractionator, the 12 fractions were clarified (to prevent clogging the trapping column) with low protein/peptide binding spin filters (0.65 micron pore size) from Millipore. The 12 peptide fractions were infused separately online to a Thermo Electron Finnigan linear ion trap mass spectrometer (LTQ) operated in positive ion mode via a Dionex U-3000 Ultimate nano LC system running Chromeleon v 6.80 SP4 Build 2361. The loading pump injected 50μL of sample onto a Dionex μ-Precolumn acclaim PepMap100 C18, 5 μm, 100A, and 300 μm × 5mm at 20 μl/min. The pre-column was washed with mobile phase (5% ACN, 0.1% formic acid) for 20 min to remove any contaminants from the OFFGEL fractionation and the flow diverted to the separation column (Agilent Zorbax 300SB-C18, 3.5μm, 75 μm × 150mm) at a flow rate of 250nL/min. The peptides were separated and eluted with a linear gradient of 10-60% solution B (95% ACN, 0.1% formic acid) in 84 min. The LTQ spray voltage was 2.0kV and the capillary temperature was set at 200°C. A survey full scan (m/z = 400-2000) and the five most intense ions were selected for a zoom scan to determine the charge state; afterwhich MS/MS was triggered in Pulsed-Q Dissociation mode (PQD) with minimum signal required (1000), isolation width 2.0, normalized collision energy 31.0, activation Q 0.600, and activation time of 0.400. PQD mode is critical in order to visualize the low m/z reporter ions from the iTRAQ reagents used for relative quantitation[22]. The dynamic exclusion list was restricted to 250 entries with duration of 120s. Mass spectrometry data were acquired with Xcalibur software version 2.0 SR2.
2.7. Data Analysis
We analyzed two analytical replicates for each of four different biological replicates. Mass spectra were converted from Xcalibur .Raw file to mzXML and MGF file formats using MassMatrix downloadable tools for conversion at http://www.massmatrix.net/mm-cgi/downloads.py. The conversion tools from MassMatrix extend ReAdW.exe developed by the Institute for Systems Biology. The MGF files from duplicate runs were concatenated and submitted as a single large MGF file (24 files = 12 OFFGEL fractions done in duplicate) and searched using a Mascot 2.2.04 search engine [23]. MassMatrix searches [24,25] were performed using identical search parameters as used for Mascot with merged files containing all 24 MGF files. Mascot and MassMatrix searches were performed against the IPI rat database (v3.65 Oct 2009) for protein identifications using automatically generated decoy databases to determine the false discovery rate for each search (FDR <5% considered significant). Fixed modifications were set for MassMatrix and Mascot as: carbamidomethyl (C), iTRAQ 4-plex (n-term), and iTRAQ 4-plex (K) and a variable modification was oxidation (M). The peptide mass tolerance was set to 2.0 Da, minimum length of peptide six amino acids, maximium of two missed cleavages allowed, trypsin as a cleavage rule, and a FDR < 5% for proteins identifications. The ratio of reporter ions for quantitation was analyzed by performing searches with the MassMatrix iTRAQ quantitation module. MassMatrix quantitation results from the four biological replicates (output as .CSV) were binned for statistical analysis into 6 catagories using an in-house Python script: 1= all four agree, 2 = three agree and one not available (N/A), -2 = three agree and one disagrees, 3= two agree and other two N/A, -3 = two agree and at least one disagrees, 0 = non-conclusive or none agreed. Only proteins that were grouped in category 1, 2, or 3 were considered for statistical comparisons of reporter ion ratios (see supplemental data Excel file Quantitation). A threshold was set to 1.2 with a p-value < 0.05 yielding at least a 20% change in abundance compared to the reference (healthy remote tissue).
Results 3
3.1. Sub-cellular fractionation
To compare rat cardiac ischemic and non-ischemic tissue the left coronary artery was ligated for an hour. The one hour ligation procedure yielded a distinct ischemic area easily distinguished from non-ischemic or remote tissue through the use of Evans Blue dye staining (Figure 1). The ischemic and non-ischemic tissue regions were separately fractionated into sub-cellular fractions through the use of differential centrifugation as summarized in Figure 2. The sarcomeric enriched fraction was used to determine relative quantitative changes in the ischemic versus non-ischemic tissue (Figure 2 dashed box) of four different animals (biological n=4). In addition to differential centrifugation, treatment with Triton X-100 helped to solubilize the membranes and remove many contaminants [16]. The protein content of each fraction is included in Figure 2 and we obtained approximately a 78% yield (n=4) of all fractions summed when compared to the whole homogenate fraction 1 in the workflow diagram. There was no significant difference in the fractions between the ischemic and non-ischemic tissue based on protein content.
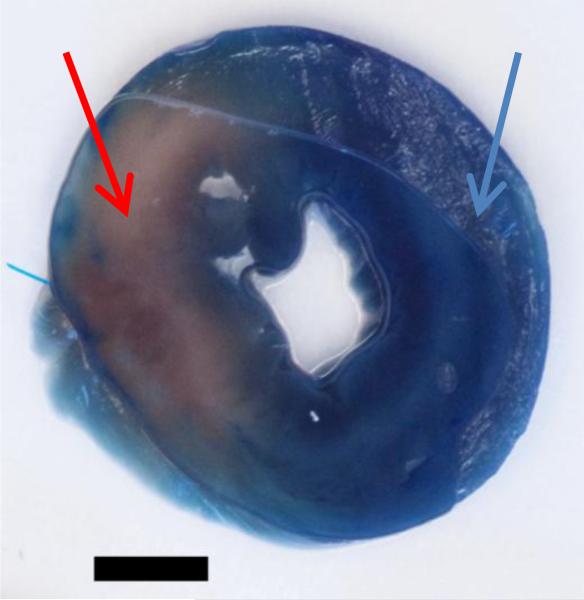
Figure 1. Cross-section of rat heart demarcating ischemic versus non-ischemic tissue.
The blue area of the heart cross-section shows the non-ischemic tissue that absorbed the Evans Blue dye indicated by the blue arrow. The pink area of the heart cross-section shows the ischemic tissue that did not absorb the Evans Blue dye indicated by the red arrow. Bar = 3mm
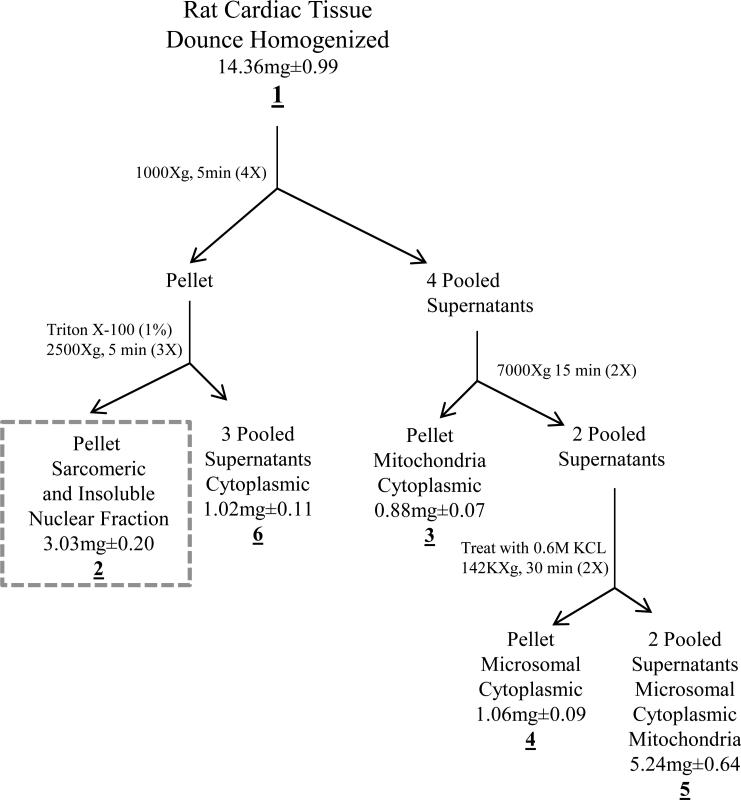
Figure 2. Sub-cellular fractionation of rat cardiac tissue.
The flowchart depicts the sub-cellular separation based mainly on differential centrifugation. The underlined numbers represent fractions taken during the procedure. The dashed box represents the sarcomeric enriched fraction used in the study. Protein concentration was determined at each of the fractions and is represented as mean (mg) ±SEM n=4. There were no significant differences between the non-ischemic and ischemic tissue protein concentrations. The fractionation yielded approximately 78% recovery when comparing the whole homogenate (fraction 1) vs fractions 2-6.
3.2. Western blot and SDS-PAGE analysis of the sub-proteomic fractions
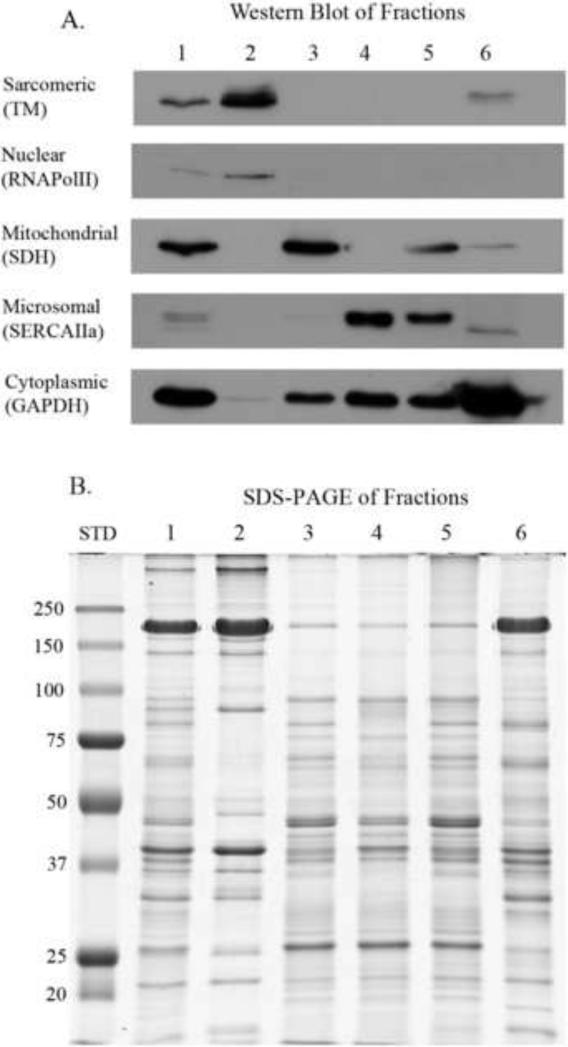
To determine the efficacy of the fractionation we ran the fractions in an SDS-PAGE gel and also performed Western blot analysis (Figure 3). Based on the Western blot, Figure 3A, the sarcomeric fraction is substantially enriched with the exception of the presence of nuclear proteins. Fraction 6 was also highly enriched for cytoplasmic proteins. The mitochondrial and microsomal fractions were enriched, and they both had some cytoplasmic proteins present (Figure 3A). Despite multiple washes there was some unavoidable carryover. Therefore, the fractionation protocol is meant as a partial fractionation method to reduce the complexity of a whole homogenate. The SDS-PAGE gel in Figure 3B shows varying band intensities and migration of the various fractions suggesting differential proteins were contained in each fraction even though there was some carryover.
Figure 3. Western blot and SDS-PAGE analysis of sub-cellular fractions.
Lanes are numbered as fractions taken during the fractionation procedure (refer to Figure 2). Samples were load equalized based on total protein. A. Western blot of the fractions. Antibodies used to determine cellular compartments: TM, tropomyosin (sarcomeric); RNApolII, RNA polymerase II (nuclear); SDH, succinate dehydrogenase (mitochondrial); SERCAIIa, sarcoplasmic or endoplasmic reticulum calcium ATPases muscle specific isoform (microsomal); and GAPDH, glyceraldehydes 3-phosphate dehydrogenase (cytoplasmic). B. SDS-PAGE of the same fractions used in the Western blot. STD, molecular weight marker in kDa.
3.3. Chromatography comparison of post labeled iTRAQ peptides
After labeling peptides with iTRAQ, it is important to remove excess reagent and reaction by-products from the labeling process in order to run the samples in the mass spectrometer. In a pilot experiment, we tested two approaches to clean up the peptides prior to MS (see supplemental data pilot experiments). The first approach as recommended by the manufacturer was to run the pooled iTRAQ labeled sample through a SCX cartridge; however, this yields a sample with high salt. To remove the high salt and other minor contaminants, we applied the sample to a reverse phase C18 column. The second approach we used was to bypass the SCX resin and apply the sample directly to the reverse phase C18 column. Figure 4A shows an overlaid chromatogram from the reverse phase C18 column of the two approaches. When we used only the C18 column, the absorbance at 214 nm was approximately twice that of the SCX as shown in Figure 4A (black line compared to the red line). The SCX resin removed excess iTRAQ reagents and other contaminants from the sample as shown by the lack of a large signal in the flow through at a retention time of 2-4 minutes (Figure 4A, red line). However, the sample applied directly to the C18 column removed all contaminants as shown in the flow through at a retention time of 2-4 minutes (Figure 4A, black line). To determine if the clean up procedure would affect the number of protein identifications, we ran the samples after C18 clean up on the LC-MS/MS without OFFGEL fractionation and searched with two search engines, MassMatrix and Mascot (Figure 4B). Figure 4B shows two Venn diagrams of the number of proteins identified at a false discovery rate of < 5%, one searched with MassMatrix and the other with Mascot. In this pilot experiment, we found that using C18 reverse phase column clean up without the SCX step, resulted in approximately twice as many protein identifications in the sarcomeric enriched fraction.
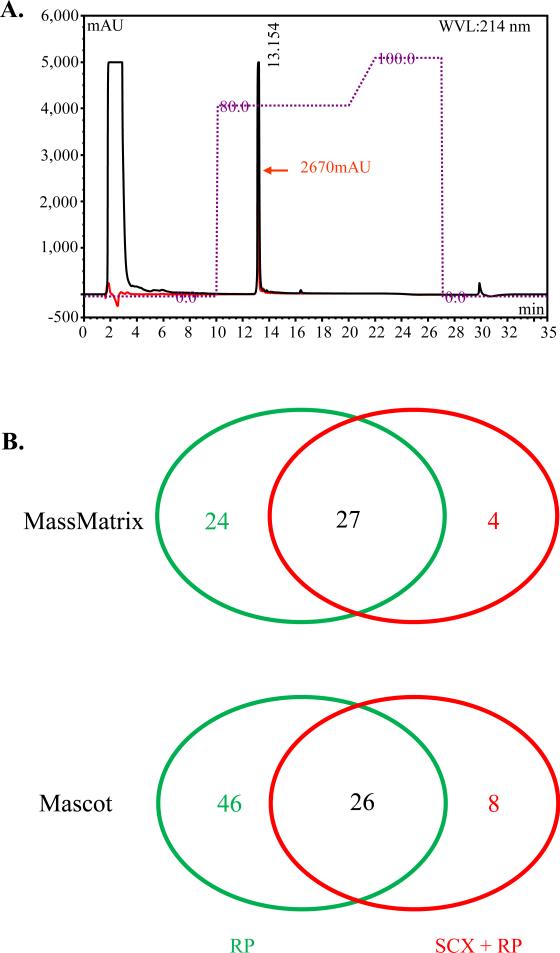
Figure 4. Post iTRAQ label clean up comparison between reverse phase HPLC only and strong cation exchange followed by reverse phase HPLC.
A. Representative HPLC chromatogram showing the amount of peptides recovered (retention time 13.154) after C18 reverse phase clean up of iTRAQ labeled peptidesThe red line indicates the sample that was first cleaned with a strong cation exchange cartridge followed with reverse phase separation. The red arrow represents the maximum absorbance for the red line at 2670 milli absorbance units (mAU). The black line represents the sample that was cleaned only with reverse phase HPLC. B. Venn diagrams showing number of protein identifications post iTRAQ label but before OFFGEL fractionation. The red numbers represent the number of unique protein identifications when peptides were cleaned with a strong cation exchange cartridge and reverse phase HPLC, and the green numbers represent the number of unique protein identifications when peptides were cleaned with reverse phase HPLC only . The black numbers are common identifications among the two cleanup methods. RP, C18 reverse phase HPLC. SCX + RP, strong cation exchange and C18 reverse phase HPLC.
3.4. OFFGEL and LC-MS/MS iTRAQ quantitation
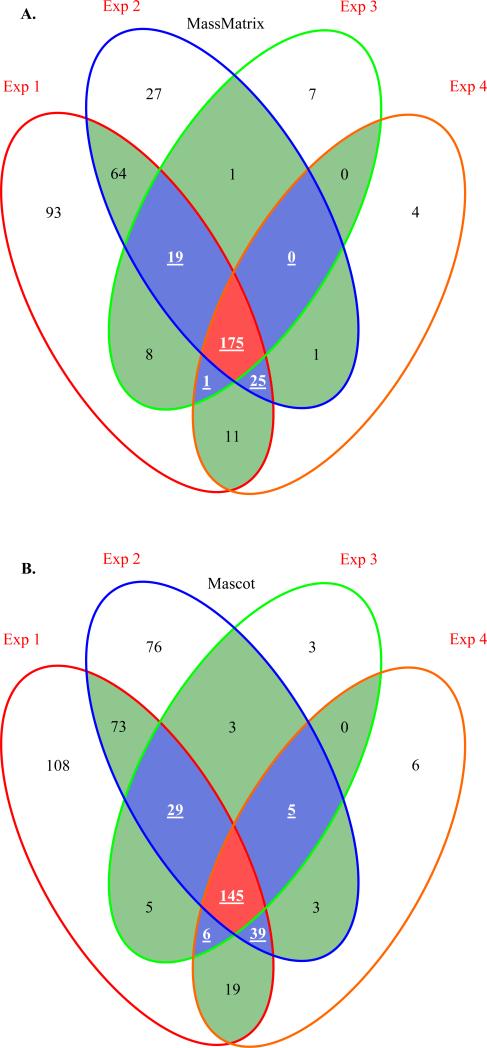
In Figure 5A using MassMatrix, we identified (FDR < 5%) 220 proteins with at least three of the four biological replicates agreeing (see supplemental data MassMatrix). Similarly when Mascot was used to analyze the same data set, 224 proteins were identified (FDR 5%) in three of the four biological replicates (Figure 5B,see supplemental data Mascot). When OFFGEL was incorporated into our fractionation procedure, we were able to approximately quadruple the number of confident protein identifications in at least three of the four biological replicates studied (compare figure 5 to 4B). Hence, incorporation of OFFGEL fractionation increased our identifications and was implemented in the experimental design.
Figure 5. Four way Venn diagrams comparing number of protein identifications in each biological replicate as identified with either Mascot or MassMatrix.
These protein identifications were derived from OFFGEL peptide fractions run on the LC-MS/MS, which were combined and searched against either Mascot or MassMatrix. The red Exp (experiment) numbers above the ovals represent four biological replicates. The white shaded area represents the number of protein identifications where only one replicate agrees, the green shaded area where two agree, the blue shaded area with white underlined bold numbers where three agree, and the red shaded area with white underlined bold numbers where all four agree. A. Four way Venn diagram where the replicates were searched with MassMatrix search engine. B. Four way Venn diagram where the replicates were searched with Mascot search engine.
From the identifications, Table 1 lists 22 unique proteins where analysis of the iTRAQ quantitation showed the abundance level changed by at least 20% (see supplemental data quantitation and MassMatrix for all data). We have additional supporting data suggesting several proteins of interest changed similarly with Western blot analysis (Figure 6) and iTRAQ (Table 1). Due to the way protein identifications were grouped Table 1 lists only a representative accession number with a description for that IPI number. In each protein grouping there could be various isoforms or truncations of the same or like proteins (see supplemental data MassMatrix or Mascot) In the case of ambiguous descriptions the rat IPI sequence that was identified was then BLAST searched [26] against the non-redundant NCBI database using Rattus norvegicus to get a meaningful description. The change was determined with a significant 20% change at a p-value < 0.05 and protein identifications from at least 3 of the 4 biological replicates had to be identified by MassMatrix. The proteins that changed were involved in regulating intracellular Ca2+, thin filament length, metabolism, cytoskeleton structure, and transport.
Table 1.
Proteins identified in LC-MS/MS with at least 20% change in abundance determined with iTRAQ
| Bin | Representative Acc. No. | Change | Description |
|---|---|---|---|
| 2 | IPI00206193 | decrease | FOUR AND A HALF LIM DOMAINS PROTEIN 2. |
| 2 | IPI00558178 | decrease | SIMILAR TO DYSTROPHIN, MUSCULAR DYSTROPHY. |
| 2 | IPI00870316 | increase | MYOSIN BINDING PROTEIN C, CARDIAC |
| 2 | IPI00869617 | increase | 52 KDA PROTEIN (Sarcalumenin glycoprotein)* |
| 2 | IPI00764107 | increase | SIMILAR TO TUBULIN ALPHA-8 CHAIN. |
| 3 | IPI00189173 | decrease | RYANODINE RECEPTOR 2 |
| 3 | IPI00208205 | decrease | HEAT SHOCK COGNATE 71 KDA PROTEIN. |
| 3 | IPI00566672 | decrease | SIMILAR TO HEAT SHOCK PROTEIN 8. |
| 3 | IPI00207355 | decrease | HEAT SHOCK-RELATED 70 KDA PROTEIN 2. |
| 3 | IPI00230941 | decrease | VIMENTIN. |
| 3 | IPI00201060 | decrease | LAMIN-A. |
| 3 | IPI00372839 | decrease | SIMILAR TO PROCOLLAGEN, TYPE VI, ALPHA 2. |
| 3 | IPI00476408 | decrease | SIMILAR TO OBSCURIN, CYTOSKELETAL CALMODULIN AND TITIN-INTERACTING RHOGEF. |
| 3 | IPI00324302 | decrease | ACETYL-COA ACETYLTRANSFERASE, MITOCHONDRIAL. |
| 3 | IPI00557049 | decrease | 39 KDA PROTEIN. (Enolase 1)* |
| 3 | IPI00765523 | decrease | SIMILAR TO SYNAPTOPODIN 2-LIKE |
| 3 | IPI00200466 | increase | ADP/ATP TRANSLOCASE 2 |
| 3 | IPI00363182 | increase | SIMILAR TO SOLUTE CARRIER FAMILY 25, MEMBER 5. |
| 3 | IPI00371236 | increase | ELONGATION FACTOR TU, MITOCHONDRIAL. |
| 3 | IPI00209115 | increase | SOLUTE CARRIER FAMILY 25 MITOCHONDRIAL CARRIER |
| 3 | IPI00390168 | increase | NEBULIN-RELATED ANCHORING PROTEIN ISOFORM C. |
| 3 | IPI00358033 | increase | NADH-UBIQUINONE OXIDOREDUCTASE 75 KDA SUBUNIT, MITOCHONDRIAL |
Change is relative to non-ischemic tissue identified in at least 3 biological replicates.
Representative assession number means more than one IPI listed for similar group of proteins.
Bin 2, three experiments agree one not available; Bin 3, two experiments agree and the other two not available.
manually annotated, See supplemental data Quantitation and MassMatrix for details.
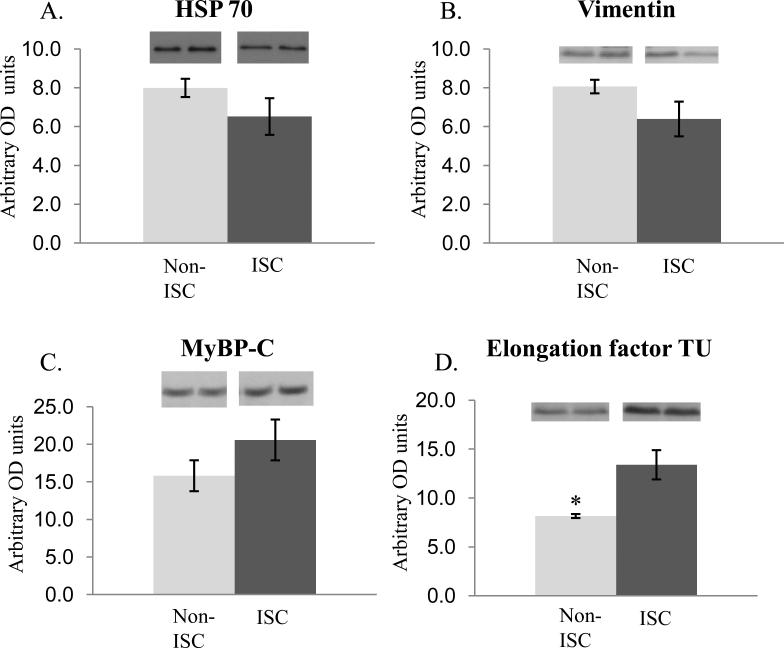
Figure 6. Western blot analysis of selected proteins from Table 1.
Densitometry quantification. Data shown as mean ± SEM, * p-value< 0.05, n=4, OD optical density, ISC = ischemic, Non-ISC = non-ischemic. Two representative bands from each group are shown above each histogram. The trends in protein abundance differences detected were similar to iTRAQ determined differences (Table 1) albeit some were not statistically significant. A. The mean heat shock protein 70 abundance levels were decreased in the ischemic samples. B. The mean vimentin abundance levels were decreased in the ischemic samples. C. The mean myosin binding protein –C abundance levels were increased in the ischemic samples. D. The mean elongation factor TU abundance levels were increased in the ischemic samples.
Discussion 4
One of the main objectives or our investigation was to develop a workflow for analysis of the sarcomeric sub-proteome. Although investigation of the other fractions was beyond the scope of the current study, the fractionation protocol might be useful for researchers to investigate the other fractions to gleam insight into what sub-cellular compartment protein pools may have translocated or changed during a perturbation event. Our data demonstrate that the workflow presented above improved protein identification over a standard approach. This workflow generated data identifying novel alterations in protein abundance in acute ischemic injury, a relevant model of a cardiac disorder. Sub-cellular fractionation has been employed for decades to decrease the complexity of proteins in cardiac tissue [16,27,28]; moreover, the application recently has been applied to proteomic approaches as a method to increase the detection limits of low abundant proteins [29-33]. Modern mass spectrometers are highly sensitive, and abundant proteins can overwhelm the system's dynamic range leading to diminished detection of low abundant proteins. In biomarker discovery and validation, fractionation is particularly important, as many biomarker candidates may not be detected due to low abundance [34]. In order to further reduce complexity, orthogonal separation of protein and or peptides is widely utilized. Some of the main orthogonal approaches are: multidimensional protein identification technology (MudPIT) [35] and SDS-PAGE followed by tryptic in-gel digestion-MS (GeLC-MS) [36]. It has been shown that the use of OFFGEL separation yields more protein identifications compared to GeLC-MS [10,21]. When comparing MudPIT to OFFGEL, one study reported similar results with membrane enriched mouse skeletal cells [11]. However, another report using membrane proteins enriched from mice brains showed similar results for SCX with a pH gradient vs OFFGEL, but more proteins were identified with reverse phase at high pH in membrane mouse brain [37]. The use of OFFGEL separation is becoming more common as a viable method for separating peptides. It may also be used to separate intact proteins even though this application is not commonly employed, perhaps due to similar limitations of protein IEF.
Underestimation of iTRAQ quantitation has been reported and some of the main causes are related to isotopic contamination and background interference [38]. The isotopic contamination can partly be corrected by incorporating the correction factors, which were available in the development kit for 114 and 117 reporter ions. In addition, the ischemic and non-ischemic samples were labeled with alternating 114 and 117 reagents to correct for labeling bias, which may also minimize the effects of manufacturing impurities of the iTRAQ reagents. Background interference is more difficult to correct however, one of the major problems is the 121 reporter ion discrimination with the immonium ions from phenylalanine having similar m/z values[38] and in the current study, we did not use this reporter ion. Another form of interference is simultaneously selecting more than one peptide for MS/MS. By reducing the complexity of the sample (sub-cellular fractionation) and peptides (OFFGEL separation along with relatively shallow separation gradient), we attempted to minimize multiple peptide fragmentation occurring at the same time. Previous rat cardiac studies utilizing iTRAQ reagents to investigate changes in hypertensive heart failure [39] and aging of the heart [40] had on average less than 2-fold differences. However, even with a possible dynamic range of approximately 2-fold, iTRAQ has been found to be more sensitive than iCAT and DIGE technologies[41].
In our data, we specified a threshold of 1.2 where 22 unique proteins had changed at least 20% in a sarcomeric enriched fraction (Table 1). In this experiment we were interested in the changes between healthy non-ischemic and ischemic cardiac tissue without the effects of reperfusion on the myocardium. It has been shown that during ischemia/reperfusion, myofilament proteins were broken down or released from the myocardium, but to a lesser extent in samples from hearts stressed by ischemia alone [1]. The difference between ischemia and ischemia/reperfusion may partially explain the lower fold changes observed along with bias of iTRAQ towards underestimation. However, biologically induced small changes in protein composition may have a large effect on function in the myocardium [42].
Our data demonstrated that with myocardial infarction, proteins involved in regulating metabolism, chaperone transport, intracellular Ca2+, thin filament length, and cytoskeleton structure changed significantly in abundance. ADP/ATP Translocase 2, Elongation factor Tu (EF-Tu), NADH-ubiquinone oxidoreductase were increased in ischemic compared to infarcted tissue. These proteins catalyze exchange of ADP and ATP across the mitochondrial membrane, mediate tRNA to free sites on the ribosome, and aid in generation of a proton gradient used in ATP synthesis respectively. In agreement with our results, EF-Tu has previously been shown in rat heart to increase in ischemia, without decreasing during reperfusion [43]. EF-Tu has been suggested to have increased levels of phosphorylation, which leads to inhibition of mitochondrial protein synthesis, which may be a way of compensating for the ischemic damage by reducing energy consumption and superoxide formation [43,44]. Acetyl-COA acetyltransferase is involved in the mevalonate pathway, which is important for biosynthesis of molecules involved in cell membrane maintenance and protein anchoring among others. The decrease in acetyl-COA acetyltransferase may be a contributing mechanism in the apoptosis of ischemic cardiac tissue.
Heat shock protein 70 (HSP 70) functions as a molecular chaperone involved in protein folding and transport [45]. We found that HSP 70 was decreased in ischemic tissue (sarcomeric fraction) in agreement with a study done with mice where heat shock cognate protein was released into the extracellular space and therefore may be decreased in the sarcomeric fraction as we observed [45]. It has been suggested that HSP 70 has a cardioprotection affect in rat by attenuation of the intracellular Ca2+ overload [46]. Hence, during ischemia the HSP 70 might be released from the myocardium in an effort to regulate cellular Ca2+ via sarcoplasmic reticulum Ca2+ -ATPase (SERCA) and the ryanodine receptor [46], which was also decreased in our ischemic samples.
The structural proteins nebulin and myosin binding protein-C (MyBP-C) were increased in the ischemic tissue and obscurin and vimentin were decreased. The reduction of obscurin may be a result of the compromised myofilament structure in ischemic samples; specifically the M-band proteins, A-band proteins, and sarcoplasmic reticulum network alignment around the contractile apparatus as previously observed in striated tissue [47]. When vimentin content was increased in rat cardiomyocytes prior to an infarct there was a cardioprotective effect against ischemia [48]. Therefore, when vimentin is decreased as in our case it may be a maladaptive effect of ischemia to breakdown the intermediate filament proteins. In a study investigating a rat model of ischemia/reperfusion, phosphorylation levels had changed in MyBP-C and nebulin in response to myocardial injury [49]. It has been proposed that MyBP-C phosphorylation may have cardioprotective effects on the myocardium by maintaining the thick filament spacing and structure [50]. The increased levels of MyBP-C may be another mechanism in addition to phosphorylation to maintain the thick filament spacing and structure. The mechanism for replacement is unclear but, would not likely be from protein turnover do to the short time span of the infarct.
Apart from our investigation of effects of MI on protein abundance, we think the methods described here provide an approach for targeted analysis of commonly studied cardiac protein subsets and reduction in their complexity for proteomic analysis. The approach can be applied to various disorders of the myocardium and with the incorporation of iTRAQ it can be multiplexed, which is an advantage to help reduce variability by running multiple (8) samples at the same time. Using this approach we revealed 22 proteins with abundance changes in ischemia that could further be investigated for potential use as a biomarker in MI injury.
Supplementary Material
Acknowledgements
This research was supported by National Institutes of Health Grants PO1 HL 62426 (project 1, core c), R01 HL22231 and Chicago Biomedical Consortium C-009. The authors are grateful to Dr. Randal C. Jaffe for help with the photography in Figure 1 and Mr. Roderick G. Davis for initial method development on the LTQ. Tropomyosin CH1 antibody was obtained from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development, maintained by the University of Iowa, Biological Sciences (Iowa City, IA). The myosin binding protein – C antibody was a kind gift from Dr. Richard Moss.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van Eyk JE, Powers F, Law W, Larue C, Hodges RS, Solaro RJ. Breakdown and release of myofilament proteins during ischemia and ischemia/reperfusion in rat hearts: Identification of degradation products and effects on the pca-force relation. Circ Res. 1998;82:261–271. doi: 10.1161/01.res.82.2.261. [DOI] [PubMed] [Google Scholar]
- 2.Westfall MV, Solaro RJ. Alterations in myofibrillar function and protein profiles after complete global ischemia in rat hearts. Circ Res. 1992;70:302–313. doi: 10.1161/01.res.70.2.302. [DOI] [PubMed] [Google Scholar]
- 3.Walker LA, Walker JS, Ambler SK, Buttrick PM. Stage-specific changes in myofilament protein phosphorylation following myocardial infarction in mice. J Mol Cell Cardiol. 2009 doi: 10.1016/j.yjmcc.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P, Hofmann PA, Li B, Malhotra A, Cheng W, Sonnenblick EH, Meggs LG, Anversa P. Myocardial infarction alters myofilament calcium sensitivity and mechanical behavior of myocytes. Am J Physiol. 1997;272:H360–370. doi: 10.1152/ajpheart.1997.272.1.H360. [DOI] [PubMed] [Google Scholar]
- 5.Arteaga GM, Martin AF, Solaro RJ. Increased cardiac myofilament ca2+ sensitivity protects murine hearts during ischemia/reperfusion injury. Biophysical Journal. 2005;88:537A–537A. [Google Scholar]
- 6.Cox B, Emili A. Tissue subcellular fractionation and protein extraction for use in mass-spectrometry-based proteomics. Nat Protoc. 2006;1:1872–1878. doi: 10.1038/nprot.2006.273. [DOI] [PubMed] [Google Scholar]
- 7.Corthals GL, Wasinger VC, Hochstrasser DF, Sanchez JC. The dynamic range of protein expression: A challenge for proteomic research. Electrophoresis. 2000;21:1104–1115. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 8.McGregor E, Dunn MJ. Proteomics of the heart: Unraveling disease. Circ Res. 2006;98:309–321. doi: 10.1161/01.RES.0000201280.20709.26. [DOI] [PubMed] [Google Scholar]
- 9.Jonsson AP, Aissouni Y, Palmberg C, Percipalle P, Nordling E, Daneholt B, Jornvall H, Bergman T. Recovery of gel-separated proteins for in-solution digestion and mass spectrometry. Anal Chem. 2001;73:5370–5377. doi: 10.1021/ac010486h. [DOI] [PubMed] [Google Scholar]
- 10.Horth P, Miller CA, Preckel T, Wenz C. Efficient fractionation and improved protein identification by peptide offgel electrophoresis. Mol Cell Proteomics. 2006;5:1968–1974. doi: 10.1074/mcp.T600037-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Elschenbroich S, Ignatchenko V, Sharma P, Schmitt-Ulms G, Gramolini AO, Kislinger T. Peptide separations by on-line mudpit compared to isoelectric focusing in an off-gel format: Application to a membrane-enriched fraction from c2c12 mouse skeletal muscle cells. J Proteome Res. 2009;8:4860–4869. doi: 10.1021/pr900318k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernoult E, Gamelin E, Guette C. Improved proteome coverage by using itraq labelling and peptide offgel fractionation. Proteome Sci. 2008;6:27. doi: 10.1186/1477-5956-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wayman NS, Ellis BL, Thiemermann C. Simvastatin reduces infarct size in a model of acute myocardial ischemia and reperfusion in the rat. Med Sci Monit. 2003;9:BR155–159. [PubMed] [Google Scholar]
- 14.Ohlendieck K, Ervasti JM, Snook JB, Campbell KP. Dystrophin-glycoprotein complex is highly enriched in isolated skeletal muscle sarcolemma. J Cell Biol. 1991;112:135–148. doi: 10.1083/jcb.112.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane LA, Yung CK, Agnetti G, Neverova I, Van Eyk JE. Optimization of paper bridge loading for 2-de analysis in the basic ph region: Application to the mitochondrial subproteome. Proteomics. 2006;6:5683–5687. doi: 10.1002/pmic.200600267. [DOI] [PubMed] [Google Scholar]
- 16.Solaro RJ, Pang DC, Briggs FN. The purification of cardiac myofibrils with triton x-100. Biochim Biophys Acta. 1971;245:259–262. doi: 10.1016/0005-2728(71)90033-8. [DOI] [PubMed] [Google Scholar]
- 17.Fritz JD, Swartz DR, Greaser ML. Factors affecting polyacrylamide gel electrophoresis and electroblotting of high-molecular-weight myofibrillar proteins. Anal Biochem. 1989;180:205–210. doi: 10.1016/0003-2697(89)90116-4. [DOI] [PubMed] [Google Scholar]
- 18.Warren CM, Arteaga GM, Rajan S, Ahmed RP, Wieczorek DF, Solaro RJ. Use of 2-d dige analysis reveals altered phosphorylation in a tropomyosin mutant (glu54lys) linked to dilated cardiomyopathy. Proteomics. 2008;8:100–105. doi: 10.1002/pmic.200700772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 20.Layland J, Cave AC, Warren C, Grieve DJ, Sparks E, Kentish JC, Solaro RJ, Shah AM. Protection against endotoxemia-induced contractile dysfunction in mice with cardiac-specific expression of slow skeletal troponin i. Faseb J. 2005;19:1137–1139. doi: 10.1096/fj.04-2519fje. [DOI] [PubMed] [Google Scholar]
- 21.Hubner NC, Ren S, Mann M. Peptide separation with immobilized pi strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics. 2008;8:4862–4872. doi: 10.1002/pmic.200800351. [DOI] [PubMed] [Google Scholar]
- 22.Griffin TJ, Xie H, Bandhakavi S, Popko J, Mohan A, Carlis JV, Higgins L. Itraq reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. J Proteome Res. 2007;6:4200–4209. doi: 10.1021/pr070291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H, Freitas MA. Massmatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Mela L, Seitz S. Isolation of mitochondria with emphasis on heart mitochondria from small amounts of tissue. Methods in enzymology. 1979;55:39–46. doi: 10.1016/0076-6879(79)55006-x. [DOI] [PubMed] [Google Scholar]
- 28.Widnell CC, Hamilton TH, Tata JR. The isolation of enzymically active nuclei from the rat heart and uterus. J Cell Biol. 1967;32:766–770. doi: 10.1083/jcb.32.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bousette N, Kislinger T, Fong V, Isserlin R, Hewel JA, Emil A, Gramolini AO. Large-scale characterization and analysis of the murine cardiac proteome. J Proteome Res. 2009;8:1887–1901. doi: 10.1021/pr800845a. [DOI] [PubMed] [Google Scholar]
- 30.Donoghue PM, Hughes C, Vissers JP, Langridge JI, Dunn MJ. Nonionic detergent phase extraction for the proteomic analysis of heart membrane proteins using label-free lc-ms. Proteomics. 2008;8:3895–3905. doi: 10.1002/pmic.200800116. [DOI] [PubMed] [Google Scholar]
- 31.Gurel E, Smeele KM, Eerbeek O, Koeman A, Demirci C, Hollmann MW, Zuurbier CJ. Ischemic preconditioning affects hexokinase activity and hkii in different subcellular compartments throughout cardiac ischemia-reperfusion. J Appl Physiol. 2009;106:1909–1916. doi: 10.1152/japplphysiol.90537.2008. [DOI] [PubMed] [Google Scholar]
- 32.Loro E, Gianazza E, Cazzola S, Malena A, Wait R, Begum S, Brizio C, Dabbeni-Sala F, Vergani L. Development and characterization of polyspecific anti-mitochondrion antibodies for proteomics studies on in toto tissue homogenates. Electrophoresis. 2009;30:1329–1341. doi: 10.1002/elps.200800576. [DOI] [PubMed] [Google Scholar]
- 33.Zlatkovic J, Arrell DK, Kane GC, Miki T, Seino S, Terzic A. Proteomic profiling of katp channel-deficient hypertensive heart maps risk for maladaptive cardiomyopathic outcome. Proteomics. 2009;9:1314–1325. doi: 10.1002/pmic.200800718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matt P, Fu Z, Fu Q, Van Eyk JE. Biomarker discovery: Proteome fractionation and separation in biological samples. Physiol Genomics. 2008;33:12–17. doi: 10.1152/physiolgenomics.00282.2007. [DOI] [PubMed] [Google Scholar]
- 35.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 36.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 37.Manadas B, English JA, Wynne KJ, Cotter DR, Dunn MJ. Comparative analysis of offgel, strong cation exchange with ph gradient, and rp at high ph for first-dimensional separation of peptides from a membrane-enriched protein fraction. Proteomics. 2009;9:5194–5198. doi: 10.1002/pmic.200900349. [DOI] [PubMed] [Google Scholar]
- 38.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. Itraq underestimation in simple and complex mixtures: “The good, the bad and the ugly”. J Proteome Res. 2009;8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 39.Jullig M, Hickey AJ, Chai CC, Skea GL, Middleditch MJ, Costa S, Choong SY, Philips AR, Cooper GJ. Is the failing heart out of fuel or a worn engine running rich? A study of mitochondria in old spontaneously hypertensive rats. Proteomics. 2008;8:2556–2572. doi: 10.1002/pmic.200700977. [DOI] [PubMed] [Google Scholar]
- 40.Grant JE, Bradshaw AD, Schwacke JH, Baicu CF, Zile MR, Schey KL. Quantification of protein expression changes in the aging left ventricle of rattus norvegicus. J Proteome Res. 2009;8:4252–4263. doi: 10.1021/pr900297f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu WW, Wang G, Baek SJ, Shen RF. Comparative study of three proteomic quantitative methods, dige, cicat, and itraq, using 2d gel- or lc-maldi tof/tof. J Proteome Res. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 42.Kirk JA, MacGowan GA, Evans C, Smith SH, Warren CM, Mamidi R, Chandra M, Stewart AF, Solaro RJ, Shroff SG. Left ventricular and myocardial function in mice expressing constitutively pseudophosphorylated cardiac troponin i. Circ Res. 2009;105:1232–1239. doi: 10.1161/CIRCRESAHA.109.205427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai J, Ishikawa H, Kojima S, Satoh H, Yamamoto S, Kanaoka M. Proteomic analysis of rat heart in ischemia and ischemia-reperfusion using fluorescence two-dimensional difference gel electrophoresis. Proteomics. 2003;3:1318–1324. doi: 10.1002/pmic.200300432. [DOI] [PubMed] [Google Scholar]
- 44.He H, Chen M, Scheffler NK, Gibson BW, Spremulli LL, Gottlieb RA. Phosphorylation of mitochondrial elongation factor tu in ischemic myocardium: Basis for chloramphenicol-mediated cardioprotection. Circ Res. 2001;89:461–467. doi: 10.1161/hh1701.096038. [DOI] [PubMed] [Google Scholar]
- 45.Zou N, Ao L, Cleveland JC, Jr., Yang X, Su X, Cai GY, Banerjee A, Fullerton DA, Meng X. Critical role of extracellular heat shock cognate protein 70 in the myocardial inflammatory response and cardiac dysfunction after global ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2008;294:H2805–2813. doi: 10.1152/ajpheart.00299.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Kam KW, Zhou JJ, Yan WY, Chen M, Wu S, Wong TM. Effects of heat shock protein 70 activation by metabolic inhibition preconditioning or kappa-opioid receptor stimulation on ca2+ homeostasis in rat ventricular myocytes subjected to ischemic insults. J Pharmacol Exp Ther. 2004;310:606–613. doi: 10.1124/jpet.104.067926. [DOI] [PubMed] [Google Scholar]
- 47.Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Sutter S, Borisov AB, Pumplin DW, Russell MW, Bloch RJ. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. Faseb J. 2006;20:2102–2111. doi: 10.1096/fj.06-5761com. [DOI] [PubMed] [Google Scholar]
- 48.Chang H, Chu XY, Zou J, Chang TH. Effect of dl-praeruptorin a on desmin and vimentin content in rat ischemia/reperfusion myocardiocytes. Chin Med J (Engl) 2007;120:2256–2259. [PubMed] [Google Scholar]
- 49.Yuan C, Guo Y, Ravi R, Przyklenk K, Shilkofski N, Diez R, Cole RN, Murphy AM. Myosin binding protein c is differentially phosphorylated upon myocardial stunning in canine and rat hearts-- evidence for novel phosphorylation sites. Proteomics. 2006;6:4176–4186. doi: 10.1002/pmic.200500894. [DOI] [PubMed] [Google Scholar]
- 50.Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Seidman CE, Seidman JG, Robbins J. Cardiac myosin binding protein c phosphorylation is cardioprotective. Proc Natl Acad Sci U S A. 2006;103:16918–16923. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.