Abstract
Inhibitors of the transmembrane protein sarco/endoplasmic reticulum calcium ATPase (SERCA) are invaluable tools for the study of the enzyme’s physiological functions and they have been recognized as a promising new class of anticancer agents. For the discovery of novel enzyme inhibitors, small molecule docking for virtual screens of large compound libraries has become increasingly important. Since the performance of various docking routines varies considerably, depending on the target and the chemical nature of the ligand, we critically evaluated the performance of four frequently used programs – GOLD, AutoDock, Surflex-Dock, and FRED – for the docking of SERCA inhibitors based on the structures of thapsigargin, di-tert-butylhydroquinone, and cyclopiazonic acid. Evaluation criteria were docking accuracy using crystal structures as references, docking reproducibility, and correlation between docking scores and known bioactivities. The best overall results were obtained by GOLD and FRED. Docking runs with conformationally flexible binding sites produced no significant improvement of the results.
Keywords: computational docking, scoring function, inhibitory potency, calcium pump, thapsigargin, di-tert-butylhydroquinone, cyclopiazonic acid, inhibitor binding site
Introduction
Since its conception in the 1980s, small ligand docking has become an increasingly important tool for the computational analysis of binding interactions between proteins and ligands [1, 2]. By predicting the most favorable position (pose) of a ligand in a protein binding site, docking is capable of revealing crucial protein/ligand interactions at the molecular level. Since docking programs can also rank-order the predicted binding affinities of multiple ligands for a given protein target and are much more rapid than time-intensive molecular dynamics calculations, they have become the method of choice for virtual high-throughput screening (vHTS) of large compound libraries for lead compounds with specific bioactivities. Therefore, docking is nowadays an integral part of many drug discovery and design programs in the pharmaceutical industry. In structure-based design projects, docking is not just a powerful tool for virtual library screens prior to experimental testing, but it can also guide the structural optimization of lead compounds. Each docking routine has two essential components, a scoring function that assigns a numerical fitness value to a computed protein/ligand conformation and a search algorithm that identifies the ligand pose with the highest fitness score in the protein binding site [3].
During the last three decades, docking routines have undergone remarkable improvements. Whereas the first docking algorithms treated both protein and ligand as rigid bodies [4], the next generation of programs permitted the ligand to be fully flexible while keeping the protein essentially static. Only with the advent of some of the most recent docking programs, partial or complete conformational flexibility of the residues in the protein binding site has become reality, allowing the simulation of a so-called “induced fit” [5–7]. Despite its widespread use, the term “induced fit” is somewhat misleading since recent studies have shown that a ligand itself cannot induce substantial conformational changes in a protein. Rather, it is thought that a ligand-free protein is dynamic and exists in an ensemble of different, low energy conformations. Ligand binding to the protein tends to selectively stabilize one of these preexisting conformers, which can differ from the conformer favored in the ligand-free state [8, 9]. Thus, we will use the term “conformational selection” instead of “induced fit” when referring to the abovementioned phenomenon throughout the remainder of this study. Despite significant improvements and remarkable overall performances, docking programs still face challenges. Examples include difficulties associated with the proper simulation of conformational selection if the changes in receptor structure are not local but encompass the movement of entire domains, the presence of interacting water molecules in the binding pocket, or metal atoms that form bonds with the ligand.
Two popular docking programs, GOLD (genetic optimisation for ligand docking; Cambridge Crystallographic Data Centre) [10, 11] and AutoDock (The Scripps Research Institute) [12–14] explore the conformational space of a ligand using genetic search algorithms in combination with various scoring functions. The most recent releases of these two programs allow for full conformational flexibility of the residues lining the binding site. This feature is believed to be an improvement over the commonly employed strategy of mimicking a conformationally flexible binding site by creating ensembles of various (rigid) protein conformations into which the ligand is docked. The program Surflex-Dock (Tripos), which is integrated into the SYBYL modeling suite, builds ligands from fragments in the binding site and evaluates their fitness with the Hammerhead scoring function [15–17]. FRED (fast rigid exhaustive docking), which is part of the OpenEye suite (OpenEye Scientific Software), implements conformational flexibility of the ligand by creating an ensemble of ligand conformers that are then rigidly docked into the binding site [18–20]. The rigid docking approach makes computations with FRED very rapid, which is an advantage when screening libraries with hundreds of thousands compounds.
The overall performance of most docking routines has been evaluated extensively using different approaches that take into account different features of the programs. So-called enrichment studies, for instance, evaluate a program’s ability to correctly identify known actives that are part of a much larger set of inactive compounds. A few examples for this widely used method include an evaluation of the program AutoDock in comparison to DOCK and FlexX using four different target receptors and about 1,000 compounds [21] as well as a comprehensive performance assessment of the programs Glide, GOLD, and DOCK with regard to the docking of sets of about 1,000 drug-like molecules containing known actives and decoys into 23 pharmacologically relevant receptors [22]. Two more examples for this type of study comprise an evaluation of five different scoring functions in conjunction with a single program, FlexX, to identify a set of known inhibitors of a specific serine protease concealed in a pool of over 25,000 drug-like compounds [23] and a critical comparison of the accuracy and speed by which the routines DOCK, DockVision, Glide, and Gold could identify 49 active molecules in a pool of more than 1,000 inactive ones [24].
Another widely used strategy assesses the accuracy of docked-predicted ligand poses, using the known ligand positions from X-ray crystal structures as a reference point. Two representative studies among many examined the ability of the programs ISE-dock, Glide, GOLD, and AutoDock to correctly reproduce the known ligand poses of 81 protein/ligand complexes [25] or applied GOLD and FlexX to predict the binding of a set of meptazinol analogs into the enzyme acetylcholinesterase [26]. A review by Cole et al comments on the difficulties associated with root-mean-square deviation-based (RMSD) and enrichment-based comparisons of docking programs [27].
Unfortunately, most studies arrived at the conclusion that their overall findings are difficult to generalize and that a particular docking protocol might work better for certain targets or compound classes than for others. Thus, when working with a particular protein and ligand class, an individual evaluation of available docking routines still remains a necessity. The lack of general guidelines regarding the performance of docking software is exacerbated for transmembrane proteins, which – due to the difficulties associated with their crystallization – are vastly underrepresented in the protein databank. As a result, most validation studies that strive to be representative of the overall content of the protein databank entries include few transmembrane proteins, despite their medicinal relevance as drug targets.
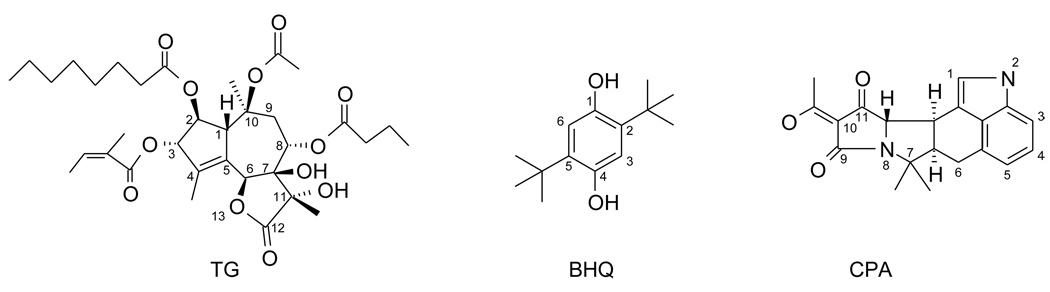
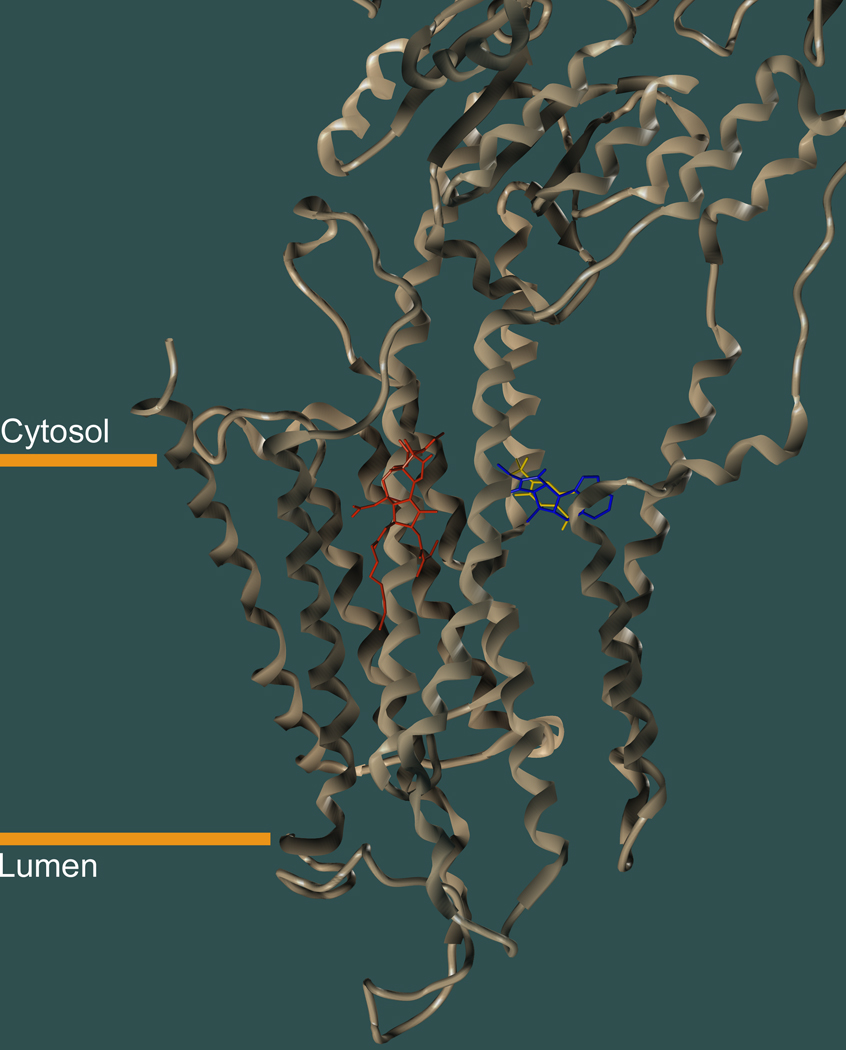
One member of this relatively small group of transmembrane proteins for which high resolution X-ray crystal structures exist is the sarco/endoplasmic reticulum calcium ATPase (SERCA), an ion transport protein present in the membranes of intracellular calcium stores [28–33]. The rapid release of calcium ions from these stores triggers a variety of physiologically important functions, such as muscle contraction. In numerous studies, the ability of small molecules to inhibit SERCA’s ion transport activity has been exploited for the investigation of the enzyme’s role in physiological processes [34]. Recently, SERCA inhibition has been suggested to be of therapeutic value in chemotherapy of prostate cancer, which has triggered new interest in the development of novel SERCA inhibitors [35, 36]. Small molecule SERCA inhibitors (Scheme 1) include thapsigargin (TG), di-tert-butylhydroquinone (BHQ), and cyclopiazonic acid (CPA), all of which bind to the transmembrane domain of the enzyme close to the membrane/cytosolic interface (Fig. 1). Although structurally quite different from each other, all three compounds possess a cyclic core carrying various substituents. TG, the most potent and most frequently used SERCA inhibitor, is a natural product produced by the Mediterranean plant Thapsia garganica [37, 38]. TG is a sesquiterpene lactone with a rigid three-membered core that bears side chains of considerable conformational flexibility. The inhibitor BHQ is a symmetric hydroquinone with two tert-butyl groups [39, 40] whereas CPA, a metabolite originally isolated from the fungus Penicillium cyclopium, is composed of five fused rings each with five or six heavy atoms [41, 42]. In contrast to TG, the conformational flexibility of BHQ and CPA is limited. All three compounds bind to the transmembrane domain of SERCA at the lipid/protein interface, which accounts for their relatively hydrophobic overall character (Fig. 1). The smaller BHQ binding site is part of the larger CPA site, which is located in close proximity of the TG binding site. For all three inhibitors, crystal structures of SERCA in complex with the inhibitor are available, thereby facilitating their study by docking-based methodologies [29, 43].
Scheme 1.
Molecular structure of the SERCA inhibitors TG, BHQ, and CPA.
Figure 1.
Location of the TG (red), BHQ (yellow), and CPA (blue) binding sites in the transmembrane domain of SERCA. The X-ray crystal structures of the TG/BHQ/SERCA (PDB entry: 2AGV [29]) and the CPA/SERCA (PDB entry: 1EAS [43]) complexes were superimposed by aligning the α-carbon atoms of the transmembrane segments.
Since systematic performance comparisons of docking routines for transmembrane proteins are rare and none to our knowledge have been conducted for SERCA inhibitors, we evaluated the performance of the abovementioned four programs: GOLD, AutoDock, Surflex-Dock, and FRED. A major criterion for the performance of the programs was their ability to both correctly and reproducibly dock the previously deleted ligand back into the X-ray crystal structures of SERCA. In the next step, analogs of TG and BHQ with known bioactivities were also docked and the correlation of docking score and bioactivity was assessed. In the case of Gold and AutoDock, the effects of conformational flexibility of the binding site on the results were explored as well.
Methods
Preparation of the SERCA receptor and the ligand structures
The coordinates of the crystal structures of SERCA/inhibitor complexes were downloaded from the Protein Databank (http://www.rcsb.org) (TG and BHQ: 2AGV [29]; CPA: 2EAS [43]) and imported into the modeling software SYBYL (version 8.0; Tripos, St. Louis, MO). All non-protein components such as water molecules, inhibitors, metal ions, and lipids were deleted and hydrogen atoms were added to the protein structures. Partial charges were assigned according to the Amber library and the positions of the added hydrogen atoms were optimized by molecular mechanics while keeping the positions of the heavy atoms static [44]. Energy minimization was performed with the Powell method in combination with the Amber7 FF99 force field, a distance-dependent dielectric constant of 4 (mimicking the hydrophobic character of SERCA’s transmembrane domain), and a convergence criterion of 0.05 kcal/(mol Å).
The molecular structures of inhibitors were also prepared in SYBYL. Since CPA has four tautomers, its structure was modeled according to the form specified in the original report of the SERCA/CPA X-ray crystal structure (Scheme 1). Prior to importing the files into the various docking programs, the conformational energy of each structure was minimized by molecular mechanics (MMFF94s force field, MMFF94 charges, and distance-dependent dielectric constant of 4) using the conjugate gradient method and a termination criterion of 0.01 kcal/(mol Å).
Docking – general procedures
To minimize user bias, all docking runs were conducted at settings as close to the defaults as possible. Docking with GOLD (version 4.0) and FRED (version 2.2.5) was performed on a Windows server equipped with an Intel Core2 processor (2.4 GHz) and 3 GB of RAM. AutoDock (version 4.0) and Surflex-Dock (version 4.0) were running on a LINUX (Red Hat) server with an Intel Pentium D processor (3.0 GHz) and 4 GB of RAM. All programs were set to conduct 30 independent repeats for each ligand under identical conditions.
Consensus sizes, defined as the number of repeats that reproduced the top-scoring pose, were determined by visual inspection of results from independent repeats. In the case of GOLD, Surflex, and FRED, this inspection was conducted with the tool VIDA (version 4.0.0; OpenEye Scientific Software), whereas AutoDockTools (version 1.5.1; The Scripps Research Institute) was employed to analyze AutoDock results. RMSD values between the heavy atoms of a top-score and the ligand pose in the crystal structure were calculated in SYBYL using a slightly modified SPL script originally authored by Jon Swanson. Execution times were ascertained from the respective docking log files.
Docking with GOLD
All three scoring functions that were available for GOLD at the time of the study (GoldScore, ChemScore, and Astex Statistical Potential (ASP)) were tested in separate runs. The genetic search algorithm was executed at the default settings and the docking sphere had a radius of 15 Å centered at the position of the following (deleted) ligand atoms: C2 in the case of TG; C2 in the case of BHQ, and C7 in the case of CPA (see Scheme 1 for numbering).
Docking with AutoDock
AutoDock runs were performed using a C-shell script that was a compilation of AutoDock virtual screening scripts released by The Scripps Research Institute. Intermediary steps, such as grid box creation, were completed using AutoDockTools. The grid box was centered at the coordinates defined for GOLD docking (see above). The number of points in each direction was set to 66, and the point spacing was set to 0.369 Å, giving a search volume comparable to that of the GOLD docking sphere (approximately 14,000 Å3).
Docking with Surflex-Dock
Prior to docking runs, Surflex-Dock requires the generation of a so-called protomol, an idealized representation of the binding site that defines the search area. For this purpose, the prepared receptor file along with the unaltered ligand from the crystal structure were loaded and the center of protomol generation was focused around the area occupied by the ligand, leaving the threshold and bloat parameters at their default values of 0.50 and 0 Å, respectively. During the subsequent docking runs, all other adjustable parameters were left at their default values.
Docking with FRED
FRED was the only program that required both a specially prepared receptor file and a ligand conformer library. The receptor file was prepared with FRED Receptor 2.2.5 (OpenEye Scientific Software). The size of the docking box centered on the co-crystallized ligand was expanded in all directions until its volume was approximately 14,000 Å3. In order to force FRED to search the entire volume of the docking box, no contours were created that would have limited the search space to the volume enclosed by them. Ligand conformer libraries were created in Omega 2.3.2 (OpenEye Scientific Software). For docking of analogs, VIDA was used to merge the structure of a parent ligand with those of the analogs (e.g. BHQ and its 10 derivatives) into a single file. Omega was then used to create conformer libraries using this file as input along with a dielectric constant of 4.0 and the search force field MMFF94s.
To be consistent with the other docking programs, FRED docking was limited to using the same scoring function throughout the entire search and for the rank-ordering of results (no rescoring with different functions was permitted). This limited the available scoring functions to Shapegauss, Piecewise Linear Potential (PLP), Chemical Gaussian Overlay (CGO), Chemgauss 2, and Chemgauss 3. The optimization scoring function was identical the search function and consensus scoring was disabled.
Results and Discussion
Comparison of accuracy and reproducibility of docking results
The accurate prediction of the pose of a ligand in its receptor’s binding site determines the value of any docking program. The accuracy of a docking result is frequently measured by the RMSD between the docking-predicted and the experimentally observed heavy atom positions of the ligand, using a somewhat arbitrary chosen RMSD cut-off value, usually somewhere between 2 and 3 Å [20, 24, 26, 27, 45]. When comparing structurally different ligands, however, the use of RMSD as the only measure for the success of a docking result can be problematic [27]. For example, RMSD calculations tend to favor smaller ligands which frequently yield lower RMSD values, even for incorrect poses as long as the center of gravity of the molecule is approximately the same. Accordingly, the RMSD threshold for successful docking should be relatively low for a molecule the size of the inhibitor BHQ whereas higher values should be acceptable for the larger molecules TG and CPA. Another issue concerning RMSD values is encountered if a ligand possesses a flexible terminal group, such as a solvent-exposed chain, whose position may fluctuate since the group is not engaged in strong interactions with the protein. RMSD evaluations disproportionally penalize binding poses that differ in the conformation of such a flexible tail, even though the overall position and crucial protein/ligand interactions in the rest of the molecule might be correct. Among the ligands docked in this study, such a scenario was encountered in the case of TG which bears a highly flexible octanoic acid residue at C2. The alkyl chain of this substituent makes hydrophobic contacts with non-polar patches on the outer surface of SERCA’s transmembrane domain and possibly with surrounding lipids. As a result, this alkyl chain can adopt a variety of orientations that are similarly favorable.
In order to avoid the abovementioned two problems of RMSD-based evaluations, we used a RMSD threshold of 2 Å for the small BHQ and of 3 Å for the larger TG and CPA molecules. Moreover, we performed RMSD calculations for TG with the flexible chain at C2 removed to avoid unreasonably high RMSD penalties. In addition, we always complemented RMSD-based assessments of the success of a particular docking run by a visual comparison of the top-ranked solution and the inhibitor position in the X-ray crystal structure. Lastly, a docking routine’s ability to reliably reproduce the top-ranked pose in repeated runs was quantified in terms of the consensus size, which was used as a further performance indicator.
Analysis of the docking results revealed considerable differences in the performance of the docking routines (Table 1–Table 3). The program GOLD performed overall very well and was able to successfully predict the poses of almost all inhibitors, with the exception of CPA if ASP was the scoring function (left column of Fig. 2 and Table 1). In the case of GoldScore, the seemingly high RMSD value of 2.1 Å obtained for BHQ was caused by a 180° rotation of the ligand around the axis formed by C2 and C5, resulting in an orientation that left the positions of the tert-butyl groups unchanged and that preserved the two hydrogen bonds between the inhibitor’s hydroxyl groups and SERCA. Thus, the prediction of the major features of BHQ binding by GoldScore was essentially correct.
Table 1.
Docking results (30 repeats) obtained from GOLD and three different scoring functions. For each set of repeats, the number of results in agreement with the top score (#) and the RMSD (Å) between the top score and the crystal structure pose (heavy atoms) are indicated.
| ASP | ChemScore | GoldScore | ||||
|---|---|---|---|---|---|---|
| # | RMSD | # | RMSD | # | RMSD | |
| TG | 28 | 3.30 | 17 | 1.54 | 23 | 2.56 |
| TG* | 1.43 | 1.53 | 1.11 | |||
| BHQ | 30 | 0.18 | 30 | 0.50 | 29 | 2.12 |
| CPA | 8 | 10.38 | 12 | 1.92 | 22 | 1.62 |
TG* refers to RMSD values after the removal of the molecule’s octanoic acid residue at C-2.
Table 3.
Docking results (30 repeats) obtained with FRED and its five scoring functions. For each set of repeats, the number of results in agreement with the top score and the RMSD (Å) between the top score (#) and the crystal structure pose (heavy atoms) are indicated.
| Shapegauss | PLP | CGO | Chemgauss 2 | Chemgauss 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| # | RMSD | # | RMSD | # | RMSD | # | RMSD | # | RMSD | |
| TG | 17 | 5.76 | 12 | 1.84 | 30 | 1.16 | 14 | 9.94 | 30 | 2.30 |
| TG* | 5.91 | 1.13 | 1.16 | 8.13 | 1.44 | |||||
| BHQ | 29 | 0.86 | 25 | 0.67 | 23 | 0.36 | 1 | 11.31 | 14 | 1.11 |
| CPA | 22 | 2.43 | 2 | 1.69 | 20 | 1.29 | 1 | 10.56 | 3 | 3.30 |
TG* refers to RMSD values after the removal of the molecule’s octanoic acid residue at C-2.
Figure 2.
Comparison of top-ranked docking poses to the known position of the inhibitors BHQ (top row), TG (middle row), and CPA (bottom row) according to crystallographic information (yellow) obtained with GOLD/ChemScore (red), AutoDock (green), Surflex-Dock (blue), and FRED/CGO (orange).
Similar to GOLD, AutoDock predicted the poses of all three inhibitors correctly. One noticeable limitation related to TG, whose pose was predicted correctly but whose reproducibility was remarkably low (left column of Fig. 2 and Table 2). Surflex-Dock, on the other hand, performed very well with TG but failed entirely in the case of BHQ and CPA (right column of Fig. 2 and Table 2).
Table 2.
Docking results (30 repeats) obtained with AutoDock and Surflex-Dock with their built-in scoring functions. For each set of repeats, the number of results in agreement with the top score (#) and the RMSD (Å) between the top score and the crystal structure pose (heavy atoms) are indicated.
| AutoDock | Surflex-Dock | |||
|---|---|---|---|---|
| # | RMSD | # | RMSD | |
| TG | 1 | 2.20 | 30 | 3.22 |
| TG* | 1.64 | 1.33 | ||
| BHQ | 30 | 0.74 | 9 | 6.41 |
| CPA | 27 | 2.52 | 2 | 5.93 |
TG* refers to RMSD values after the removal of the molecule’s octanoic acid residue at C-2.
The performance of FRED was highly dependent on the scoring function used (right column of Fig. 2 and Table 3). Among the five functions tested, the best results with regard to accuracy and reproducibility were obtained with CGO. PLP also gave excellent accuracy, but low reproducibility in the case of CPA.
Interestingly, most of the docking difficulties were encountered with CPA, which was surprising since rigid molecules are usually more reliably docked than flexible ones. One possible explanation could relate to the resolution of the crystal structure of the SERCA/CPA complex, which was of lower quality (3.4 Å) than that of the SERCA/BHQ/TG complex (2.4 Å) [29, 43]. Since certain types of protein/ligand interactions are strongly distance-dependent (London dispersion interactions, for example), even slight inaccuracies in the binding site geometry can have a significant impact on computationally predicted binding poses, which might account for the observed shortcomings.
In an attempt to identify characteristics of docking routines and fitness functions that produce favorable results for SERCA inhibitors, the properties of the best-performing routines and functions were examined. With regard to the search algorithm, the programs GOLD and AutoDock are similar since they both utilize stochastic, genetic search algorithms to flexibly dock the entire ligand into the binding site [11, 13]. In contrast, Surflex-Dock builds a given ligand by assembling it from conformationally optimized fragments in the binding site and maximizing its similarity to that of a previously defined idealized ligand [15]. FRED, on the other hand, is non-stochastic and finds the most favorable ligand pose via a systematic exploration of conformational and rotational space [19].
The majority of the more successful scoring functions – ChemScore [46, 47], the Hammerhead scoring function Surflex-Dock [15, 17], and AutoDock’s built-in scoring function [48] – are empirical and have been calibrated by regression against measured binding affinities for ligand/receptor complexes with known structures. They take into account a sum of terms that contribute to the overall free energy of binding, such as dispersion interactions, hydrogen bonds, electrostatic interactions, desolvation energies, hydrophobic interactions, and entropy terms. A quite different approach is implemented in ASP, which employs statistical potentials that reflect the frequency of interactions between ligand and receptor atoms in crystal structures of ligand/protein complexes. CGO, on the other hand, focuses solely on molecular shape of the ligand and uses Gaussian functions to compute how well a given ligand’s pose overlaps with that of the original ligand in the crystal structure [18]. One might therefore argue that CGO has somewhat of an “unfair” advantage since it explicitly utilizes the position of the co-crystallized ligand whereas the other functions do not make use of that information, other than for defining the location of the binding site.
Given that the best results were obtained by two fundamentally different algorithms (GOLD versus FRED) using two unrelated scoring functions (ChemScore versus CGO), we were unable to generalize our observations with regard to what general type of algorithm or fitness functions is likely to generate best results for SERCA inhibitors. Rather, we concur with the majority of previous studies that concluded that the performance of different docking programs and scoring functions is specific for a given receptor and type of ligand.
Correlation between docking score and bioactivity
In addition to correctly predicting binding poses, a useful docking program should provide a good measure for a ligand’s affinity for the target protein. By design, the value of the scoring function for a given ligand directly relates to its affinity for the target, often in a linear fashion [3, 16–18, 46, 47, 49].
Using a set of TG and BHQ analogs with known bioactivities [50, 51], we evaluated the ability of the four docking programs to predict inhibitor affinities for SERCA. The inhibitory potencies of these compounds had been determined by the same type of assay and covered an activity range of almost four orders of magnitude. The BHQ analogs differed from each other with regard to the chemical structure and position of the hydroxyl and alkyl groups at the central phenyl ring. Within the set of TG analogs, structural variations related to the shortening of the aliphatic chain at C2, the reduction of the carbonyl group at C12, the introduction of a five-membered ring about C11 and C12, and the hydrolysis of the acetate group at C10. It should be noted that with the exception of two remotely related compounds [52], no comparable experimental data sets are available for CPA analogs, which is why this inhibitor class was not included in this part of the study.
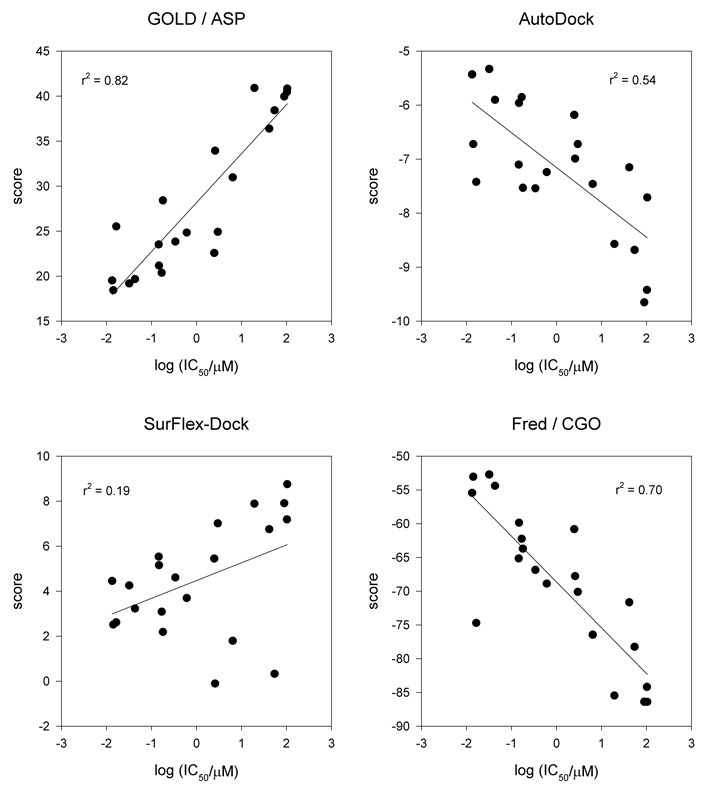
Both for GOLD and FRED, only the scoring function that had performed best for BHQ and TG (not CPA) according to the criteria for accuracy defined in the previous section was employed (ASP and CGO, respectively). Graphical analysis of semi-logarithmic plots of docking score (top-ranked result) versus bioactivity (Fig. 3) showed that the best correlation was obtained by the program GOLD in conjunction with the scoring function ASP (r2 = 0.82). This value signifies a noticeable improvement over results obtained in earlier studies that focused either on BHQ or TG analogs alone [50, 53]. In the case of FRED, the scoring function CGO was able to establish a good correlation as well (r2 = 0.70). Whereas the correlation for AutoDock was somewhat lower but still at an acceptable level (r2 = 0.54), the poor correlation obtained with Surflex-Dock (r2 = 0.19) prohibited a reliable prediction of bioactivity. In the latter case, the correlation could have been notably improved by the removal of three poorly predicted outliers, but such action would have violated the premise of minimizing user input.
Figure 3.
Correlation between docking score and bioactivity for TG and BHQ analogs. For programs with multiple scoring functions, only results for the best-performing function for these two compound classes (GOLD: ASP; FRED: CGO) are displayed.
Overall, the results indicated that the best-performing docking routines, in particular GOLD/ASP and FRED/CGO, permitted the prediction of inhibitory potency within one to two orders of magnitude. Whereas this level of accuracy might be inadequate for the development of detailed structure-activity relationship studies at the molecular level, it is certainly sufficient for the development of docking protocols suited for virtual screens of large compound libraries for the identification of lead compounds that can serve as the starting point for the design of novel SERCA inhibitors with improved properties [54].
Effect of conformational flexibility of the binding site
At the time of this study, the only programs allowing for full conformational flexibility of residues lining the protein binding site were GOLD (only in conjunction with the GoldScore scoring function) and AutoDock. In principle, this feature represents a significant improvement over previous program versions since it can simulate – within certain limits – conformational selection, a phenomenon describing the stabilization of the protein in a conformation that is different from the conformation in the ligand-free state. Originally proposed by Koshland under the term “induced fit” [7], the concept of conformational selection has been observed in some proteins (the glycolytic enzyme hexokinase, for example [55]), whereas others are known to not appreciably alter their binding site structure upon ligand binding [56]. Flexible docking becomes most valuable if the docked ligand’s structure differs significantly from that of the parent ligand in the crystal structure because under these circumstances, the likelihood of a conformational selection is greatest. It should be noted, though, that the docking-based simulations are restricted to local conformational changes confined to the immediate proximimity of the binding site and cannot properly mimic some of the large-scale conformational changes that involve the movement of the protein backbone or even entire protein domains.
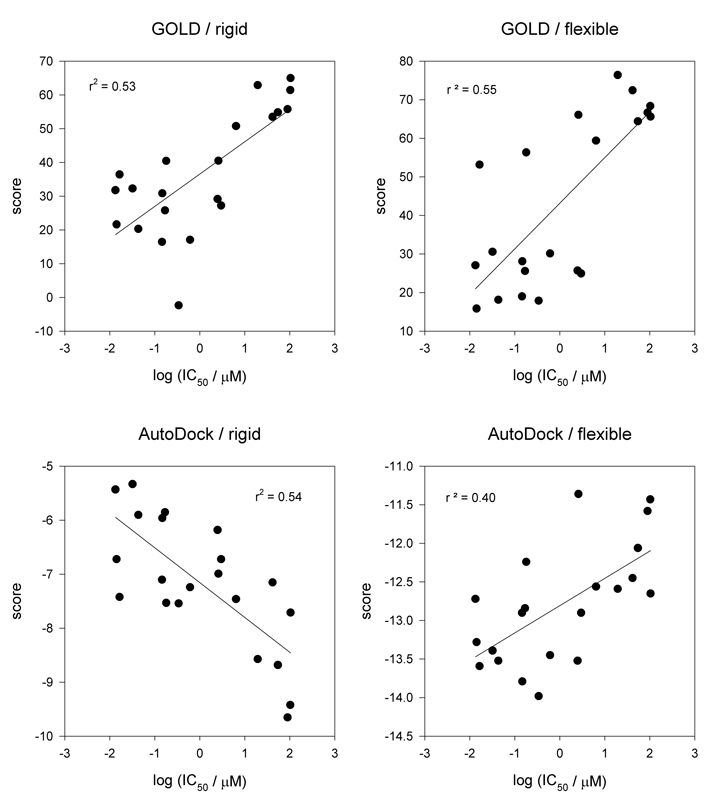
It is well-known that SERCA undergoes significant conformational changes as it moves through its catalytic cycle [28, 31, 57]. Almost all SERCA inhibitors, including the ones examined in the present study, bind to SERCA in its so-called E2 conformation and prevent it from reverting into the E1 conformation, a step necessary for maintaining catalytic activity. Whereas no major differences between the ligand-free and ligand-bound E2 forms have been observed, it is presently unknown if smaller changes occur that would be confined to residues in the immediate proximity of the inhibitor. In order to explore this possibility, we used the same series of SERCA inhibitors described above to conduct flexible docking runs with GOLD (GoldScore) and AutoDock, allowing the side chains of ten amino acids that were closest to the parent inhibitor to be fully flexible. These were Asp59, Leu61, Val62, Leu65, Asn10, Leu253, Phe256, Ile307, Glu309, and Leu311 in the case of BHQ and Phe256, Gln259, Leu260, Val263, Ile765, Val769, Leu828, Ile829, Phe834, and Met838 in the case of TG. Whereas the two docking protocols (GOLD/GoldScore and AutoDock) achieved almost identical correlation coefficients if docking was conducted in rigid fashion, this was clearly not the case if the binding site was allowed to flex (Fig. 4). In the case of GOLD/GoldScore, the improvement of the correlation was insignificant (from r2 = 0.53 to r2 = 0.55) and in the case of AutoDock, having a flexible binding site was detrimental to the correlation (from r2 = 0.54 to r2 = 0.40). Clearly, there was no obvious benefit of flexible docking for these two SERCA inhibitor types.
Figure 4.
Effect of flexible docking. Correlation of docking score and bioactivity for TG and BHQ analogs. Results were obtained with GOLD (GoldScore) and AutoDock using a rigid (left panels) and flexible (right panels) binding site comprised of ten residues (see Results and Discussion).
This observed lack of improvement could have several reasons. The most straightforward explanation would be that the SERCA inhibitor binding sites are rather rigid and do not undergo appreciable conformational changes upon inhibitor binding. This would be consistent with the finding that rigid docking produced results of similar quality as flexible docking in the case of GOLD, but it does not account for the decrease in correlation in the case of AutoDock. Alternatively, the structural differences within the set of analogs might not have been sufficient to stabilize the binding site in a substantially altered conformation. Whereas the BHQ and TG analogs considered in this study exhibited systematic structural variations, they still resembled each other in a sense that their main structural scaffolds (i.e. the cyclic core) were preserved. Provided that bioactivities become available, it would be interesting to conduct similar simulations with a structurally highly diverse set of analogs to see if the observed trend persists.
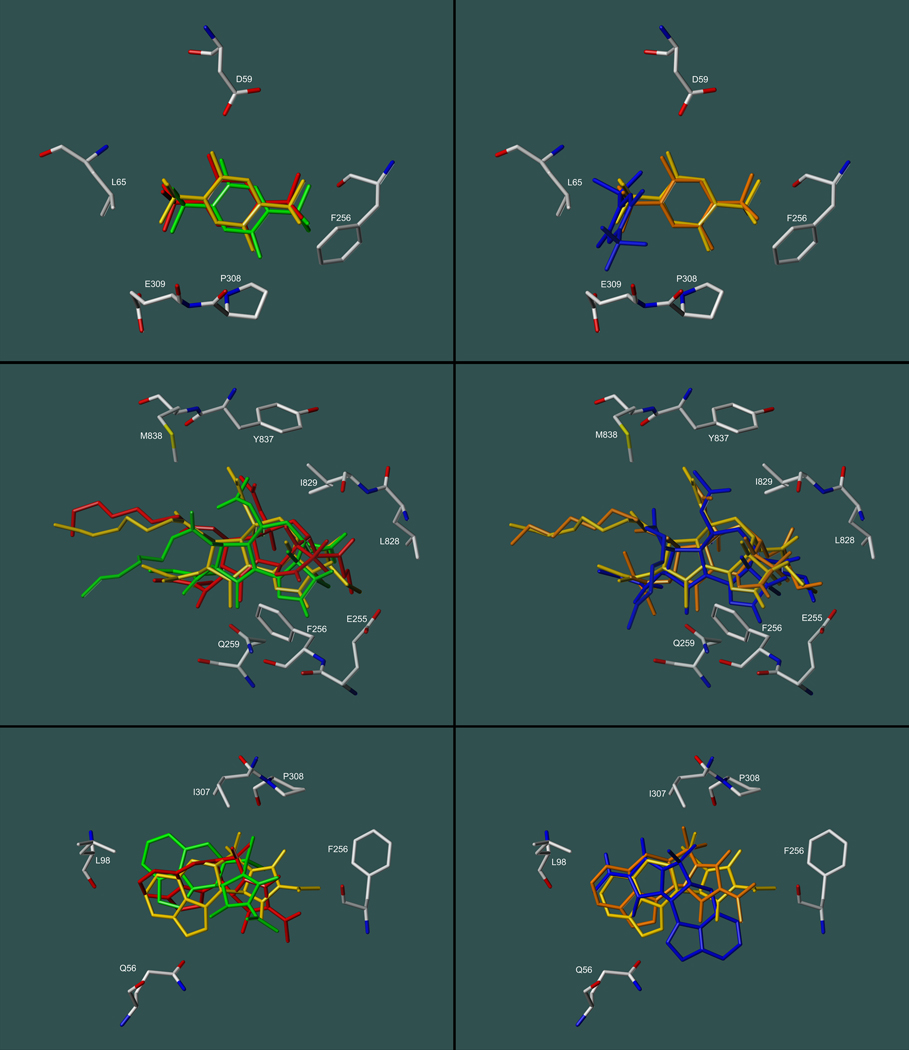
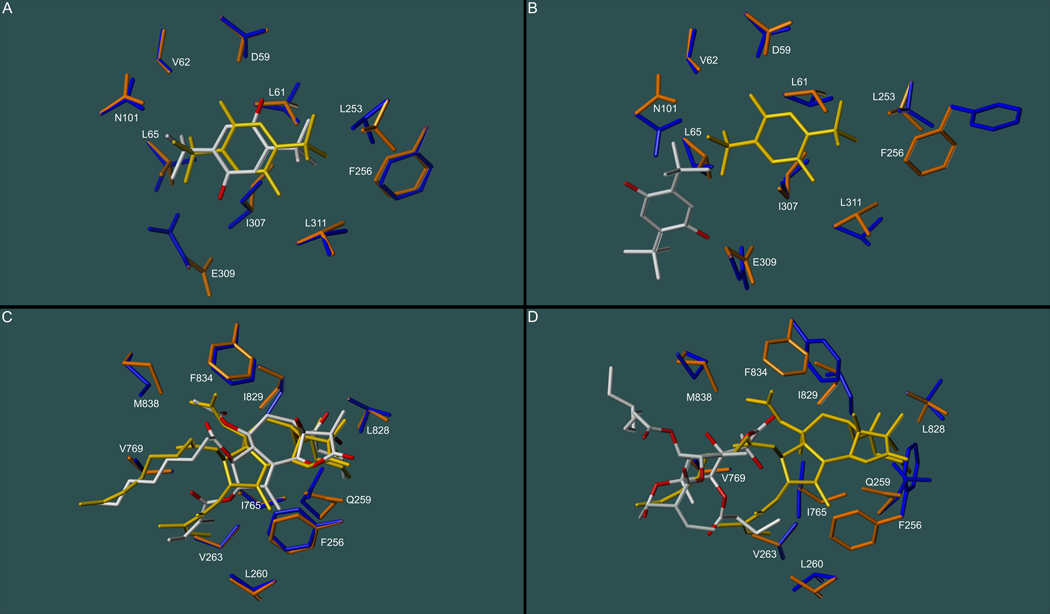
Another possibility would be a scenario in which the added conformational flexibility of the SERCA side chains may lead to hypothetical binding site conformations that might be energetically favorable from a computational aspect, but that are not assumed by the protein in reality. The challenges associated with the calculation of protein conformations by the relatively simplistic methodologies employed by the docking algorithms (such as molecular force fields) are well known. These difficulties may very well be accentuated by the fact that the SERCA inhibitor binding sites reside in the transmembrane domain at the protein/lipid interface of the enzyme with their openings exposed to the hydrophobic parts of the lipid bilayer, thereby creating a heterogeneous environment that cannot be approximated by continuum models with sufficient accuracy. Whether docking-predicted binding site conformations are correct or not can only be determined if the structure of the protein/ligand complex is known. Therefore, we compared flexible docking results for the compounds BHQ and TG to their corresponding crystal structure counterparts. Ideally, the docking routines should have left the residues in their original positions as defined by the crystal structure (Fig. 5). Whereas GOLD preserved the overall structure of the binding sites and the BHQ and TG positions, AutoDock predicted drastic changes both in the conformation of side chains (Phe256, Leu311, and Asn101, for instance) and in the position of the two inhibitors, which were severely dislodged from their original positions and thus presumed unrealistic. The observed trends became even more pronounced if analogs of BHQ and GOLD were docked (supplementary materials). Consistent with the correlations depicted in Fig. 4, GOLD’s performance with regard to flexible docking appears to be better than AutoDock’s, but certainly not better that rigid docking. Overall, the findings suggest that flexible docking simulations for the two SERCA inhibitor classes can lead to erroneous ligand and receptor conformations and are in need of further improvement to be of practical use.
Figure 5.
Docking-predicted conformational changes in the binding sited upon binding of BHQ (panels A and B) and TG (panels C and D). Docking was performed with GOLD/GoldScore (panel A for BHQ and panel C for TG) and AutoDock (panel B for BHQ and panel D for TG), showing noticeable deviations of computed side chain positions (blue) from the X-ray crystal structure (orange). Ligand positions are docking-predicted (CPK colored) and according to the crystallographic information (yellow).
One final issue pertinent to flexible docking is the inevitable increase in computation time to explore the additional conformational space. On average, the time to complete a flexible docking run for a BHQ analog was about three (GOLD) to six (AutoDock) times greater than for rigid docking. For the larger TG, the increase was proportionally smaller (a factor of 1.5 for GOLD and of 3 for AutoDock), but still evident. The additional time (several minutes per compound) is certainly not of concern if only a small number of compounds is under investigation, but might impose restrictions if large compound libraries are screened. Given the lack of improvement in docking performance and the added computation time, simulating a flexible SERCA binding site does presently not offer any real advantage, at least not for the two inhibitor classes investigated in this study.
Conclusion and Future Directions
The findings of our comparative study revealed substantial differences in the performance of commonly used programs for docking of SERCA inhibitors. This observation underscored the need for an individual evaluation of available software for a given inhibitor class and receptor type. The best overall results were obtained with GOLD and FRED, as far as docking accuracy and reproducibility were concerned. AutoDock and Surflex-Dock performed well with some inhibitors but not with others. Using a flexible binding site did not improve the quality of the docking results, suggesting a conformationally rigid inhibitor binding site and/or a need for further improvement of this relatively new feature. Notwithstanding the problems encountered, the best performing routines were able to accurately and reliably predict the binding pose of all three inhibitors in the transmembrane domain of SERCA and can therefore be the basis for future vHTS studies aimed at the discovery of novel SERCA inhibitors with improved properties.
Supplementary Material
Acknowledgements
This work was supported by a summer student fellowship to M.L. from the Greaves Foundation at Northern Kentucky University and by grants from the Kentucky Biomedical Research Infrastructure Network (P20RR016481-09), Research Corporation (Award 6843), and the National Institutes of Health (1R15GM084431-01) to S.P. We are grateful to OpenEye Scientific Software and The Scripps Research Institute (Arthur Olson) for making their docking software available. We also acknowledge Jon Swanson, formerly of Tripos, for writing the script for the calculation of RMSD values.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taylor RD, Jewsbury PJ, Essex JW. A review of protein-small molecule docking methods. Journal of computer-aided molecular design. 2002;16:151–166. doi: 10.1023/a:1020155510718. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer T, Rarey M. Computational methods for biomolecular docking. Current opinion in structural biology. 1996;6:402–406. doi: 10.1016/s0959-440x(96)80061-3. [DOI] [PubMed] [Google Scholar]
- 3.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: An overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 4.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromolecule-ligand interactions. Journal of molecular biology. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 5.Carlson HA, McCammon JA. Accommodating protein flexibility in computational drug design. Molecular pharmacology. 2000;57:213–218. [PubMed] [Google Scholar]
- 6.B-Rao C, Subramanian J, Sharma SD. Managing protein flexibility in docking and its applications. Drug discovery today. 2009;14:394–400. doi: 10.1016/j.drudis.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nature chemical biology. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valente AP, Miyamoto CA, Almeida FC. Implications of protein conformational diversity for binding and development of new biological active compounds. Current medicinal chemistry. 2006;13:3697–3703. doi: 10.2174/092986706779026147. [DOI] [PubMed] [Google Scholar]
- 10.Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. Journal of molecular biology. 1995;245:43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- 11.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. Journal of molecular biology. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 12.Goodsell DS, Olson AJ. Automated docking of substrates to proteins by simulated annealing. Proteins. 1990;8:195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- 13.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using Lamarckian genetic algorithm and an empirical bidning free energy function. J. Comp. Chem. 1998;19:1639–1662. [Google Scholar]
- 14.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. Journal of computer-aided molecular design. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 15.Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. Journal of medicinal chemistry. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 16.Welch W, Ruppert J, Jain AN. Hammerhead: fast, fully automated docking of flexible ligands to protein binding sites. Chemistry & biology. 1996;3:449–462. doi: 10.1016/s1074-5521(96)90093-9. [DOI] [PubMed] [Google Scholar]
- 17.Jain AN. Scoring noncovalent protein-ligand interactions: a continuous differentiable function tuned to compute binding affinities. Journal of computer-aided molecular design. 1996;10:427–440. doi: 10.1007/BF00124474. [DOI] [PubMed] [Google Scholar]
- 18.McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Gaussian docking functions. Biopolymers. 2003;68:76–90. doi: 10.1002/bip.10207. [DOI] [PubMed] [Google Scholar]
- 19.McGaughey GB, Sheridan RP, Bayly CI, Culberson JC, Kreatsoulas C, Lindsley S, Maiorov V, Truchon JF, Cornell WD. Comparison of topological, shape, and docking methods in virtual screening. Journal of chemical information and modeling. 2007;47:1504–1519. doi: 10.1021/ci700052x. [DOI] [PubMed] [Google Scholar]
- 20.Vigers GP, Rizzi JP. Multiple active site corrections for docking and virtual screening. Journal of medicinal chemistry. 2004;47:80–89. doi: 10.1021/jm030161o. [DOI] [PubMed] [Google Scholar]
- 21.Park H, Lee J, Lee S. Critical assessment of the automated AutoDock as a new docking tool for virtual screening. Proteins. 2006;65:549–554. doi: 10.1002/prot.21183. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Felts AK, Friesner RA, Levy RM. Comparative performance of several flexible docking programs and scoring functions: enrichment studies for a diverse set of pharmaceutically relevant targets. Journal of chemical information and modeling. 2007;47:1599–1608. doi: 10.1021/ci7000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xing L, Hodgkin E, Liu Q, Sedlock D. Evaluation and application of multiple scoring functions for a virtual screening experiment. Journal of computer-aided molecular design. 2004;18:333–344. doi: 10.1023/b:jcam.0000047812.39758.ab. [DOI] [PubMed] [Google Scholar]
- 24.Cummings MD, DesJarlais RL, Gibbs AC, Mohan V, Jaeger EP. Comparison of automated docking programs as virtual screening tools. Journal of medicinal chemistry. 2005;48:962–976. doi: 10.1021/jm049798d. [DOI] [PubMed] [Google Scholar]
- 25.Gorelik B, Goldblum A. High quality binding modes in docking ligands to proteins. Proteins. 2008;71:1373–1386. doi: 10.1002/prot.21847. [DOI] [PubMed] [Google Scholar]
- 26.Xie Q, Tang Y, Li W, Wang XH, Qiu ZB. Investigation of the binding mode of (-)-meptazinol and bis-meptazinol derivatives on acetylcholinesterase using a molecular docking method. Journal of molecular modeling. 2006;12:390–397. doi: 10.1007/s00894-005-0058-y. [DOI] [PubMed] [Google Scholar]
- 27.Cole JC, Murray CW, Nissink JW, Taylor RD, Taylor R. Comparing protein-ligand docking programs is difficult. Proteins. 2005;60:325–332. doi: 10.1002/prot.20497. [DOI] [PubMed] [Google Scholar]
- 28.Moller JV, Juul B, le Maire M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim Biophys Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 29.Obara K, Miyashita N, Xu C, Toyoshima I, Sugita Y, Inesi G, Toyoshima C. Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+ Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14489–14496. doi: 10.1073/pnas.0506222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 31.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 32.Toyoshima C, Nomura H, Sugita Y. Structural basis of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. FEBS Lett. 2003;555:106–110. doi: 10.1016/s0014-5793(03)01086-x. [DOI] [PubMed] [Google Scholar]
- 33.Olesen C, Picard M, Winther AM, Gyrup C, Morth JP, Oxvig C, Moller JV, Nissen P. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–1042. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 34.Treiman M, Caspersen C, Christensen SB. A tool coming of age: thapsigargin as an inhibitor of sarco-endoplasmic reticulum Ca2+-ATPases. Trends Pharmacol Sci. 1998;19:131–135. doi: 10.1016/s0165-6147(98)01184-5. [DOI] [PubMed] [Google Scholar]
- 35.Isaacs J. New strategies of the medical treatment of prostate cancer. BJU International. 2005;96:35–40. doi: 10.1111/j.1464-410X.2005.05945.x. [DOI] [PubMed] [Google Scholar]
- 36.Sohoel H, Jensen AM, Moller JV, Nissen P, Denmeade SR, Isaacs JT, Olsen CE, Christensen SB. Natural products as starting materials for development of second-generation SERCA inhibitors targeted towards prostate cancer cells. Bioorganic & medicinal chemistry. 2006;14:2810–2815. doi: 10.1016/j.bmc.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen U, Broogger Christensen S, Sandberg F. Thapsigargine and thapsigargicine, two new histamine liberators from Thapsia garganica L. Acta Pharm Suec. 1978;15:133–140. [PubMed] [Google Scholar]
- 38.Sagara Y, Inesi G. Inhibition of the sarcoplasmic reticulum Ca2+ transport ATPase by thapsigargin at subnanomolar concentrations. The Journal of biological chemistry. 1991;266:13503–13506. [PubMed] [Google Scholar]
- 39.Floreani M, Forlin A, Bellin S, Carpenedo F. Cardiac sarcoplasmic reticulum Ca2+ pump as a target for benzoquinones. Gen Pharmacol. 1996;27:873–878. doi: 10.1016/0306-3623(95)02130-2. [DOI] [PubMed] [Google Scholar]
- 40.Khan YM, Wictome M, East JM, Lee AG. Interactions of dihydroxybenzenes with the Ca2+-ATPase: separate binding sites for dihydroxybenzenes and sesquiterpene lactones. Biochemistry. 1995;34:14385–14393. doi: 10.1021/bi00044a015. [DOI] [PubMed] [Google Scholar]
- 41.Plenge-Tellechea F, Soler F, Fernandez-Belda F. On the inhibition mechanism of sarcoplasmic or endoplasmic reticulum Ca2+-ATPases by cyclopiazonic acid. The Journal of biological chemistry. 1997;272:2794–2800. doi: 10.1074/jbc.272.5.2794. [DOI] [PubMed] [Google Scholar]
- 42.Soler F, Plenge-Tellechea F, Fortea I, Fernandez-Belda F. Cyclopiazonic acid effect on Ca2+-dependent conformational states of the sarcoplasmic reticulum ATPase. Implication for the enzyme turnover. Biochemistry. 1998;37:4266–4274. doi: 10.1021/bi971455c. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi M, Kondou Y, Toyoshima C. Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5800–5805. doi: 10.1073/pnas.0700979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paula S, Ball WJ., Jr Molecular determinants of thapsigargin binding by SERCA Ca2+-ATPase: a computational docking study. Proteins. 2004;56:595–606. doi: 10.1002/prot.20105. [DOI] [PubMed] [Google Scholar]
- 45.Perola E, Walters WP, Charifson PS. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins. 2004;56:235–249. doi: 10.1002/prot.20088. [DOI] [PubMed] [Google Scholar]
- 46.Baxter CA, Murray CW, Clark DE, Westhead DR, Eldridge MD. Flexible docking using Tabu search and an empirical estimate of binding affinity. Proteins. 1998;33:367–382. [PubMed] [Google Scholar]
- 47.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. Journal of computer-aided molecular design. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 48.Huey R, Morris GM, Olson AJ, Goodsell DS. A Semiempirical Free Energy Force Field with Charge-Based Desolvation. J. Computational Chemistry. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 49.Mooij WT, Verdonk ML. General and targeted statistical potentials for protein-ligand interactions. Proteins. 2005;61:272–287. doi: 10.1002/prot.20588. [DOI] [PubMed] [Google Scholar]
- 50.Lape M, Elam C, Versluis M, Kempton R, Paula S. Molecular determinants of sarco/endoplasmic reticulum calcium ATPase inhibition by hydroquinone-based compounds. Proteins. 2008;70:639–649. doi: 10.1002/prot.21542. [DOI] [PubMed] [Google Scholar]
- 51.Nielsen SF, Thastrup O, Pedersen R, Olsen CE, Christensen SB. Structure-activity relationships of analogues of thapsigargin modified at O-11 and O-12. Journal of medicinal chemistry. 1995;38:272–276. doi: 10.1021/jm00002a009. [DOI] [PubMed] [Google Scholar]
- 52.Riley RT, Goeger DE, Yoo H, Showker JL. Comparison of three tetramic acids and their ability to alter membrane function in cultured skeletal muscle cells and sarcoplasmic reticulum vesicles. Toxicol Appl Pharmacol. 1992;114:261–267. doi: 10.1016/0041-008x(92)90076-5. [DOI] [PubMed] [Google Scholar]
- 53.Singh P, Mhaka AM, Christensen SB, Gray JJ, Denmeade SR, Isaacs JT. Applying linear interaction energy method for rational design of noncompetitive allosteric inhibitors of the sarco- and endoplasmic reticulum calcium-ATPase. Journal of medicinal chemistry. 2005;48:3005–3014. doi: 10.1021/jm049319a. [DOI] [PubMed] [Google Scholar]
- 54.Deye J, Elam C, Lape M, Ratliff R, Evans K, Paula S. Structure-based virtual screening for novel inhibitors of the sarco/endoplasmic reticulum calcium ATPase and their experimental evaluation. Bioorganic & medicinal chemistry. 2009;17:1353–1360. doi: 10.1016/j.bmc.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 55.Steitz TA, Shoham M, Bennett WS., Jr Structural dynamics of yeast hexokinase during catalysis. Philosophical transactions of the Royal Society of London. 1981;293:43–52. doi: 10.1098/rstb.1981.0058. [DOI] [PubMed] [Google Scholar]
- 56.Gunasekaran K, Nussinov R. How different are structurally flexible and rigid binding sites? Sequence and structural features discriminating proteins that do and do not undergo conformational change upon ligand binding. Journal of molecular biology. 2007;365:257–273. doi: 10.1016/j.jmb.2006.09.062. [DOI] [PubMed] [Google Scholar]
- 57.Moller JV, Nissen P, Sorensen TL, le Maire M. Transport mechanism of the sarcoplasmic reticulum Ca2+-ATPase pump. Current opinion in structural biology. 2005;15:387–393. doi: 10.1016/j.sbi.2005.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.