Abstract
Trytophan Hydroxylase Type I (TPH1), most abundantly expressed in the gastrointestinal tract, initiates the synthesis of serotonin by catalyzing hydroxylation of tryptophan in the presence of biopterin and oxygen. We have previously described three series of novel, periphery-specific TPH1 inhibitors that selectively deplete serotonin in the gastrointestinal tract. We have now determined co-crystal structures of TPH1 with three of these inhibitors at high resolution. Analysis of the structural data showed that each of the three inhibitors fills the tryptophan binding pocket of TPH1 without reaching into the binding site of the cofactor pterin, and induces major conformational changes of the enzyme. The enzyme-inhibitor complexes assume a compact conformation that is similar to the one in tryptophan complex. Kinetic analysis showed that all three inhibitors are competitive versus the substrate tryptophan, consistent with the structural data that the compounds occupy the tryptophan binding site. On the other hand, all three inhibitors appear to be uncompetitive versus the cofactor 6-methyltetrahydropterin, which is not only consistent with the structural data but also indicate that the hydroxylation reaction follows an ordered binding mechanism in which a productive complex is formed only if tryptophan binds only after pterin, similar to the kinetic mechanisms of tyrosine and phenylalanine hydroxylase.
Keywords: Serotonin, structure, kinetics, gastrointestinal disorder, carcinoid, monooxygenase.
INTRODUCTION
Serotonin (5-hydroxytryptamine, 5-HT) has a plethora of functions in both the central nervous system and the periphery. It is synthesized from tryptophan by the sequential action of two enzymes, tryptophan hydroxylase (TPH) and aromatic amino acid decarboxylase with TPH catalyzing the rate-limiting step. Two forms of TPH have been identified in mammals: TPH1, expressed highly in the gastrointestinal tract and the pineal gland, is responsible for >90% of 5-HT synthesis in the periphery, while TPH2, expressed in neuronal cells located in the dorsal raphe nucleus of the brain and the myenteric plexus of the gut, is responsible for the vast majority of 5-HT synthesis in the central nervous system [1-4]. The two enzymes, sharing an overall identity of ~80% at the amino acid level, are homologous to the other two aromatic amino acid hydroxylases, phenylalanine hydroxylase (PAH) and tyrosine hydroxylase (TH), with approximately 50% amino acid sequence identity [3, 5]. The four enzymes use the same cofactors, tetrahydrobiopterin, iron, and oxygen to catalyze the hydroxylation of their respective substrates [6].
X-ray crystal structures have been determined for various forms of TPH1, PAH and TH [7-13]. The catalytic domains of all three enzymes have a very similar fold with a mixture f α and β structures. The active site is formed by two long channels, one occupied by the amino acid substrate, the other one by pterin, with iron sitting at the intersection of the two channels. For PAH, binding of 5,6,7,8-tetrahydro-L-biopterin (BH4) caused small structural changes to the catalytic domain [8]. In contrast, binding of a phenylalanine analogue, such as L-norleucine or 3-(2-thienyl)-L-alanine (THA) in the presence of BH4 and iron ion led to large structural changes in the catalytic domain [9, 14]. Specifically, the loop containing residues 133-141 of PAH moved much closer to the catalytic site. For TPH1, a structure of the catalytic domain of the human enzyme with 7,8-dihydrobiopterin (BH2) and ferric iron ion bound was solved first, which showed remarkable similarity to the structure of the catalytic domain of PAH with BH4 bound [11]. More recently, a structure of the catalytic domain of chicken TPH1 with tryptophan bound was solved [12]. Compared with the BH2-bound structure of human TPH1, binding of tryptophan caused major structural changes with two loops (Leu124-Asp139 and Ile367-Thr369) brought closer to the catalytic site, as in the corresponding regions of PAH after substrate analogue binding [12].
The kinetic mechanism of mammalian TH and bacterial PAH has been determined. For both enzymes, all three substrates, i.e., amino acid, pterin, and oxygen, have to be bound before catalysis can happen [6]. For rat TH, the order of substrate binding is pterin first, followed by oxygen, and then tyrosine [15]. For bacterial PAH, one study reported oxygen binding first, followed by pterin and amino acid in random order [16]. A more recent study, however, conclusively showed that pterin binds first, followed by phenylalanine and then oxygen [17]. Such an ordered binding mechanism with pterin binding first is consistent with the observations that both enzymes are inhibited by their amino acid substrates at high concentrations [6]. TPH1 is also inhibited by high concentrations of Trp [18], implicating a similar kinetic mechanism , even though no detailed mechanism studies have been reported.
Dysregulation of the serotonergic system in the periphery is associated with various kinds of maladies, including chemotherapy-induced nausea and vomiting [19], gastrointestinal disorders [20, 21], and pulmonary hypertension [22]. Inhibition of TPH1 will lead to decrease or total abolishment of serotonin synthesis, and thus may provide effective treatment for such disorders that are caused by excessive synthesis or release of serotonin. Previously, we reported the discovery and characterization as well as structure-activity relationship of three series of novel TPH1 inhibitors that are able to deplete peripheral but not central serotonin [4, 23, 24]. We have now determined X-ray structures of the catalytic domain of human TPH1 co-crystallized with one compound from each of the three series of inhibitors. Hereby, we report the structure and analysis of the co-crystals. We also carried out kinetic studies of the three inhibitors in attempt to understand their mechanism of inhibition.
MATERIALS AND METHODS
Materials
Compounds LP-521834, LP-533401, and LP-534193 were synthesized in-house as described before [23, 24]. All other reagents were purchased from Sigma-Aldrich.
Protein Purification, Crystallization, and Data Collection
The polypeptide encompassing amino acid sequence 102 to 402 of human TPH1 with a C-terminal six-histidine tag was produced in E. coli BL21(DE3) cells using a modified pET28 expression vector. The protein was purified by Ni2+ affinity, anion exchange, and gel filtration chromatography essentially as described previously [11]. The final protein buffer contained 50 mM Tris-Cl, pH 8.0, 0.5 mM EDTA, 1 mM DTT and 200 mM NaCl. For co-crystallization, inhibitor (from a 100 mM DMSO stock) was added to the purified enzyme to a final concentration of 0.5 mM and the resulting complex was concentrated to a final protein concentration of 10-15 mg/ml with a Centricon centrifugal concentrator. To obtain crystals, hanging drop vapor diffusion was employed (20º C), using drops containing a 1:1 ratio of the concentrated protein solution and a reservoir comprising 24-28% (w/v) PEG 6000, and 100 mM Tris-Cl, pH 8.5. Crystals were cryoprotected by brief immersion in the reservoir solution supplemented with 20% (v/v) ethylene glycol, harvested in nylon loops and frozen in liquid nitrogen for data collection. X-ray data were collected at beamline 4.2.2 at the ALS, Berkeley, at 10000 eV on a Noir1 CCD detector. Data were analyzed and reduced using d*TREK [25] and CCP4 [26]. The complex structures were solved by molecular replacement using the structure of human TPH1 bound with BH2 and Fe(III) [11] as a probe. The molecular replacement calculations were carried using PHASER in the CCP4 suite2. The structures were refined using REFMAC in the CCP4 suite2. All electron density map visualization and manual model rebuilding was carried out using the XtalView/Xfit package [27].
Enzyme Kinetic Studies
Full-length human TPH1 was expressed and purified as described before [4], to a specific activity of approximately 60 nmole/min/(mg of protein). Enzyme assays were carried out at room temperature with atmosphere oxygen in a volume of 0.1 ml containing 50 mM 3-(N-morpholino)-propanesulfonate (MOPS), pH 7.2, 100 mM (NH4)2SO4, 0.05 mg/ml of catalase, 1 mg/ml of bovine serum albumin, 0.05 mM (NH4)2Fe(SO4)2 , various concentrations of tryptophan and 6-methyltetrahydropterin, and 0.5 μg of TPH1. The reactions were started with the addition of pterin and were generally linear with time up to 10 min. For kinetic studies, the reactions proceeded for 5 min with 0.5 μg of protein per reaction in the presence of the inhibitors at the indicated concentrations and were then terminated immediately with 0.1 ml of 1 M trichloroacetic acid. The reaction mixtures were filtered through GF/B filter plates; five ul of each filtrate was then analyzed for 5-HTP using in-line fluorescence-coupled HPLC as described before [4]. Double reciprocal plots were used to determine type of inhibition. Competitive Ki’s of the inhibitors were calculated using the global fit formula (GraphPad Prism 4.03): v = Vmax*[S]/(KmObs + [S]), where v = initial velocity, Vmax = maximum velocity, [S] = substrate concentration, [I] = inhibitor concentration, KmObs = Km*(1+[I]/Ki), and Km = Michaelis-Menten constant [28]. Uncompetitive Ki’s of the inhibitors were calculated using the global fit formula v = [S]*Vmax*KmObs/Km/ (KmObs+[S]), where v, [S], Km, Vmax, and [I] are as defined above except that KmObs = Km/(1+[I]/Ki) [28].
RESULTS
Crystal Structures of Inhibitor-TPH1 Complexes
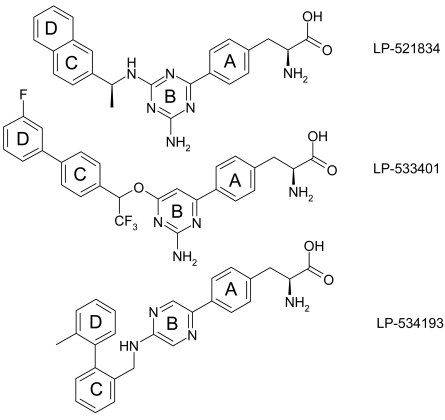
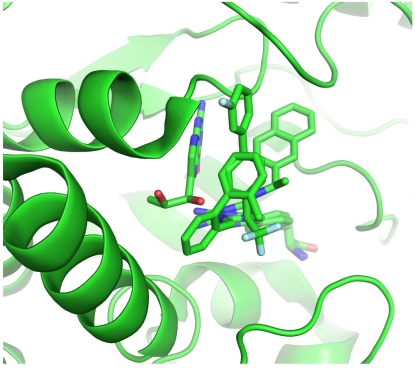
To gain a better understanding of the interactions between TPH1 and its inhibitors, we solved co-crystal structures of the catalytic domain of human TPH1 with three of our TPH1 inhibitors, LP-521834, LP-533401, and LP-534193 at a resolution of 1.80, 1.85, and 1.92 Å, respectively. All three crystal structures were determined with one iron ion plus one inhibitor molecule but without pterin. The coordinates are deposited in Protein Data Bank and designated as 3HF6 (TPH1 +LP-521834), 3HF8 (TPH1 + LP-533401), and 3HFB (TPH1 + LP-534193). Due to poor electron density, loops 122-137 in 3HF8 and 3HFB, and 119-128 in 3HF6 were not resolved. The three inhibitor molecules share a phenylalanine moiety but represent three distinct subtypes on the basis of substitutions on the phenylalanine moiety (Fig. 1). They all contain four ring structures which, for the purpose of clarity, are designated A to D (Fig. 1).
Fig. (1).
Structure and ring designation of the three inhibitors: from top to bottome, LP-521834, LP-533401, and LP-534193.
Analysis of the three X-ray structures shows that TPH1 has a catalytic core composed of mixed α and β structures, similar to that of human PAH and rat TH, as noted previously [11]. The active site is formed by a large cavity divided into a ~13 Å long channel, where the amino acid part of the inhibitors is bound, and a ~9 Å long chamber previously identified as the BH2 binding site [11]. Approximately at the conjunction of the two areas, the active site is exposed to the solvent through a roughly circular, ~12Å wide opening. The iron ion is situated at the intersection of the two areas and coordinated to His272, His277, three water molecules, and to Glu317 with a monodentate interaction in 3HF6 (LP-521834) and 3HFB (LP-534193). This pattern of iron coordination is identical to that observed in human PAH-BH4-Fe(II) and TPH1-BH2-Fe(III) complexes [8, 11]. In contrast, the iron ion in 3HF8 (LP-533401) is coordinated to His272, His277, one water molecule and to Glu317 with a bidentate interaction. Such a bidentate arrangement of the metal ligand was only observed in TPH1-Trp-Fe(III) and PAH-BH4-THA-Fe(II) complexes [9, 12]. The oxidation state of the iron in the three structures was not resolved, but it can be assumed to be ferric since the enzyme purification and crystallization were carried out under aerobic conditions.
The Tyr235 residue showed different orientations in the three structures: in 3HF8 and 3HFB, its side chain is directed inward and forms a hydrogen-bond with the backbone carbonyl of Pro268 which, through a bridging water molecule, interacts with Thr367 backbone nitrogen. In this conformation, the Tyr side chain is almost orthogonal to the equivalent Tyr236 in the chicken TPH1 X-ray structure 3E2T [12]. In 3HF6, the Tyr235 side chain is rotated outward approximately 90 o, bringing the aromatic ring to an almost coplanar position with that of Ty236 in the chicken TPH1 X-ray structure accompanied by a ~5 Å shift toward the solvent. An additional change observed in all three structures is that the loop Ile336-Phe369 moved toward the loop Phe263-Pro266 when compared to the TPH1-BH2-Fe(III) structure, as in the corresponding loop of chicken TPH1 in the Trp complex [12].
Analysis of the Binding Modes of the Ligands
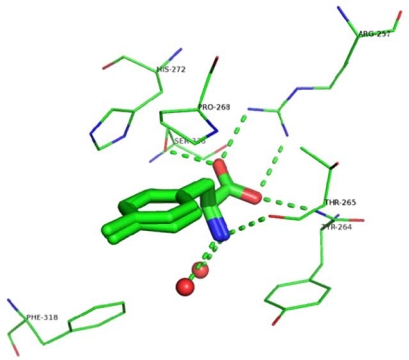
Analysis of the ligands’ binding mode reveals that the three compounds form similar interactions with TPH1. The 2-amino-3-phenyl-propionic acid (phenylalanine) part is a common substructure across the ligands and, not surprisingly, the interactions between this moiety and the protein are almost the same in the three structures, as shown in Fig. (2). The carboxylate group interacts with Arg257 through a salt-bridge and accepts two hydrogen bonds: one from the backbone nitrogen of Thr265 and another one from the hydroxyl group of the side chain of Ser336. Similar interactions occur between the carboxylate group of tryptophan and Arg258, Thr266, and Ser337 in the chicken TPH1-Trp-Fe(III) structure, and between the carboxylate group of THA and Arg270, Thr278, Ser349 in PAH-BH4-THA-Fe(II) structure. The primary amine group, protonated at physiological pH, forms a hydrogen-bond with the carbonyl oxygen of Thr265, and two interactions with water molecules in 3HF6 and 3HF8 but only one in 3HFB, also similar to the interaction between the primary amine of Trp with the carbonyl oxygen of Thr266 and two water molecules in the chicken TPH1-Trp-Fe(III) structure. In addition, similar to the structure of the chicken TPH1 (3E2T), the aromatic surface of Tyr264 closes this section of the binding pocket (Fig. 2).
Fig. (2).
Interactions between TPH1 and the amino acid portion of the ligands.
Ring A of the inhibitors is ~3.75 Å from the almost coplanar side chain of Pro268 and interacts with the iron ion-coordinating His272 (Fig. 2). Distinct from what was observed in the X-ray structure of PAH co-crystallized with THA where the thiophene ring interacted with His285 through a face-to-face-π-π interaction, no stacking interaction was observed between Ring A and His272. In the structures presented here, in fact, Ring A is rotated ~70° away from the plane of the imidazole and makes hydrophobic contacts with the side chain of Pro268 (Fig. 2). Overall, the data indicate that the phenylalanine moiety in all three compounds binds to the enzyme in a way that is highly similar to the way in which the substrate tryptophan binds in the chicken TPH1-Trp-Fe(III) complex.
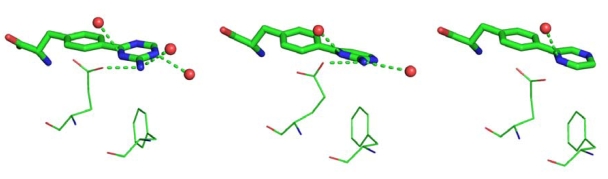
The three ligands have different moieties as Ring B: 2-amino-triazine in LP-521834, 2-amino-pyrimidine in LP-533401, and pyrazine in LP-534193. Two interactions between Ring B and the enzyme are shared among the three structures: the edge-to-face π-π interaction with Phe313 and the contact between the nitrogen atoms in the ortho position relative to phenylalanine and one of the iron ion-coordinating water molecules. The two ligands with a 2-amino group, i.e., LP-521834 and LP-533401, show two additional hydrogen bonds: one with the side chain of Glu317 and the other with a water molecule (Fig. 3). In addition, LP-521834 accepts a hydrogen bond from water 449 (Fig. 3).
Fig. (3).
Interactions between TPH1 and Ring B of the inhibitors. The contact with one of the iron-coordinating water and the edge-to-face π-π interaction with Phe313 are common to the three complexes. LP-521834, left panel; LP-533401, middle panel; LP-534193, right panel.
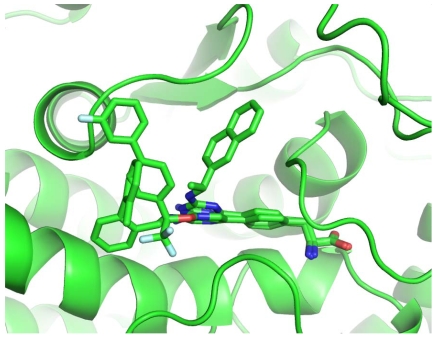
The remaining parts of the ligands (linker and Rings C and D) establish less specific interactions with TPH1: no hydrogen bond was observed between the protein and these substructures; only hydrophobic and π-π interactions are found. This part of the ligands is the least analogous, and thus showed different conformations and interactions with TPH1 in the three complexes (Fig. 4). In 3HF6, LP-521834, most distinct from the other two in the ring C/D moiety, exploits a peculiar conformation of Tyr235 by having its naphthyl structure sandwiched between the Tyr235 side chain (π-π-face-to-face stacking) and Pro268 (hydrophobic interaction). In 3HF8, the CF3 group of LP-533401 is placed in a small hydrophobic pocket located at the entrance to the catalytic site defined by Ile336, Leu365, Cys364 and Phe313. Ring C makes hydrophobic contact with Pro238 and Tyr235 while Ring D is placed on the protein's external surface at the entrance to the binding pocket in an area defined by the Pro238 side chain and backbone atoms of Ser237 and Leu236. In 3HFB, LP-534193 has Ring D interacting with Pro238 and Ring C in a hydrophobic pocket defined by Ala309, Cys364 and Phe313.
Fig. (4).
Different conformations of Ring C and D in the three complexes.
The human TPH1-BH2-Fe(III) structure [11] shows that Tyr235 and Pro238 interact with pterin. Superposition of this X-ray structure on 3HF6, 3HFB and 3HF8 reveals that the three inhibitors do not occupy the pterin binding pocket (Fig. 5). Whether a pterin molecule and any of the inhibitors can bind to the enzyme simultaneously remains to be tested experimentally.
Fig. (5).
Superposition of 3HF6, 3HF8, 3HFB and 1MLW shows that the three inhibitors do not occupy the pterin binding pocket.
Kinetic Analysis of Inhibition
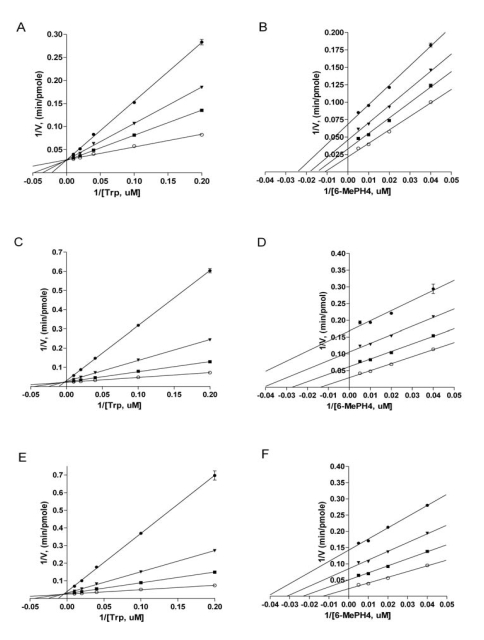
We also studied mechanism of inhibition of the three inhibitors with purified recombinant full-length TPH1. When tryptophan concentration was varied at a fixed concentration of the pterin cofactor 6-methyltetrahydropterin (6-MePH4), all three inhibitors displayed characteristics of competitive inhibition, as shown by the lines converging on the Y axis in the double reciprocal plots of the data (Figs. 6A, 6C, and 6E). The Ki values of LP-533401, LP-521834, and LP-534193, determined using non-linear regression based on competitive inhibition, are 0.31, 0.036, and 0.03 μM, respectively (Table 1). The finding that all three compounds are competitive versus tryptophan is entirely consistent with the observations from the X-ray structures that all three inhibitors occupy the binding site of tryptophan. On the other hand, when 6-MePH4 concentration was varied at a fixed concentration of tryptophan, all three compounds appeared to be uncompetitive, as shown by the lines being nearly parallel in the double reciprocal plots (Figs. 6B, 6D, 6F). The Ki values for LP-533401, LP-521834, and LP-534193, determined using non-linear regression fitting based on uncompetitive inhibition, are 0.81, 0.19, and 0.17 μM, respectively. The finding that all three inhibitors are not competitive versus pterin is consistent with the structural data that they don’t occupy the pterin binding site. Since uncompetitive behavior is typically displayed by inhibitors that can bind to the enzyme only after the varied substrate is bound [28], the data suggest that the inhibitors can only bind to the enzyme after pterin is bound.
Fig. (6).
Double reciprocal plots of initial velocities at various concentration of the inhibitors: LP-533401 (A, B), LP-521834 (C, D), and LP-534193 (E, F). A, C, E, varying tryptophan concentration at a fixed concentration of 6-MePH4 (200 uM). B, D, F, varying 6-MePH4 at fixed concentration of tryptophan (50 uM). The inhibitor concentrations used here are: A, LP-533401 at 0 uM (◯), 0.25 uM (■), 0.5 uM (▼), and 1.0 uM (●); B, LP-533401 at 0 uM (◯), 0.5 uM (■), 1.0 uM (▼), and 2.0 uM (●); C, LP-521834 at 0 uM (◯), 0.05 uM (■), 0.15 uM (▼), and 0.5 uM (●); D, LP-521834 at 0 uM (◯), 0.2 uM (■), 0.5 uM (▼), and 1.0 uM (●); E, LP-534193 at 0 uM (◯), 0.05 uM (■), 0.15 uM (▼), and 0.5 uM (●); F, LP-534193 at 0 uM (◯), 0.2 uM (■), 0.5 uM (▼), and 1.0 uM (●).
Table 1.
Inhibition Patterns of the Three Compounds for TPH1
| Inhibitor | Varied Substrate | Fixed Substrate | Inhibition Patterns | Ki ( mean ± S.E.M, uM)a |
|---|---|---|---|---|
| LP-533401 | Trp | 200 uM 6-MePH4 | Cb | 0.31 ± 0.01 |
| LP-533401 | 6-MePH4 | 50 uM Trp | UCc | 0.81± 0.03 |
| LP-521834 | Trp | 200 uM 6-MePH4 | C | 0.036 ± 0.001 |
| LP-521834 | 6-MePH4 | 50 uM Trp | UC | 0.19 ± 0.01 |
| LP-534193 | Trp | 200 uM 6-MePH4 | C | 0.030 ± 0.001 |
| LP-534193 | 6-MePH4 | 50 uM Trp | UC | 0.17 ± 0.01 |
Values are calculated based on two independent experiments with each experiment carried out in triplicates.
C, competitive.
UC, uncompetitive.
DISCUSSION
We determined the crystal structures of the catalytic domain of human TPH1 in complex with three novel TPH inhibitors. The structures of TPH1 in these complexes resemble the structure of the chicken TPH1 (3E2T) co-crystallized with tryptophan and iron ion (3E2T) more than that of human TPH1 bound with BH2 and iron ion (1MLW). Superposition of all five structures shows that the enzyme adopts a more compact structure when bound with either the phenylalanine-based inhibitors or tryptophan than by the pterin analogue BH2. In all three inhibitor-TPH1 structures, the loop Ile336-Phe369 moved toward the loop Phe263-Pro266, closing around the amino acid binding pocket when compared to the TPH1-BH2-Fe(III) structure. Such a movement was also observed in the chicken TPH1-Trp-Fe(III) structure. Another large structural change observed in the chicken TPH1-Trp-Fe(III) structure is the movement of the loop Leu124-Asp139 toward the catalytic site [12]. Unfortunately, this loop was not resolved in any of the three inhibitor-TPH1 structures due to poor electron density. Such a movement is suggested to have also occurred in these structures, given the confirmed movement of the Ile336-Phe369 loop and the similarity in interactions between the phenylalanine moiety of the inhibitors and tryptophan with the enzyme. Since phenylalanine is also an effective substrate of TPH1 with its catalytic domain showing comparable V/K and Vmax for the two amino acids [18], it is not surprising that the structures of the enzyme bound with the phenylalanine-based inhibitors are more similar to the one bound with Trp.
The pattern of the iron ion coordination is different among the three complexes. Two of the structures (3HF6 and 3HFB) show a monodentate coordination by Glu317, identical to the pattern in TPH1-BH2-Fe(III) and PAH-BH4-Fe(II). In contrast, in the structure of LP-533401-TPH1 (3HF8), Glu317 has a bidentate interaction with the metal, making the coordination geometry very distorted. Such a bidentate interaction was also observed in the structures of the complexes PAH-BH4-THA-Fe(II) and TPH1-Trp-Fe(III) [9, 12, 14]. In the bidentate interaction mode, iron ion is only coordinated to five sites, allowing one open site for oxygen [12]. Interestingly, the mechanism of catalysis by these aromatic amino acid hydroxylases is hypothesized to have a step that involves the formation of a Fe(II)-O-O-BH4 intermediate for oxygen activation followed by the formation of Fe(IV)O for hydroxylation [6]. Therefore, it is tempting to suggest that of the three inhibitors, LP-533401 binds to the enzyme in a conformation that is closest to its catalytic state.
The other major difference among the three structures is the conformation of Tyr235. This residue is conserved in all known TPH1 and TPH2 sequences. In PAH and TH, this position is leucine. When Tyr235 was changed to an alanine or leucine, the mutant enzyme showed much lower specific activity (~5% remaining) and increased Km for Trp but decreased Km for BH4 [29], implying a role in both pterin and Trp binding. Tyr235 was indeed found to interact not only with pterin in the TPH1-BH2-Fe(III) structure but also with Trp in the TPH1-Trp-Fe(III) structure [11, 12]. In the TPH1-LP-521834-Fe(III) structure, Tyr235 has a conformation similar to the one in Trp- or BH2-bound structure. However, in the TPH1 structures containing either LP-533401 or LP-534193, it is rotated inward by ~90o and forms a new hydrogen bond with Pro268. These results suggest that Tyr235 has considerable conformation freedom that may be critical to the reaction mechanism of TPH1.
The results of kinetic analysis with the three inhibitors clearly showed that they are all competitive versus tryptophan but predominantly uncompetitive versus pterin. Being competitive with tryptophan is entirely consistent with the structural data that the phenylalanine moiety of the inhibitors occupies the tryptophan binding site. Being uncompetitive with pterin is also consistent with the structural data that the side chains of the three compounds are not in the space where pterin binds. More importantly, the uncompetitive behavior of the three inhibitors strongly suggests that the substrate tryptophan and the inhibitors bind only after pterin is bound [28]. For TH and PAH, similar analyses were used to demonstrate that pterin binds before the amino acid substrate does [15, 17]. Intriguingly, these co-crystal structures were determined in the absence of pterin, indicating that the inhibitors can bind to the enzyme before pterin does, at least at high concentrations (inhibitors were at >500 uM during crystallization). Similarly, when the crystal structure of chicken TPH1 was solved in the absence of pterin, it was also found to contain tryptophan, even though the authors suggested that tryptophan binds after biopterin during catalysis [12]. No detailed kinetic studies were reported for TPH1 before. One study showed that the product 5-HTP inhibits TPH1 in a mechanism that is competitive versus phenylalanine but uncompetitive versus pterin [30], analogous to the behavior of the three inhibitors examined here. Given its structural similarity to tryptophan, 5-HTP is expected to occupy the tryptophan site, and thus to be competitive versus trytophan but uncompetitive versus pterin. Taken together, the data showed that, similar to PAH and TH, TPH1 also follows an ordered binding mechanism with pterin binding first. Our data don’t address the binding order of tryptophan and oxygen.
In summary, we have determined co-crystal structures of the human TPH1 catalytic domain with three inhibitors. The three inhibitors all have a phenylalanine moiety which occupies the tryptophan binding site of the enzyme. All three complexes showed an overall structure that is more similar to that of chicken TPH1 bound with tryptophan rather than that of human TPH1 bound with pterin, reflecting the structural similarity between the inhibitors and tryptophan, and the major conformational change induced by the binding of the amino acid substrate and its analogue. Kinetic analysis showed that all three inhibitors are competitive versus tryptophan but largely uncompetitive with pterin, consistent with the structural data and suggestive of an ordered binding mechanism in which tryptophan must bind after pterin in order for a productive complex to form. The data shed important light on the mechanism of TPH1 catalysis and inhibition.
ABBREVIATIONS
- 6-MePH4
= 6-methyl-5,6,7,8-tetrahydropterin
- BH2
= 7,8-tetrahydrobiopterin
- BH4
= 5,6,7,8-tetrahydrobiopterin
- PAH
= phenylalanine hydroxylase
- Phe
= L-phenylalanine
- TH
= tyrosine hydroxylase
- THA
= 3-(2-thienyl)-L-alanine
- TPH1
= tryptophan hydroxylase type I
- Trp
= L-tryptophan
REFERENCES
- 1.Cote F, Thevenot E, Fligny C, et al. Disruption of the nonneuronal tph1 gene demonstrates the importance of peripheral serotonin in cardiac function. Proc Natl Acad Sci USA. 2003;100:13525–30. doi: 10.1073/pnas.2233056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel PD, Pontrello C, Burke S. Robust and tissue-specific expression of TPH2 versus TPH1 in rat raphe and pineal gland. Biol Psychiatry. 2004;55:428–33. doi: 10.1016/j.biopsych.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Walther DJ, Peter JU, Bashammakh S, et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- 4.Liu Q, Yang Q, Sun W, et al. Discovery and characterization of novel tryptophan hydroxylase inhibitors that selectively inhibit serotonin synthesis in the gastrointestinal tract. J Pharmacol Exp Ther. 2008;325:47–55. doi: 10.1124/jpet.107.132670. [DOI] [PubMed] [Google Scholar]
- 5.Flatmark T, Stevens RC. Structural insight into the aromatic amino acid hydroxylases and their disease-related mutant forms. Chem Rev. 1999;99:2137–60. doi: 10.1021/cr980450y. [DOI] [PubMed] [Google Scholar]
- 6.Fitzpatrick PF. Mechanism of aromatic amino acid hydroxylation. Biochemistry. 2003;42:14083–91. doi: 10.1021/bi035656u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erlandsen H, Fusetti F, Martinez A, Hough E, Flatmark T, Stevens RC. Crystal structure of the catalytic domain of human phenylalanine hydroxylase reveals the structural basis for phenylketonuria. Nat Struct Biol. 1997:995–1000. doi: 10.1038/nsb1297-995. [DOI] [PubMed] [Google Scholar]
- 8.Andersen OA, Flatmark T, Hough E. High resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin. J Mol Biol. 2001;314:279–91. doi: 10.1006/jmbi.2001.5061. [DOI] [PubMed] [Google Scholar]
- 9.Andersen OA, Stokka AJ, Flatmark T, Hough E. 2.0A resolution crystal structures of the ternary complexes of human phenylalanine hydroxylase catalytic domain with tetrahydrobiopterin and 3-(2-thienyl)-L-alanine or L-norleucine: substrate specificity and molecular motions related to substrate binding. J Mol Biol. 2003;333:747–57. doi: 10.1016/j.jmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Daubner SC, McGinnis JT, Gardner M, Kroboth SL, Morris AR, Fitzpatrick PF. A flexible loop in tyrosine hydroxylase controls coupling of amino acid hydroxylation to tetrahydropterin oxidation. J Mol Biol. 2006;359:299–307. doi: 10.1016/j.jmb.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Erlandsen H, Haavik J, Knappskog PM, Stevens RC. Three-dimensional structure of human tryptophan hydroxylase and its implications for the biosynthesis of the neurotransmitters serotonin and melatonin. Biochemistry. 2002;41:12569–74. doi: 10.1021/bi026561f. [DOI] [PubMed] [Google Scholar]
- 12.Windahl MS, Petersen CR, Christensen HE, Harris P. Crystal structure of tryptophan hydroxylase with bound amino acid substrate. Biochemistry. 2008;47:12087–94. doi: 10.1021/bi8015263. [DOI] [PubMed] [Google Scholar]
- 13.Goodwill KE, Sabatier C, Marks C, Raag R, Fitzpatrick PF, Stevens RC. Crystal structure of tyrosine hydroxylase at 2.3 A and its implications for inherited neurodegenerative diseases. Nat Struct Biol. 1997;4:578–85. doi: 10.1038/nsb0797-578. [DOI] [PubMed] [Google Scholar]
- 14.Andersen OA, Flatmark T, Hough E. Crystal structure of the ternary complex of the catalytic domain of human phenylalanine hydroxylase with tetrahydrobiopterin and 3-(2-thienyl)-L-alanine, and its implications for the mechanism of catalysis and substrate activation. J Mol Biol. 2002;320:1095–108. doi: 10.1016/s0022-2836(02)00560-0. [DOI] [PubMed] [Google Scholar]
- 15.Fitzpatrick PF. Steady-state kinetic mechanism of rat tyrosine hydroxylase. Biochemistry. 1991;30:3658–62. doi: 10.1021/bi00229a010. [DOI] [PubMed] [Google Scholar]
- 16.Pember SO, Johnson KA, Villafranca JJ, Benkovic SJ. Mechanistic studies on phenylalanine hydroxylase from Chromobacterium violaceum. Evidence for the formation of an enzyme-oxygen complex. Biochemistry. 1989;28:2124–30. doi: 10.1021/bi00431a024. [DOI] [PubMed] [Google Scholar]
- 17.Volner A, Zoidakis J, Abu-Omar MM. Order of substrate binding in bacterial phenylalanine hydroxylase and its mechanistic implication for pterin-dependent oxygenases. J Biol Inorg Chem. 2003;8:121–8. doi: 10.1007/s00775-002-0395-6. [DOI] [PubMed] [Google Scholar]
- 18.Moran GR, Daubner SC, Fitzpatrick PF. Expression and characterization of the catalytic core of tryptophan hydroxylase. J Biol Chem. 1998;273:12259–66. doi: 10.1074/jbc.273.20.12259. [DOI] [PubMed] [Google Scholar]
- 19.Andrews PL, Davis CJ, Bingham S, Davidson HI, Hawthorn J, Maskell L. The abdominal visceral innervation and the emetic reflex: pathways, pharmacology, and plasticity. Can J Physiol Pharmacol. 1990;68:325–45. doi: 10.1139/y90-047. [DOI] [PubMed] [Google Scholar]
- 20.Gershon MD. Nerves, reflexes, and the enteric nervous system: pathogenesis of the irritable bowel syndrome. J Clin Gastroenterol. 2005;39:S184–93. doi: 10.1097/01.mcg.0000156403.37240.30. [DOI] [PubMed] [Google Scholar]
- 21.Spiller R. Serotonin and GI clinical disorders. Neuropharmacology. 2008;55:1072–80. doi: 10.1016/j.neuropharm.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Maclean MR, Dempsie Y. Serotonin and pulmonary hypertension-from bench to bedside? Curr Opin Pharmacol. 2009;9:281–6. doi: 10.1016/j.coph.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Shi ZC, Devasagayaraj A, Gu K, et al. Modulation of peripheral serotonin levels by novel tryptophan hydroxylase inhibitors for the potential treatment of functional gastrointestinal disorders. J Med Chem. 2008;51:3684–7. doi: 10.1021/jm800338j. [DOI] [PubMed] [Google Scholar]
- 24.Jin H, Cianchetta G, Devasagayaraj A, et al. Substituted 3-(4-(1, 3, 5-triazin-2-yl) phenyl)-2-aminopropanoic Acids as Novel Tryptophan Hydroxylase Inhibitors. Bioorg Med Chem. 2009;19:5229–32. doi: 10.1016/j.bmcl.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. 1999;55:1718–25. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 26.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 27.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–65. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 28.Segel IH. Enzyme Kinetics. USA: John Wiley & Sons; 1993. [Google Scholar]
- 29.Jiang GC, Yohrling GJT, Schmitt JD, Vrana KE. Identification of substrate orienting and phosphorylation sites within tryptophan hydroxylase using homology-based molecular modeling. J Mol Biol. 2000;302:1005–17. doi: 10.1006/jmbi.2000.4097. [DOI] [PubMed] [Google Scholar]
- 30.Ichiyama A, Hasegawa H, Tohyama C, Dohmoto C, Kataoka T. Some properties of bovine pineal tryptophan hydroxylase. Adv Exp Med Biol. 1976;74:103–17. doi: 10.1007/978-1-4684-3270-1_9. [DOI] [PubMed] [Google Scholar]