Abstract
Purpose
The epidermal growth factor receptor (EGFR) is overexpressed in several tumor types, and its expression is influenced by the length of a 5′-end microsatellite repeat (CA)n: the longer the repeat, the lower the expression. Dinucleotide repeats accumulate insertion/deletion types of mutations in tumors with microsatellite instability. We designed this study to estimate the occurrence of these mutations in EGFR(CA)n and their relevance in carcinogenesis of microsatellite instability – positive colon and gastric tumors.
Experimental Design
We analyzed the frequency of EGFR(CA)n mutations in vivo in 55 colorectal and 14 gastric microsatellite instability – positive cancers, and in vitro in single-cell clone cultures of microsatellite instability – positive colon tumor cell line LS174. Single-cell clone cultures with different repeat lengths were analyzed by fluorescent-activated cell sorter for EGFR cell-surface expression. A correlation analysis was done between EGFR(CA)n mutations and mutations in KRAS, BRAF, and p53.
Results
Unlike single-cell clone cultures, which exhibited higher rate of deletions compared with insertions, most of EGFR(CA)n mutations in colon and gastric tumors were insertions. Longer EGFR(CA)n correlated with lower EGFR cell-surface expression in single-cell clone cultures. In colon cancers, the elongation of the repeat was associated negatively with mutations in KRAS and BRAF, but not in p53.
Conclusions
The EGFR(CA)n elongation observedin tumors cannot be explained by anintrinsic property of this repeat favoring insertions versus deletions. Instead, a selection for repeat elongation occurs in microsatellite instability – positive tumors, leading to EGFR down-regulation. These findings suggest that in microsatellite instability – positive tumors current therapies targeting EGFR overexpression may have either no effect or an opposite to the expected effect.
The epidermal growth factor receptor (EGFR), a member of the erbB gene family, encoding a tyrosine kinase receptor, regulates cell growth and survival by binding specific ligands such as epidermal growth factor, transforming growth factor α (TGF-α), and heparin-binding epidermal growth factor (1, 2). Over-expression of EGFR, which has been detected in a variety of cancer types, is believed to contribute to neoplastic transformation and is associated with a poor cancer prognosis (3, 4). Overexpression of EGFR in glioblastomas can be explained by gene amplification (5), but amplification of EGFR in other types of cancer occurs only in a small percentage of tumors. Therefore, it is widely accepted that EGFR overexpression is regulated at the transcriptional level.
Transcription of EGFR starts at several initiation sites within a GC-rich promoter region (6). Two enhancer elements have been identified for EGFR: one upstream of the promoter and another in intron 1 (7). It has been also shown that the basal EGFR gene expression is influenced by the length of a microsatellite repeat sequence (CA)n located in the proximity of the intron 1 enhancer: the longer the repeat length, the lower the gene expression (8, 9).
Microsatellites are unstable repetitive sequences and for this reason are not only “naturally” highly polymorphic in the human population but also may be abnormally mutated in tumors with compromised mismatch repair. Genetic or epigenetic inactivation of mismatch repair genes causes a mutator phenotype, which was discovered by the ubiquitous presence of slippage-induced insertion/deletion mutations in microsatellite sequences (10–12). When microsatellite instability takes place in a tumor precursor cell, it leads to the accumulation of many mutations, some of which occur in the coding regions of cancer genes, contributing to neoplastic growth (reviewed in ref. 13).
Translational Relevance
EGFR is overexpressed in different tumor types, and its overexpression correlates with malignant progression. Thus, we were intrigued by finding elongation of a microsatellite repeat at the 5′ untranslated region in gastrointestinal cancers with microsatellite instability because it had been reported that elongation reduced expression. Our study indicates that, indeed, this tendency for elongation led to down-regulation of the gene product. Therefore, the selection for EGFR down-regulation during tumorigenesis implies that the gene works as a tumor suppressor and not as an oncogene in gastric and colon cancer of the microsatellite mutator phenotype.
The immediate consequence of these findings has translational potential related to EGFR-targeted cancer therapies. Numerous cancer therapies, ranging from therapeutic and imaging antibodies to toxin-linked ligands for enhancement of targeting for gene therapy vectors, have used up-regulated EGFR as a tumor-specific target. Our findings predict that some of these therapies may have no effect or an opposite to the expected effect on microsatellite instability – positive colon tumors. It also remains to be determined whether this opposite effect of epidermal growth factor signaling in microsatellite instability – positive colon cancer compared with breast cancer, forinstance, is restricted to the mutator pathway or is extensive to other cancers.
In this study, we have analyzed somatic mutations in the EGFR(CA)n microsatellite in microsatellite instability–positive gastric and colon tumors. We have further studied the relationship between the mutations in the 5′EGFR(CA)n and 3′EGFR (A) n noncoding microsatellite sequences, and the mutations in the KRAS, BRAF, and p53 cancer genes.
Materials and Methods
Tumor samples, cell lines, and microsatellite instability analysis
Co-lorectal and gastric tumors were obtained from the Cooperative Human Tissue Network (University of Alabama, Birmingham, AL). From a consecutive series of >500 unselected colorectal normal-tumor matched pairs, we selected for EGFR repeat analysis the microsatellite instability–positive cases for which DNA was available (n = 55). Sixty-one MSS colorectal cancers were randomly selected for comparative purposes. From the 52 microsatellite instability – positive cases for which mutation data was available, some of the cases could be classified as HNPCC (n = 5) or familial cases (n = 4), some had no family history (i.e., sporadic, n = 8), and for the rest, no family history information was available (n = 36). Fourteen microsatellite instability and 53 MSS gastric cancers were also analyzed. Genomic DNA from frozen specimens was extracted with phenol-chloroform. The LS174T colon cancer cell line was obtained from the American Type Culture Collection and was grown in DMEM supplemented with 15% of fetal bovine serum (Tissue Culture Biologicals). Microsatellite instability status in primary tumors was analyzed by PCR, as described previously (8, 14).
PCR analysis of the EGFR (CA)n length
The following primers were used for PCR amplification of the EGFR (CA)n-containing region within the first intron of EGFR: 5′-GTTTGAAGAATTTGAGCCAACC-3′ and 5′-TTCTTCTGCACACTTGGCAC-3′. PCR was carried out using a mix of Taq (Perkin Elmer) and Pfu DNA polymerases (Stratagene; ratio, 1:0.01) in the presence of 0.2 mCi of [α-P32]dCTP as follows: incubation at 94°C for 4 min, followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and one last incubation at 72°C for 10 min. PCR products were resolved on a 6% Sequencing gel (National Diagnostics) and subjected to autoradiography.
Isolation of the LS174T subclones with different numbers of CA repeats
Subclones with different (CA)n repeat numbers of LS174T cells were isolated as described (15). Cells were lysed in 100 μL of Tris-EDTA buffer (10 mmol/L Tris, 1 mmol/L EDTA; pH 7.4) supplemented with 150 μg/mL proteinase K and heated at 65°C for 1 h. The lysates were used for PCR amplification of the (CA)n region of EGFR. Two or three independent subclones with the same allele length were collected for further fluorescent-activated cell sorter analysis.
Frequency of insertion/deletion mutations in EGFR (CA)n in LS174T colon cancer cell line
To measure the frequency of mutations in EGFR (CA)n, we followed the Luria and Delbruck approach (16). The LS174T subclone culture was used to start 48 independent single cell cultures, which were grown separately for 25 cell replications. Each independent culture was then subcloned to obtain 96 single-cell clones, which were used for PCR analysis of the (CA)n length. A change in the repeat length by one CA unit was considered a single mutational event. Hence, a (CA)n+2 allele was scored as having undergone two insertions, and a (CA)n-2 allele was scored as having two deletions. After 25 cell replications, the total number of insertions and deletions was calculated in every one of the 48 independent cultures, analyzing at least 30 subclones per culture. The total number of insertions and deletions was then divided by the number of subclones analyzed in that particular culture to obtain the frequency of elongation and shortening, respectively. These independently estimated frequencies were subsequently averaged to account for fluctuations in the mutational frequency among the different 48 cultures.
Analysis of EGFR cell surface expression in LS174T subclones
Cells from two or three LS174T independent subclones with the same allele length were harvested with a nonenzymatic cell dissociation reagent (Specialty Media) and used for fluorescent-activated cell sorter analysis. Cells (5 × 105) from each subclone were incubated with EGFR-specific monoclonal antibodies (mouse immunoglobulin G; clone EGFR.1 from BD Pharmingen) at 5 μg/μL for 30 min at 4°C (fluorescent-activated cell sorter buffer: 1% bovine serum albumin, 1 mmol/L Ca2+, 1 mmol/L Mg2+, 0.02% NaN3 in PBS). Isotype-matched immunoglobulin G were used as negative control. Primary antibody incubation was followed by washing with fluorescent-activated cell sorter buffer and incubating with FITC-labeled secondary antibodies at 5 μg/μL for 30 min at 4°C (BD Biosciences). Fluorescence was detected on a FACScan flow cytometer (BD Biosciences) using standard procedures. The experiment was repeated twice from different independent clone cultures.
KRAS, p53, and BRAF mutation analysis
KRAS mutations at codons 12 and 13 and p53 mutations in exons 4 to 9 were analyzed by single-strand conformational polymorphism and sequencing, as described previously (18, 19). BRAF mutations in exons 11 and 15 were determined by sequencing, as described (20).
Analysis of the polyA tract of the 3′ untranslated region (UTR) of EGFR
Specific primers were designed to amplify the polyA tract in the 3′ UTR of EGFR (forward, 5′-GAAACGCATCCAGCAAGAAT-3′; reverse, 5′-ACTCCAAGATCCCCAATCAA-3′). Reverse primer was labeled with [γ-32P]ATP using T4-polynucleotide kinase (Promega). PCRs were done in standard conditions (Roche Applied Science) as follows: a single incubation at 94°C for 3 min followed by 30 cycles at 94°C for 45s, 58°C for 45 s, and 72°C for 45 s, and one final incubation at 72°C for 10 min. PCR products were resolved on a 6% Sequencing gel (National Diagnostics) and autoradiographed. The allele size was assessed by direct DNA sequencing using an automated DNA Analyzer (Applied Biosystems).
Results
Elongation of EGFR (CA)n in microsatellite instability–positive gastric and colon tumors
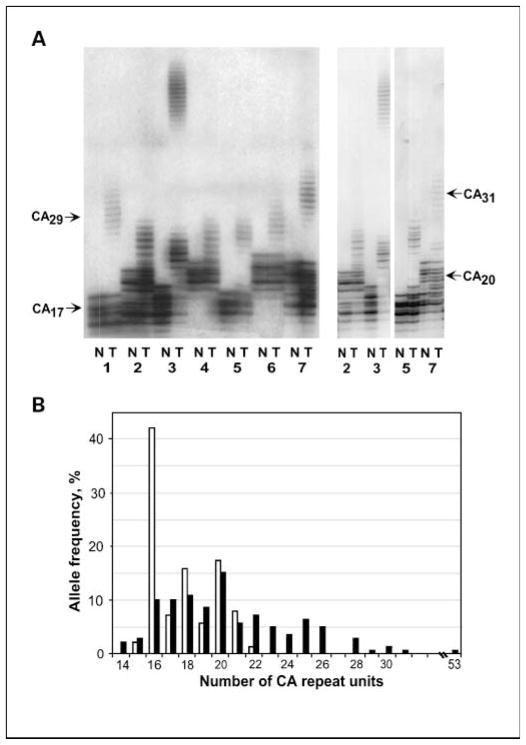
We examined EGFR (CA)n for somatic insertion/deletion mutations in microsatellite instability–positive stomach and colon cancers. The (CA)n sequence was PCR amplified from pairs of tumor and matched normal tissue DNA and their length determined by PAGE (Fig. 1A). The most frequent (CA)n alleles in normal tissues contained 16, 18, and 20 CA units with frequencies 42%, 16%, and 17%, respectively (Fig. 1B). This was in accordance with previously published data (21). As for cancer tissues, 82% of the colon and 62% of the stomach exhibited mutations in at least one of the two (CA)n alleles.
Fig. 1.
EGFR (CA)n mutations in microsatellite instability – positive tumors. A, electrophoregrams of (CA)n-containing PCR fragments amplified from seven (1–7) microsatellite instability – positive colon tumor and matched normal tissues. Arrows, positions of bands that correspond to alleles with 17, 20, 29, and 31CA units. Right, a lower exposure of some of the samples at left from an independent experiment. B, distributionof EGFR allele length in microsatellite instability – positive gastrointestinal (52 colorectal and 14 gastric) tumors and matchednormal tissues. Openbars, normal tissue; closedbars, tumor tissue.
Surprisingly, most of these mutations were insertions, resulting in elongation of the repeat (Fig. 1). Overall, 60 alleles with insertions versus 13 with deletions and 22 alleles with insertions versus 0 with deletions were detected in colon and gastric cancers, respectively (Table 1). The insertions ranged from 1 to 35 repeat units for colon and from 1 to 9 for gastric cancer. Length distribution of the CA alleles in tumor and normal tissues is shown in Fig. 1B. Eight alleles with repeats from 15 to 22 CA units were found in normal tissues of 69 gastrointestinal cancer patients, whereas 17 alleles with 14 to 53 CA repeats were found in the matched tumors. The allele with 16 CA repeats was the most prevalent in normal tissues (42.0%, 58 of 138). Its frequency was significantly lower in the tumor tissues (10.1%, 14 of 138; P = 1.62 × 10−9).
Table 1.
Frequency of insertion and deletion types of mutations in EGFR (CA)n repeat in tumor tissues of gastrointestinal cancer patients
| Mutations | Insertions | Deletions | No change |
|---|---|---|---|
| Colon cancer | 54.6% (60/110) | 11.8%(13/110) | 33.6% (37/110) |
| Stomach cancer | 78.6% (22/28) | 0% (0/28) | 21.4% (6/28) |
| Total | 59.4% (82/138) | 9.4% (13/138) | 31.2% (43/138) |
Expression of EGFR in microsatellite instability–positive and –negative colon tumors
EGFR expression was estimated by quantitative reverse transcription-PCR in 14 colon micro-satellite instability and 38 MSS cancers for which RNA was available. Although there was a slight increase in the overall expression levels of EGFR mRNA in MSS tumors relative to matched normal tissues (108%), the average level of expression was about half (56%) in the microsatellite instability tumors (P = 0.008; data not shown).
Frequency of insertion and deletion mutations in EGFR (CA)n in vitro
To compare insertion and deletion mutation rates for the (CA)n repeat in vitro, we used the Luria and Delbruck approach (16). For this purpose, 48 single-cell clone subcultures of the LS174T colon cancer cell line all with the same (CA)n allele sizes 20/28 were started. The subcultures were cultivated separately for 25 cell divisions, and the frequency of the CA repeat mutations were determined for each subculture. This was done by isolating 96 single-cell clones from each of the 48 independent cultures and analyzing their CA repeat length by PCR. A change in the repeat length by one CA unit was considered a single mutational event.
Average frequencies of deletions and insertions for the short (CA)20 and the long (CA)28 alleles were calculated over 48 independently grown cultures (Table 2) to accommodate for fluctuation of mutation frequency. Average frequencies of short and long allele mutations were used to compare their mutation rate. Short (1626) and long (1508) alleles were analyzed. The long allele had significantly higher rate of deletions versus insertions in vitro as measured by a higher average frequency of deletions versus insertions (χ2 = 19.89; P = 0.001). The short allele also had more deletions than insertions, although the difference was not statistically significant.
Table 2.
Frequency of deletions and insertions in (CA)20 and (CA)28 alleles of EGFR in vitro as determined by Luria and Delbrück approach
| Repeat | Frequency of shortening | Frequency of elongation |
|---|---|---|
| (CA)20 | 0.147 ± 0.020 | 0.144 ± 0.018 |
| (CA)28 | 0.313 ± 0.025 | 0.193 ± 0.021 |
These data illustrate that the expansion of EGFR (CA)n observed in microsatellite instability–positive tumors cannot be explained by an intrinsic property of this repeat favoring insertions versus deletions. Instead, the expansion of the repeat observed in the tumors must be the result of selection in favor of insertions due to clonal advantage in vivo.
Modulation of EGFR expression by the number of CA units in EGFR (CA)n
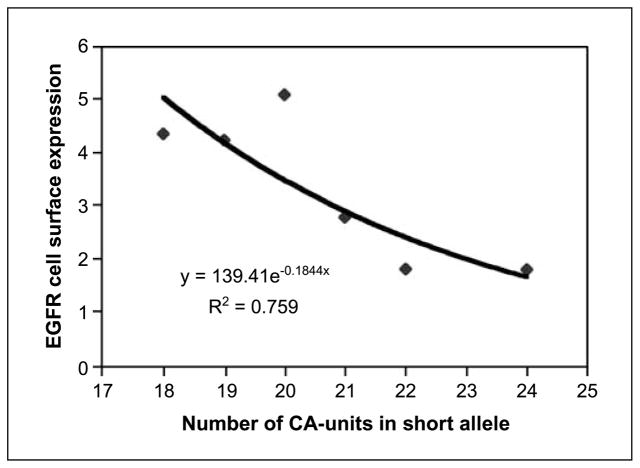
We examined EGFR cell surface expression by fluorescent-activated cell sorter analysis using monoclonal EGFR-specific antibodies. Binding to primary antibodies was visualized with the secondary FITC-labeled antibodies. The experiment was done with selected subclones of LS174T cell line, all sharing the same long allele of 28 CA units but different in the length of the short allele: 18, 19, 20, 21, 22, and 24 CA units.
Figure 2 shows that EGFR cell surface expression decreased almost 3-fold with the increase of the length of the short allele from 18 to 24 CA units. The expression level could be approximated by a formula y = 139.41e−0.1844X, where X is the number of CA units and y is the relative EGFR cell surface expression. We used this trend line formula to calculate EGFR expression for allele sizes outside the 18 to 24 CA size range.
Fig. 2.
EGFR cell-surface expression in LS174T subclones with alleles of different (CA)n lengths. Each dot on the graph, the average value of the expression of at least two independent clones with the same allele length from two independent repeated experiments. The trend line was automatically drawn by Microsoft Excel software using y = α × exp(β × X) formula for approximation of the experimental results. The exact formula is shown inside the chart area.
Our experimentally estimated relative levels of EGFR expression for different allele sizes agreed well with previously published in vitro transcription data for repeat sizes <20 CA units (8, 9). This result illustrates that the modulation of EGFR expression occurs continuously throughout a wide range of repeat lengths in cells of different tissue origin.
Correlation between EGFR (CA)n repeat expansion and mutations in KRAS, BRAF, and p53
To explore the possibility that the expansions of the EGFR repeat could be related to the mutational status of KRAS, BRAF, and p53, we did correlation studies between the EGFR (CA)n length and the presence of mutations in these genes in microsatellite instability–positive colon cancers. We used the results of previous screening of these tumors for mutations in codons 12 and 13 of KRAS (exon 1), in exon 15 of BRAF, and in the coding region of p53 from exons 4 to 9 (Table 3).
Table 3.
EGFR (CA)n, KRAS, BRAF, and p53 genotypes for 52 microsatellite instability–positive colorectal cancers
| Case no. | EGFR(CA)n† | EGFR(CA)n‡ | KRAS§ | BRAF|| | p53¶ |
|---|---|---|---|---|---|
| 1 | +3/+6 | +9 | wt | wt | wt |
| 2 | +1/−1 | 0 | wt | mut | wt |
| 3 | 0/+12 | +12* | wt | mut | Ala159Thr |
| 4 | 0/+6 | +6* | wt | wt | wt |
| 5 | +1/+3 | +4 | wt | mut | wt |
| 6 | 0/0 | 0 | mut | wt | Arg283Cys |
| 7 | 0/+2 | +2* | wt | wt | Glu326Gly |
| 8 | 0/−1 | −1 | wt | wt | wt |
| 9 | +4/+1 | +5 | wt | mut | wt |
| 10 | 0/+1 | +1 | mut | wt | Met246Val |
| 11 | −2/−2 | −4 | wt | mut | wt |
| 12 | +2/+9 | +11 | wt | wt | wt |
| 13 | +1/+2 | +3 | mut | wt | wt |
| 14 | 0/+5 | +5* | wt | mut | wt |
| 15 | +4/+6 | +10 | wt | wt | wt |
| 16 | 0/+8 | +8* | mut | wt | wt |
| 17 | +7/+9 | +16 | wt | wt | mut |
| 18 | 0/+1 | +1 | wt | mut | wt |
| 19 | 0/0 | 0 | wt | wt | wt |
| 20 | 0/+1 | +1 | wt | wt | Val73Met |
| 21 | +2/+5 | +7 | wt | mut | wt |
| 22 | +4/+4 | +8 | wt | mut | wt |
| 23 | 0/+5 | +5 | wt | wt | wt |
| 24 | −2/+2 | 0 | wt | mut | Arg175Cys |
| 25 | −1/+1 | 0 | wt | mut | His168Arg |
| 26 | −1/0 | −1 | wt | wt | wt |
| 27 | −2/+3 | +1 | wt | wt | wt |
| 28 | −3/0 | −3* | wt | mut | wt |
| 29 | +1/+10 | +11 | mut | wt | wt |
| 30 | −2/+3 | +1 | wt | mut | wt |
| 31 | −2/0 | −2* | wt | mut | Thr155Asp |
| 32 | 0/0 | 0 | wt | wt | wt |
| 33 | 0/0 | 0 | mut | wt | wt |
| 34 | −2/0 | −2 | wt | mut | wt |
| 35 | 0/+3 | +3* | wt | wt | Gly245Asp |
| 36 | +1/+1 | +2 | mut | wt | wt |
| 37 | 0/0 | 0 | wt | mut | wt |
| 38 | +1/0 | +1 | mut | wt | wt |
| 39 | 0/+1 | +1 | mut | wt | wt |
| 40 | +4/+6 | +10 | wt | mut | Arg181Cys |
| 41 | +8/+35 | +43 | wt | wt | wt |
| 42 | 0/0 | 0 | mut | wt | wt |
| 43 | 0/0 | 0 | wt | mut | wt |
| 44 | +1/+3 | +4 | wt | wt | wt |
| 45 | 0/0 | 0 | wt | mut | wt |
| 46 | 0/+1 | +1 | wt | mut | wt |
| 47 | +1/−1 | 0 | wt | mut | wt |
| 48 | +6/+4 | +10 | wt | wt | Gly154Ser |
| 49 | +3/+4 | +7 | wt | wt | wt |
| 50 | +1/+3 | +4 | wt | mut | wt |
| 51 | +2/+6 | +8 | wt | wt | wt |
| 52 | 0/0 | 0 | wt | mut | wt |
Abbreviations: wt, wild type; mut, mutations.
Asterisks denote noninformative cases in regards to whether one or two alleles underwent insertion/deletions. These 8 samples were excluded from the correlation study (Fig. 3). The result was not affected if the values of these cases were included. The interpretation of these cases was difficult due to the length similarity of constitutional and/or mutated alleles, together with the presence of significant contamination by normal tissue in the tumor tissue samples.
Number of added (+) or deleted (−) CA repeat units in the two alleles (shorter/longer) in the tumor compared with the constitutional alleles.
Accumulative value in the number of mutated (CA) repeat units from the two EGFR alleles.
KRAS mutations were Gly12Asp (cases 6, 10, 29, and 38), Gly12Ala (case 16), and Gly13Asp (cases 13, 33, 36, 39 and 613).
BRAF mutations were exclusively the Val600Glu at exon 15. No mutations were identified in exon 11. Two mutations at exon 11 were identified among 72 microsatellite instability–negative colon cancers.
p53 missense mutations. Case 7 has an additional silent nucleotide substitution in codon Ser96; the mutation in case 17 was only characterized by single-strand conformational polymorphism analysis.
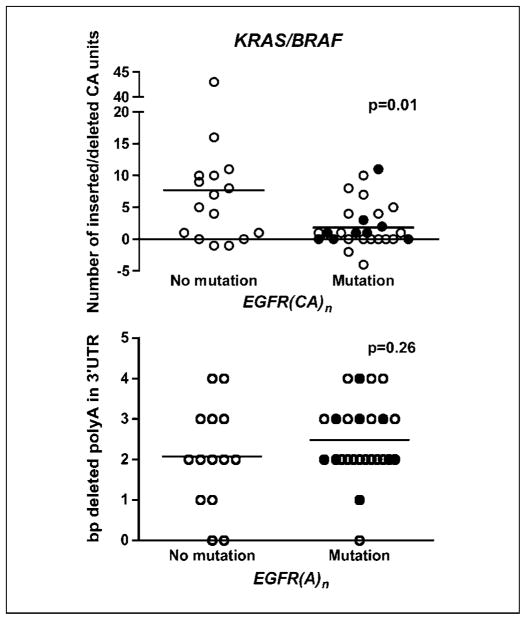
There was a negative association between the level of EGFR (CA)n expansion and mutations in either KRAS or BRAF (t test; P = 0.01). Tumors with mutations in either one of these two genes had less frequent insertions than the tumors without (Fig. 3, top). On the other hand, no correlation between mutations in p53 and EGFR (CA)n repeat elongation was found (t test; P = 0.74).
Fig. 3.
Association analysis between insertions in EGFR (CA)n (top) and deletions in EGFR 3′ UTR (A)n (bottom), and mutations in KRAS (solid dots) and BRAF (empty dots). Similar trend was observed when analyzing separately microsatellite instability tumors with mutations in K-ras (P = 0.13; n = X) or in B-raf (P = 0.03; n = X) versus microsatellite instability tumors with no mutations in these genes. No cases were found with concomitant K-ras and B-raf mutations.
Analysis of the polyA tract of the EGFR 3′ UTR
The 3′ UTR of EGFR harbors a polyadenine tract that is susceptible to mutation in microsatellite instability–positive tumors. Previous studies have shown that mutations in the 3′ UTR could modulate gene expression (22, 23). We determined the length of this polyA tract in 40 microsatellite instability–positive colon tumors and corresponding normal tissues. EGFR (A)n is polymorphic in the normal samples: the most common alleles contain 13 or 14 adenines. As expected, most tumors (92.5%) showed a shortening in this sequence (12). There was no correlation between the number of deletions in the 3′-end poly(A)n and the number of CA insertions in the (CA)n of the first intron of EGFR. No correlation between the deletions in the poly(A)n of EGFR and mutations in the KRAS and BRAF oncogenes was found (Fig. 3, bottom).
We also examined the length of EGFR (A)n in the LS174T subclones with different numbers of CA repeats and found that all of the subclones used in the fluorescent-activated cell sorter analysis were haploidentical. Therefore, the difference in EGFR expression observed in these clones was due solely to the changes in the number of (CA)n units in the 5′ end of the gene and not to alterations in the 3′-end noncoding repeat.
Discussion
EGFR is overexpressed in several tumors, and its over-expression correlates with malignant progression (24). Thus, we were intrigued by finding elongation of a microsatellite repeat at the 5′ UTR in gastrointestinal cancers with micro-satellite instability because it had been reported that elongation reduced expression (8, 9). Our study indicates that indeed this tendency for elongation that leads to down-regulation of the gene product is not due to an intrinsic tendency of the DNA sequence. Therefore, the selection for EGFR down-regulation during tumorigenesis implies that, in gastric and colon cancer of the microsatellite mutator phenotype, the gene works as a tumor suppressor and not as an oncogene. These observations may not be contradictory to each other, considering the differences between tumorigenic pathways of microsatellite instability–positive and microsatellite instability–negative cancers, and the pleiotropy of the biological responses to EGFR activation.
The research history of TGFs is an excellent example of divergent growth roles of proteins in different cell contexts. TGFs were initially detected and isolated by their transforming activities in normal anchorage-dependent cells (25, 26). It was only later that it became apparent that the common effects of these TGFs were of a negative nature. Thus, TGF-β plays a preferentially repressor function, despite its misleading name (27).
Although overexpression of EGFR was detected in several tumor types, particularly brain and breast, there is also a report that the EGFR level is lower in carcinomatous than in normal colorectal tissues (28). Because one of the EGFR ligands is TGF-α, down-regulation of EGFR in microsatellite instability–positive tumors may have an effect similar, additive or synergistic, to the mutational inactivation of the TGF-β receptor type II gene, which is a major event in the tumorigenesis of microsatellite instability – positive tumors (29).
In contrast to the accepted positive growth role of EGFR in breast and brain tumorigenesis, ligand stimulation of cells that overproduce EGFR may result in apoptosis, and apoptosis is a predictable outcome of EGFR overexpression in a variety of cell types (30, 31). To explain this apparent paradox, the latter authors proposed that during the clonal evolution of a tumor, low levels of receptor activation may enhance survival and indirectly promote mitogenesis and confer growth advantage to cells.
It has been shown that the induction of apoptosis by exposure of EGFR-overexpressing cells to epidermal growth factor is enhanced by inhibition of RAS signaling. The Raf–MAP/ERK kinase–extracellular signal-regulated kinase (mitogen-activated protein kinase) signal transduction is an important mediator in cell growth, proliferation, and survival. BRAF is activated by oncogenic RAS, leading to cooperative effects in cells responding to growth factor signals. The cells that expressed a dominant negative RAS/BRAF mutant exhibited a significant enhancement of EGFR-induced apoptosis, suggesting that RAS activation is a key survival signal generated by EGFR (31). Microsatellite instability–positive tumors are characterized by a paradoxically low incidence of RAS mutations (6, 32). Thus, microsatellite instability–positive tumor cells that carry normal RAS and BRAF proteins may be under selective pressure to down-regulate EGFR expression to escape apoptosis. These circumstances might cause a negative association between the presence of RAS/BRAF mutations and the EGFR (CA)n repeat expansion.
We failed to find any EGFR (CA)n mutations in 53 stomach and 61 colon microsatellite instability–negative tumors. The negative result is not surprising because the rate of insertion/deletion mutations in microsatellite instability – positive tumors is three orders of magnitude higher than in tumors without microsatellite instability. In this context, expression of the EGFR gene in vivo, analyzed by immunohistochemistry using a specific antibody, showed that EGFR expression was inhibited in some of the microsatellite instability–positive colon cancers analyzed, whereas staining was generally strong in microsatellite instability–negative tumors (data not shown).
The complex and pleiotropic role of the EGFR signaling system may dictate an alternative use of the EGFR signaling pathway during tumorigenesis, depending on the cancer organ site and the mechanism of cancer development. A microsatellite instability–positive tumor precursor cell produces many mutations without yet manifesting a neoplastic phenotype. Inactivation of DNA mismatch repair and the resulting unrepaired genomic errors may serve as apoptotic signals. The elongation of EGFR (CA)n occurs with high frequency because of the intrinsic instability of microsatellite sequences. The immediate consequential down-regulation of the gene expression may hamper the apoptotic response of the cell and help its survival until more powerful mechanisms of apoptosis inhibition are triggered, for example, an inactivating frameshift mutation in the mononucleotide stretch of the BAX gene (33).
If mutational inactivation of the apoptotic machinery occurs, the down-regulation of EGFR may no longer be required and the elongated EGFR (CA)n sequence could shorten to a stable size during further growth of the tumor or tumor precursor cell. However, it is possible that the partial inhibition of apoptosis by EGFR down-regulation early in tumor development may never lose its selective advantage. In contrast to microsatellite instability–negative tumors, for which the two-hit tumor suppressor gene inactivation model (34) applies so well, microsatellite instability–positive tumors are characterized by the presence of a plethora of monoallelic mutations in cancer-related genes (8, 35, 36). Such monoallelic mutations can cause haploinsufficiency and partial inactivation of gene function, which, when occurring simultaneously in many genes of the same oncogenic signaling networks, may bring down the normal homeostasis in cell growth or survival.
In microsatellite instability–positive cells, such mechanism would lead to a cascade of inactivation of tumor suppressor function analogous to the cascade increase of mutator phenotype, which we also observed for these tumors (15, 37, 38). In this case, each of these small effects, such as down-regulation of EGFR gene expression, heterozygous frameshift mutations in BAX, and other apoptotic genes (33, 35), can presumably cause altogether a significant inhibition of apoptosis and may remain under a selective pressure during tumor development and progression.
Thus, although EGFR was one of the first discovered factors implicated in the process of malignant transformation, its complex role in tumorigenesis may be far from having been completely understood. As an illustrative example, a recent report described EGFR to be frequently hypermethylated and silenced in several cancers, including breast, lung, and head and neck carcinomas (39).
The immediate consequence of these findings has translational potential related to EGFR-targeted cancer therapies. Numerous cancer therapies, ranging from therapeutic and imaging antibodies to toxin-linked ligands for enhancement of targeting for gene therapy vectors, have used up-regulated EGFR as a tumor-specific target (reviewed in ref. 40). Our findings predict that some of these therapies may have no effect or an opposite to the expected effect in microsatellite instability–positive gastrointestinal tumors.
Acknowledgments
Grant support: NIH grants R01CA098818 and R37 CA63585.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Savage CR, Jr, Cohen S. Epidermal growth factor and a new derivative. Rapid isolation procedures and biological and chemical characterization. J Biol Chem. 1972;247:7609–11. [PubMed] [Google Scholar]
- 2.Carpenter G. Receptors for epidermal growth factor and other polypeptide mitogens. Annu Rev Biochem. 1987;56:881–914. doi: 10.1146/annurev.bi.56.070187.004313. [DOI] [PubMed] [Google Scholar]
- 3.Huang SM, Harari PM. Epidermal growth factor receptor inhibition in cancer therapy: biology, rationale and preliminary clinical results. Invest New Drugs. 1999;17:259–69. doi: 10.1023/a:1006384521198. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37:S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 5.Wang X-Y, Smith DI, Frederick L, James CD. Analysis of EGF receptor amplicons reveals amplification of multiple expressed sequences. Oncogene. 1998;16:191–5. doi: 10.1038/sj.onc.1201476. [DOI] [PubMed] [Google Scholar]
- 6.Ishii S, Xu YH, Stratton RH, Roe BA, Merlino GT, Pastan I. Characterization and sequence of the promoter region of the human epidermal growth factor receptor gene. Proc Natl Acad Sci U S A. 1985;82:4920–4. doi: 10.1073/pnas.82.15.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maekawa T, Itoh F, Okamoto T, Kurimoto M, Imamoto F, Ishii S. Identification and purification of the enhancer-binding factor of human immunodeficiency virus-1. Multiple proteins and binding to other enhancers. J Biol Chem. 1989;264:2826–31. [PubMed] [Google Scholar]
- 8.Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274:13176–80. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 9.Gebhardt F, Burger H, Brandt B. Modulation of EGFR gene transcription by secondary structures, a polymorphic repetitive sequence and mutations—a link between genetics and epigenetics. Histol Histopathol. 2000;15:929–36. doi: 10.14670/HH-15.929. [DOI] [PubMed] [Google Scholar]
- 10.Aaltonen LA, Peltomäki P, Leach FS, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–6. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 11.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–9. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 12.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–61. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 13.Woerner SM, Benner A, Sutter C, et al. Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative real common target genes. Oncogene. 2003;22:2226–35. doi: 10.1038/sj.onc.1206421. [DOI] [PubMed] [Google Scholar]
- 14.Perucho M. Correspondence re: C.R. Boland, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 15.Baranovskaya S, Soto JL, Perucho M, Malkhosyan SR. Functional significance of concomitant inactivation of hMLH1 and hMSH6 in tumor cells of the micro-satellite mutator phenotype. Proc Natl Acad Sci U S A. 2001;98:15107–12. doi: 10.1073/pnas.251234498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krajewska M, Kim H, Shin E, et al. Tumor-associated alterations in caspase-14 expression in epithelial malignancies. Clin Cancer Res. 2005;11:5462–71. doi: 10.1158/1078-0432.CCR-04-2527. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto H, Perez-Piteira J, Yoshida T, et al. Gastric cancers of the microsatellite mutator phenotype display characteristic genetic and clinical features. Gastroenterology. 1999;116:1348–57. doi: 10.1016/s0016-5085(99)70499-3. [DOI] [PubMed] [Google Scholar]
- 19.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–3. [PubMed] [Google Scholar]
- 20.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Innocenti F, Chen P, Das S, Cook EH, Jr, Ratain MJ. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1polymorphism. Clin Cancer Res. 2003;9:1009–12. [PubMed] [Google Scholar]
- 22.Ruggiero T, Olivero M, Follenzi A, Naldini L, Calogero R, Di Renzo MF. Deletion in a (T)8 microsatellite abrogates expression regulation by 3′-UTR. Nucleic Acids Res. 2003;31:6561–9. doi: 10.1093/nar/gkg858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puga I, Lainez B, Fernandez-Real JM, et al. A polymorphism in the 3′ untranslated region of the gene for tumor necrosis factor receptor 2 modulates reporter gene expression. Endocrinology. 2005;146:2210–20. doi: 10.1210/en.2004-1366. [DOI] [PubMed] [Google Scholar]
- 24.Aaronson SA. Growth factors and cancer. Science. 1991;254:1146–53. doi: 10.1126/science.1659742. [DOI] [PubMed] [Google Scholar]
- 25.De Larco JE, Todaro GJ. Transforming growth factors produced by certain human tumor cells: poly-peptides that interact with epidermal growth factor receptors. Proc Natl Acad Sci U S A. 1978;75:4001–5. doi: 10.1073/pnas.77.9.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquardt H, Todaro GJ. Human transforming growth factor. Production by a melanoma cell line, purification, and initial characterization. J Biol Chem. 1982;257:5220–5. [PubMed] [Google Scholar]
- 27.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–78. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 28.Koenders PG, Peters WH, Wobbes T, Beex LV, Nagengast FM, Benraad TJ. Epidermal growth factor receptor levels are lower in carcinomatous than in normal colorectal tissue. Br J Cancer. 1992;65:189–92. doi: 10.1038/bjc.1992.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong DK, Kaufmann SH, Ottaviano YL, et al. Epidermal growth factor-mediated apoptosis of MDA-MB-468 human breast cancer cells. Cancer Res. 1994;54:5280–3. [PubMed] [Google Scholar]
- 31.Hognason T, Chatterjee S, Vartanian T, Ratan RR, Ernewein KM, Habib AA. Epidermal growth factor receptor induced apoptosis: potentiation by inhibition of Ras signaling. FEBS Lett. 2001;491:1–3. doi: 10.1016/s0014-5793(01)02166-4. [DOI] [PubMed] [Google Scholar]
- 32.Konishi M, Kikuchi-Yanoshita R, Tanaka K, et al. Molecular nature of colon tumors in hereditary nonpolyposis colon cancer, familial polyposis, and sporadic colon cancer. Gastroenterology. 1996;111:307–17. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- 33.Rampino N, Yamamoto H, Ionov Y, et al. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–9. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 34.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto H, Gil J, Schwartz S, Jr, Perucho M. Frameshift mutations in Fas, Apaf-1, and Bcl-10 in gastro-intestinal cancer of the microsatellite mutator phenotype. Cell Death Differ. 2000;7:238–9. doi: 10.1038/sj.cdd.4400651. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto H, Yamashita K, Perucho M. Somatic mutation of the beta2-microglobulin gene associates with unfavorable prognosis in gastrointestinal cancer of the microsatellite mutator phenotype. Gastroenterology. 2001;120:1565–7. doi: 10.1053/gast.2001.24497. [DOI] [PubMed] [Google Scholar]
- 37.Malkhosyan S, Rampino N, Yamamoto H, Perucho M. Frameshift mutator mutations. Nature. 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- 38.Ohmiya N, Matsumoto S, Yamamoto H, Baranovskaya S, Malkhosyan SR, Perucho M. Germline and somatic mutations in hMSH6 and hMSH3 in gastrointestinal cancers of the microsatellite mutator phenotype. Gene. 2001;272:301–13. doi: 10.1016/s0378-1119(01)00517-0. [DOI] [PubMed] [Google Scholar]
- 39.Montero AJ, Diaz-Montero CM, Mao L, et al. Epigenetic inactivation of EGFR by CpG island hypermethylation in cancer. Cancer Biol Ther. 2006;5:1494–501. doi: 10.4161/cbt.5.11.3299. [DOI] [PubMed] [Google Scholar]
- 40.Voldborg BR, Damstrup L, Spang-Thomsen M, Poulsen HS. Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann Oncol. 1997;8:1197–206. doi: 10.1023/a:1008209720526. [DOI] [PubMed] [Google Scholar]