Abstract
The development of complex multispecies communities such as biofilms is controlled by interbacterial communication systems. We have previously reported an intergeneric communication between two oral bacteria, Streptococcus cristatus and Porphyromonas gingivalis, that results in inhibition of fimA expression. Here, we demonstrate that a surface protein, arginine deiminase (ArcA), of S. cristatus serves as a signal that initiates intergeneric communication. An ArcA-deficient mutant of S. cristatus is unable to communicate with P. gingivalis. Furthermore, arginase activity is not essential for the communication, and ArcA retains the ability to repress expression of fimA in the presence of arginine deiminase inhibitors. These results present a novel mechanism by which intergeneric communication in dental biofilms is accomplished.
INTRODUCTION
Human dental plaque is a multispecies microbial biofilm that is associated with two common oral diseases, dental caries and periodontal disease. More than 700 bacterial species have been detected in the oral cavity, over 50 % of which are identified by culture-independent molecular techniques (Aas et al., 2005). Formation of dental plaque is a highly organized developmental process involving a specific sequence of colonization that results in spatially and temporally organized structures (Kolenbrander et al., 2006). Formation of dental plaque is initiated by Gram-positive species, including streptococci and Actinomyces spp., which recognize salivary receptors exposed on the tooth surfaces (Gibbons et al., 1991; Li et al., 2000; Scannapieco et al., 1995). These early colonizers in turn provide new surfaces that attract and recruit succeeding organisms including Gram-negative potential pathogens, such as Porphyromonas gingivalis and Aggregatibacter (Actinobacillus) actinomycetemcomitans (Kolenbrander et al., 2002). Therefore, the early colonizers play a key role in the development of the dental plaque biofilm.
It is recognized that cell–cell communication occurs between bacterial strains, species and genera. A universal language for interspecies bacterial communication is autoinducer-2 (AI-2). LuxS, the AI-2 synthase, has been discovered in many oral bacteria, including Streptococcus mutans, S. oralis, S. gordonii, P. gingivalis and A. actinomycetemcomitans (Chung et al., 2001; James et al., 2006; Merritt et al., 2005; Rickard et al., 2006). LuxS-dependent intercellular communication appears to play an important role in biofilm formation in the oral cavity. McNab et al. (2003) found that a S. gordonii luxS mutant was unable to form normal biofilms with a LuxS-deficient strain of P. gingivalis, and complementation of the luxS mutation in S. gordonii restored normal biofilm formation with the luxS-deficient P. gingivalis. In addition to communication mediated through soluble extracellular signalling molecules, interspecies crosstalk can occur through direct cell-to-cell contact (Aoki et al., 2005). We reported earlier that expression of the P. gingivalis fimA gene, encoding the long fimbrial major subunit protein, is repressed by surface extracts of Streptococcus cristatus (Xie et al., 2000). As the long fimbriae of P. gingivalis are required to initiate heterotypic biofilm formation with oral streptococci, substrata of S. cristatus do not support the development of a mixed biofilm with P. gingivalis (Xie et al., 2000). We show here that arginine deiminase (ArcA) is the inhibitory molecule of S. cristatus. The ability of S. cristatus to communicate with P. gingivalis is diminished in an arcA mutant. We also provide evidence that the ability of ArcA to repress expression of the fimA in P. gingivalis is not correlated with its enzymic activity. This work presents a novel inter-species contact-dependent communication system between P. gingivalis and S. cristatus.
METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids are listed in Table 1. Streptococcus strains were grown in Trypticase peptone broth (TPB) supplemented with 0.5 % glucose at 37 °C under aerobic conditions. S. cristatus CC5A was used as the parental strain for mutant construction. P. gingivalis ATCC 33277 was grown from frozen stocks in Trypticase soy broth (TSB) or on TSB blood agar plates, supplemented with 1 mg yeast extract ml−1, 5 μg haemin ml−1 and 1 μg menadione ml−1, at 37 °C in an anaerobic chamber (85 % N2, 10 % H2, 5 % CO2). Escherichia coli DH5α was used as the host for plasmids. E. coli strains were grown in L broth at 37 °C. Antibiotics were used when appropriate, at the following concentrations: 100 μg gentamicin ml−1 for P. gingivalis, 200 μg erythromycin ml−1 for E. coli and 10 μg erythromycin ml−1 for S. cristatus, 2 μg tetracycline ml−1 for E. coli and S. cristatus, 50 μg ampicillin ml−1 and 50 μg kanamycin ml−1 for E. coli.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics* | Source or reference |
|---|---|---|
| S. cristatus | ||
| CC5A | Low-passage plaque isolate | Lab collection |
| ArcAE | Derivative of CC5A containing an insertional mutation in the arcA gene; Emr | This study |
| cArcAE | A complemented strain of ARCE harbouring pT-ARCA | This study |
| P. gingivalis | ||
| ATCC 33277 | Type strain from ATCC | Lab collection |
| UPF | Derivative of ATCC 33277 containing fimA–lacZ gene fusion in its chromosomal DNA; Emr | Xie et al. (1997) |
| Mflac | Derivative of ATCC 33277 containing mfa1–lacZ gene fusion in its chromosomal DNA; Emr | This study |
| E. coli | ||
| DH5α | F−φ80dlacZΔ(lacZYA–argF)U169 endA1 supE44recA1 relA1 | BRL |
| Plasmids | ||
| pVA3000 | A suicide vector for Bacteroides; Emr, 5.3 kb | Lee et al. (1996) |
| pDN19lac | Contains a promoterless lacZ gene | Xie et al. (1997) |
| pJRD215 | A wide-host-range plasmid | Xie et al. (1997) |
| pPGS749 | E. coli–Streptococcus shuttle plasmid with Emr | Kuramitsu & Wang (2006) |
| pSF143 | Suicide vector for streptococci with Tetr; replicates only in E. coli | Tao et al. (1992) |
| pTet | Shuttle plasmid derived from both pPGS749 and pSF143 with Tetr; replicates in both E. coli and streptococci | This study |
| pT-ARCA | pTet plasmid carrying the arcA gene of S. cristatus CC5A | This study |
| pCRII-TOTO | A linearized plasmid with single 3′ dT residues; Kmr Amr | Invitrogen |
*Kmr, Tetr, Emr, Amr, resistance to kanamycin, tetracyline, erythromycin and ampicillin, respectively.
Partial purification of the S. cristatus inhibitory protein.
Surface extracts of S. cristatus CC5A were collected by sonication and centrifugation (13 000 g for 30 min) followed by filtration (0.2 μm pore size). The crude extract of CC5A was partially purified by ammonium sulfate fractionation as described earlier (Xie et al., 2004). The fractions precipitated with 33, 42, 50, 55, 60 and 66 % saturated ammonium sulfate were designated AS1, AS2, AS3, AS4, AS5 and AS6, respectively. For further purification, the AS6 fraction (1 ml) was dialysed overnight against Tris buffer (50 mM, pH 7.3). The dialysed sample was then applied to a Blue Sepharose column (GE Healthcare), which was pre-washed with the same Tris buffer (Nelson et al., 2001). The non-bound proteins were collected from the column. Bound proteins were eluted with Tris buffer supplemented with 1 mM NAD+.
Proteomic analysis.
Samples were separated by SDS-PAGE (12 % gel) along with prestained size standards (Bio-Rad). Coomassie-stained protein bands of interest were excised and reduced with 10 μl 45 mM dithiothreitol for 20 min at 37 °C. The gel pieces were then digested with trypsin overnight. The peptides were extracted and reconstituted in 20 μl 0.1 % trifluoroacetic acid. Approximately 0.4 μl of the peptides were spotted onto a MALDI plate. For each individual sample, the MALDI-TOF mass spectrum and the corresponding MS/MS fragmentation spectra were collectively searched against the SWISS-PROT database using GPS Explorer software (Applied Biosystems) running the MASCOT database search engine (Matrix-Science). MALDI-TOF peptide mass maps were internally calibrated to within 20 p.p.m. mass accuracy using trypsin autolytic peptides (m/z 842.51 and 2211.10).
Sequencing of the S. cristatus arcA gene.
The entire arcA gene of S. cristatus CC5A was amplified by the primers 5′-GTACCGATGGTCTTGTTTGA-3′ and 5′-AGGTATTCTAACTCTGCACG-3′, which were designed based on the completely conserved regions among Streptococus suis flps (AF546864), Streptococcus equi subsp. zooepidemicus arcA (AB210842) and the Streptococcus gordonii DL1 arc operon (AF534569). The PCR product was cloned into pCRII-TOPO vector (Invitrogen) and sequenced by using an ABI capillary sequencer (Perkin-Elmer). The sequence is deposited in GenBank (accession number EF435044).
Construction of the S. cristatus arcA mutant and arcA-complemented strains.
An insertional arcA mutant was generated by using ligation-independent cloning of PCR-mediated mutagenesis (LIC-PCR) (Aslanidis & de Jong, 1990). This procedure involved three steps of PCR to introduce a 2.1 kb ermF-ermAM cassette (Fletcher et al., 1995) into the arcA gene. First, the upstream DNA fragment (549 bp) of the arcA gene was amplified by using Taq RNA polymerase (1 U, Invitrogen) and chromosomal DNA of S. cristatus CC5A (0.1 μg) as template with specific primers (5′-ATGTCTACACATCCAATTC-3′ and 5′-GATGTTGCAAATACCGATGAGCATCTGCATACATGTGGTTGA-3′) containing the sequence (underlined) corresponding to the 5′ end of the ermF-ermAM cassette. The downstream DNA fragment (549 bp) of the arcA gene was amplified with specific primers (5′-ACAACGAGGTCCACCACG-3′and 5′-CCTCTAGAGTCGACCTGCAGATCGAAGGTGGAGATGAGTT-3′) containing the sequence (underlined) corresponding to the 3′ end of the ermF-ermAM cassette. Primers 5′-GCTCATCGGTATTTGCAACA-3′ and 5′-CTGCAGGTCGACTCTAGAGG-3′ were used to amplify the ermF-ermAM cassette. Each PCR product of the arcA gene was then ligated with the ermF-ermAM cassette by the second PCR step with primers arcAF and ermR or primers ermF and arcAR, respectively. The second-step PCR products (100 ng) were then mixed and used as template with arcAF and arcAR as primers in the third PCR step to create the fragment arcA-erm-arcA containing the ermF-ermAM cassette flanked with upstream and downstream fragments of arcA.
The arcA-erm-arcA fragment was introduced into S. cristatus CC5A cells by DNA transformation (Wang & Kuramitsu, 2005). arcA-deficient mutants were constructed via a double-crossover event that introduces the arcA-erm-arcA fragment into the CC5A chromosome. The mutants were selected on TPB plates supplemented with erythromycin (10 μg ml−1). The mutations were confirmed by PCR analysis, and the one selected for study was designated S. cristatus ArcAE.
An E. coli–Streptococcus shuttle vector was used to construct a complemented strain of ArcAE. To create the E. coli–Streptococcus shuttle vector, plasmid pSF143 (obtained from L. Tao, University of Illinois, Chicago, IL, USA), which replicates only in E. coli, was digested with HincII and BamHI to obtain a 5.4 kb fragment containing a tetracycline-resistance gene (Tobian et al., 1984). Plasmid pPGS749 (Kuramitsu & Wang, 2006) was digested with SmaI and BglII, and a 2.2 kb fragment that contains a Rep origin which replicates in streptococci was purified using a QIAEX II Gel Extraction kit (Qiagen). The two fragments were ligated using T4 ligase to generate pTet, a shuttle plasmid with tetracycline resistance that replicates in both E. coli and streptococci. pTet was then used for complementation of the arcA gene. The encoding region of CC5A arcA along with 330 bp of upstream sequence from the potential start codon was amplified by PCR with primers 5′-GCGGTACCTCAGCTATGAGCACAAACAG (KpnI site underlined), and 5′-GCCCATGGACAACGAGGTCCACCACG (NcoI site underlined). The PCR product was cloned into pTet vector. The recombinant plasmid, pT-ARCA, was introduced by transformation into the arcA-deficient mutant, S. cristatus ArcAE, to create S. cristatus cArcAE. After transformation, erythromycin- and tetracycline-resistant transconjugants were selected, and plasmid identity was confirmed by PCR analysis.
Cloning and expression of the arcA gene in E. coli.
arcA, encoding arginine deiminase, was amplified by PCR with primers 5′-GCGGTACCTATGTCTACACATCCAATTC-3′ (KpnI site underlined) and 5′-GCGAGCTCACAACGAGGTCCACCACG-3′ (SacI site underlined), which produced a 1200 bp PCR product. The PCR product was then cloned into pCRII-TOPO (Invitrogen). Recombinant arginine deiminase (rArcA) was expressed in E. coli by using a pThiohis protein expression system (Invitrogen). The arcA DNA fragment was subcloned into pThiohis-A downstream of a His tag. The recombinant ArcA was expressed in E. coli DH5α cells carrying the pThiohis-A/arcA plasmid in the presence of IPTG and kanamycin. His-tagged rArcA was purified with ProBond resin (Invitrogen). The His-tag on the recombinant protein was cleaved with enterokinase and removed by His-bind resin. Enterokinase was then removed by using Ekapture agarose.
Arginine deiminase assay.
The arginine deiminase assay was performed in 96-well microplates as described by Thirkill et al. (1983). S. cristatus CC5A protein samples were adjusted with PBS to a constant 100 μl volume in each well, and mixed with 50 μl 0.1 M l-arginine. The mixtures were allowed to react for 1 h at 37 °C and the reactions were then terminated by the addition of 50 μl 20 % sulfuric acid. Finally, 1 % 2,3-butanedione monoxime (Sigma) was added to each well, and the reaction was developed by incubation in the dark for 1 h at 56 °C. The peach colour was quantified with a Benchmark plus microplate spectrophotometer (Bio-Rad) at 492 nm.
Construction of P. gingivalis Mflac strain.
A P. gingivalis strain carrying an mfa1 promoter–lacZ fusion was generated by the method described before (Xie et al., 1997). Briefly, the mfa1 promoter region was amplified by PCR with primers 5′-ACCCATCCTCTGTCTTCTGC-3′ and 5′-CTCGTTATCACATATCCGAACC-3′, and cloned into pDN19lac to generate the mfa1 promoter–lacZ fusion. The recombinant plasmid was introduced into P. gingivalis ATCC 33277 by conjugation. The P. gingivalis transconjugants (Mflac) were selected on TSB plates containing 10 μg erythromycin ml−1.
β-Galactosidase assays.
S. cristatus protein fractions (25 μg) were mixed with 105 cells of P. gingivalis UPF, which contains a chromosomal fimA promoter–lacZ reporter construct, and spotted onto a TSB blood agar plate. The ability of the fractions to inhibit fimA expression in P. gingivalis was determined with a β-galactosidase assay. Expression of the lacZ gene under control of the fimA promoter was measured by the standard spectrophotometric β-galactosidase assay with ONPG as the substrate, as described by Xie et al. (1997).
RESULTS
Identification of S. cristatus inhibitory protein
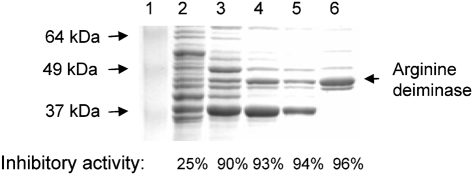
We reported previously that the expression of the fimA gene is repressed in the presence of surface extracts of S. cristatus, but not in the culture medium, indicating the presence of a LuxS-independent intergeneric communication system (Xie et al., 2000). To purify the signal molecule, we first fractionated S. cristatus CC5A surface extracts by ammonium sulfate precipitation (Xie et al., 2004). For further purification, the active fraction (AS6, 1 ml) was then applied to a Blue Sepharose column to remove glyceraldehyde-3-phosphate dehydrogenase, one of the major proteins in the AS6 fraction. The non-bound proteins were collected from the column and the fractions were analysed by SDS-PAGE. To test their ability to repress fimA expression in P. gingivalis, each fraction was mixed with P. gingivalis UPF, a strain carrying a fimA promoter–lacZ fusion. Expression of the lacZ gene under the control of the fimA promoter was measured by β-galactosidase assay (Xie et al., 1997). The results shown in Fig. 1 reveal that a protein band of approximately 47 kDa had a high correlation with the inhibitory activity. The ability to repress fimA expression was enhanced as the purity of this 47 kDa protein increased. The expression of fimA in P. gingivalis was inhibited by as much as 96 % by the unbound fraction after Blue Sepharose column chromatography (Fig. 1). These data suggested the involvement of the 47 kDa protein in intergeneric communication between S. cristatus and P. gingivalis.
Fig. 1.
SDS-PAGE analysis and inhibitory activity of the fractions of S. cristatus surface proteins. Surface extracts of S. cristatus were precipitated with ammonium sulfate at increasing concentrations, separated by SDS-PAGE and stained with Coomassie blue. Lane 1, molecular size standards; lane 2, ammonium sulfate (AS) fraction AS3; lane 3, AS4; lane 4, AS5; lane 5, AS6; lane 6, Blue Sepharose unbound fraction of AS6. Molecular sizes and the ArcA band are denoted by arrows. Inhibitory activity is expressed as % reduction (compared to buffer control) of LacZ activity in P. gingivalis UPF, a strain carrying the transcriptional fusion of a promoterless lacZ and the fimA promoter region.
To identify the 47 kDa protein, we performed in-gel digestion followed by MALDI-TOF mass spectrometry, as described in Methods. Searches against the SWISS-PROT database identified the protein as arginine deiminase (ArcA). Identification was accepted based on the significant molecular weight search (MOWSE) scores. Protein scores greater than 66 are significant (P<0.05). The score for the 47 kDa protein was 604. The molecular mass (46 752 Da) of streptococcal ArcA is consistent with the corresponding region of the gel as determined by the molecular mass markers.
Activity of the arcA mutant and complemented strains
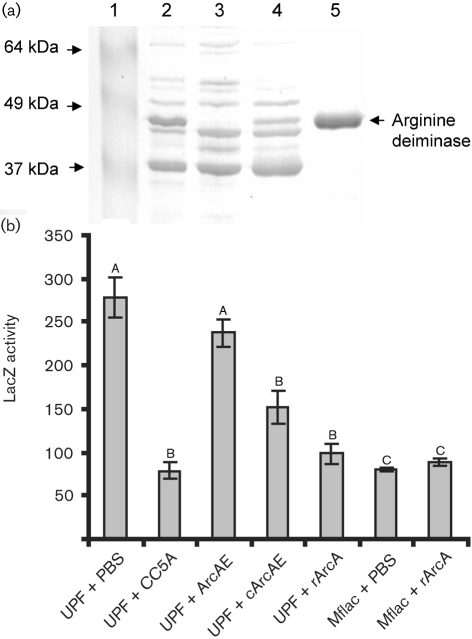
To confirm the role of ArcA in regulation of fimA expression, we constructed an arcA mutant of S. cristatus. Insertional inactivation of the S. cristatus arcA gene resulted in a prolonged lag period under the standard growth conditions for streptococci (Fig. 2). This is not surprising since the arginine deiminase pathway is partly responsible for ATP regeneration in bacteria (Crow & Thomas, 1982). Comparison of the ammonium sulfate precipitation fractions AS6 between wild-type CC5A and the mutant strain ArcAE showed that a 47 kDa band was missing from the mutant (Fig. 3a). Furthermore, mutation of arcA abrogated the inhibitory activity toward P. gingivalis fimA expression (Fig. 3b), indicating that arginine deiminase is indeed an effector molecule mediating communication between S. cristatus and P. gingivalis.
Fig. 2.
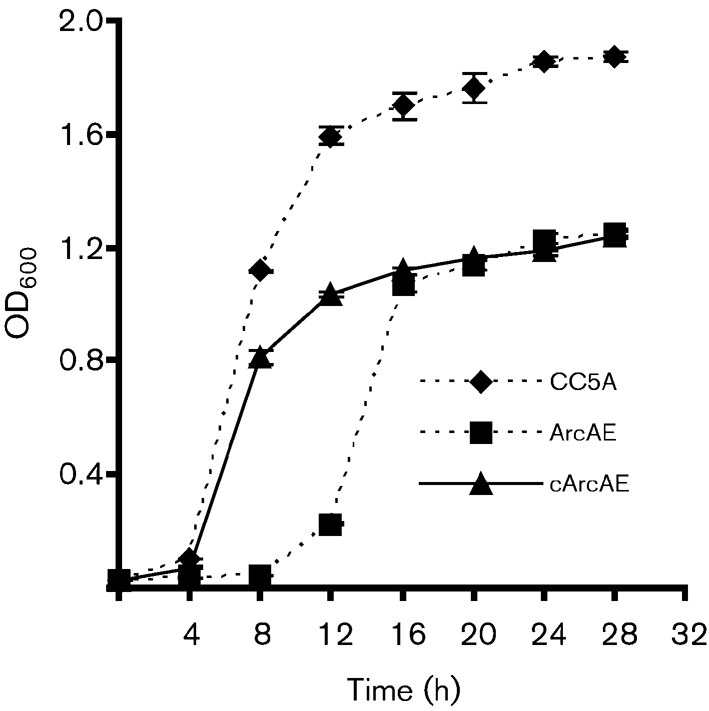
Comparison of the growth curves of S. cristatus strains in TSB medium. The data points are means±sem of four samples (error bars not shown where smaller than symbols). Samples of 1 ml were taken and the OD600 was measured over a period of 30 h.
Fig. 3.
Inhibition of fimA expression in P. gingivalis by ArcA. (a) S. cristatus surface proteins were subjected to SDS-PAGE (12 %) and stained with Coomassie blue. Lane 1, molecular mass markers; lane 2, ammonium sulfate fraction AS6 of CC5A; lane 3, ammonium sulfate fraction AS6 of ArcAE (arcA mutant); lane 4, ammonium sulfate fraction AS6 of cArcAE (arcA mutant complemented with the wild-type allele); lane 5, recombinant ArcA purified from E. coli. (b) P. gingivalis UPF carrying a fimA promoter–lacZ fusion and P. gingivalis Mflac carrying an mfa1 promoter–lacZ fusion were tested for LacZ activity in the presence or absence of surface extracts (50 μg) isolated from the S. cristatus strains indicated, or rArcA. The results are means±sem (n=3). Means with different letters are significantly different (P<0.05; Bonferroni test); means with the same letter are not significantly different.
The arginine deiminase operon has been extensively studied in S. gordonii DL1 (Caldelari et al., 2000; Dong et al., 2002; Zeng et al., 2006) and consists of five genes that encode enzymes involved in the conversion of arginine to ornithine, ammonia and CO2 with the concomitant production of ATP (Dong et al., 2002). arcA is the first gene in this operon. To eliminate the possibility that a polar effect plays a role in abolishing inhibitory activity in the arcA mutant, we complemented the mutant with the wild-type allele in trans. As shown in Fig. 3(a), production of ArcA was restored in the complemented strain cArcAE, although the expression level was lower compared to the parental CC5A strain. Complementation of the arcA mutant with the arcA gene partially restored the wild-type phenotype, since surface extracts isolated from the complemented strain cArcAE inhibited 50 % of fimA expression in P. gingivalis (Fig. 3b).
Activity of recombinant ArcA protein
We further confirmed the role of arginine deiminase in the repression of fimA expression in P. gingivalis by cloning and expressing arcA in E. coli. The fimA expression was repressed 2.5- to 3-fold in the presence of the recombinant protein (rArcA) (Fig. 3b), although the inhibitory activity was not as high as that of the natural protein, which was able to inhibit 96 % of the fimA expression (Fig. 1). This could be due to incorrect folding or post-translational modification in the heterologous host. The role of rArcA in expression of the short fimbriae (mfa1) was also examined by using a P. gingivalis strain carrying an mfa1–lacZ fusion. In the presence of rArcA, the promoter activity of mfa1 was not modulated in P. gingivalis (Fig. 3b), suggesting a specific role of S. cristatus ArcA in fimA expression. As a control, a major surface protein, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), of S. cristatus CC5A was also cloned and expressed in E. coli. The rGAPDH had no effect on fimA expression (data not shown).
Dual function of arginine deiminase
While the arginine deiminase system is found in many bacteria (Burne & Marquis, 2000), relatively few arginine deiminase-positive bacteria are found in oral biofilms (Zeng et al., 2006). Arginine deiminase catalyses the hydrolysis of l-arginine to l-citrulline and ammonia, and the latter is believed to be important for oral biofilm pH homeostasis and caries prevention (Burne & Marquis, 2000). Besides arginase activity, ArcA can also function as an inhibitor of angiogenesis and tumour growth, which may be due to the depletion of arginine (Gong et al., 2000; Kang et al., 2000; Park et al., 2003). In addition, arginine deiminase plays an important role in the regulation of the level of nitric oxide that is synthesized by NO synthase from arginine, a substrate of arginine deiminase (Gotoh & Mori, 1999). Since these two enzymes compete for the same substrate, antiangiogenic activity may result from the suppression of nitric oxide generation. To address whether the inhibitory activity of ArcA depends on enzyme activity, we examined each fraction for its arginase activity. Relatively high arginine hydrolytic activity was detected in the surface extract of S. cristatus (Table 2). Arginine hydrolytic activity was abolished in the arcA mutant, but was partially restored in the surface extracts of the arcA-complemented strain, which is consistent with production of arginine deiminase. Surprisingly, the purified fraction of arginine deiminase (the unbound fraction of the Blue Sepharose column) did not show an increased hydrolytic activity, despite the fact that at least 10 times more inhibitory activity was found in the purified fraction than in the surface extracts (Table 2). We speculated that the arginase activity is not required for intergeneric communication between S. cristatus and P. gingivalis. To test this hypothesis, communication was tested in the presence of aminoguanidine (20 μM) and l-lysine (5 mM), both of which are arginine deiminase inhibitors (Ulisse et al., 2001). These agents completely inhibited the arginase activity in CC5A fractions, but had little effect on the inhibitory activity of the fractions on fimA expression in P. gingivalis (Table 2). These data suggest that the catalytic activity of ArcA is not required for the mechanism of inhibition of fimA expression. It appears that ArcA now joins a growing list of bacterial proteins that can have multiple functions, possibly depending on their location (Jeffery, 1999).
Table 2.
Comparison of arginase activity and the inhibitory activity in protein fractions of S. cristatus
| Protein fraction | Arginine deiminase activity* | LacZ acivity† |
|---|---|---|
| PBS | 0.04±0.00 | 278±21 |
| S. cristatus CC5A surface extract (50 μg) | 2.12±0.06 | 78±10 |
| S. cristatus ArcAE surface extract (50 μg) | 0.18±0.01 | 237±15 |
| S. cristatus cArcAE surface extract (50 μg) | 1.46±0.10 | 152±18 |
| CC5A surface extract (50 μg)+10 mM aminoguanidine | 0.32±0.04 | nd |
| CC5A surface extract (50 μg)+5 mM lysine | 0.14±0.01 | nd |
| Purified fraction AS6 (25 μg) | 0.8±0.10 | 12±2 |
| Purified fraction AS6 (25 μg)+10 mM aminoguanidine | 0.15±0.02 | 19±3 |
| Purified fraction AS6 (25 μg)+5 mM lysine | 0.15±0.01 | 21±4 |
*Arginine deiminase levels are means±sd (n=3).
†Expression of the fimA gene was determined by measuring LacZ activity, expressed in Miller units. Results are means±SEM (n=3). nd, Not determined.
DISCUSSION
P. gingivalis is a secondary colonizer of dental plaque, and is significantly more prevalent in both supra- and subgingival plaque samples from periodontitis subjects in comparison with healthy subjects (Ximenez-Fyvie et al., 2000). The surface attachment of P. gingivalis is promoted by adhesive molecules including fimbriae. The long fimbriae, composed of the FimA subunit, mediate adherence of P. gingivalis to a variety of oral substrates and molecules, including proline-rich proteins and glycoproteins, statherin, fibrinogen, fibronectin and lactoferrin (Lamont & Jenkinson, 1998). The fimbriae are also important effector molecules in coaggregation interaction with various early plaque-forming bacteria, such as Actinomyces viscosus (Goulbourne & Ellen, 1991), Streptococcus gordonii (Lamont et al., 1993) and Streptococcus oralis. Amano et al. (1997) also demonstrated that the FimA C-terminal region is involved in coaggregation with S. oralis, with functional domains located in regions spanning amino acids 266–286 and 287–337. FimA is also a specific adhesin mediating coaggregation of P. gingivalis and Treponema denticola, another secondary colonizer (Hashimoto et al., 2003). This specific coaggregation ability with other oral bacteria suggests that the P. gingivalis long fimbriae contribute to bacterial integration into dental plaque by interacting with the early and secondary colonizers of dental plaque. Evidently, dental plaque colonization is beneficial to P. gingivalis survival in their optimum ecological niche, periodontal pockets. Our earlier finding demonstrated that S. cristatus is able to repress expression of the fimA gene in P. gingivalis and thus prevent subsequent heterotypic biofilm formation (Xie et al., 2000). The present results provide evidence for the first time that the surface protein arginine deiminase of S. cristatus is a signal molecule responsible for cell–cell communication between S. cristatus and P. gingivalis. As a result of this signal, P. gingivalis shuts down expression of the fimA gene. Communication between Gram-positive and Gram-negative bacteria as observed here may be fundamental to bacteria in multispecies biofilms. Interspecies cooperation and competition play important roles in biofilm development and organization. The identification of the molecular basis for an intergeneric contact-dependent communication system provides a molecular basis to begin to understand the differentiation of oral microbial communities from commensal to pathogenic. The study presented here could ultimately lead to the development of novel means to inhibit oral colonization of periodontal pathogens. Oral streptococci are some of the predominant early colonizers of oral plaque (Li et al., 2004), and this unique communication system of sensing foreign species via a surface protein may have been developed to uphold a dominant position in this specialized niche. The consequent inhibition of P. gingivalis biofilm formation suggests that susceptibility to periodontal disease may depend to some extent on the microbial composition of the early plaque biofilm and, moreover, that production of arginine deiminase by the oral streptococci may be significant in protection against periodontitis.
Acknowledgments
This work was supported by Public Health Service grants DE014699 (H. X.) and DE12505 (R. J. L.) from the National Institute of Dental and Craniofacial Research.
Footnotes
The GenBank/EMBL/DDBJ accession number for the arcA gene sequence of S. cristatus is EF435044.
References
- Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I. & Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43, 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano, A., Fujiwara, T., Nagata, H., Kuboniwa, M., Sharma, A., Sojar, H. T., Genco, R. J., Hamada, S. & Shizukuishi, S. (1997). Prophyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res 76, 852–857. [DOI] [PubMed] [Google Scholar]
- Aoki, S. K., Pamma, R., Hernday, A. D., Bickham, J. E., Braaten, B. A. & Low, D. A. (2005). Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248. [DOI] [PubMed] [Google Scholar]
- Aslanidis, C. & de Jong, P. J. (1990). Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18, 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne, R. A. & Marquis, R. E. (2000). Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett 193, 1–6. [DOI] [PubMed] [Google Scholar]
- Caldelari, I., Loeliger, B., Langen, H., Glauser, M. P. & Moreillon, P. (2000). Deregulation of the arginine deiminase (arc) operon in penicillin-tolerant mutants of Streptococcus gordonii. Antimicrob Agents Chemother 44, 2802–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, W. O., Park, Y., Lamont, R. J., McNab, R., Barbieri, B. & Demuth, D. R. (2001). Signaling system in Porphyromonas gingivalis based on a LuxS protein. J Bacteriol 183, 3903–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, V. L. & Thomas, T. D. (1982). Arginine metabolism in lactic streptococci. J Bacteriol 150, 1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., Chen, Y. Y., Snyder, J. A. & Burne, R. A. (2002). Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl Environ Microbiol 68, 5549–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, H. M., Schenkein, H. A., Morgan, R. M., Bailey, K. A., Berry, C. R. & Macrina, F. L. (1995). Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun 63, 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons, R. J., Hay, D. I. & Schlesinger, D. H. (1991). Delineation of a segment of adsorbed salivary acidic proline-rich proteins which promotes adhesion of Streptococcus gordonii to apatitic surfaces. Infect Immun 59, 2948–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, H., Zolzer, F., von Recklinghausen, G., Havers, W. & Schweigerer, L. (2000). Arginine deiminase inhibits proliferation of human leukemia cells more potently than asparaginase by inducing cell cycle arrest and apoptosis. Leukemia 14, 826–829. [DOI] [PubMed] [Google Scholar]
- Gotoh, T. & Mori, M. (1999). Arginase II downregulates nitric oxide (NO) production and prevents NO-mediated apoptosis in murine macrophage-derived RAW 264.7 cells. J Cell Biol 144, 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulbourne, P. A. & Ellen, R. P. (1991). Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J Bacteriol 173, 5266–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M., Ogawa, S., Asai, Y., Takai, Y. & Ogawa, T. (2003). Binding of Porphyromonas gingivalis fimbriae to Treponema denticola dentilisin. FEMS Microbiol Lett 226, 267–271. [DOI] [PubMed] [Google Scholar]
- James, D., Shao, H., Lamont, R. J. & Demuth, D. R. (2006). The Actinobacillus actinomycetemcomitans ribose binding protein RbsB interacts with cognate and heterologous autoinducer 2 signals. Infect Immun 74, 4021–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery, C. J. (1999). Moonlighting proteins. Trends Biochem Sci 24, 8–11. [DOI] [PubMed] [Google Scholar]
- Kang, S. W., Kang, H., Park, I. S., Choi, S. H., Shin, K. H., Chun, Y. S., Chun, B. G. & Min, B. H. (2000). Cytoprotective effect of arginine deiminase on taxol-induced apoptosis in DU145 human prostate cancer cells. Mol Cells 10, 331–337. [PubMed] [Google Scholar]
- Kolenbrander, P. E., Andersen, R. N., Blehert, D. S., Egland, P. G., Foster, J. S. & Palmer, R. J., Jr (2002). Communication among oral bacteria. Microbiol Mol Biol Rev 66, 486–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander, P. E., Palmer, R. J., Rickard, A. H., Jakubovics, N. S., Chalmers, N. I. & Diaz, P. I. (2006). Bacterial interactions and successions during plaque development. Periodontol 2000 42, 47–79. [DOI] [PubMed] [Google Scholar]
- Kuramitsu, H. K. & Wang, B. Y. (2006). Virulence properties of cariogenic bacteria. BMC Oral Health 6 (Suppl. 1), S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont, R. J. & Jenkinson, H. F. (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 62, 1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont, R. J., Bevan, C. A., Gil, S., Persson, R. E. & Rosan, B. (1993). Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol 8, 272–276. [DOI] [PubMed] [Google Scholar]
- Lee, S. W., Hillman, J. D. & Progulske-Fox, A. (1996). The hemagglutinin genes hagB and hagC of Porphyromonas gingivalis are transcribed in vivo as shown by use of a new expression vector. Infect Immun 64, 4802–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., Bratt, P., Jonsson, A. P., Ryberg, M., Johansson, I., Griffiths, W. J., Bergman, T. & Stromberg, N. (2000). Possible release of an ArgGlyArgProGln pentapeptide with innate immunity properties from acidic proline-rich proteins by proteolytic activity in commensal streptococcus and actinomyces species. Infect Immun 68, 5425–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Helmerhorst, E. J., Leone, C. W., Troxler, R. F., Yaskell, T., Haffajee, A. D., Socransky, S. S. & Oppenheim, F. G. (2004). Identification of early microbial colonizers in human dental biofilm. J Appl Microbiol 97, 1311–1318. [DOI] [PubMed] [Google Scholar]
- McNab, R., Ford, S. K., El-Sabaeny, A., Barbieri, B., Cook, G. S. & Lamont, R. J. (2003). LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol 185, 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, J., Kreth, J., Shi, W. & Qi, F. (2005). LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol Microbiol 57, 960–969. [DOI] [PubMed] [Google Scholar]
- Nelson, D., Goldstein, J. M., Boatright, K., Harty, D. W., Cook, S. L., Hickman, P. J., Potempa, J., Travis, J. & Mayo, J. A. (2001). pH-regulated secretion of a glyceraldehyde-3-phosphate dehydrogenase from Streptococcus gordonii FSS2: purification, characterization, and cloning of the gene encoding this enzyme. J Dent Res 80, 371–377. [DOI] [PubMed] [Google Scholar]
- Park, I. S., Kang, S. W., Shin, Y. J., Chae, K. Y., Park, M. O., Kim, M. Y., Wheatley, D. N. & Min, B. H. (2003). Arginine deiminase: a potential inhibitor of angiogenesis and tumour growth. Br J Cancer 89, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard, A. H., Palmer, R. J., Jr, Blehert, D. S., Campagna, S. R., Semmelhack, M. F., Egland, P. G., Bassler, B. L. & Kolenbrander, P. E. (2006). Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol 60, 1446–1456. [DOI] [PubMed] [Google Scholar]
- Scannapieco, F. A., Torres, G. I. & Levine, M. J. (1995). Salivary amylase promotes adhesion of oral streptococci to hydroxyapatite. J Dent Res 74, 1360–1366. [DOI] [PubMed] [Google Scholar]
- Tao, L., LeBlanc, D. J. & Ferretti, J. J. (1992). Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120, 105–110. [DOI] [PubMed] [Google Scholar]
- Thirkill, C. E., Song, D. Y. & Gregerson, D. S. (1983). Application of monoclonal antibodies to detect intraocular mycoplasma antigens in Mycoplasma arthritidis-infected Sprague-Dawley rats. Infect Immun 40, 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobian, J. A., Cline, M. L. & Macrina, F. L. (1984). Characterization and expression of a cloned tetracycline resistance determinant from the chromosome of Streptococcus mutans. J Bacteriol 160, 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulisse, S., Gionchetti, P., D'Alo, S., Russo, F. P., Pesce, I., Ricci, G., Rizzello, F., Helwig, U., Cifone, M. G. & other authors (2001). Expression of cytokines, inducible nitric oxide synthase, and matrix metalloproteinases in pouchitis: effects of probiotic treatment. Am J Gastroenterol 96, 2691–2699. [DOI] [PubMed] [Google Scholar]
- Wang, B. Y. & Kuramitsu, H. K. (2005). Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol 71, 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, H., Cai, S. & Lamont, R. J. (1997). Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect Immun 65, 2265–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, H., Cook, G. S., Costerton, J. W., Bruce, G., Rose, T. M. & Lamont, R. J. (2000). Intergeneric communication in dental plaque biofilms. J Bacteriol 182, 7067–7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, H., Kozlova, N. & Lamont, R. J. (2004). Porphyromonas gingivalis genes involved in fimA regulation. Infect Immun 72, 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenez-Fyvie, L. A., Haffajee, A. D. & Socransky, S. S. (2000). Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J Clin Periodontol 27, 722–732. [DOI] [PubMed] [Google Scholar]
- Zeng, L., Dong, Y. & Burne, R. A. (2006). Characterization of cis-acting sites controlling arginine deiminase gene expression in Streptococcus gordonii. J Bacteriol 188, 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]