Abstract
The linoleic acid isomerase enzyme from Propionibacterium acnes responsible for bioconversion of linoleic acid to trans-10, cis-12 conjugated linoleic acid (t10, c12 CLA) was cloned and overexpressed in Lactococcus lactis and Escherichia coli, resulting in between 30 and 50 % conversion rates of the substrate linoleic acid to t10, c12 CLA. The anti-proliferative activities of the fatty acids produced following isomerization of linoleic acid by L. lactis and E. coli were assessed using the human SW480 colon cancer cell line. Fatty acids generated from both L. lactis and E. coli contained a mixture of linoleic acid and t10, c12 CLA at a ratio of ∼1.35 : 1. Following 5 days of incubation of SW480 cells with 5–20 μg ml−1 (17.8–71.3 μM) of the t10, c12 CLA, there was a significant (P<0.001) reduction in growth of the SW480 cancer cells compared with the linoleic acid control. Cell viability after treatment with the highest concentration (20 μg ml−1) of the t10, c12 CLA was reduced to 7.9 % (L. lactis CLA) and 19.6 % (E. coli CLA), compared with 95.4 % (control linoleic acid) and 31.7 % (pure t10, c12 CLA). In conclusion, this is believed to represent the first report in which recombinant strains are capable of producing CLA with an anti-proliferative potential.
INTRODUCTION
Conjugated linoleic acids (CLAs) are a family of positional and geometric isomers of linoleic acid with conjugated double bonds. Biological activities associated with the cis-9, trans-11 CLA (c9, t11 CLA) and the trans-10, cis-12 CLA (t10, c12 CLA) isomers include anti-cancer, anti-atherosclerotic, anti-diabetic and immune-enhancing properties, and positive effects on body composition and bone formation (Belury, 2002; Pariza et al., 1999, 2000). A number of studies have shown that the t10, c12 CLA isomer is the most potent isomer for prevention of cell proliferation and induction of apoptosis in cancer cells (Cho et al., 2005, 2006; Lee et al., 2006; Kim et al., 2002a; Ochoa et al., 2004). Furthermore, this isomer has the ability to alter body composition by reducing the fat content and increasing the lean body tissue in animal models, and also in some studies with humans (Smedman & Vessby, 2001; Blankson et al., 2000; Thom et al., 2001).
c9, t11 CLA is formed as an intermediate during biohydrogenation of linoleic acid by rumen bacteria, and consequently natural sources of this isomer are milk and fats from ruminants. The main CLA isomer in milk fat is c9, t11 CLA, which accounts for 80–90 % of total milk fat CLA, whereas the t10, c12 CLA isomer is only present at about 1 % (Jensen, 2002). A range of cultures with the ability to convert linoleic acid to c9, t11 CLA are known, in addition to the rumen microflora. We have previously shown that some human-derived Bifidobacterium species can produce CLA (Coakley et al., 2003; Rosberg-Cody et al., 2004). The predominant isomer formed after 48–72 h incubation with 0.5 mg linoleic acid ml−1 is c9, t11 CLA, with some of this isomer further converted to the t9, t11 CLA isomer (Coakley et al., 2006). Other species reported to biosynthesize CLA, mainly the c9, t11 CLA isomer, are propionibacteria used as dairy starter cultures (Jiang et al., 1998), strains of the intestinal flora of rats (Chin et al., 1994), Lactobacillus (Lin et al., 1999), Corynebacterium (Fairbank et al., 1988, 1989), Lactococcus (Kim & Liu, 2002) and some common bacterial lung pathogens (Jack et al., 1994). The only known species which can perform a c9 to t10 isomerization are Propionibacterium acnes isolated from mouse caecum (Verhulst et al., 1987) and the rumen bacterium Megasphaera elsdenii (Kim et al. 2000, 2002b). P. acnes has been shown to convert ∼85 % of linoleic acid into t10, c12 CLA after 24 h of incubation and ∼10 % into t10 octadecenoic acid (Verhulst et al., 1987). Since both reaction products have proved to be derived from linoleic acid, the results indicate that the c9 double bond of linoleic acid is first isomerized into a t10 double bond, partly followed by reduction of the c12 double bond (Verhulst et al., 1987). The t10, c12 CLA could be formed from linoleic acid alone by P. acnes and M. elsdenii, and not from linolenic acid, as with the c9, t11 CLA isomer (Verhulst et al., 1987; Kim et al., 2000).
The mechanisms by which CLA exerts all its physiological effects are not yet fully understood, but at least two different pathways have been proposed. The first suggests that CLA reduces the arachidonate pool, leading to a reduced production of downstream eicosanoid products, which modulates cytokine production involved in inflammation and cancer. The other model includes the regulation of expression of genes known to control the cell cycle and apoptosis, but also lipid oxidation, adipocyte differentiation, energy balance and atherogenesis (Belury, 2002; Pariza et al., 2000). Recently, it has been proposed that different CLA isomers produce different effects. The t10, c12 CLA isomer seems to work preferentially through modulation of apoptosis and cell cycle control, while c9, t11 CLA appears to affect arachidonic acid metabolism. The t10, c12 CLA isomer has been shown to control the cell cycle and induce apoptosis by decreasing bcl-2 gene expression and increasing p21WAF1/Cip1 mRNA levels in prostate cancer cells (Ochoa et al., 2004). It has been shown to activate caspase-3, and -9, translocate the anti-apoptotic protein Bax into the mitochondrial membrane and cleave the anti-apoptotic protein Bid in rat hepatoma cells (Yamasaki et al., 2005), along with induction of caspase-dependent apoptosis in MIP-101 and PC-3 cells (Palombo et al., 2002). This isomer has also been reported to decrease cell growth due to a decrease in the secretion of insulin-like growth factor II (IGF-II) in Caco-2 colon cancer cells (Kim et al., 2002a), and to cause a decrease of expression of the oncogene ErbB3 in HT-29 human colon cancer cells (Cho et al., 2005). Lee et al. (2006) have reported that t10, c12 CLA induces pro-apoptotic and anti-tumorigenic NAG-1 in human colorectal cancer cells. Moreover, t10, c12 CLA induces p53, p21 and p27, and reduces the levels of cyclins D1 and E and hyperphosphorylated retinoblastoma protein (required for G1 to S phase transition) in human breast cancer cells (Kemp et al., 2003). Cho et al. (2006) have reported the induction by t10, c12 CLA of G1 cell cycle arrest through increased expression of p21, as well as decreased CDK2 activity, decreased levels of hyperphosphorylated retinoblastoma (Rb) protein and increased hypophosphorylated Rb protein in HT-29 colon cancer cells. However, an integrated model of the mechanism of action of t10, c12 CLA has not been developed to date.
The aims of this study were to clone and overexpress the linoleic acid isomerase capable of producing t10, c12 CLA from P. acnes (Hornung et al., 2005) in Lactococcus lactis and Escherichia coli, and to investigate the anti-proliferative activity of the generated t10, c12 CLA using the human SW480 colon cancer cell line.
METHODS
Cultures and media.
L. lactis NZ9800 (an L. lactis NZ9700 derivative which, due to a deletion in the nisA gene, does not produce nisin, and which contains the nisRK signal transduction genes integrated on the chromosome) was cultured at 30 °C in M17 (Difco) broth and/or agar containing glucose (0.5 %, w/v). L. lactis carrying the plasmid pNZ44 was routinely cultured in the presence of chloramphenicol (5 μg ml−1) as a selective marker. E. coli DH5α (TOP 10, Invitrogen) harbouring the plasmid pNZ44 was cultured in Luria–Bertani (LB) media supplemented with chloramphenicol (20 μg ml−1) with shaking at 37 °C.
Human colon cancer cells were obtained from the American Type Culture Collection (ATCC). The SW480 cell line was originally established from a primary adenocarcinoma of the colon (Leibovitz et al., 1976). The cell line is somewhat dedifferentiated, lacking many of the phenotypic markers of normal colon epithelial cells. SW480 cells do retain an epithelial-like morphology, with regular polygonal cells predominating in culture, with some round cells. The t10, c12 CLA isomer (>98 % purity) was obtained from Matreya and linoleic acid was purchased from Sigma Aldrich. Cell culture media and supplements were purchased from Sigma, unless otherwise stated. SW480 cells were maintained in Dulbecco's Minimum Essential Medium (DMEM) supplemented with 5 % (v/v) fetal bovine serum, 0.2 mM l-glutamine, 1 mM HEPES, and 1 U ml−1 penicillin and streptomycin. SW480 cells were grown in 75 ml flasks and maintained at 37 °C in a humidified atmosphere at pH 7.2–7.4 with a flow of 95 % air/5 % CO2, as required.
DNA manipulation.
Two oligonucleotide primers were designed to amplify the complete linoleic acid isomerase (coPAI) for production of t10, c12 CLA from the original construct pC33.1-coPAI (linoleic acid isomerase gene in a plant vector; BASF). The forward primer, designated ERcoPAI1, contained a PstI restriction site and a ribosome-binding site (RBS), four extra bases at the 5′ end and seven extra bases between the RBS and the gene start: 5′-aaaactgcagaggaggaaaaaaaATGGGTTCCATTTCCAAGGA-3′, where bold capitals represent part of the DNA sequence and small capitals represent artificial sequence (e.g. restriction sites and added bases). The reverse primer, designated ERcoPAI2, contained a KpnI restriction site and three extra bases at the 5′ end: 5′-cggggtaccTCACACGAAGAACCGCGTCA-3′. The 1278 bp coPAI gene was amplified in an Eppendorf Mastercycler Gradient with High Fidelity Expand polymerase, as described by the supplier (Roche Diagnostics) using 200 ng plasmid DNA (pC33.1-coPAI) as template. PCR reactions were performed in a total volume of 50 μl containing 1 μl of each primer, 3 mM MgCl2, 5 μl 10× Expand buffer, 1 μl dNTPs and 0.75 μl Expand polymerase. PCR conditions were as follows: one cycle of 2 min denaturation (94 °C), 10 cycles of 15 s denaturation (94 °C), 30 s annealing (55 °C), 2 min elongation (72 °C), followed by 20 cycles of 15 s (94 °C), 30 s (55 °C), 2 min+5 s per cycle (72 °C), and finally one 7 min cycle at 72 °C. The PCR reaction mixture was analysed in a 1 % (w/v) agarose gel to visualize the resulting PCR fragment.
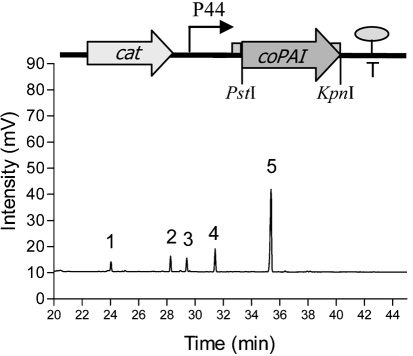
The Qiagen Plasmid Mini kit was used to isolate plasmid DNA from E. coli and L. lactis NZ9800, with one minor modification for L. lactis, i.e. 40 mg lysozyme ml−1 was added to buffer P1 and incubated for 20 min at 37 °C. PCR products were purified using a Qiaquick PCR Purification kit (Qiagen). The two plasmids pNZ8048 (nisin-inducible plasmid containing PnisA promoter, initially used for cloning of the coPAI gene) and pNZ44 (a derivative of pNZ8048 in which the PnisA promoter is replaced by P44, a constitutive L. lactis chromosomal promoter), and the coPAI gene fragment, were restricted with PstI and KpnI, followed by a ligation reaction at 15 °C with T4 DNA ligase, as described by the supplier (New England Biolabs), resulting in the construct pEV01, shown in Fig. 1. Recombinant plasmids were double-digested with the same enzymes to verify the correct clone and then electroporated into L. lactis NZ9800. Electrocompetent L. lactis was prepared and transformed according to the method described by de Ruyter et al. (1996). Chemically competent E. coli DH5α (TOP 10) was purchased from Invitrogen. The authenticity of the clones was verified by sequencing (MWG-Biotech) and sequence analysis was performed using dnastar software.
Fig. 1.
The pEV01 construct (above) and a representative GLC chromatogram (below) of supernatant following 72 h incubation of L. lactis pEV01 in media containing 0.5 mg linoleic acid ml−1. Peaks: 1, C16 : 0 (palmitic acid); 2, C18 : 0 (stearic acid); 3, C18 : 1 (oleic acid); 4, C18 : 2 (linoleic acid); 5, C18 : 2 (t10, c12 CLA).
Screening for CLA production.
The L. lactis pEV01 and E. coli pEV01 clones were tested for their ability to convert free linoleic acid (0.1–0.5 mg ml−1) to t10, c12 CLA as follows. Inoculum (1 %) of an overnight culture was transferred to 10 ml of the appropriate broth and incubated until the culture reached OD600 ∼0.5. Then, linoleic acid (0.1–0.5 mg ml−1) (added from a 30 mg ml−1 stock solution, sterile-filtered through a 0.45 μm filter, Sarstedt) was added to cultures, followed by further incubation to 72 h. Following incubation, the culture was centrifuged (4345 g) for 20 min and fatty acids were extracted from the supernatant and cell pellets, followed by methylation and analysis by GLC, as described by Coakley et al. (2003). All conversion rates (%) were related to the amount of linoleic acid that was recovered and extracted from the media following incubation without culture for the same time as with culture, which represents 100 % of available linoleic acid. All experiments were done at least in triplicate.
Preparation of fatty acids.
The E. coli and L. lactis pEV01 clones, as well as L. lactis and E .coli vector controls (empty pNZ44) were inoculated (1 % overnight culture) into 500 ml of the respective media and grown to OD600 ∼0.5, after which linoleic acid (0.5 mg ml−1) was added and incubation continued to 72 h. A linoleic acid control consisting of medium containing linoleic acid (0.5 mg ml−1) but without culture was also prepared and incubated at 37 °C for 72 h. After 72 h, fatty acids (fermented oils) were co-extracted from the supernatant and the bacterial pellet, as described for the supernatant (Coakley et al., 2003), but with the difference that the supernatant was not separated from the growing culture by centrifugation prior to fatty acid extraction. Control samples from fermentations were also prepared, methylated and analysed by GLC in triplicate to calculate the amounts of fatty acids and the CLA : linoleic acid ratio for the samples.
Anti-proliferative activity of fatty acids on human SW480 colon cancer cells.
To examine the anti-proliferative activity of the fatty acids extracted following fermentation of cultures of L. lactis and E. coli pEV01 in the respective media containing linoleic acid (0.5 mg ml−1), SW480 human colon cancer cells were cultured in the presence of different concentrations of the extracted fatty acids. The fermented oils generated from both L. lactis and E. coli (pEV01) expressing coPAI contained a mixture of linoleic acid and t10, c12 CLA at a ratio of ∼1.35 : 1. All samples, except for the pure t10, c12 CLA standard, contained smaller amounts of palmitic, stearic and oleic acids. E. coli fermented oil also contained 1.6 % t9, t11 CLA (Table 1). Since the recombinantly produced t10, c12 CLA samples contained even more linoleic acid as well as other fatty acids, all calculated concentrations added to the SW480 cancer cells were based on the amount of t10, c12 CLA present in the samples (or linoleic acid in the control sample, and the fermented oil samples from the L. lactis and E. coli vector controls). Initially, 1×104 cells were seeded per well in 96-well plates and cultured for 24 h at 37 °C to allow the cells to adhere to the surface prior to treatment with 5–20 μg (ml medium)−1 t10, c12 CLA (bacterial t10, c12 CLA/linoleic acid mix and pure Matreya standard in ethanol) and 5–25 μg (ml medium)−1 linoleic acid (control fatty acid extracted from broth). All fatty acids were sterile-filtered through a 0.2 μm Millex FG filter unit (Millipore) for non-aqueous solvents. Control flasks were supplemented with ethanol to a final concentration of 0.1 % (v/v). Following incubation for 5 days, cell viability was measured and relative cell numbers were determined using the MTS method (Promega), a colorimetric method for determining the number of viable cells in proliferation or cytotoxicity assays, subsequent to incubation with MTS tetrazolium compound. Following incubation with MTS for 2 h, A492 was recorded with a 96-well plate reader. Cell viability (%) after treatment was expressed relative to the ethanol control (100 %). Three independent experiments were performed on separate occasions, each time in triplicate for each fatty acid/fermented oil treatment of SW480. Student's t test was used to determine significant differences between treatments (P<0.001).
Table 1.
Fatty acid composition of the various fatty acid treatments (% of total fatty acids in samples)
Values represent the mean of triplicate analyses±sd.
| Fatty acid treatment | Fatty acid composition (percentage of total fatty acids) | |||||
|---|---|---|---|---|---|---|
| C16 : 0 | C18 : 0 | C18 : 1 c9 | C18 : 2 c9, c12 | C18 : 2 t10, c12 CLA | C18 : 2 t9, t11 CLA | |
| Control linoleic acid | 0.7±0.06 | 1.2±0.15 | 5.4±0.06 | 92.7±0.2 | − | − |
| Pure t10, c12 CLA | − | − | − | − | 100 | − |
| E. coli (pEV01) fermented oils | 4.2±0.0 | 1.6±0.07 | 6.2±0.0 | 49.2±0.07 | 37.3±0.28 | 1.6±0.42 |
| L. lactis (pEV01) fermented oils | 1.1±0.06 | 1.8±1.5 | 4.3±3.29 | 53.7±1.8 | 39.0±1.5 | − |
| E. coli (pNZ44) vector control fermented oils | 4.0±0.28 | 1.2±0.07 | 4.8±0.14 | 90.1±0.49 | − | − |
| L. lactis (pNZ44) vector control fermented oils | 0.8±0.07 | 0.9±0.07 | 4.9±0.07 | 93.3±0.07 | − | − |
RESULTS
Sequence analysis
The 1278 bp gene from P. acnes (Hornung et al., 2005) encodes a linoleic acid isomerase protein of 425 aa. The molecular mass of the isomerase is 49 077 Da. Comparison with sequences in the database revealed that the cloned isomerase protein showed significant homology with flavin-containing amino oxidases over most of the sequence (approx. amino acids 25–400; NCBI Conserved Domain Database; http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). The isomerase showed 96 % identity to a putative amino oxidase from P. acnes (GenBank accession no. Q6A8X5_PROAC; EXPASY/UniProtKB database), but only 26 % identity to the next best match, a protein from the plant Oryza sativa (japonica cultivar group; accession no. Q7XR12_ORYSA; EXPASY/UniProtKB database) (http://www.expasy.ch/cgi-bin/blast.pl) spanning amino acids 145–423. The aligned region in these proteins includes a flavin binding site. The flavin-containing amine oxidase family also contains phytoene hydrogenases and related enzymes. An NAD/FAD binding domain located in the region between amino acid residues 10 and 39 was identified by the Prosite database (http://www.expasy.org/prosite/). The crystal structure of the isomerase protein PAI (P. acnes isomerase) has recently been determined (Liavonchanka et al., 2006), which has confirmed the protein to be an FAD-containing monomer consisting of three intricately connected domains. Structural comparisons also show that the closest homologues of the isomerase protein domain 1, identified by the DALI program (www.ebi.ac.uk/dali) are FAD-containing oxidoreductases and isomerases, such as yeast polyamine oxidases. No structural homologues of PAI domains 2 and 3 have been found (DALI database) (Liavonchanka et al., 2006). The isomerase protein is soluble (http://www.psort.org/psortb/) and the predicted location of the protein is cytoplasmic (http://bp.nuap.nagoya-u.ac.jp/sosui/sosuimenu0E.html).
Bioconversion of linoleic acid
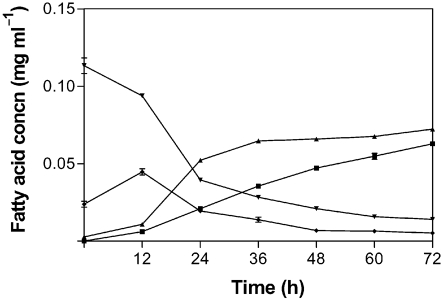
L. lactis carrying the construct pEV01 was shown to convert free linoleic acid into t10, c12 CLA, while the control L. lactis culture containing the vector pNZ44 alone produced no CLA isomers. L. lactis pEV01 converted >50 % of the free linoleic acid to t10, c12 CLA (Figs 1 and 2). Interestingly, L. lactis pEV01 accumulated a large amount of t10, c12 CLA in the cell pellets. After 72 h incubation, up to 60 % of total t10, c12 CLA was recovered in the bacterial membranes. Nearly all the t10, c12 CLA that was recovered in the membranes was already incorporated after half the incubation time (36 h). At this time point, the concentration of t10, c12 CLA in the supernatant was only about half of the final concentration (Fig. 2). Given that L. lactis did not grow well when initially incubated in the presence of linoleic acid (0.4–0.5 mg ml−1), the fatty acid was instead added to the growing culture at OD600 0.5. At this growth stage, the culture still showed sensitivity to linoleic acid, resulting in a reduced growth rate at this linoleic acid concentration. Greater rates of conversion to t10, c12 CLA were observed at lower concentrations of free linoleic acid (0.1 and 0.2 mg ml−1). E. coli cells carrying the construct pEV01 converted about 40 % of linoleic acid after 72 h incubation in the presence of the fatty acid (0.5 mg ml−1), whereas E. coli pNZ44 (vector control) did not produce any CLA. Compared with L. lactis pEV01, E. coli pEV01 accumulated much less CLA in the membranes: only about 10 % of total t10, c12 CLA.
Fig. 2.
t10, c12 CLA production versus linoleic acid usage and accumulation of the fatty acids in the cell pellets by L. lactis pEV01 incubated with 0.2 mg linoleic acid ml−1 for 72 h. The culture was incubated in linoleic acid at OD600 0.5. Samples were analysed in triplicate. ▪, t10, c12 CLA supernatant; ▾, linoleic acid supernatant; ▴, t10, c12 CLA bacterial pellets; ⧫, linoleic acid bacterial pellets.
Anti-proliferative activity of microbially generated fatty acids on human SW480 colon cancer cells
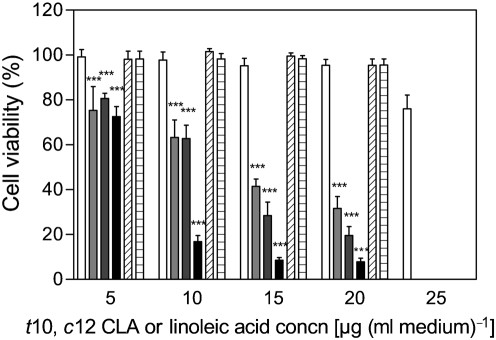
To investigate the anti-proliferative effects of fatty acids produced following isomerization of linoleic acid by L. lactis pEV01 and E. coli pEV01, human colon cancer cells (SW480) were cultured in the presence of the extracted fermented oils (consisting of a mixture of linoleic acid and t10, c12 CLA at a ratio of ∼1.35 : 1, and smaller amounts of other fatty acids; Table 1). Controls of linoleic acid extracted from LB broth after 72 h incubation at 37 °C and of the pure synthetic t10, c12 CLA standard, but also controls of fermented oils from L. lactis and E. coli transformed with the empty vector, were inoculated with SW480 cells. Since linoleic acid has been shown to have an anti-proliferative effect on SW480 cancer cells at higher concentrations [42.8 μg (ml medium)−1; 152.5 μM], and a slightly proliferative effect at a lower concentration [16.9 μg (ml medium)−1; 60.2 μM] (Miller et al., 2003), concentrations of t10, c12 CLA (microbially generated fatty acid samples) between 5 and 20 μg (ml medium)−1 [equivalent to 6.7–27 μg linoleic acid (ml medium)−1 in the same fatty acid sample] were chosen so as not to exceed the threshold concentration at which linoleic acid inhibits cell growth. Therefore, the highest linoleic acid concentration added to the cancer cells was 25 μg (ml medium)−1 from the control samples, to examine the effect of linoleic acid alone, given that the fermented oil samples contained approximately this concentration of linoleic acid together with 20 μg (ml medium)−1 t10, c12 CLA. Following 5 days of incubation with t10, c12 CLA between 5 and 20 μg ml−1, there was a significant (P<0.001) reduction in growth of the SW480 cancer cells compared with control linoleic acid at all concentrations. Average cell viability following treatment with the highest concentration (20 μg ml−1) of t10, c12 CLA was reduced to 7.9 % (L. lactis fermented oil) and 19.6 % (E. coli fermented oil), compared with 95.4 % (control linoleic acid+L. lactis vector control fermented oils) and 95.6 % (E. coli vector control fermented oils) (Fig. 3). Viable cell numbers following treatment with the highest concentration (25 μg ml−1) of linoleic acid were decreased to 76 %, which showed that this concentration had a slight anti-proliferative effect on the SW480 cells and therefore must have contributed to the killing effect of the highest concentrations of the fermented oils. All figures are related to ethanol controls (100 % cell viability). Significant differences in cell viability were observed at all concentrations between the control linoleic acid and the t10, c12 CLA from L. lactis and E. coli (P<0.001). There was no significant difference in cell viability after treatment between E. coli t10, c12 CLA and the pure t10, c12 CLA standard at any concentration. However, there was a significant difference in cell viability between treatments with L. lactis t10, c12 CLA and the pure t10, c12 CLA standard at concentrations between 10 and 20 μg ml−1 (P<0.001) (Fig. 3).
Fig. 3.
Cell viability of SW480 cells after 5 days of incubation with 5–20 μg (ml medium)−1 t10, c12 CLA (fermented oils from CLA-producing L. lactis and E. coli or pure t10, c12 CLA) or 5–25 μg (ml medium)−1 linoleic acid (control linoleic acid extracted from broth and fermented oils of L. lactis and E. coli vector controls). Data represent cell viability expressed as a percentage of the ethanol control, which was taken as 100 %. White bars, control linoleic acid (extracted from broth after 72 h incubation); light-grey bars, E. coli pEV01 fermented oil; dark-grey bars, pure t10, c12 CLA standard (Matreya); black bars, L. lactis pEV01 fermented oil; diagonal-hatched bars, L. lactis vector control fermented oil; horizontal-hatched bars, E. coli vector control fermented oil. Since the recombinantly produced t10, c12 CLA samples also contained other fatty acids, all calculated concentrations were based on the amount of t10, c12 CLA (or linoleic acid in the control samples) present in the samples. Three independent experiments were performed on separate occasions and each time in triplicate for each fatty acid treatment, and Student's t test was used to determine significant differences between treatments. Error bars represent sd. ***Values significantly different (P<0.001) from that of the control (linoleic acid extracted from broth).
DISCUSSION
We have successfully cloned and overexpressed the t10, c12 CLA isomerase from P. acnes in L. lactis and E. coli. The recombinant isomerase protein was functionally active, with conversion rates of >50 % of the added linoleic acid to t10, c12 CLA when expressed in L. lactis, and 40 % in E. coli. The isomerase protein was also expressed in the probiotic strain Lactobacillus paracasei subsp. paracasei NFBC 338, which was also capable of converting linoleic acid to t10, c12 CLA (data not shown). The enzyme has previously been characterized and expressed in bacteria, yeast and tobacco seeds (Rosson et al., 2001; Hornung et al., 2005). However, not many sequences of polyunsaturated fatty acid isomerases are known, and the homology between coPAI and other fatty acid isomerase sequences in the database was not significant. Recently, Liavonchanka et al. (2006) have described six crystal structures of PAI, which has revealed a unique gating mechanism for substrate specificity due to conformational changes in a hydrophobic channel toward the active site. The length preference for C18 fatty acids can thus be explained by the geometry of the substrate binding site.
Interestingly, higher conversion rates of linoleic acid to t10, c12 CLA were achieved with recombinant lactococci than with recombinant E. coli, while the former were more sensitive to free linoleic acid. For this reason, the linoleic acid was added to the medium only after the cells had reached OD600 ∼0.5. Another notable feature of L. lactis CLA production was that approximately half of the produced t10, c12 CLA was cell associated and could be extracted from pelleted cells. The reason for this is unknown, but could be related to the fact that polyunsaturated fatty acids are antibacterial (Kankaanpää et al., 2001), and the demonstrated incorporation into the membranes might be part of a detoxification mechanism (Jiang et al., 1998) used by L. lactis. A positive correlation between the inhibitory activity of a fatty acid and the level to which the fatty acid is incorporated into the cell membrane has been reported (Khulusi et al., 1995). In general, a long-chain fatty acid with a higher degree of unsaturation is more inhibitory than a fatty acid of the same chain length with fewer double bonds. Also, Gram-negative bacteria are less susceptible than Gram-positive bacteria to long-chain unsaturated fatty acids (Jenkins & Courtney, 2003). In particular, linoleic acid is known to exhibit an inhibitory effect towards some bacteria, e.g. Lactobacillus (Alonso et al., 2003) and propionic acid bacteria (Jiang et al., 1998).
The anti-proliferative effect of the fatty acids produced by L. lactis and E. coli on human SW480 cancer cells was confirmed in this study. Cell growth inhibition by t10, c12 CLA was dose-dependent, with the highest growth inhibitory effect at concentrations of 20 μg ml−1 t10, c12 CLA. Most effective was L. lactis fermented oil, which left only 8 % viable cells (compared with the ethanol control). E. coli fermented oil caused a reduction of viable cell numbers to 20 %. Linoleic acid alone also showed a slight anti-proliferative effect on the cancer cells following incubation with media containing 25 μg ml−1, and therefore must have contributed to the killing effect of the highest concentrations of the fermented oils.
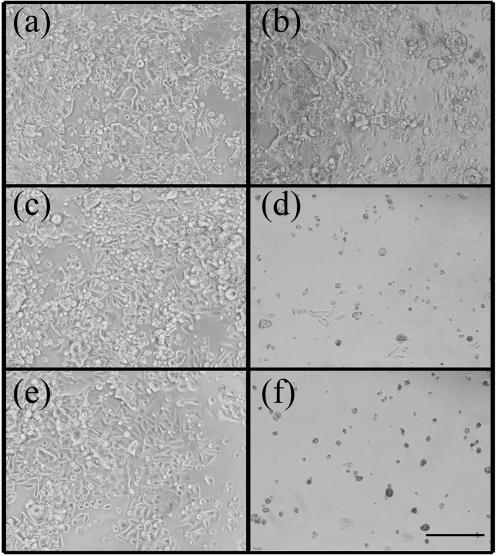
Microscopic examination of the SW480 colon cancer cells following 5 days of incubation with the various fatty acids treatments (Fig. 4) confirmed the results from the MTS cell viability assay (Fig. 3). SW480 cells incubated with 20 μg linoleic acid ml−1 were largely unaffected, in terms of either cell numbers or morphology (Fig. 4a, b). In contrast, in the samples incubated with 20 μg ml−1 L. lactis and E. coli (pEV01) fermented oils, there was a noticeable reduction in cell numbers presenting a viable epithelial-like morphology. Cells undergoing cell death and associated morphological changes were also evident (Fig. 4c–f).
Fig. 4.
Micrographs of human SW480 colon cancer cells following 5 days of incubation with the different fatty acid treatments. (a, b) Linoleic acid control (extracted from broth); (a) 5 and (b) 20 μg (ml medium)−1. (c, d) E. coli (pEV01) fermented oils containing (c) 5 and (d) 20 μg (ml medium)−1 t10, c12 CLA. (e, f) L. lactis (pEV01) fermented oils containing (e) 5 and (f) 20 μg (ml medium)−1 t10, c12 CLA. All panels are ×100 magnification; bar, ** μm.
An interesting observation was the increased inhibition of SW480 cell numbers in the samples treated with the L. lactis (pEV01)-generated fermented oil, compared with the samples treated with the same concentration of pure CLA standard or E. coli (pEV01) fermented oil. The added anti-proliferative effect of lactococcal-derived oil compared with that from E. coli may be due to other hydrophobic constituents which were co-extracted with the fatty acids. These putative constituents do not appear to have anti-proliferative activity on their own, given that the vector controls did not exhibit significant anti-proliferative activity. It is possible that such hydrophobic constituents act synergistically with the t10, c12 CLA isomer to give the observed increased killing with L. lactis-derived oils.
The t10, c12 CLA isomer has been shown to inhibit growth and induce apoptosis in a number of cell lines, such as HT-29 human colon cancer cells (Cho et al., 2005, 2006), Caco-2 colon cancer cells (Kim et al., 2002b), human colorectal cancer (CRC) cells (Lee et al., 2006), dRLh-84 rat hepatoma cells (Yamasaki et al., 2005) and the PC-3 human prostatic carcinoma cell line (Ochoa et al., 2004), while similar effects have not been reported for the c9, t11 CLA isomer at the same concentrations.
Both c9, t11 CLA and t10, c12 CLA have been shown to inhibit growth of MCF-7 cells (Scultz et al., 1992; O'Shea et al., 1999). The human SW480 colon cancer cell line was chosen in the present study, as it has previously been shown to be efficient as an in vitro cancer model (Miller et al., 2001, 2002, 2003). In the study of Miller et al. (2002), the t10, c12 CLA isomer was the most potent isomer examined, reducing cell viability by 47–61 % at a concentration of 16 μg ml−1, compared with a 40–52 % reduction by c9, t11 CLA at the same concentration. The same concentration of t10, c12 CLA reduces viability by 50–60 % in both SW480 and the MCF-7 breast cancer cell line following 4 days of incubation (Miller et al., 2001). In contrast, 16 μg linoleic acid ml−1 increases viability of SW480 cells by 23 % (Miller et al., 2001). Linoleic acid, however, has also been shown to inhibit growth of SW480 cells at higher concentrations. Incubation with 42.8 μg ml−1 (152.5 μM) linoleic acid reduces viability by 40 % after 3 days of incubation (Miller et al., 2003).
In order to be a valuable therapeutic agent for cancer, CLA or any other anti-proliferative agent must demonstrate selective growth inhibition of cancer cells. A number of studies have presented evidence supporting such a claim for CLA for a range of tissue types. No cytotoxic or apoptotic effect of CLA (as a blend of isomers and specific isomers) has been found on human osteoblast-like cells (SaOS2 and MG63) in culture at a concentration range of 0–50 μM (Cusack et al., 2005). Similarly, neither apoptosis nor any of the apoptosis regulatory proteins was affected by CLA in normal mammary gland alveoli or terminal end buds, whereas CLA inhibited the formation of pre-malignant lesions of rat mammary gland by ∼50 % and significantly increased apoptosis, reducing the expression of bcl-2 in these lesions (Ip et al., 2000). Yamasaki et al. (2002) have demonstrated a decrease in viable cell numbers of normal rat RLN-10 hepatocytes by the t10, c12 CLA isomer only at 25 μM, compared with rat hepatoma dRLh-84 cells, which are much more sensitive to t10, c12 CLA: less than 25 % viable cells persisted following treatment with 1 μM of this isomer.
CONCLUSIONS
In this study, we have heterologously expressed linoleic acid isomerase in L. lactis and E. coli, and in so doing generated cultures which can produce t10, c12 CLA. Moreover, this is believed to be the first instance in which the CLA produced by recombinant strains has been shown to exhibit an anti-proliferative effect. These results demonstrate the potential for using recombinant strains to deliver bioactive CLAs to (or produce within) the mammalian intestine, where they exert an anti-proliferative activity.
Acknowledgments
This work was supported by BASF Plant Science GmbH, and funds supplied by the Irish Government under the National Development Plan 2000–2006, EU Project QLK1-2002-02362 and Science Foundation Ireland (SFI) are gratefully acknowledged. The technical assistance of Seamus Aherne is gratefully acknowledged.
Abbreviations
CLA, conjugated linoleic acid
c9, t11 CLA, cis-9, trans-11 CLA
t10, c12 CLA, trans-10, cis-12 conjugated linoleic acid
PAI, P. acnes isomerase
References
- Alonso, L., Cuesta, E. P. & Gilliland, S. E. (2003). Production of free conjugated linoleic acid by Lactobacillus acidophilus and Lactobacillus casei of human intestinal origin. J Dairy Sci 86, 1941–1946. [DOI] [PubMed] [Google Scholar]
- Belury, M. A. (2002). Dietary conjugated linoleic acid in health: physiological effects and mechanisms of action. Annu Rev Nutr 22, 505–531. [DOI] [PubMed] [Google Scholar]
- Blankson, H., Stakkestad, J. A., Fagertun, H., Thom, E., Wadstein, J. & Gudmundsen, O. (2000). Conjugated linoleic acid reduces body fat mass in overweight and obese humans. J Nutr 130, 2943–2948. [DOI] [PubMed] [Google Scholar]
- Chin, S. F., Storksson, J. M., Liu, W., Albright, K. J. & Pariza, M. W. (1994). Conjugated linoleic acid (9,11- and 10,12-octa-decadienoic acid) is produced in conventional but not germ-free rats fed linoleic acid. J Nutr 124, 694–701. [DOI] [PubMed] [Google Scholar]
- Cho, H. J., Kim, W. K., Jung, J. I., Kim, E. J., Lim, S. S., Kwon, D. Y. & Park, J. H. Y. (2005). Trans-10,cis-12, not cis-9,trans-11, conjugated linoleic acid decreases ErbB3 expression in HT-29 human colon cancer cells. World J Gastroenterol 11, 5142–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. J., Kim, E. J., Lim, S. S., Kim, M. K., Sung, M. K., Kim, J. S. & Park, J. H. Y. (2006). Trans-10,cis-12, not cis-9,trans-11, conjugated linoleic acid inhibits G1-S Progression in HT-29 human colon cancer cell. J Nutr 136, 893–898. [DOI] [PubMed] [Google Scholar]
- Coakley, M., Ross, R. P., Nordgren, M., Fitzgerald, G., Devery, D. & Stanton, C. (2003). Conjugated linoleic acid biosynthesis by human-derived Bifidobacterium species. J Appl Microbiol 94, 138–145. [DOI] [PubMed] [Google Scholar]
- Coakley, M., Johnson, M. C., McGrath, E., Rahman, S., Ross, R. P., Fitzgerald, G. F., Devery, R. & Stanton, C. (2006). Intestinal bifidobacteria that produce trans-9, trans-11 CLA: a fatty acid with anti-proliferative activity against SW480 and HT-29 colon cancer cells. Nutr Cancer 56, 95–102. [DOI] [PubMed] [Google Scholar]
- Cusack, S., Jewell, C. & Cashman, K. D. (2005). The effect of conjugated linoleic acid on the viability and metabolism of human osteoblast-like cells. Prostaglandins Leukot Essent Fatty Acids 72, 29–39. [DOI] [PubMed] [Google Scholar]
- de Ruyter, P. G., Kuipers, O. P. & de Vos, W. M. (1996). Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol 62, 3662–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbank, J., Ridgway, L., Griffin, J., Wickens, D., Singer, A. & Dormandy, T. L. (1988). Octadeca-9-11-dienoic acid in diagnosis of cervical intraepithelial neoplasia. Lancet 2, 329–330. [DOI] [PubMed] [Google Scholar]
- Fairbank, J., Hollingworth, A., Griffin, J., Ridgway, E., Wickens, D., Singer, A. & Dormandy, T. (1989). Octadeca-9,11-dienoic acid in cervical intraepithelial neoplasia: a colposcopic study. Clin Chim Acta 186, 53–58. [DOI] [PubMed] [Google Scholar]
- Hornung, E., Krueger, C., Pernstich, C., Gipmans, M., Porzel, A. & Feussner, I. (2005). Production of (10E,12Z)-conjugated linoleic acid in yeast and tobacco seeds. Biochim Biophys Acta 1738, 105–114. [DOI] [PubMed] [Google Scholar]
- Ip, C., Ip, M., Loftus, T., Shoemaker, S. & Shea-Eaton, W. (2000). Induction of apoptosis by conjugated linoleic acid in cultured mammary tumor cells and premalignant lesions of the rat mammary gland. Cancer Epidemiol Biomarkers Prev 9, 689–696. [PubMed] [Google Scholar]
- Jack, C. I. A., Ridgway, F., Jackson, M. J. & Hind, C. R. K. (1994). Serum octadeca-9,11-dienoic acid – an assay of free radical activity or a result of bacterial population?. Clin Chim Acta 224, 139–146. [DOI] [PubMed] [Google Scholar]
- Jenkins, J. K. & Courtney, P. D. (2003). Lactobacillus growth and membrane composition in the presence of linoleic or conjugated linoleic acid. Can J Microbiol 49, 51–57. [DOI] [PubMed] [Google Scholar]
- Jensen, R. G. (2002). The composition of bovine milk lipids. J Dairy Sci 85, 295–350. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Björck, L. & Fonden, R. (1998). Production of conjugated linoleic acid by dairy starter cultures. J Appl Microbiol 85, 95–102. [DOI] [PubMed] [Google Scholar]
- Kankaanpää, P. E., Salminen, S. J., Isolauri, E. & Lee, Y. K. (2001). The influence of polyunsaturated fatty acids on probiotic growth and adhesion. FEMS Microbiol Lett 194, 149–153. [DOI] [PubMed] [Google Scholar]
- Kemp, M. Q., Jeffy, B. D. & Romagnolo, D. F. (2003). Conjugated linoleic acid inhibits cell proliferation through a p53-dependent mechanism: effects on the expression of G1-restriction points in breast and colon cancer cells. J Nutr 133, 3670–3677. [DOI] [PubMed] [Google Scholar]
- Khulusi, S., Ahmed, H. A., Patel, P., Mendall, M. A. & Northfield, T. C. (1995). The effects of unsaturated fatty acids on Helicobacter pylori in vitro. J Med Microbiol 42, 276–282. [DOI] [PubMed] [Google Scholar]
- Kim, Y. J. & Liu, R. H. (2002). Increase of conjugated linoleic acid content in milk by fermentation with lactic acid bacteria. J Food Sci 67, 1731–1737. [Google Scholar]
- Kim, Y. J., Liu, R. H., Bond, D. R. & Russell, J. B. (2000). Effect of linoleic acid concentration on conjugated linoleic acid production by Butyrivibrio fibrisolvens A38. Appl Environ Microbiol 66, 5226–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E. J., Holthuizen, P. E., Park, H. S., Ha, Y. L., Jung, K. C. & Park, J. H. (2002a). Trans-10, cis-12-conjugated linoleic acid inhibits Caco-2 colon cancer cell growth. Am J Physiol Gastrointest Liver Physiol 283, G357–G367. [DOI] [PubMed] [Google Scholar]
- Kim, Y. J., Liu, R. H., Rychlik, J. L. & Russell, J. B. (2002b). The enrichment of a ruminal bacterium (Megasphaera elsdenii YJ-4) that produces the trans-10, cis-12 isomer of conjugated linoleic acid. J Appl Microbiol 92, 976–982. [DOI] [PubMed] [Google Scholar]
- Lee, S. H., Yamaguschi, K., Kim, J. S., Eling, T. E., Safe, S., Park, Y. & Baek, S. J. (2006). Conjugated linoleic acid stimulates an anti-tumorigenic protein NAG-1 in an isomer specific manner. Carcinogenesis 27, 972–981. [DOI] [PubMed] [Google Scholar]
- Leibovitz, A., Stinson, J. C., McCombs, W. B., III, McCoy, C. E., Mazur, K. C. & Mabry, N. D. (1976). Classification of human colorectal adenocarcinoma cell lines. Cancer Res 36, 4562–4569. [PubMed] [Google Scholar]
- Liavonchanka, A., Hornung, E., Feussner, I. & Rudolph, M. G. (2006). Structure and mechanism of the Propionibacterium acnes polyunsaturated fatty acid isomerase. Proc Natl Acad Sci U S A 103, 2576–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, T. Y., Lin, C. W. & Lee, C. H. (1999). Conjugated linoleic acid concentration as affected by lactic cultures and added linoleic acid. Food Chem 67, 1–5. [Google Scholar]
- Miller, A., Stanton, C. & Devery, R. (2001). Modulation of arachidonic acid distribution by conjugated linoleic acid isomers and linoleic acid in MCF-7 and SW480 cancer cells. Lipids 36, 1161–1168. [DOI] [PubMed] [Google Scholar]
- Miller, A., Stanton, C. & Devery, R. (2002). Cis 9,trans 11- and trans 10, cis 12-conjugated linoleic acid isomers induce apoptosis in cultured SW480 cells. Anticancer Res 22, 3879–3888. [PubMed] [Google Scholar]
- Miller, A., Stanton, C., Murphy, J. & Devery, R. (2003). Conjugated linoleic acid (CLA)-enriched milk fat inhibits growth and modulates CLA-responsive biomarkers in MCF-7 and SW480 human cancer cell lines. Br J Nutr 90, 877–885. [DOI] [PubMed] [Google Scholar]
- Ochoa, J. J., Farquharson, A. J., Grant, I., Moffat, L. E., Heys, S. D. & Wahle, K. W. J. (2004). Conjugated linoleic acids (CLAs) decrease prostate cancer cell proliferation: different molecular mechanisms for cis-9, trans-11 and trans-10, cis-12 isomers. Carcinogenesis 25, 1185–1191. [DOI] [PubMed] [Google Scholar]
- O'Shea, M., Stanton, C. & Devery, R. (1999). Antioxidant enzyme defence responses of human MCF-7 and SW480 cancer cells to conjugated linoleic acid. Anticancer Res 19, 1953–1959. [PubMed] [Google Scholar]
- Palombo, J. D., Gangulu, A., Bistrian, B. R. & Menard, M. P. (2002). The antiproliferative effects of biologically active isomers of conjugated linoleic acid on human colorectal and prostatic cancer cells. Cancer Lett 177, 163–172. [DOI] [PubMed] [Google Scholar]
- Pariza, M. W., Park, Y. & Cook, M. E. (1999). Conjugated linoleic acid and the control of obesity and cancer. Toxicol Sci 52, 107–110. [DOI] [PubMed] [Google Scholar]
- Pariza, M. W., Park, Y. & Cook, M. E. (2000). Mechanism of action of conjugated linoleic acid: evidence and speculation. Proc Soc Exp Biol Med 223, 8–13. [DOI] [PubMed] [Google Scholar]
- Rosberg-Cody, E., Ross, R. P., Hussey, S., Ryan, C. A., Murphy, B. P., Fitzgerald, G. F., Devery, R. & Stanton, C. (2004). Mining the microbiota of the neonatal gastrointestinal tract for CLA-producing bifidobacteria. Appl Environ Microbiol 70, 4635–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosson, R. A., Deng, M., Grund, A. D. & Peng, S. S. (2001). Nucleotide encoding a propionibacterium linoleate isomerase and uses thereof. Patent WO 01/00846.
- Shultz, T. D., Chew, B. P. & Seaman, W. R. (1992). Differential stimulatory and inhibitory responses of human MCF-7 breast cancer cells to linoleic acid and conjugated linoleic acid in culture. Anticancer Res 12, 2143–2145. [PubMed] [Google Scholar]
- Smedman, A. & Vessby, B. (2001). Conjugated linoleic acid supplementation in humans – metabolic effects. Lipids 36, 773–781. [DOI] [PubMed] [Google Scholar]
- Thom, E., Wadstein, J. & Gudmundsen, O. (2001). Conjugated linoleic acid reduces body fat in healthy exercising humans. J Int Med Res 29, 392–396. [DOI] [PubMed] [Google Scholar]
- Verhulst, A., Janssen, G., Parmentier, G. & Eyssen, H. (1987). Isomerization of polyunsaturated long chain fatty acids by Propionibacteria. Syst Appl Microbiol 9, 12–15. [Google Scholar]
- Yamasaki, M., Chujo, H., Koga, Y., Oishi, A., Rikimaru, T., Shimada, M., Sugimachi, K., Tachibana, H. & Yamada, K. (2002). Potent cytotoxic effect of the trans10, cis12 isomer of conjugated linoleic acid on rat hepatoma dRLh-84 cells. Cancer Lett 188, 171–180. [DOI] [PubMed] [Google Scholar]
- Yamasaki, M., Miyamoto, Y., Chujo, H., Nishiyama, K., Tachibana, H. & Yamada, K. (2005). Trans10, cis12-conjugated linoleic acid induces mitochondria-related apoptosis and lysosomal destabilization in rat hepatoma cells. Biochim Biophys Acta 1735, 176–184. [DOI] [PubMed] [Google Scholar]