Abstract
Clumping factor B (ClfB) of Staphylococcus aureus binds to cytokeratin 10 and to fibrinogen. In this study the binding site in human fibrinogen was localized to a short region within the C terminus of the Aα-chain. ClfB only bound to the Aα-chain of fibrinogen in a ligand-affinity blot and in solid-phase assays with purified recombinant fibrinogen chains. A variant of fibrinogen with wild-type Bβ- and γ-chains but with a deletion that lacked the C-terminal residues from 252–610 of the Aα-chain did not support adherence of S. aureus Newman expressing ClfB. A series of truncated mutants of the recombinant Aα-chain were tested for their ability to support adherence of S. aureus Newman ClfB+, which allowed the binding site to be localized to a short segment of the unfolded flexible repeated sequence within the C terminus of the Aα-chain. This was confirmed by two amino acid substititions within repeat 5 of the recombinant Aα-chain which did not support adherence of Newman ClfB+. Lactococcus lactis expressing ClfB mutants with amino acid substitutions (N256 and Q235) located in the putative ligand-binding trench between domains N2 and N3 of the A-domain were defective in adherence to immobilized fibrinogen and cytokeratin 10, suggesting that both ligands bind to the same or overlapping regions.
INTRODUCTION
Staphylococcus aureus is an important opportunistic pathogen of humans that is responsible for a wide range of infections ranging from superficial skin infections to more serious invasive diseases such as endocarditis, osteomyelitis and septicaemia. The primary habitat of S. aureus is the moist squamous epithelium of the anterior nares (Cole et al., 2001; Peacock et al., 2001). The success of S. aureus as a pathogen is due in part to its ability to adhere to a wide range of host tissues including host extracellular matrix proteins such as fibrinogen (Fg), fibronectin and collagen. Adhesion to host proteins is mediated by bacterial cell-wall-associated proteins called MSCRAMMs (microbial surface components recognizing adhesive matrix molecules). S. aureus can express up to 20 different potential MSCRAMMs that are covalently anchored by sortase to peptidoglycan (Mazmanian et al., 1999; Navarre & Schneewind, 1994).
S. aureus expresses several different proteins that can bind specifically to Fg, including clumping factors A and B (ClfA and ClfB) and the bifunctional fibronectin- (and Fg-) binding proteins A and B, FnbpA and FnbpB (McDevitt et al., 1994; Ni Eidhin et al., 1998; Perkins et al., 2001; Wann et al., 2000). ClfA, FnbpA and FnbpB bind to the extreme C terminus of the γ-chain protruding from domain D of Fg. In contrast, the SdrG protein from Staphylococcus epidermidis binds to the fibrinopeptide-B protruding from domain E (Davis et al., 2001). S. aureus also secretes several proteins that bind Fg, notably coagulase, the extracellular Fg-binding protein (Efb) and MHC class II analogue protein (Map) (Jonsson et al., 1995; McGavin et al., 1993; Palma et al., 1998; Phonimdaeng et al., 1990).
ClfB is only expressed on the cell surface during the exponential phase of growth (McAleese et al., 2001). Ligand binding is specified by the A region, which is divided into three independently folded subdomains N1, N2 and N3 (Perkins et al., 2001). The S. aureus metalloprotease aureolysin cleaves ClfB between N1 and N2, resulting in the loss of Fg-binding activity at the end of the exponential phase of growth (McAleese et al., 2001; Perkins et al., 2001). ClfB is a bifunctional MSCRAMM. It binds to cytokeratin 10 (CK10) exposed on the surface of desquamated epithelial cells in addition to Fg. It is a major determinant of the ability of S. aureus to adhere to squamous cells and to colonize the anterior nares (O'Brien et al., 2002). The binding domain in CK10 was shown to be quasi-repeats of glycine and serine residues that occur as unfolded loops located at the C terminus of the protein, which likely protrude from keratin filaments (Walsh et al., 2004).
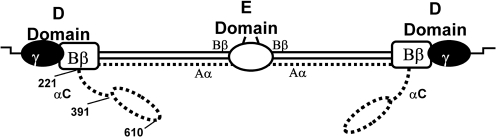
Fg is a 340 kDa plasma protein that plays a crucial role in haemostasis. It is composed of two identical disulfide-linked subunits, each of which is composed of three non-identical polypeptide chains, Αα, Bβ and γ (Doolittle, 1984; Henschen & MCDonagh, 1986; Herrick et al., 1999). The removal of C termini of the Aα-chains (residues 220–610) by proteolysis results in generation of αC fragments, representing the whole or parts of the αC-domain (Weisel & Medved, 2001). The αC-domains are involved in fibrin assembly and clot formation (Cierniewski & Budzynski, 1992; Gorkun et al., 1994; Medved et al., 1985) and control activation of factor XIII (Credo et al., 1981). The αC-domains and the N-terminal portions of the Bβ-chains are the parts of the Fg molecule for which the 3D structure has not been established (Yang et al., 2001). However, recent studies have shown that the αC-domain consists of two structurally distinct regions, a flexible connector region from residues 221–391 and an independently folded, compact portion from residues 392–610 (Fig. 1) (Burton et al., 2006; Tsurupa et al., 2002). In human Fg the flexible NH2-connector region is unordered and is composed of a 43-residue segment followed by ten 13-residue tandem repeats.
Fig. 1.
Structure of fibrinogen. Fg consists of two identical disulfide-linked subunits, each of which is composed of three non-identical polypeptide chains, Αα, Bβ and γ. Fg can be divided into four major regions, the central E region, two identical terminal D regions and the αC-domains. The αC-domains (residues 221–610) contain two distinct regions, a compact C-terminal half and an unordered NH2-terminal half or connector region.
In this study we investigated the binding of ClfB to human Fg and have localized the binding site to one of the tandem repeats within the flexible connector region of the αC-domain.
METHODS
Bacterial strains and growth conditions.
Escherichia coli strain XL-1 Blue was used as the host for plasmid cloning and was routinely grown in L-broth or agar with ampicillin (100 μg ml−1) and tetracycline (10 μg ml−1) as appropriate. Strain TOPP3 (Stratagene) was used as the host for recombinant ClfB A-domain (rClfB 45–542) or rClfA A-domain (rClfA 221–559) protein expression. E. coli strain JM101 was used for expression of recombinant Fg proteins.
The S. aureus strains are mutants of strain Newman (Duthie & Lorenz, 1952) defective in ClfA (DU5876 clfA2 : : Tn917) (McDevitt et al., 1994) and a double mutant defective in ClfA and ClfB (DU5944 clfA2 : : Tn917 clfB : : Tcr) (Ni Eidhin et al., 1998). Bacteria were routinely cultured in trypticase soy broth or on agar. For optimum expression of ClfB, S. aureus was grown to exponential phase (OD600 0.6) in 50 ml brain heart infusion broth in a 250 ml conical flask shaken at 200 r.p.m. at 37 °C (McAleese et al., 2001; Ni Eidhin et al., 1998).
Lactococcus lactis strain NZ9800 carrying the nisin-inducible expression plasmid pNZ8037 expressing clfB or clfB Q235A was described previously (Miajlovic et al., 2007). L. lactis pNZ8037clfB N526A was constructed by site-directed mutagenesis as described for L. lactis pNZ8037clfB Q235A (Miajlovic et al., 2007) using the primers shown in Table 1. L. lactis strains were grown statically at 28 °C in M17 (Difco) broth incorporating 0.5 % (w/v) glucose, chloramphenicol (Sigma, 10 μg ml−1) with nisin at 3.2 ng ml−1 to stimulate maximum induction (Miajlovic et al., 2007).
Table 1.
Synthetic oligonucleotides used to amplify Aα-chain fragments and for site-directed and deletion mutants of the α-chain
Restriction endonuclease sites are underlined. The nucleotides in boldface indicate the location of the desired mutation. F corresponds to forward primer; R corresponds to reverse primer.
| Expression constructs | Primer name | Primer sequence |
|---|---|---|
| Truncated proteins | Aα-1F | CGG GGA TCC GCA GAT AGT GGT GAA GGT G |
| Aα-232F | GAC GGA TCC TTA ACA GAC ATG CCG CAG ATG AG | |
| Aα-315F | CGG GGA TCC TGG AAC TCT GGG AGC TCT GG | |
| Aα-367F | CGG GGA TCC TGG CAC TCT GAA TCT GGA AG | |
| Aα-283R | CGG AAG CTT AGG TCC AGA GCT CCC AGA GTT C | |
| Aα-574R | CGG AAG CTT TTA GTC TCC TCT GTT GTA ACT CGT G | |
| Aα-625R | CGG AAG CTT TTA GGG GGA CAG GGA AGC CTT C | |
| Deletion mutagenesis | Aα-329F | GCG CTG ATA TCG GAA ACC AAA ACC CTG GGA G |
| Aα-341F | GCG CTG ATA TCG GTA GTA CCG GAA CCT GG | |
| Aα-344F | GCG CTG ATA TCT GGA ATC CTG GCA GCT CTG | |
| Aα-373F | CGC TGA TAT CGG AAG TTT TAG GCC AGA TAG | |
| Aα-286R | ATC TCG GTT TCC AGT ACT TCC AG | |
| Aα-315R | ATC CCA GCT TCC AGT ACT TCC AG | |
| Aα-331R | ATC GTC CCC AGG GTT TTG GTT CTT | |
| Aα-341R | ATC CCA GGT TCC GGT ACT ACC AG | |
| Site-directed mutagenesis | Aα-S317PF | GCTGGAACCCTGGGAGCTCTGGAAC |
| Aα-S317PR | GTTCCAGAGCTCCCAGGGTTCCAGC | |
| Aα-T322PF | TGGGAGCTCTGGACCTGGAAGTACTGGAAAC | |
| Aα-T322PR | GTTTCCAGTACTTCCAGGTCCAGAGCTCCCA | |
| ClfB mutant | N526F | GTT GGA ATA ATG AGG CTG TTG TAC GTT ATG GTG G |
| N526R | CCA CCA TAA CGT ACA ACA GCC TCA TTA TTC CAA C |
Manipulation of DNA.
Restriction and DNA modification enzymes were purchased from New England Biolabs or Roche Molecular Biochemicals and were used according to the manufacturers' instructions. DNA procedures were carried out according to standard protocols (Sambrook et al., 1989).
Cloning and PCR amplification of Fg constructs.
E. coli strains expressing human Fg Aα-, Bβ- and γ-chains were provided by Professor Susan Lord, Department of Pathology, University of North Carolina at Chapel Hill. A 1878 bp fragment from plasmid p166.9 (Lord, 1985) and a 1236 bp fragment from p253 (Bolyard & Lord, 1988) were subcloned into the plasmid pQE30 (Qiagen) to produce recombinant mature Aα- and γ-chains with N-terminal His tags. Subcloning of the mature recombinant Bβ-chain was described previously (Davis et al., 2001). The recombinant plasmids were transformed into E. coli strain JM101 for protein expression.
Segments of the Aα-chain were amplified by PCR from the plasmid containing the full-length Aα-chain (residues 1–625 in the common Fg α-chain, NCBI accession number AAA17055). Although Fg α-chain DNA encodes 625 residues, in plasma the Aα-chain is only 610 residues long due to a post-translational modification. The oligonucleotide primers used for PCR are listed in Table 1. Restriction sites were incorporated at the 5′ ends of the primers to facilitate directional cloning. PCR amplification was carried out in a DNA thermal cycler (Perkin-Elmer Cetus) with Phusion DNA-polymerase (Bioline). Reactions were carried out with a 30 s denaturation step at 98 °C, a 10 s annealing step (the temperature of which depended on the individual primers) and elongation at 72 °C depending on the length of the PCR product. This standard cycle was repeated 25 times followed by incubation at 72 °C for 10 min. PCR products were purified using the Favourgen Gel/PCR Purification Kit (Favourgen Biotech), cleaved with the appropriate restriction enzyme and cloned into plasmid pQE30.
Mutagenesis of Aα-chain of Fg.
Site-directed mutagenesis was carried out on pQE30 carrying the gene encoding Aα-1–625 using Quickchange (Stratagene). The primers used in the mutagenesis protocol are listed in Table 1. Two mutants were created: Aα-S317P and Aα-T322P.
Tandem repeat 5 of Aα-S317P was made to resemble repeat 3. Repeat 5 of Aα-T322P resembles repeats 1 and 2, which have proline residues in the middle of their repeat sequences.
Construction of recombinant α-chain mutants.
Sections of the Aα-chain were deleted by inverse PCR using oligonucleotide primers in Table 1. The forward primers incorporated a complete EcoRV site and the reverse primers contained half an EcoRV site at their 5′ ends, respectively. The PCR products were digested with EcoRV, religated and transformed into E. coli XL-1 Blue cells. Amino acid residues D and I were inserted in place of the region of DNA that was deleted from the αC-domain.
Sequencing of recombinant plasmids.
All recombinant plasmids were sequenced by GATC Biotech.
Expression and purification of recombinant proteins.
Recombinant rClfA 221–559, rClfB 45–542 and rClfB 197–542 were purified by Ni2+ affinity chromatography as described previously (O'Connell et al., 1998; Perkins et al., 2001). Purification of the recombinant Fg Aα-, Bβ- and γ-chains, and each of the recombinant Aα-chain deletion and truncated proteins, was carried out by Ni2+ affinity chromatography (Perkins et al., 2001) with the addition of 6 M urea (Sigma) to the bacterial cell suspension prior to lysis.
SDS-PAGE and Western immunoblotting.
Recombinant proteins were analysed by SDS-PAGE by standard procedures (Laemmli, 1970) on gels containing 10–15 % acrylamide. Gels were stained with Coomassie blue. Human Fg (Calbiochem) was separated by SDS-PAGE and transferred electrophoretically to PVDF Western blotting membranes (Roche Applied Science) by the wet system (Bio-Rad) in Tris/HCl (0.02 M), glycine (0.15 M) and methanol (20 %, v/v). Membranes were blocked for 15 h at 4 °C in 10 % (v/v) non-dry fat milk and then incubated with recombinant ClfA or rClfB (10 μg ml−1) for 1 h with shaking. The membranes were washed three times with PBS containing Tween 20 (0.01 %, w/v) and then incubated with polyclonal antisera to ClfA (1 : 2000) (McDevitt et al., 1995) or ClfB (1 : 2000) (McAleese et al., 2001) as appropriate. Horseradish peroxidase (HRP)-labelled goat anti-rabbit IgG (Dako, 1 : 2000) was used to detect bound antibody. Membranes were developed using LumiGLO chemiluminescent substrate (New England BioLabs) according to the manufacturer's instructions and exposed to X-ray film.
Fibrinogen.
Native human Fg was from Calbiochem. Recombinant Fg Aα251 contains Aα-chains truncated at residue 251 but is otherwise identical to normal human Fg. It was purified from CHO cells media (Gorkun et al., 1998). Plasmin digestion of native Fg was performed to isolate fragment D-domains (Rudchenko et al., 1996). Briefly, Fg (70 mg) in Tris-buffered saline (TBS) pH 7.4 was treated with plasmin [0.0015 unit (mg Fg)−1] for 6 h at room temperature. The reaction was stopped by the addition of phenylmethylsulfonyl fluoride (Sigma) to 0.5 mM final concentration and dialysed overnight at 4 °C into TBS pH 7.4. The sample was then applied to a MonoQ HP anion-exchange column and eluted with a gradient of 0–1 M NaCl. SDS-PAGE and Western blotting with anti-Fg domain D and anti-Fg domain E polyclonal antibodies (Cambio) showed the D- and E-domains eluting at 250 mM and 350 mM NaCl, respectively.
Adherence of bacteria to immobilized proteins.
Adherence of S. aureus or L. lactis to immobilized proteins was performed as described previously (Walsh et al., 2004). Nunc-Immuno MaxiSorb microtitre plates were coated with the protein in carbonate buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) and incubated overnight at 4 °C. Wells were washed with PBS, BSA (5 mg ml−1) was added and the plates were incubated for 2 h at 37 °C. The plates were washed three times with PBS. A bacterial cell suspension (OD600 1.0 in PBS) was added (100 μl per well) and the plates were incubated for 1 h at room temperature. Plates were washed three times with PBS and bound cells were fixed with formaldehyde (25 %, v/v) for 30 min and stained with crystal violet (0.5 %, v/v 100 μl per well) for 1 min. Following three washes with PBS, acetic acid (5 %, v/v) was added (100 μl per well) for 10 min at room temperature. The absorbance was measured at 570 nm in an ELISA plate reader (Labsystems Multiskan Plus). Inhibition of bacterial adherence by rClfB 197–542 was performed as described by Perkins et al. (2001).
Binding of rClfB 45–542 to immobilized proteins.
ELISA plates were coated with the appropriate protein in carbonate buffer overnight at 4 °C. Wells were washed twice with PBS and incubated at 37 °C with BSA in PBS for 2 h at 37 °C. They were then washed with PBS and varying concentrations of rClfB 45–542 in PBS with Tween 20 (0.01 %, w/v) were added. The plates were then incubated for 1 h at room temperature. Any unbound protein was removed by washing with PBS, and plates were incubated with rabbit anti-ClfB 45–452 antibodies for 1 h at room temperature. Wells were washed and HRP-labelled goat anti-rabbit IgG (1 : 2000) was added for 1 h at room temperature. After washing, 1 mg ml−1 tetramethylbenzidine chromogenic substrate and 0.006 % (v/v) H2O2 in 0.05 M phosphate citrate buffer pH 5.0) was added (100 μl per well) and plates developed for 10 min in the dark. The reaction was stopped by the addition of 2 M H2SO4 (50 μl per well), and plates were read at 450 nm.
To check that native and mutant Fg Aα251 were coating the ELISA plates efficiently, HRP-labelled anti-human Fg antibody (Dako 1 : 4000) was added to the plates following the initial blocking step. The plates were incubated at room temperature for 1 h with shaking. After washing with PBS, bound antibody was detected by adding tetramethylbenzidine and H2SO4 and reading the absorbance at 450 nm.
RESULTS
ClfB binds to the α-chain of human Fg
Native whole human Fg was separated into individual Aα-, Bβ- and γ-chains by denaturing SDS-PAGE, transferred to PVDF membranes and probed with recombinant (r) ClfB region A-domain (rClfB 45–542). Binding of rClfB was detected with anti-ClfB region A antibodies. The rClfB protein bound specifically to the Aα-chain (Fig. 2a). In contrast, rClfA region A-domain rClfA 40–559 reacted with the γ-chain of human Fg (Fig. 2a), confirming previous reports (McDevitt et al., 1997).
Fig. 2.
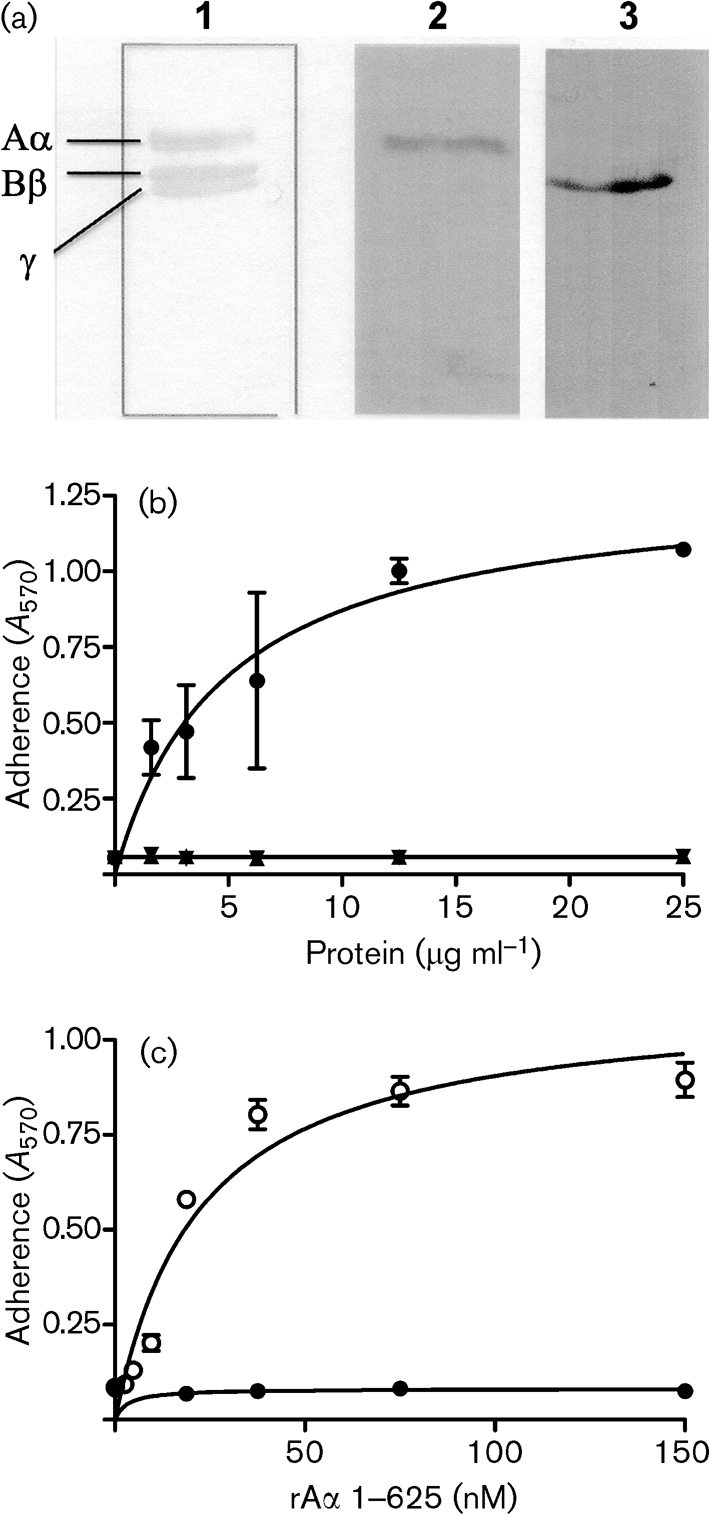
ClfB binds to Fg Aα-chain. (a) Ligand affinity blotting. Human Fg was fractionated by SDS-PAGE and stained with Coomassie blue (lane 1), or transferred to a PVDF-membrane and probed with rClfB 45–542 (lane 2) or rClfA 220–559 (lane 3). Binding of rClf proteins was detected with anti-Clf polyclonal antisera. (b) Adherence of S. aureus Newman ClfA− to immobilized recombinant Aα- (•), Bβ- (▴) or γ- (▾) chains of Fg. (c) Adherence of S. aureus Newman ClfA− (○) and Newman ClfA− ClfB− (•) to immobilized recombinant Aα-chain of Fg. Cells were grown to exponential phase and suspensions (1×108 c.f.u.) were added to wells. Adherent bacteria were detected by crystal violet staining. The experiment was performed twice with similar results and values represent means±sd of triplicate wells.
Purified recombinant Fg Aα-, Bβ- and γ- chains were immobilized and tested for their ability to support the adherence of ClfB-expressing S. aureus cells. The ability of S. aureus Newman cells defective in ClfA to adhere to Fg is exclusively due to expression of ClfB. Newman does not express the bifunctional fibronectin- and Fg-binding proteins FnBPA and FnBPB (Wann et al., 2000). Newman clfA adhered to recombinant Aα-chain (rAα 1–625) in a dose-dependent and saturable manner but did not adhere to either the recombinant Bβ-chain or the γ-chain (Fig. 2b). The double clfA clfB mutant did not adhere to Fg or the Aα-chain (Fig. 2c). These data clearly show that ClfB binds only to the Aα-chain of Fg and not to the Bβ-chain as was reported previously (Ni Eidhin et al., 1998). A truncated but functional form of rClfB 197–542 (Perkins et al., 2001) inhibited adherence of Newman clfB to rAα 1–625 in a dose-dependent manner (Fig. 3), indicating that recombinant ClfB and ClfB expressed by S. aureus compete for the same binding site(s).
Fig. 3.
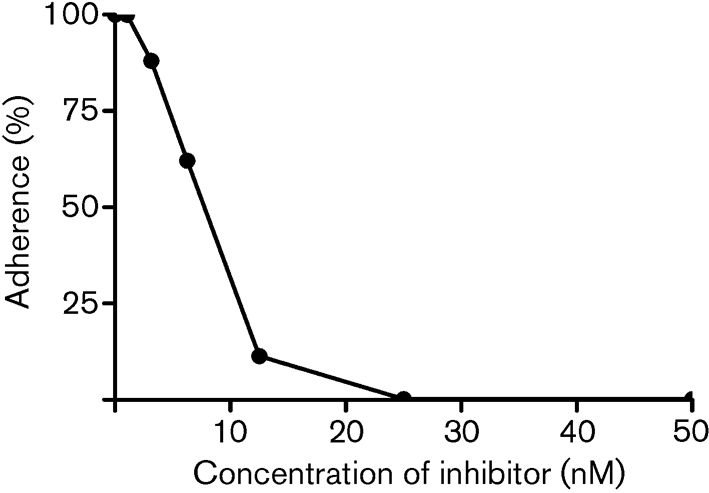
Inhibition of S. aureus Newman ClfA− adherence to recombinant Aα-chain by rClfB 197–542. ELISA plates coated with Fg rAα-chain 1–625 were incubated with increasing concentrations of rClfB 197–542 for 1 h at room temperature. Newman ClfA− cells grown to exponential phase (1×108 c.f.u.) were added to wells and adherent bacteria were detected by crystal violet staining. The values represent means of triplicate wells.
ClfB binds to a site within the αC-domain of human Fg
Mutant recombinant human Fg with a deletion of residues 252–625 comprising the C terminus of the Aα-chain (Fg Aα251) was purified from CHO cells (Gorkun et al., 1998). Native human Fg and Fg Aα251 were coated onto the wells of a microtitre dish and tested for their ability to support adherence of S. aureus cells and binding by rClfB protein. S. aureus Newman clfA adhered strongly to native Fg but did not bind to Fg Aα251 (Fig. 4a). Also rClfB 45–542 bound very weakly to Fg Aα251 in an ELISA-based assay (Fig. 4b). Plasmin treatment of Fg cleaves the protein into an E-fragment, two D-fragments and two αC-domains. Purified D- and E-domains of Fg did not inhibit the adherence of S. aureus cells to Fg (data not shown). These data suggest that the binding site for ClfB in Fg lies in the C-terminal region of the Aα-chain of Fg between residues T251 and V610.
Fig. 4.
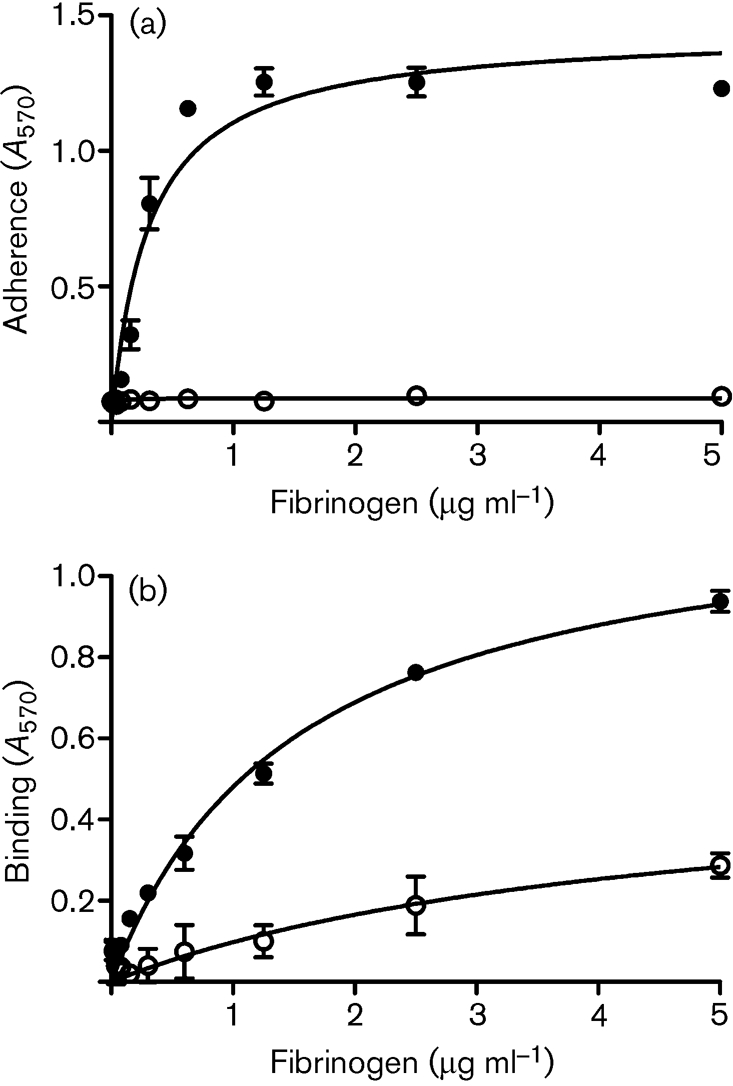
Binding to native and mutant Fg. (a) Adherence of S. aureus Newman ClfA− cells to immobilized native Fg (•), or mutant Fg Aα251 (○). Exponential-phase cells (1×108 c.f.u.) were added to wells and adherent bacteria were detected by crystal violet staining. (b) Binding of rClfB 45–542 to native Fg (•), or mutant Fg Aα251 (○). The experiment was performed twice with similar results and the values represent means±sd of triplicate wells.
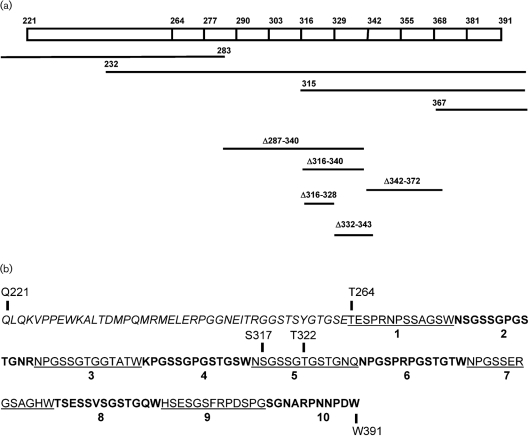
To investigate further the binding site for ClfB within the C terminus of the Aα-chain, a series of truncated forms of the recombinant α-chain were expressed (Fig. 5a). The proteins were purified, immobilized in ELISA wells and tested for their ability to support the adherence of S. aureus Newman clfA. Bacteria adhered to the protein corresponding to the C-terminal half of the Aα-chain (rAα 232–625) but did not adhere to the N-terminal region (rAα 1–283, Fig. 5a), confirming that the ClfB-binding site in the Fg α-chain is in the C-terminal domain. The C-terminal α-chain construct (rAα 232–625) was further truncated in order to locate the binding site for ClfB (Fig. 5b). The truncated proteins were tested for their ability to support the adherence of ClfB+ S. aureus cells. The smallest truncate that supported binding of ClfB was rAα 315–574, whereas rAα 367–625 did not, suggesting that ClfB binds to a site between residues W315 and W367 in the αC-domain.
Fig. 5.
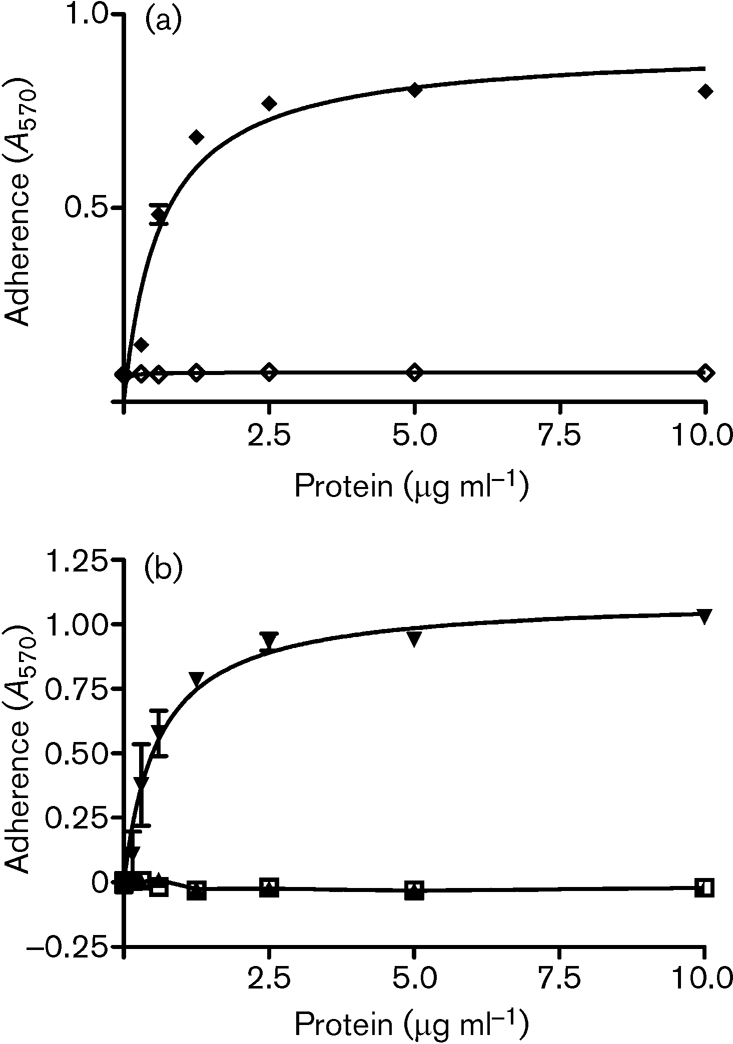
Localization of the ClfB-binding site in Fg using recombinant Aα-chain truncates. (a) Adherence of S. aureus Newman ClfA− to immobilized recombinant Fg Aα-chain truncates, rAα 232–625 (⧫) and rAα 1–283 (□). (b) Adherence of Newman ClfA− to rAα 367–574 (□), rAα 367–625 (▴) and rAα 315–574 (▾). Increasing concentrations of recombinant α-chain constructs were immobilized on ELISA plates and exponential-phase cells (1×108 c.f.u.) were added to the wells. Adherent bacteria were detected by crystal violet staining. The values represent the means±sd of triplicate wells.
ClfB binds to repeat region 5 within the flexible connector region of the Aα-chain
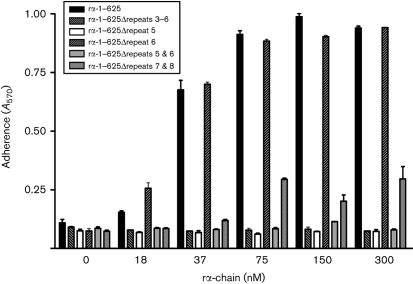
The structural organization of the Aα C-terminal domain (residues 221–610) of human Fg has not yet been fully established. It has been shown that each αC-domain consists of two structurally distinct regions, a compact C-terminal half connected to the rest of the molecule via an unordered NH2-terminal connector region (Fig. 1) (Burton et al., 2006; Tsurupa et al., 2002). In human Fg the flexible connector region starts with a 43-residue segment followed by ten 13-residue tandem repeats (Fig. 6) (Tsurupa et al., 2002). Using rAα-chain truncates we located the binding site for ClfB to between residues W315 and W367 (Fig. 5), corresponding to tandem repeats 5–8. Deletion mutants that had lost one or more of the tandem repeats were created in the plasmid expressing Aα 1–625. The ability of S. aureus ClfB+ cells to adhere to the recombinant mutant proteins was investigated (Fig. 7). S. aureus cells adhered in a dose-dependent and saturable manner both to the wild-type recombinant Aα-chain and to an Aα-chain construct that contained a deletion of most of repeat 6 (Aα 1–625 Δ332–343). Cells also adhered to a construct with deletion of repeats 7 and 8 (Aα 1–625 Δ342–372), although the binding appeared to be weaker (Fig. 7). However, ELISA with anti-Fg antibodies showed that rAα 1–625 Δ342–372 coated the plates poorly compared to all of the other proteins (data not shown), which might explain the lower adherence. Crucially, a deletion lacking repeat 5 (rAα 1–625 Δ316–328) failed to support adherence, suggesting that the ClfB-binding site is located in this region of the α-chain of Fg.
Fig. 6.
Schematic diagram showing the organization of the αC-domain of human Fg. (a) The unordered NH2-terminal half (residues 221–391) is composed of a 43 amino acid segment followed by ten 13-residue tandem repeats. The recombinant Aα-chain proteins used in this study are shown by lines and correspond to truncated or deleted variants. (b) Amino acid sequence of the N-terminal part of the αC-domain. The ten tandem repeats begin at T264 and are shown in alternating normal and bold type.
Fig. 7.
Localization of the ClfB-binding site in the tandem repeat region of the αC-domain of Fg using recombinant Aα-chain deletions. Increasing concentrations of recombinant α-chain constructs were immobilized on ELISA plates. Exponential-phase Newman ClfA− cells (1×108 c.f.u.) were added to the wells and adherent bacteria were detected by staining with crystal violet. Error bars represent standard deviations for three replicates. The experiment was performed twice with similar results.
Altering residues in repeat 5 of the Aα-chain disrupts binding to ClfB
The possibility that an additional ClfB-binding site occurs in repeats 1–3 (residues 264–302) was investigated by isolating two proline substitutions within repeat 5 of rFg Aα 1–625. Purifed recombinant Aα 1–625 chain and two mutants (S317P and T322P) were tested for their ability to support adherence of Newman clfA. ELISA with anti-6xHis antibody (Roche) showed that each protein coated the microtitre plates equally well (data not shown). Newman clfA bound to the wild-type Aα-chain in a dose-dependent and saturable manner (Fig. 8). Newman clfA was not able to adhere to the Aα-T322P mutant, while adherence to the Aα-S317P mutant was reduced. The inability of Aα-T322P to support binding to ClfB suggests that there is a single binding site for ClfB in the α-chain of Fg located in repeat 5.
Fig. 8.
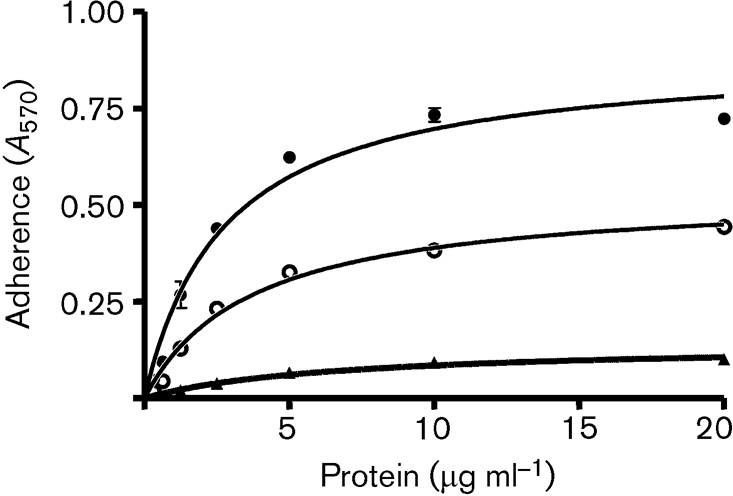
Adherence of S. aureus Newman ClfA− cells to immobilized recombinant Fg Aα- (•), AαS317P- (○) and Aα-T322P- (▴) chains. Increasing concentrations of recombinant α-chain constructs were immobilized on ELISA plates. Exponential-phase Newman ClfA− cells (1×108 c.f.u.) were added to the wells and adherent bacteria were detected by staining with crystal violet. Bars represent standard deviations for three replicates. The experiment was performed twice with similar results.
Expression of ClfB mutants by L. lactis
In order to investigate if the trench located between domains N2 and N3 of ClfB is important in binding to the Fg Aα-chain, several residues with side chains that were predicted to be located close to or within the trench were converted to alanine and expressed on the surface of L. lactis NZ9800 from the nisin-inducible vector pNZ8037. L. lactis-expressing mutants Q235A and N256A were defective in adherence to immobilized Fg (Fig. 9) and CK10 (data not shown) compared to the wild-type control. Western immunoblotting indicated that the proteins were expressed at the same level as the wild-type and were intact. This suggests that CK10 and Fg likely bind to the same region of ClfB.
Fig. 9.
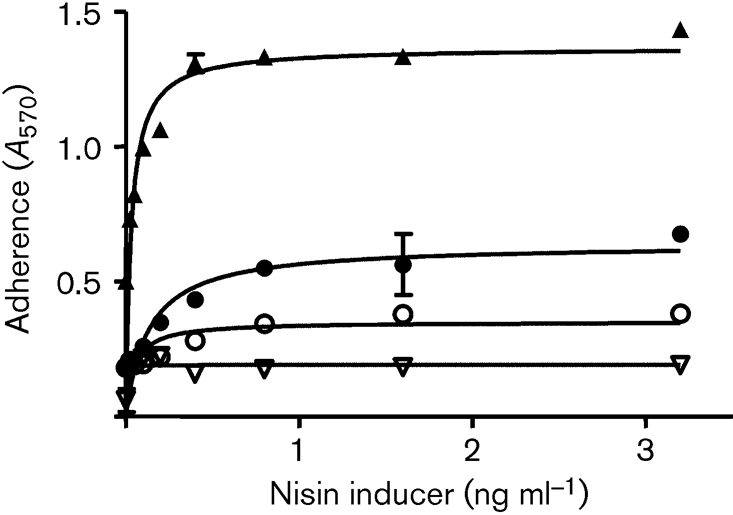
Fg binding by L. lactis ClfB+ and L. lactis ClfB mutants. L. lactis ClfB+ (▴), L. lactis ClfB Q235A (○), L. lactis ClfB N526A (•) and L. lactis pNZ8037 (▿) were induced with nisin and grown to stationary phase. Adherence of washed cultures to ELISA plates coated with Fg (10 μg ml−1) was assessed by crystal violet staining; the values represent the means±sd of triplicate wells.
DISCUSSION
Many aspects of haemostasis and wound healing involve the αC-domains of Fg playing a central role. The two self-interacting αC-domains are formed by the C-terminal two-thirds of the two Aα-chains (residues 220–610) (Burton et al., 2006; Medved et al., 1983). Compared to the rest of the Fg molecule, the αC-domains are very sensitive to proteases and are readily cleaved into smaller fragments (Doolittle, 1984; Henschen & MCDonagh, 1986; Weisel & Medved, 2001), and they are the first portions of fibrin to be removed upon fibrinolysis (Tsurupa & Medved, 2001).
This paper presents data that define the binding site in Fg for ClfB as being located in repeat 5 of the flexible region of the Aα-chain. Only the Aα-chain and not the Bβ- or γ-chains could support binding of ClfB. Fg with a deletion of the entire αC region and a recombinant Aα-chain mutant lacking the C-terminal αC region did not support binding. A series of recombinant truncated proteins narrowed down a binding domain to repeat 5 located between residues 316 and 328.
In human Fg each individual tandem repeat is composed of 13 amino acids (Fig. 6). Up to eight residues in the repeats are glycine or serine. Repeat 5 (NSGSSGTGSTGNQ) may form a loop similar to the Tyr-(Gly/Ser)n Ω loops present in the tail region of CK10, to which ClfB also binds (Walsh et al., 2004). Since the repeats that apparently do not support ClfB binding contain proline and/or arginine residues (Fig. 6), it is possible that these residues interfere with the potential of ClfB to bind to other parts of the repeat region. To examine this, two mutants in repeat 5 mimicking the presence of proline residues located in other putative non-ClfB-binding repeats were isolated in a recombinant Fg α-chain construct that contained each of repeats 1–8. The S317P mutant had reduced affinity for ClfB whereas the T322P mutant was unable to bind ClfB. This suggests that repeat 5 is the only site in the Fg α-chain that ClFB binds to, and that T322 is crucially important for this. The presence of a P in the centre of the putative Ω loops in other repeats, in particular repeat 2, might explain their apparent inability to support binding in the T322 mutant. The S317P substitution creates a sequence in repeat 5 that resembles repeat 3. It is unclear why wild-type repeat 3 did not support reduced ClfB binding in the S317 mutant similar to the T322P substitution in repeat 5. Perhaps the sequences at the C terminus of repeat 3 that differ from repeat 5 are responsible.
This study suggests that Fg and CK10 have the same or overlapping binding sites on ClfB (Walsh et al., 2004) and that the mechanism of ClfB binding to Fg is likely to be similar to K10 binding. Amino acid substitution mutants Q235A and N256A located in the putative binding trench in ClfB between domains N2 and N3 were defective in binding to both Fg and CK10.
A common theme is emerging concerning the nature of ligands recognized by surface proteins of staphylococci of the Clf-Sdr family. Each binds flexible unfolded peptides, with ClfA and FnBPA binding to the short flexible γ-chain C-terminal peptide that protrudes from domain D of Fg (McDevitt et al., 1997) while SdrG binds to peptides at the N-terminal end of the β-chain that extend from domain E (Davis et al., 2001; Ponnuraj et al., 2003). ClfB binds to glycine- and serine-rich loops at the C terminus of CK10 (Walsh et al., 2004) and this study reveals that ClfB also binds to the flexible connector region of the αC region of Fg. It is logical to postulate that each of these MSCRAMMs binds their ligand(s) by the dock latch and lock mechanism (Ponnuraj et al., 2003) but this can only be shown for certain by solving the X-ray crystal structure of the MSCRAMM with the ligands bound.
Acknowledgments
This work was supported by the Wellcome Trust and the Health Research Board of Ireland.
Abbreviations
CK10, cytokeratin 10
Fg, fibrinogen
HRP, horseradish peroxidase
MSCRAMMs, microbial surface components recognizing adhesive matrix molecules
References
- Bolyard, M. G. & Lord, S. T. (1988). High-level expression of a functional human fibrinogen gamma chain in Escherichia coli. Gene 66, 183–192. [DOI] [PubMed] [Google Scholar]
- Burton, R. A., Tsurupa, G., Medved, L. & Tjandra, N. (2006). Identification of an ordered compact structure within the recombinant bovine fibrinogen alphaC-domain fragment by NMR. Biochemistry 45, 2257–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cierniewski, C. S. & Budzynski, A. Z. (1992). Involvement of the alpha-chain in fibrin clot formation. Effect of monoclonal-antibodies. Biochemistry 31, 4248–4253. [DOI] [PubMed] [Google Scholar]
- Cole, A. M., Tahk, S., Oren, A., Yoshioka, D., Kim, Y. H., Park, A. & Ganz, T. (2001). Determinants of Staphylococcus aureus nasal carriage. Clin Diagn Lab Immunol 8, 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credo, R. B., Curtis, C. G. & Lorand, L. (1981). Alpha-chain domain of fibrinogen controls generation of fibrinoligase (coagulation factor XIIIa). Calcium ion regulatory aspects. Biochemistry 20, 3770–3778. [DOI] [PubMed] [Google Scholar]
- Davis, S. L., Gurusiddappa, S., McCrea, K. W., Perkins, S. & Hook, M. (2001). SdrG, a fibrinogen-binding bacterial adhesin of the microbial surface components recognizing adhesive matrix molecules subfamily from Staphylococcus epidermidis, targets the thrombin cleavage site in the Bbeta chain. J Biol Chem 276, 27799–27805. [DOI] [PubMed] [Google Scholar]
- Doolittle, R. F. (1984). Fibrinogen and fibrin. Annu Rev Biochem 53, 195–229. [DOI] [PubMed] [Google Scholar]
- Duthie, E. S. & Lorenz, L. L. (1952). Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol 6, 95–107. [DOI] [PubMed] [Google Scholar]
- Gorkun, O. V., Veklich, Y. I., Medved, L. V., Henschen, A. H. & Weisel, J. W. (1994). Role of the alpha C domains of fibrin in clot formation. Biochemistry 33, 6986–6997. [DOI] [PubMed] [Google Scholar]
- Gorkun, O. V., Henschen-Edman, A. H., Ping, L. F. & Lord, S. T. (1998). Analysis of A alpha 251 fibrinogen: the alpha C domain has a role in polymerization, albeit more subtle than anticipated from the analogous proteolytic fragment X. Biochemistry 37, 15434–15441. [DOI] [PubMed] [Google Scholar]
- Henschen, A. & McDonagh, J. (1986). In Fibrinogen, Fibrin and Factor XIII in Blood Coagulation, pp. 171–241. Edited by R. F. A. Zwaal & H. C. Hemker. Amsterdam: Elsevier Science Publishers.
- Herrick, S., Blanc-Brude, O., Gray, A. & Laurent, G. (1999). Fibrinogen. Int J Biochem Cell Biol 31, 741–746. [DOI] [PubMed] [Google Scholar]
- Jonsson, K., McDevitt, D., McGavin, M. H., Patti, J. M. & Hook, M. (1995). Staphylococcus aureus expresses a major histocompatibility complex class II analog. J Biol Chem 270, 21457–21460. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lord, S. T. (1985). Expression of a cloned human fibrinogen cDNA in Escherichia coli: synthesis of an A alpha polypeptide. DNA 4, 33–38. [DOI] [PubMed] [Google Scholar]
- Mazmanian, S. K., Liu, G., Ton-That, H. & Schneewind, O. (1999). Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763. [DOI] [PubMed] [Google Scholar]
- McAleese, F. M., Walsh, E. J., Sieprawska, M., Potempa, J. & Foster, T. J. (2001). Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J Biol Chem 276, 29969–29978. [DOI] [PubMed] [Google Scholar]
- McDevitt, D., Francois, P., Vaudaux, P. & Foster, T. J. (1994). Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol 11, 237–248. [DOI] [PubMed] [Google Scholar]
- McDevitt, D., Francois, P., Vaudaux, P. & Foster, T. J. (1995). Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol Microbiol 16, 895–907. [DOI] [PubMed] [Google Scholar]
- McDevitt, D., Nanavaty, T., House-Pompeo, K., Bell, E., Turner, N., McIntire, L., Foster, T. & Hook, M. (1997). Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur J Biochem 247, 416–424. [DOI] [PubMed] [Google Scholar]
- McGavin, M. H., Krajewska-Pietrasik, D., Ryden, C. & Hook, M. (1993). Identification of a Staphylococcus aureus extracellular matrix-binding protein with broad specificity. Infect Immun 61, 2479–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medved, L. V., Gorkun, O. V. & Privalov, P. L. (1983). Structural organization of C-terminal parts of fibrinogen A alpha-chains. FEBS Lett 160, 291–295. [DOI] [PubMed] [Google Scholar]
- Medved, L. V., Gorkun, O. V., Manyakov, V. F. & Belitser, V. A. (1985). The role of fibrinogen alpha C-domains in the fibrin assembly process. FEBS Lett 181, 109–112. [DOI] [PubMed] [Google Scholar]
- Miajlovic, H., Loughman, A., Brennan, M., Cox, D. & Foster, T. J. (2007). Both complement- and fibrinogen-dependent mechanisms contribute to platelet aggregation mediated by Staphylococcus aureus clumping factor B. Infect Immun 75, 3335–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre, W. W. & Schneewind, O. (1994). Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol Microbiol 14, 115–121. [DOI] [PubMed] [Google Scholar]
- Ni Eidhin, D., Perkins, S., Francois, P., Vaudaux, P., Hook, M. & Foster, T. J. (1998). Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol 30, 245–257. [DOI] [PubMed] [Google Scholar]
- O'Brien, L. M., Walsh, E. J., Massey, R. C., Peacock, S. J. & Foster, T. J. (2002). Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol 4, 759–770. [DOI] [PubMed] [Google Scholar]
- O'Connell, D. P., Nanavaty, T., McDevitt, D., Gurusiddappa, S., Hook, M. & Foster, T. J. (1998). The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J Biol Chem 273, 6821–6829. [DOI] [PubMed] [Google Scholar]
- Palma, M., Wade, D., Flock, M. & Flock, J. I. (1998). Multiple binding sites in the interaction between an extracellular fibrinogen-binding protein from Staphylococcus aureus and fibrinogen. J Biol Chem 273, 13177–13181. [DOI] [PubMed] [Google Scholar]
- Peacock, S. J., de Silva, I. & Lowy, F. D. (2001). What determines nasal carriage of Staphylococcus aureus? Trends Microbiol 9, 605–610. [DOI] [PubMed] [Google Scholar]
- Perkins, S., Walsh, E. J., Deivanayagam, C. C., Narayana, S. V., Foster, T. J. & Hook, M. (2001). Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J Biol Chem 276, 44721–44728. [DOI] [PubMed] [Google Scholar]
- Phonimdaeng, P., O'Reilly, M., Nowlan, P., Bramley, A. J. & Foster, T. J. (1990). The coagulase of Staphylococcus aureus 8325-4. Sequence analysis and virulence of site-specific coagulase-deficient mutants. Mol Microbiol 4, 393–404. [DOI] [PubMed] [Google Scholar]
- Ponnuraj, K., Bowden, M. G., Davis, S., Gurusiddappa, S., Moore, D., Choe, D., Xu, Y., Hook, M. & Narayana, S. V. (2003). A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115, 217–228. [DOI] [PubMed] [Google Scholar]
- Rudchenko, S., Trakht, I. & Sobel, J. H. (1996). Comparative structural and functional features of the human fibrinogen alpha C domain and the isolated alpha C fragment. Characterization using monoclonal antibodies to defined COOH-terminal A alpha chain regions. J Biol Chem 271, 2523–2530. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Labratory.
- Tsurupa, G. & Medved, L. (2001). Identification and characterization of novel tPA- and plasminogen-binding sites within fibrin(ogen) alpha C-domains. Biochemistry 40, 801–808. [DOI] [PubMed] [Google Scholar]
- Tsurupa, G., Tsonev, L. & Medved, L. (2002). Structural organization of the fibrin(ogen) alpha C-domain. Biochemistry 41, 6449–6459. [DOI] [PubMed] [Google Scholar]
- Walsh, E. J., O'Brien, L. M., Liang, X., Hook, M. & Foster, T. J. (2004). Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. J Biol Chem 279, 50691–50699. [DOI] [PubMed] [Google Scholar]
- Wann, E. R., Gurusiddappa, S. & Hook, M. (2000). The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J Biol Chem 275, 13863–13871. [DOI] [PubMed] [Google Scholar]
- Weisel, J. W. & Medved, L. (2001). The structure and function of the αC domains of fibrinogen. Ann N Y Acad Sci 936, 312–327. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Kollman, J. M., Pandi, L. & Doolittle, R. F. (2001). Crystal structure of native chicken fibrinogen at 2.7 Å resolution. Biochemistry 40, 12515–12523. [DOI] [PubMed] [Google Scholar]