Abstract
The homothallic ascomycete fungus Gibberella zeae (anamorph: Fusarium graminearum) is a major toxigenic plant pathogen that causes head blight disease on small-grain cereals. The fungus produces the mycotoxins deoxynivalenol (DON) and zearalenone (ZEA) in infected hosts, posing a threat to human and animal health. Despite its agricultural and toxicological importance, the molecular mechanisms underlying its growth, development and virulence remain largely unknown. To better understand such mechanisms, we studied the heterotrimeric G proteins of G. zeae, which are known to control crucial signalling pathways that regulate various cellular and developmental responses in fungi. Three putative Gα subunits, GzGPA1, GzGPA2 and GzGPA3, and one Gβ subunit, GzGPB1, were identified in the F. graminearum genome. Deletion of GzGPA1, a homologue of the Aspergillus nidulans Gα gene fadA, resulted in female sterility and enhanced DON and ZEA production, suggesting that GzGPA1 is required for normal sexual reproduction and repression of toxin biosynthesis. The production of DON and ZEA was also enhanced in the GzGPB1 mutant, suggesting that both Gα GzGPA1 and Gβ GzGPB1 negatively control mycotoxin production. Deletion of GzGPA2, which encodes a Gα protein similar to A. nidulans GanB, caused reduced pathogenicity and increased chitin accumulation in the cell wall, implying that GzGPA2 has multiple functions. Our study shows that G. zeae heterotrimeric G protein subunits can regulate vegetative growth, sexual development, toxin production and pathogenicity.
INTRODUCTION
Gibberella zeae (anamorph: Fusarium graminearum) is an important fungal pathogen of small-grain cereal crops, such as barley, wheat and rice, and is distributed worldwide (Desjardins, 2006; Goswami & Kistler, 2004; Leslie & Summerell, 2006). This homothallic ascomycete reproduces both asexually and sexually, resulting in the production of macroconidia and ascospores, respectively, as major inoculum sources for plant infection (Goswami & Kistler, 2004). G. zeae produces a variety of toxic secondary metabolites, notably deoxynivalenol (DON) and zearalenone (ZEA), that threaten human and animal health (Desjardins, 2006). Diverse virulence factors such as mycotoxins, cyclic peptides, amino acids and a lipase have been shown to play crucial roles during pathogenesis in G. zeae (Han et al., 2004; Kim et al., 2007; Lu et al., 2003; Oide et al., 2006; Proctor et al., 1995; Seong et al., 2005; Voigt et al., 2005). However, the complex signalling mechanisms that regulate virulence in G. zeae are at best poorly understood. To develop innovative control strategies for Fusarium head blight, a better understanding is needed of the molecular mechanisms that underpin virulence gene regulation in G. zeae.
One conserved signalling pathway in filamentous fungi that needs further investigation in relation to G. zeae virulence is the heterotrimeric G protein-mediated signalling pathway. Heterotrimeric G protein-mediated signal perception and propagation are conserved from lower eukaryotes to humans (Dohlman & Thorner, 2001; Lengeler et al., 2000). This signalling pathway plays a central role in controlling cell growth, development, virulence and secondary metabolite production in fungi (Bölker, 1998; Lengeler et al., 2000). The basic units of this pathway are a G protein-coupled receptor, G proteins (consisting of Gα, Gβ and Gγ subunits) and downstream effectors such as cAMP-dependent protein kinase A (PKA), phospholipases, mitogen-activated protein kinases and ion channels (Yu, 2006). A number of Gαi subunit genes of filamentous fungi have been characterized (Choi et al., 1995; Gronover et al., 2001; Horwitz et al., 1999; Jain et al., 2002; Turner & Borkovich, 1993; Yu et al., 1996), with gna1 in Neurospora crassa (Turner & Borkovich, 1993) and fadA in Aspergillus nidulans (Yu et al., 1996) amongst the first to be described. In plant-pathogenic fungi, such as Magnaporthe grisea, Cryphonectria parasitica, Colletotrichum trifolii and Botrytis cinerea, some Gα subunits positively regulate signalling mechanisms important for fungal vegetative growth and pathogenesis (Gao & Nuss, 1996; Gronover et al., 2001; Liu & Dean, 1997; Truesdell et al., 2000). In the chestnut blight pathogen C. parasitica, deletion of CPG-1 results in reduced growth and pigmentation, female infertility, loss of virulence and asexual sporulation, and altered gene expression, which are highly similar to the phenotypic changes caused by hypovirus infection (Dawe & Nuss, 2001). Deletion of CPG-2, the gene encoding a second Gα subunit in C. parasitica, causes only a slight reduction in growth rate and asexual development, and has no effect on either pathogenicity or hypovirulence-related phenotypes (Dawe & Nuss, 2001), suggesting that these Gα subunits have different functions in C. parasitica. In the rice blast fungus M. grisea, three Gα subunit genes have been identified and characterized (Liu & Dean, 1997). Deletion of MAGB, the gene orthologous to C. parasitica CPG-1, A. nidulans fadA and N. crassa gna1, results in significantly reduced hyphal growth, sporulation, appressorium formation and pathogenicity, whereas disruptions of the other Gα genes, MAGA and MAGC, do not affect growth or appressorium formation.
While substantial basic knowledge regarding the functional role of heterotrimeric G protein signalling complexes has been obtained from other fungi, such as A. nidulans and N. crassa, our understanding of the biological significance of these genes in Gibberella species (anamorph: Fusarium species) is very limited. However, given the very different biological life styles found in Gibberella species, e.g. host range, sexual development and virulence mechanisms, we can anticipate greater functional diversity for members of the G protein complex. For instance, Gβ subunits in Fusarium oxysporum and Gibberella moniliformis differ only in one amino acid, even though their functional roles in plant pathogenesis are quite different. Jain et al. (2003) showed that F. oxysporum FGB1, a gene encoding a Gβ subunit, is involved in virulence, while G. moniliformis GBB1 has no impact on maize stalk rot pathogenesis (Sagaram & Shim, 2007). The availability of three Fusarium genomes enables comparative functional analysis of heterotrimeric G protein subunits in G. zeae, G. moniliformis and F. oxysporum.
Our working hypothesis is that proper functioning of the G protein complex is required for sexual reproduction, mycotoxin production and scab virulence in G. zeae. We tested this hypothesis by deleting the G. zeae genes that encode heterotrimeric G protein Gα and Gβ subunits, and evaluating strains carrying the deletion mutations. We also compared structural and functional properties of the Gβ subunits from G. zeae, G. moniliformis and F. oxysporum in order to better understand the divergence of gene function in the three species.
METHODS
Fungal strains, media and culture.
G. zeae strain GZ3639 [obtained from Robert L. Bowden of the Plant Science and Entomology Research Unit, United States Department of Agriculture–Agricultural Research Service (USDA–ARS), Manhattan, KS, USA] was used as the wild-type strain. G. zeae ΔMAT1-2 deletion strain T39ΔM2-1 (Lee et al., 2003), derived from GZ3639, was used as the tester strain for outcrosses. All strains used in this study were stored in 25 % (v/v) glycerol at −80 °C and reactivated on potato dextrose agar (PDA; Difco Laboratories). Carrot agar plates (Leslie & Summerell, 2006) were used for induction of sexual development. For genomic DNA isolation, fungal strains were grown in 50 ml liquid complete medium (CM; Leslie & Summerell, 2006) at 25 °C, 150 r.p.m. for 3 days, and subsequently the hyphal mass was harvested and lyophilized. To produce macrospores, some mycelial agar blocks were inoculated into CMC liquid medium (15 g carboxymethyl cellulose l−1, 1 g yeast extract l−1, 0.5 g MgSO4 l−1, 1 g NH4NO3 l−1, and 1 g KH2PO4 l−1 in distilled water) and incubated at 25 °C, 150 r.p.m. for 4–5 days. Macroconidia were collected by filtering through several layers of sterile cheese cloth and were used as an inoculum source. For total RNA extraction, wild-type and mutant strains were grown in 25 ml liquid starch–glutamate (SG) medium (Bacon et al., 1977). For ZEA production and total RNA extraction, all strains were grown on SG medium for 15 days at 25 °C with brief shaking twice a day for the first 3 days. Rice grains (30 g) were used as the solid medium for DON and ZEA production. Wild-type and mutant strains were inoculated and incubated for 3 weeks at 25 °C in the dark, as described previously (Lee et al., 2002).
Nucleic acid manipulations, PCR primers and sequence analysis.
Fungal genomic DNA and total RNA were extracted as previously described (Han et al., 2004). Standard procedures were used for restriction endonuclease digestion, agarose gel electrophoresis and gel blot hybridization (Sambrook & Russell, 2001). PCRs were performed as previously described (Lee et al., 2002). Nucleotide sequences were assembled and analysed using the dnastar software package. Sequences were compared with the Fusarium group genome databases (http://www.broad.mit.edu/annotation/genome/fusarium_group/MultiHome.html) by using the blast algorithm (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov).
Double-joint PCR.
For targeted gene deletion, a gene disruption construct carrying a geneticin-resistance gene (gen) flanked by DNA sequences homologous to the sequences located at the 5′ and 3′ ends of the genomic target region was amplified by using a previously described double-joint PCR method with slight modifications (Yu et al., 2004). Briefly, DNA fragments corresponding to regions 5′ and 3′ of the target ORF were amplified from wild-type genomic DNA with specific primer pairs (Table 1). The 5′ (982 bp) and 3′ (992 bp) flanking regions of GzGPA1 were amplified with primer pairs GPA1-5F and GPA1-5R and GPA1-3F and GPA1-3R, respectively. The 5′ (974 bp) and 3′ (958 bp) flanking regions of GzGPA2 were amplified with GPA2-5F and GPA2-5R and GPA2-3F and GPA2-3R, respectively. The 5′ (953 bp) and 3′ (920 bp) flanking regions of GzGPA3 were amplified with GPA3-5F and GPA3-5R and GPA3-3F and GPA3-3R, respectively. The 5′ (928 bp) and 3′ (928 bp) flanking regions of GzGPB1 were amplified with primer pairs GPB1-5F and GPB1-5R and GPB1-3F and GPB1-3R, respectively. A 1.9 kb DNA fragment containing gen was amplified from the vector pII99 (Kim et al., 2005) with primer pair Gen-F and Gen-R. The reverse primers for amplifying the 5′ region and the forward primers for the 3′ region contained ∼20 bp tail sequences that overlapped with the 5′ and 3′ end of the gen cassette (Table 1). Three amplicons (the 5′ flanking region, the gen cassette and the 3′ flanking region) were mixed at 1 : 2 : 1 molar ratio and used as the template for a second round of PCR with new primer pairs, which were nested in the primers used for the first round of PCR (GPA1-5N and GPA1-3N, GPA2-5N and GPA2-3N, GPA3-5N and GPA3-3N, and GPB1-5N and GPB1-3N). Following purification, PCR products were mixed with fungal protoplasts for transformation, as previously described (Kim et al., 2007).
Table 1.
Primers used in this study
| Primer | Sequence* (5′→3′) | Base-pair positions† at contig |
|---|---|---|
| GPA1-5F | CATCTCGGATCTGCCTCTGATT | 1439–1460 at 1.228 |
| GPA1-5R | GCACAGGTACACTTGTTTAGAGAGTGACGGTAGTTTGGCTTTGCT | 2420–2399 at 1.228 |
| GPA1-3F | CCTTCAATATCACTTCTGTCGAAATGATTTTTTTCTTG | 3691–3706 at 1.228 |
| GPA1-3R | GTCTATCGACAGGTCACCGTGT | 4683–4662 at 1.228 |
| GPA1-5N | ATTTCTCACTCCCAGCTC | 1462–1479 at 1.228 |
| GPA1-3N | GCATGTCTCATCACCAGA | 4622–4605 at 1.228 |
| GPA2-5F | GAACGGTCGAGGTCTCTTTGTA | 5674–5653 at 1.398 |
| GPA2-5R | GCACAGGTACACTTGTTTAGAGATTGAGAATTAGAAA AA AGCGGC | 4700–4721 at 1.398 |
| GPA2-3F | CCTTCAATATCATCTTCTGTCGATAGTGGAACGCTGCTTTTTTCT | 3344–3323 at 1.398 |
| GPA2-3R | GCCAACAAGCCATATACCGATA | 2386–2407 at 1.398 |
| GPA2-5N | CGCTCCAATTAGTCCATC | 5611–5594 at 1.398 |
| GPA2-3N | CACATCATTCGTTCCCAA | 2440–2457 at 1.398 |
| GPA3-5F | CCTAGAGAGGTACGCTCCAAAG | 55614–55635 at 1.415 |
| GPA3-5R | GCACAGGTACACTTGTTTAGAGAATTGTGTGTTGTTGAAGCGACT | 56564–56543 at 1.415 |
| GPA3-3F | CCTTCAATATCATCTTCTGTCGAGATATACGGCGTTTGGACATTT | 57978–57999 at 1.415 |
| GPA3-3R | CTTTTGCAGTTTCGTTGTGTGT | 58898–58877 at 1.415 |
| GPA3-5N | CCACTGCCAAAAAAACCT | 55644–55661 at 1.415 |
| GPA3-3N | GAGCATCAAGCAGACCAG | 58872–58855 at 1.415 |
| GPB1-5F | TTTTTGCAGATGCGAAATAGGT | 66401–66422 at 1.179 |
| GPB1-5R | GCACAGGTACACTTGTTTAGAGAGCGACAGGAGAGATTAAACGAG | 67328–67307 at 1.179 |
| GPB1-3F | CCTTCAATATCATCTTCTGTCGACGTTCACGACCATGAAACTATT | 68794–68815 at 1.179 |
| GPB1-3R | TCATCAATGGGATATGCTAAGC | 69722–69701 at 1.179 |
| GPB1-5N | GTACATCTAGGCCAGCCA | 66469–66486 at 1.179 |
| GPB1-3N | AAAAGACCGAGCAACAGA | 69688–69671 at 1.179 |
| Gen-F | CGACAGAAGATGATATTGAAGG | For amplification of gen cassette from pII99 vector |
| Gen-R | CTCTAAACAAGTGTACCTGTGC |
*The underlined sequences are matched with the primer sequences of either Gen-F or Gen-R.
†Based on the F. graminearum genome database.
Fungal transformation.
Protoplasts of the wild-type strain were prepared by treatment of fresh mycelia grown on YPG liquid medium (3 g yeast extract l−1, 10 g peptone l−1, 20 g glucose l−1 in distilled water) for 12 h at 25 °C with 1 M NH4Cl containing 1 % Driselase (1 mg ml−1 in NH4Cl; InterSpex Products), as described previously (Lee et al., 2002). Approximately 5 μg of fusion PCR product obtained by double-joint PCR was directly added along with 1.2 ml 60 % (w/v) PEG (MW 3350) to protoplast suspensions. Fungal transformants expressing gen were selected on regeneration medium containing 75 p.p.m. geneticin. For complementation analyses, the intact copies of four genes amplified from genomic DNA of the wild-type strain were directly added into fungal protoplasts along with the vector pIGPAPA (Horwitz et al., 1999) carrying the hygromycin-resistance gene (hygB) as a selective marker.
Pathogenicity and sexual cross test.
Macroconidia were collected from cultures grown on carrot agar plates for 2 weeks at 25 °C and were suspended in sterile water at a concentration of 1×106 conidia ml−1. This conidial suspension was sprayed onto heads of barley at early anthesis. For each treatment, 10 replicate heads were sprayed. The plants were incubated for 2 days in a growth chamber at 25 °C with 100 % relative humidity and then were transferred to a greenhouse. Both selfing and outcrossing of the G. zeae strains were performed as previously described (Han et al., 2004).
Mycelial staining.
Young mycelia of G. zeae strains grown in YPG medium were harvested by centrifugation at 12 000 g for 5 min, washed twice with distilled water and resuspended in distilled water. A 20 μl volume of the mycelial suspension was placed on a glass slide and 2 μl Calcofluor white stock solution (10 mg ml−1) was added to it. After incubation for 15 min at 4 °C, the mycelia were observed under an LSM5-Duo (Carl Zeiss) laser scanning microscope.
ZEA and DON analyses.
To detect ZEA in liquid culture, the fungal culture was filtered through Whatman number 2 filter paper. The filtrate (20 ml) was defatted with 20 ml n-hexane and extracted twice with 20 ml ethyl acetate. The ethyl acetate layer was collected, dried and dissolved in 2 ml methanol. A Shimadzu LC-10 AD HPLC equipped with an RF-10A fluorescence detector (Shimadzu) was used for ZEA analysis. The column was a Symmetry C18 column (15 mm × 4.6 mm; particle size, 5 μm; Waters). The detection wavelength was from 274 to 466 nm. The mobile phase was 65 % aqueous methanol at a flow rate of 1 ml min−1.
To quantify ZEA and DON production on rice substrate, rice cultures were harvested and extracted with a method from a previously described protocol (Lee et al., 2002). A portion of each extract was injected into the HPLC to analyse ZEA, as described above. Each chemical analysis of toxins was replicated three times.
For DON analysis, 500 μl extract was concentrated to dryness and dissolved in 50 μl trimethylsilylating (TMS) reagent [BSA/trimethylchlorosilane (TMCS)/TMS1, 3 : 2 : 3, Supelco], mixed gently and heated at 60 °C for 5 min. After the reaction, 200 μl n-hexane was added to the solution, which was vortexed with 20 μl distilled water and allowed to stand until the two layers separated. The upper layer (1 μl) was analysed with a Hewlett Packard 5890 series II gas chromatograph with a DB-5 fused silica capillary column [0.25 mm (inside diameter)× 30 mm; 0.25 μm film] (J&W Scientific). The column temperature was maintained at 120 °C for 5 min and then increased to 275 °C at 8 °C min−1. The injector and detector temperatures were 280 and 300 °C, respectively.
RESULTS
Identification of heterotrimeric G proteins in G. zeae
The major heterotrimeric G proteins and downstream effectors are all present in the G. zeae genome. We selected components of G protein signalling in A. nidulans, i.e. FadA (AAC49476), GanB (AAF12813), GanA (AAD34893), SfaD (AAC33436), GpgA (ABG73391), PkaA (Ni et al., 2005), PkaB (Ni et al., 2005) and PkaR (O59922) (Chang et al., 2004; Rosén et al., 1999; Seo et al., 2005; Yu et al., 1996), and performed a comparison against the F. graminearum genome (http://www.broad.mit.edu/annotation/genome/fusarium_group/MultiHome.html) with the tblastn algorithm. We identified G. zeae genes corresponding to the major heterotrimeric G proteins and downstream effectors, which we designated GzGPA1, GzGPA2, GzGPA3, GzGPB1, GzGPG1, GzPKA1, GzPKA2 and GzPKAR (Table 2).
Table 2.
Genes encoding putative G protein signalling components in G. zeae
| Gene (linkage group)* | Protein used for tblastn/protein (A. nidulans)† | Pairwise blastp‡ | Gene coding and physical information | ||
|---|---|---|---|---|---|
| E-value | Contig | ORF region in contig defined in this study/coding strand | Locus number | ||
| GzGPA1 (III) | FadA/Gα subunit | 0.0 | 1.228 | 2422–3691/+ | FG05535.1 |
| GzGPA2 (VI) | GanB/Gα subunit | 1e−153 | 1.398 | 4698–3345/− | FG09614.1 |
| GzGPA3 (VII) | GanA/Gα subunit | 2e−98 | 1.415 | 56 563–57 911/− | FG09988.1 |
| GzGPB1 (II) | SfaD/Gβ subunit | 8e−144 | 1.179 | 67 367–68 789/+ | FG04104.1 |
| GzGPG1 (IV) | GpgA/Gγ subunit | 1e−25 | 1.303 | 158 138–157 801/− | FG07235.1 |
| GzACY1 (I) | CyaA/adenylyl cyclase | 0.0 | 1.62 | 29 627–22 382/− | FG01234.1 |
| GzPKA1 (IV) | PkaA/cAMP-dependent protein kinase | 2e−146 | 1.303 | 212 185–214 114/+ | FG07251.1 |
| GzPKA2 (V) | PkaB/cAMP-dependent protein kinase catalytic subunit | 2e−110 | 1.353 | 83 645–84 932/+ | FG08729.1 |
| GzPKAR (VII) | PkaR/cAMP-dependent protein kinase regulatory subunit | 9e−123 | 1.411 | 91 620–90 371/− | FG09908.1 |
*Linkage groups are determined based on the contig and supercontig alignment with the genetic map and supplied by the website (http://www.broad.mit.edu/annotation/genome/fusarium_group/MultiHome.html).
†tblastn between the proteins in A. nidulans and F. graminearum genome databases was performed.
‡Results of the pairwise blastp analysis between putative G. zeae proteins and orthologous A. nidulans proteins.
GzGPA1 (FG05535.1, annotated as a conserved hypothetical protein in the F. graminearum genome database) encodes a 353 aa protein with high similarity (93 % identity) to A. nidulans FadA. GzGPA1, like the other members of the Gαi family, contains conserved myristylation (Buss et al., 1987, MGXXXS) and pertussis toxin-labelling sites (West et al., 1985, CXXX) in the N- and C-terminal regions, respectively. GzGPA1 and FadA can be grouped in the Gαi family or subgroup I, which inhibit adenylyl cyclase activity. The GzGPA2 gene (FG09614.1, annotated as G protein alpha-3 subunit in the genome database), located on contig 1.398 with five introns, encodes a protein with high similarities to A. nidulans GanB, M. grisea MagA and C. parasitica Cpg-2, which are classified as the Gαs family or subgroup III (Bölker, 1998). GzGPA3 (FG09988.1, annotated as G protein alpha-2 subunit in the genome database), located on contig 1.415, is interrupted by four introns, and encodes a 354 aa protein similar to GanA of A. nidulans, MagC of M. grisea and Cpg-3 of C. parasitica, which are classified as subgroup II (Bölker, 1998). GzGPA2 contains an N-myristylation site at the N-terminus, but GzGPA3 has neither N-myristylation nor pertussis toxin-labelling sites. All three Gα subunits of G. zeae contain a conserved GTPase domain.
A tblastn search with A. nidulans SfaD and GpgA led to the identification of the Gβ subunit gene, GzGPB, and Gγ subunit gene, GzGPG1, in the F. graminearum genome. GzGPB1 (FG04104.1, annotated as G protein subunit beta in the genome database) encodes a 359 aa protein that contains six conserved WD40 domains, whose proposed biochemical function is to coordinate multi-protein assemblies and protein–protein interactions that lead to signal transduction and transcriptional regulation (Lengeler et al., 2000).
Targeted deletion of Gα and Gβ orthologues in G. zeae
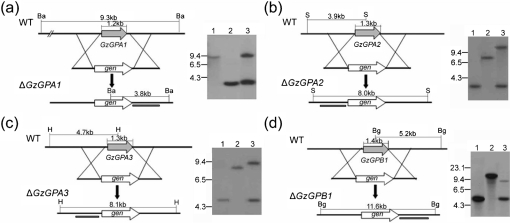
The deletion of the 1.2 kb GzGPA1 gene was confirmed by the presence of a single 3.8 kb hybridizing band in a blot of BamHI-digested genomic DNA of the GzGPA1 deletion (ΔGzGPA1) strain, rather than the 9.3 kb band that hybridized to the probe in the wild-type strain (Fig. 1a). Deletion of the three other orthologous genes was also confirmed by Southern blot analyses. The size differences in the hybridizing bands between the wild-type and the mutant strains, i.e. a single 8.0 kb band in the ΔGzGPA2 strains instead of a 3.6 kb wild-type band (Fig. 1b), an 8.1 kb band in the ΔGzGPA3 strains instead of a 4.7 kb wild-type band (Fig. 1c), and a 11.6 kb band in the ΔGzGPB1 strains rather than a 5.2 kb wild-type band (Fig. 1d), were as predicted and provided molecular evidence of successful homologous gene replacement events.
Fig. 1.
Targeted deletion of GzGPA1 (a), GzGPA2 (b), GzGPA3 (c) and GzGPB1 (d) from the genome of G. zeae wild-type strain GZ3639 (WT). Left of each panel: schematic representation of homologous gene recombination strategy resulting in gene-deletion strains. Ba, BamHI; S, SalI; H, HindIII; Bg, BglII restriction sites; gen, geneticin-resistance gene. The transcription direction of each gene is indicated by an arrow. Right of each panel: Southern blot analysis of WT (lane 1), a deletion strain (lane 2), and a transformant carrying the disruption construct at an ectopic site (lane 3). Genomic DNAs were digested with the restriction enzymes whose recognition sites are indicated on the left. The blot was hybridized with a 32P-labelled DNA fragment, indicated by a solid bar in the deletion scheme. The sizes of standards (in kb) are indicated to the left of each blot.
GzGPA2 and GzGPB1 are required for pathogenicity and normal growth
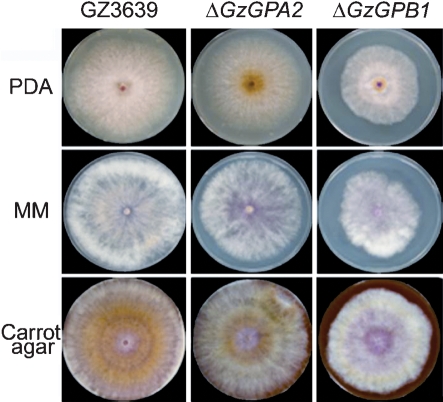
The ΔGzGPA1 and ΔGzGPA3 strains had the same level of pathogenicity towards barley as did the wild-type strain. ΔGzGPA2 and ΔGzGPB1 mutants were much less virulent than the wild-type strain (Fig. 2). When intact copies of GzGPA2 or GzGPB1 genes were introduced into deletion mutants, their virulence was fully restored, suggesting that both GzGPA2 and GzGPB1 are essential for the virulence of G. zeae. Deletion of GzGPA1, GzGPA2 and GzGPA3 slightly affected vegetative growth on PDA, with the growth rates of the mutants being similar to that of the wild-type strain, while deletion of GzGPB1 resulted in 75 % of the hyphal growth of the wild-type strain (Fig. 3).
Fig. 2.
Fungal virulence assay on barley. GZ3639, wild-type strain; ΔGzGPA2, GzGPA2-deleted strain; GPA2com, GzGPA2-complemented strain derived from ΔGzGPA2; ΔGzGPB1, GzGPB1-deleted strain; GPB1com, GzGPB1-complemented strain derived from ΔGzGPB1.
Fig. 3.
Hyphal growth of GZ3639 (wild-type strain), ΔGzGPA2 and ΔGzGPB1 strains on PDA, minimal medium (MM) and carrot agar at 4 days after inoculation.
GzGPA1 is required for sexual reproduction
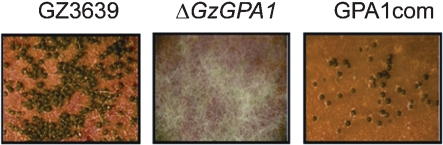
We compared sexual reproduction between the wild-type strain and its mutants on carrot agar medium. Deletion mutants of GzGPA2, GzGPA3 and GzGPB1 produced normal sexual fruiting bodies (perithecia). However, ΔGzGPA1 mutants failed to develop perithecia (Fig. 4). When we introduced an intact copy of the GzGPA1 gene back into a ΔGzGPA1 mutant, sexual reproduction was partially restored (Fig. 4). This result implies that Gα GzGPA1 plays an important role in G. zeae sexual development. The ΔGzGPA1 mutant did not show significant changes in vegetative growth or asexual sporulation on PDA or carrot agar medium. The pathogenicity of the ΔGzGPA1 mutants was comparable to that of the wild-type strain on barley.
Fig. 4.
Perithecia formation on carrot agar medium. GZ3639, wild-type strain; ΔGzGPA1, GzGPA1-deleted strain; GPA1com, GzGPA1-complemented strain derived from ΔGzGPA1.
GzGPA2 negatively regulates cell wall chitin content
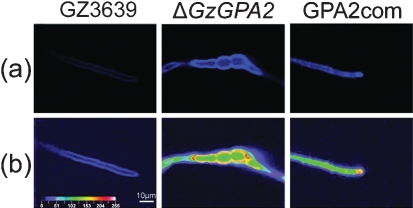
In addition to reduced virulence, the ΔGzGPA2 mutants exhibited other different phenotypes compared with their wild-type progenitor. We could not produce protoplasts from the ΔGzGPA2 mutants even when we added five times as much Driselase, a commonly used cell wall-degrading enzyme for G. zeae, and incubated for a longer time (up to 15 h). However, when the lysing enzyme solution for Aspergillus spp. (Sigma–Aldrich) was added to the Driselase solution, we were able to obtain as many protoplasts from the ΔGzGPA2 mutants as from the wild-type strain using only 1 % Driselase (data not shown). The cell wall chitin level in the ΔGzGPA2 mutant, as measured by the intensity of emitted fluorescence from Calcofluor white-stained mycelia, was much higher than that of the wild-type strain (Fig. 5). These results strongly indicate that the GzGPA2 deletion led to higher chitin accumulation in the ΔGzGPA2 cells. Moreover, the outcross of the ΔGzGPA2 mutant to the MAT1-2 deletion strain (T39ΔM2-1) produced no perithecia, although the self cross of the ΔGzGPA2 mutant formed as many normal perithecia as that of the wild-type strain (data not shown). All of these altered phenotypes were recovered in transgenic GzGPA2 mutants expressing an intact GzGPA2 copy but the chitin content in the complemented cells was still higher than that in the wild-type strain (Fig. 5).
Fig. 5.
Chitin accumulation in G. zeae ΔGzGPA2 strain. GZ3639, wild-type strain; ΔGzGPA2, GzGPA2-deleted strain; GPA2com, GzGPA2-complemented strain derived from ΔGzGPA2. (a) Chitin in the mycelia of each strain was stained with a Calcofluor white solution and observed under UV (420 nm) light. (b) Quantification of fluorescence by laser scanning microscopy. The intensity scale for quantification is indicated on the bottom left, ranging from violet for a weak signal to white for strong fluorescence.
Gα GzGPA1 and Gβ GzGPB1 negatively regulate toxin production
ΔGzGPA1 and ΔGzGPB1 mutants produced significantly more DON and ZEA than did the wild-type strain. The wild-type strain produced 460 p.p.m. ZEA; ΔGzGPA1 and ΔGzGPB1 mutants produced 800 and 1180 p.p.m., which was ∼60 and ∼250 % higher, respectively, than the wild-type strain (Fig. 6a). Moreover the ΔGzGPB1strain produced 50 % more ZEA than the ΔGzGPA1 strain. The wild-type strain produced about 10 p.p.m. DON, whereas ΔGzGPA1 and ΔGzGPB1 mutants produced 270 and 280 p.p.m., respectively (Fig. 6b). Consistent with increased ZEA production, the mRNA level of ZEB2, a gene encoding the transcription factor involved in ZEA biosynthesis, was upregulated in ΔGzGPA1 and ΔGzGPB1 mutants relative to the wild-type strain (Fig. 6c). When we introduced an intact copy of each gene back into the deletion mutants, the level of toxin production returned to approximately that of the wild-type strain, confirming that deletion of the genes was responsible for enhanced toxin production (Fig. 6a, b).
Fig. 6.
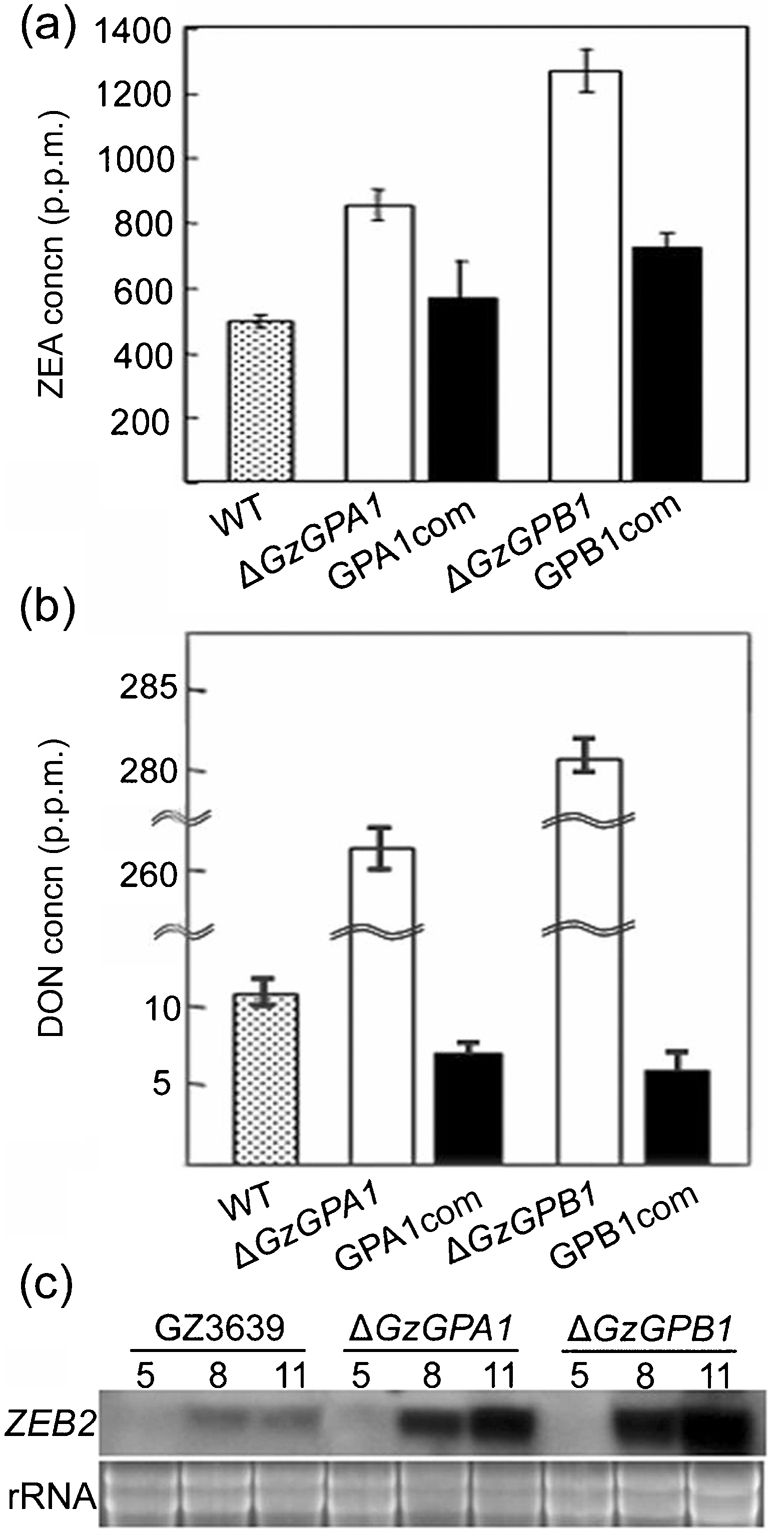
Mycotoxin production in the wild-type (WT), ΔGzGPA1 and ΔGzGPB1 strains. Quantification of ZEA (a) and DON (b) produced by the ΔGzGPA1, ΔGzGPB1 and complemented strains on rice medium. (c) Northern blot analysis of wild-type (GZ3639) and mutant strains probed with ZEB2. Total RNA samples were extracted from WT, ΔGzGPA1 and ΔGzGPB1 grown on SG medium. The probe and the incubation time (days) are indicated at the left and above the blot, respectively. The ethidium bromide-stained rRNAs used as a loading control are indicated.
DISCUSSION
Heterotrimeric G proteins are highly conserved in model filamentous and plant-pathogenic fungi. The key units of the G protein signalling complex are G protein-coupled receptors, G proteins (consisting of Gα, Gβ and Gγ subunits) and downstream effectors. We identified and characterized three putative Gα subunits and one Gβ subunit from the F. graminearum genome in this study. These G protein subunits are essential for fungal development, secondary metabolism and virulence (Bölker, 1998; Lengeler et al., 2000). One of the most extensively studied G protein signalling models in filamentous fungi is A. nidulans, in which G protein subunits control asexual/sexual development, vegetative growth and sterigmatocystin (ST) biosynthesis (Yu & Keller, 2005).
In Gibberella (Fusarium) species, however, the picture is far from clear. Two Gα subunits from F. oxysporum, FGA1 and FGA2, and the Gβ subunit from F. oxysporum, FGB1, and G. moniliformis, GBB1, have been functionally characterized (Delgado-Jarana et al., 2005; Jain et al., 2002, 2003, 2005; Sagaram & Shim, 2007). However, they do not have direct parallels in Gibberella (Fusarium) species. Given the number of phenotypic variations known in Gibberella (Fusarium) species, e.g. in host range, reproductive strategy and secondary metabolite production, there is a need to unambiguously characterize the functional role of the G protein components in G. zeae.
Heterotrimeric G protein signalling controls sexual reproduction in model and plant-pathogenic fungi (Liu & Dean, 1997; Seo et al., 2005). In G. zeae, the Gα and Gβ subunits seem to regulate either sexual reproduction or pathogenic processes, but not both. Deletion of GzGPA1 results in female sterility, whereas deletion of GzGPB1 does not alter sexual fertility. Our results suggest that GzGPA1 alone controls sexual reproduction in G. zeae. Similar results have been observed in G. moniliformis (Sagaram & Shim, 2007). Considering the genetic relatedness of G. zeae and G. moniliformis, we hypothesize that the GzGPA1 orthologue (FVEG_06962.3) in G. moniliformis, which is 100 % identical at the amino acid level, serves as the primary, if not exclusive, regulator of sexual mating in G. moniliformis.
The G. zeae G protein subunits also have an important and complicated role in plant pathogenesis. Deletion of GzGPA2 or GzGPB1 significantly reduced fungal virulence toward host plants. Pathogenicity tests were done with 1×106 conidia ml−1 on barley. Lower concentrations should be tested to exclude the possibility that GzGPA1 and GzGPA3 mutants show a more subtle phenotype. The reduced virulence in the ΔGzGPA2 mutant may be attributed to its higher level of chitin, which may elicit plant defence mechanisms. This is plausible, since chitin has been shown to be, or implicated as, a signal in plant defence (Barber et al., 1989; Tsutsui et al., 2006; Wan et al., 2004). The fungal G proteins are known to regulate cell wall composition in several cases, but their roles may not be directly comparable to those in G. zeae. In A. nidulans, FadA and SfaD (orthologous to GzGPA1 and GzGPB1, respectively) seem to negatively regulate cell wall chitin content (Coca et al., 2000), while only ΔGzGPA2 is responsible for the hyperaccumulation of chitin in G. zeae. The deletion of Trichoderma atroviride tga3, orthologous to GzGPA2, results in resistance to protoplasting by cell wall-lysing enzymes, as did ΔGzGPA2, but no changes in cell wall chitin content (Zeilinger et al., 2005). Final confirmation of this hypothesis, however, awaits further evidence supporting the hypothesis that the chitin-overproducing ΔGzGPA2 mutant elicits plant defence responses, and that GzGPA2 directly controls some chitin synthesis-related genes.
The role of fungal Gβ subunits in plant pathogenesis is not conserved. Deletion of the M. grisea Gβ subunit, mgb1, and Cochliobolus heterostrophus CGB1 leads to defective appressorium formation that prevents host penetration and infection (Ganem et al., 2004; Nishimura et al., 2003). Disruption of the C. parasitica and F. oxysporum Gβ subunit genes also results in significantly reduced virulence (Jain et al., 2003; Kasahara & Nuss, 1997). However, in Ustilago maydis, deletion of the Gβ subunit gene, BPP1, does not block tumour formation, although a slight reduction in virulence does occur (Muller et al., 2004). Deletion of the Gβ subunit gene in G. moniliformis does not alter the ability of the strain to infect and colonize maize stalk tissue (Sagaram & Shim, 2007). Similar ranges of chitin accumulation in the ΔGzGPB1 mutant compared to the wild-type strain indicate that the Gβ protein may control fungal virulence in a different manner to the Gα GzGPA2 protein in G. zeae.
In A. nidulans, both activated Gα (FadA) and Gβγ (SfadA : GpgA) propagate the vegetative growth signal through the cAMP–PKA signalling pathway (Rosén et al., 1999; Seo et al., 2005; Shimizu & Keller, 2001; Yu et al., 1996). However, independent gene deletion of the three Gα subunits slightly affects hyphal growth (∼10 % reduction), implying that Gα proteins have minor roles in hyphal growth of G. zeae.
Another important role of the heterotrimeric G protein signalling complex in filamentous fungi is to regulate secondary metabolite production. This complicated biosynthetic process is closely associated with physiological and morphological development (Calvo et al., 2002). FadA-mediated vegetative growth signalling represses ST biosynthesis in A. nidulans and asexual and sexual development, but enhances penicillin production (Yu et al., 1996). Expression of the A. nidulans fadAG42R dominant activating mutant allele in Fusarium sporotrichioides increases production of T-2 toxin (Tag et al., 2000), but inhibits production of aflatoxin in Aspergillus parasiticus (Hicks et al., 1997) and Aspergillus flavus (McDonald et al., 2005). Deletion of the Gβ subunit gene sfaD of A. nidulans drastically reduces ST biosynthesis. In G. zeae, deletion of either GzGPA1 or GzGPB1 results in increased mycotoxin production, suggesting that both G protein subunits have a role in the negative regulation of mycotoxin production. The significant upregulation of ZEB2 observed in the ΔGzGPA1 and ΔGzGPB1 mutants is consistent with this conclusion (Fig. 6c). In G. moniliformis, however, disruption of the Gβ subunit results in significant down-regulation of FUM1, a polyketide synthase gene essential for fumonisin biosynthesis, and severely curtails fumonisin production (Sagaram & Shim, 2007). The role of Gα subunits of G. moniliformis in the regulation of fumonisin biosynthesis remains unknown.
Even at the DNA level, the Gβ gene, including its location and the number of introns, is highly conserved amongst Gibberella species (data not shown). When analysed at the amino acid level, there is only a single amino acid difference between F. oxysporum (FOXG_11532.2) and G. zeae (FGSG_04104.3) or G. moniliformis (FVEG_10291.3). The Gβ proteins from the Gibberella species all contain six WD40 repeats, whereas the homologous proteins from A. nidulans and N. crassa contain seven and four WD40 repeats, respectively. When compared to previous studies of G. moniliformis and F. oxysporum, the Gβ protein from G. zeae regulates fungal development, virulence and secondary metabolism in a manner different from that in the other two species. We hypothesize that certain biological traits, e.g. host adaptation and/or sexual life style, may have altered gene function during evolution, but the basis for this functional divergence by such highly conserved proteins remains to be tested and explained.
The orthologues of the Gγ subunit GzGPG1, the protein kinase catalytic subunits GzPKA1 and GzPKA2, and the regulatory subunit GzPKAR are all present in G. zeae. These genes are the components of a cAMP-dependent PKA signalling pathway, one of the major downstream regulatory cascades in heterotrimeric G protein-mediated regulatory mechanisms in fungi (Shimizu & Keller, 2001). Identification of these components in the G. zeae genome suggests that the G protein–cAMP–PKA signalling pathway is involved in controlling G. zeae growth, development, pathogenicity and toxin production. Further functional characterization of the G protein complex and the downstream signalling pathways is needed to understand the molecular mechanisms of virulence and mycotoxin production and their regulation in G. zeae.
Acknowledgments
This study was supported by grant CG 1412 from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Korean Ministry of Science and Technology and the Korea Research Foundation Grant funded by the Korean Government [Ministry of Education and Human Resources Development (MOEHRD), Basic Research Promotion Fund, KRF-2006-005-JO4701]. H.-Y. Y. and J.-E. K. were supported by graduate fellowships from the Korean Ministry of Education through the Brain Korea 21 project. W.-B. S. was supported in part by the United States Department of Agriculture–National Research Initiative (USDA–NRI) Food Safety Program (2005-35201-16233) and Texas A&M University International Research Travel Assistance Grant (IRTAG) Program.
Abbreviations
DON, deoxynivalenol
PKA, protein kinase A
ST, sterigmatocystin
ZEA, zearalenone
References
- Bacon, C. W., Robins, J. D. & Porter, J. K. (1977). Media for identification of Gibberella zeae and production of F-2 (zearalenone). Appl Environ Microbiol 33, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber, M. S., Bertram, R. E. & Ride, J. P. (1989). Chitin oligosaccharides elicit lignification in wounded wheat leaves. Physiol Mol Plant Pathol 34, 3–12. [Google Scholar]
- Bölker, M. (1998). Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet Biol 25, 143–156. [DOI] [PubMed] [Google Scholar]
- Buss, J. E., Mumby, S. M., Casey, P. J., Gilman, A. G. & Sefton, B. M. (1987). Myristoylated α subunits of guanine nucleotide-binding regulatory proteins. Proc Natl Acad Sci U S A 84, 7493–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo, A. M., Wilson, R. A., Bok, J. W. & Keller, N. P. (2002). Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev 66, 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, M. H., Chae, K. S., Han, D. M. & Jahng, K. Y. (2004). The GanB Gα-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans. Genetics 167, 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, G. H., Chen, B. & Nuss, D. L. (1995). Virus-mediated or transgenic suppression of a G-protein α subunit and attenuation of fungal virulence. Proc Natl Acad Sci U S A 92, 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coca, M. A., Damsz, B., Yun, D.-J., Hasegawa, O. M., Bressan, R. A. & Narasimhan, M. L. (2000). Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J 22, 61–69. [DOI] [PubMed] [Google Scholar]
- Dawe, A. L. & Nuss, D. L. (2001). Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu Rev Genet 35, 1–29. [DOI] [PubMed] [Google Scholar]
- Delgado-Jarana, J., Martinez-Rocha, A. L., Roldan-Rodriguez, R., Roncero, M. I. & Di Pietro, A. (2005). Fusarium oxysporum G-protein β subunit Fgb1 regulates hyphal growth, development, and virulence through multiple signalling pathways. Fungal Genet Biol 42, 61–72. [DOI] [PubMed] [Google Scholar]
- Desjardins, A. E. (2006). Fusarium Mycotoxins. Chemistry, Genetics, and Biology. St Paul, MN: The American Phytopathological Society.
- Dohlman, H. G. & Thorner, J. W. (2001). Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem 70, 703–754. [DOI] [PubMed] [Google Scholar]
- Ganem, S., Lu, S.-W., Lee, B.-N., Chou, D. Y.-T., Hadar, R., Turgeon, B. G. & Horwitz, B. A. (2004). G-protein β subunit of Cochliobolus heterostrophus involved in virulence, asexual and sexual reproductive ability, and morphogenesis. Eukaryot Cell 3, 1653–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, S. & Nuss, D. L. (1996). Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc Natl Acad Sci U S A 93, 14122–14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, R. S. & Kistler, H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Gronover, C. S., Kasulke, D., Tudzynski, P. & Tudzynski, B. (2001). The role of G protein α subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol Plant Microbe Interact 14, 1293–1302. [DOI] [PubMed] [Google Scholar]
- Han, Y.-K., Lee, T., Han, K.-H., Yun, S.-H. & Lee, Y.-W. (2004). Functional analysis of the homoserine O-acetyltransferase gene and its identification as a selectable marker in Gibberella zeae. Curr Genet 46, 205–212. [DOI] [PubMed] [Google Scholar]
- Hicks, J. K., Yu, J. H., Keller, N. P. & Adams, T. H. (1997). Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J 16, 4916–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz, B. A., Sharon, A., Lu, S. W., Ritter, V., Sandrock, T. M., Yoder, O. C. & Turgeon, B. G. (1999). A G protein α subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet Biol 26, 19–32. [DOI] [PubMed] [Google Scholar]
- Jain, S., Akiyama, K., Kan, T., Ohguchi, T. & Takata, R. (2002). Targeted disruption of a G protein α subunit gene results in reduced pathogenicity in Fusarium oxysporum. Curr Genet 41, 407–413. [DOI] [PubMed] [Google Scholar]
- Jain, S., Akiyama, K., Kan, T., Ohguchi, T. & Takata, R. (2003). The G protein β subunit FGB1 regulates development and pathogenicity in Fusarium oxysporum. Curr Genet 43, 79–86. [DOI] [PubMed] [Google Scholar]
- Jain, S., Akiyama, K., Takata, R. & Ohguchi, T. (2005). Signaling via the G protein α subunit FGA2 is necessary for pathogenesis in Fusarium oxysporum. FEMS Microbiol Lett 243, 165–172. [DOI] [PubMed] [Google Scholar]
- Kasahara, S. & Nuss, D. L. (1997). Targeted disruption of a fungal G-protein β subunit gene results in increased vegetative growth but reduced virulence. Mol Plant Microbe Interact 10, 984–993. [DOI] [PubMed] [Google Scholar]
- Kim, J.-E., Han, K.-H., Jin, J., Kim, H., Kim, J.-C., Yun, S.-H. & Lee, Y.-W. (2005). Putative polyketide synthase and laccase genes for biosynthesis of aurofusarin in Gibberella zeae. Appl Environ Microbiol 71, 1701–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.-E., Myong, K., Shim, W.-B., Yun, S.-H. & Lee, Y.-W. (2007). Functional characterization of acetylglutamate synthase and phosphoribosylamine-glycine ligase genes in Gibberella zeae. Curr Genet 51, 99–108. [DOI] [PubMed] [Google Scholar]
- Lee, T., Han, Y. K., Kim, K. H., Yun, S. H. & Lee, Y. W. (2002). Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl Environ Microbiol 68, 2148–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Lee, T., Lee, Y. W., Yun, S. H. & Turgeon, B. G. (2003). Shifting fungal reproductive mode by manipulation of mating-type genes: obligatory heterothallism of Gibberella zeae. Mol Microbiol 50, 145–152. [DOI] [PubMed] [Google Scholar]
- Lengeler, K. B., Davidson, R. C., D'souza, C., Harashima, T., Shen, W. C., Wang, P., Pan, X., Waugh, M. & Heitman, J. (2000). Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev 64, 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, J. F. & Summerell, B. A. (2006). The Fusarium Laboratory Manual. Ames, IA: Blackwell.
- Liu, S. & Dean, R. A. (1997). G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol Plant Microbe Interact 10, 1075–1086. [DOI] [PubMed] [Google Scholar]
- Lu, S.-W., Kroken, S., Lee, B.-N., Robbertse, B., Churchill, A. C. L., Yodor, O. C. & Turgeon, B. G. (2003). A novel class of gene controlling virulence in plant pathogenic ascomycete fungi. Proc Natl Acad Sci U S A 100, 5980–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, T., Brown, D., Keller, N. P. & Hammond, T. M. (2005). RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol Plant Microbe Interact 18, 539–545. [DOI] [PubMed] [Google Scholar]
- Muller, P., Leibbrandt, A., Teuissen, H., Cubasch, S., Aichinger, C. & Kahmann, R. (2004). The Gβ-subunit-encoding gene bpp1 controls cyclic-AMP signaling in Ustilago maydis. Eukaryot Cell 3, 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M., Rierson, S., Seo, J.-A. & Yu, J.-H. (2005). The pkaB gene encoding the secondary protein kinase A catalytic subunit has a synthetic lethal interaction with pkaA and plays overlapping and opposite roles in Aspergillus nidulans. Eukaryot Cell 4, 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M., Park, G. & Xu, J. R. (2003). The G-beta subunit MGB1 is involved in regulating multiple steps of infection-related morphogenesis in Magnaporthe grisea. Mol Microbiol 50, 231–243. [DOI] [PubMed] [Google Scholar]
- Oide, S., Moeder, W., Krasnoff, S., Gibson, D., Haas, H., Yoshioka, K. & Turgeon, B. G. (2006). NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 18, 2836–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor, R. H., Hohn, T. M. & McCormick, S. P. (1995). Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe Interact 8, 593–601. [DOI] [PubMed] [Google Scholar]
- Rosén, S., Yu, J. H. & Adams, T. H. (1999). The Aspergillus nidulans sfaD gene encodes a G protein β subunit that is required for normal growth and repression of sporulation. EMBO J 18, 5592–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaram, U. S. & Shim, W.-B. (2007). Fusarium verticillioides GBB1, a gene encoding heterotrimeric G protein β subunit, is associated with fumonisin B1 biosynthesis and hyphal development but not with fungal virulence. Mol Plant Pathol 8, 375–384. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. W. (2001). Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Seo, J. A., Han, K. H. & Yu, J. H. (2005). Multiple roles of a heterotrimeric G-protein γ-subunit in governing growth and development of Aspergillus nidulans. Genetics 171, 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, K., Hou, Z. M., Tracy, M., Kistler, H. C. & Xu, J.-R. (2005). Random insertional mutagenesis identifies genes associated with virulence in the wheat scab fungus Fusarium graminearum. Phytopathology 95, 744–750. [DOI] [PubMed] [Google Scholar]
- Shimizu, K. & Keller, N. P. (2001). Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tag, A., Hicks, J., Garifullina, G., Ake, C., Jr, Phillips, T. D., Beremand, M. & Keller, N. (2000). G-protein signalling mediates differential production of toxic secondary metabolites. Mol Microbiol 38, 658–665. [DOI] [PubMed] [Google Scholar]
- Truesdell, G. M., Yang, Z. H. & Dickman, M. B. (2000). A Gα subunit gene from the phytopathogenic fungus Colletotrichum trifolii is required for conidial germination. Physiol Mol Plant Pathol 56, 131–140. [Google Scholar]
- Tsutsui, T., Morta-Yamamuro, C., Asada, Y., Minami, E., Shibuya, N., Ikeda, A. & Yamaguchi, J. (2006). Salicylic acid and a chitin elicitor both control expression of the CAD1 gene involved in the plant immunity of Arabidopsis. Biosci Biotechnol Biochem 70, 2042–2048. [DOI] [PubMed] [Google Scholar]
- Turner, G. E. & Borkovich, K. A. (1993). Identification of a G protein α subunit from Neurospora crassa that is a member of the Gi family. J Biol Chem 268, 14805–14811. [PubMed] [Google Scholar]
- Voigt, C. A., Schafer, W. & Salomon, S. (2005). A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J 42, 364–375. [DOI] [PubMed] [Google Scholar]
- Wan, J., Zhang, S. & Stacey, G. (2004). Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol Plant Pathol 5, 125–135. [DOI] [PubMed] [Google Scholar]
- West, R. E., Moss, J., Jr, Vaughan, M., Liu, T. & Liu, T. Y. (1985). Pertussis toxin-catalyzed ADP-ribosylation of transducin. Cysteine 347 is the ADP-ribose acceptor site. J Biol Chem 260, 14428–14430. [PubMed] [Google Scholar]
- Yu, J. H. (2006). Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J Microbiol 44, 145–154. [PubMed] [Google Scholar]
- Yu, J. H. & Keller, N. P. (2005). Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol 43, 437–458. [DOI] [PubMed] [Google Scholar]
- Yu, J. H., Wieser, J. & Adams, T. H. (1996). The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J 15, 5184–5190. [PMC free article] [PubMed] [Google Scholar]
- Yu, J. H., Hamari, Z., Han, K. H., Seo, J. A., Reyes-Dominguez, Y. & Scazzocchio, C. (2004). Double-joint PCR, a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41, 973–981. [DOI] [PubMed] [Google Scholar]
- Zeilinger, S., Barbara, R., Scala, V., Peissl, I., Lorito, M. & Mach, R. L. (2005). Signal transduction by Tga3, a novel G protein α subunit of Trichoderma atroviride. Appl Environ Microbiol 71, 1591–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]