Abstract
Agrobacterium tumefaciens is a Gram-negative plant-pathogenic bacterium that causes crown gall disease by transferring and integrating its transferred DNA (T-DNA) into the host genome. We characterized the chromosomally encoded alpha-crystallin-type small heat-shock protein (α-Hsp) HspL, which was induced by the virulence (vir) gene inducer acetosyringone (AS). The transcription of hspL but not three other α-Hsp genes (hspC, hspAT1, hspAT2) was upregulated by AS. Further expression analysis in various vir mutants suggested that AS-induced hspL transcription is not directly activated by the VirG response regulator but rather depends on the expression of VirG-activated virB genes encoding components of the type IV secretion system (T4SS). Among the 11 virB genes encoded by the virB operon, HspL protein levels were reduced in strains with deletions of virB6, virB8 or virB11. VirB protein accumulation but not virB transcription levels were reduced in an hspL deletion mutant early after AS induction, implying that HspL may affect the stability of individual VirB proteins or of the T4S complex directly or indirectly. Tumorigenesis efficiency and the VirB/D4-mediated conjugal transfer of an IncQ plasmid RSF1010 derivative between A. tumefaciens strains were reduced in the absence of HspL. In conclusion, increased HspL abundance is triggered in response to certain VirB protein(s) and plays a role in optimal VirB protein accumulation, VirB/D4-mediated DNA transfer and tumorigenesis.
INTRODUCTION
Agrobacterium tumefaciens is a soil-borne plant-pathogenic bacterium causing crown gall disease in a wide range of plants through an interkingdom DNA delivery system. A. tumefaciens is capable of sensing plant-released wound signal molecules such as sugars and phenolic compounds to activate a signal transduction pathway for infection. This event is regulated by the VirA/VirG two-component system encoded by the tumour-inducing (Ti) plasmid in conjunction with ChvE, a chromosomally encoded periplasmic galactose/glucose-binding protein to activate the expression of virulence (vir) genes and operons, including virA, B, G, C, D and E (McCullen & Binns, 2006). The transferred DNA (T-DNA) is then processed, followed by transfer of the T-complex and effector proteins via the Ti plasmid-encoded Vir type IV secretion system (T4SS) from bacteria into the host plant cells (Baron, 2006; Christie et al., 2005).
The Ti Vir T4SS is a transmembrane complex consisting of VirD4 and 11 VirB proteins that also assembles T pili (Christie, 2001; Lai & Kado, 2000). Accumulating biochemical and genetic data suggest a model of an ordered VirB/D4 T4SS assembly pathway (Baron, 2006; Christie et al., 2005; Ward et al., 2002). First, VirB8 initiates T4SS assembly and targets VirB1 to the cell pole, where it may locally lyse the cell wall to facilitate T4SS assembly across the double membranes (Judd et al., 2005; Yuan et al., 2005). VirB6, VirB4, VirB7, VirB8, VirB9 and VirB10 then assemble a core complex, which is followed by recruitment of the subunits important for pilus assembly, including VirB2, VirB3 and VirB5 (Krall et al., 2002). The VirB11 homohexameric ATPase may supply energy for VirB2 polymerization across the periplasm to form the T pilus (Atmakuri et al., 2004; Rashkova et al., 2000). Finally, VirB4 and VirD4 are required for substrate translocation, which may be mechanistically linked to a conformational change of VirB10 (Cascales & Christie, 2004a). The T-DNA/substrate is translocated via four discrete steps of sequential interactions with VirD4, VirB11, VirB6/VirB8 and VirB2/VirB9, as demonstrated by T-DNA immunoprecipitation assay (Cascales & Christie, 2004b). Biochemical approaches have identified subassemblies of VirB proteins constituting the ‘core’ components believed to form the translocation channel and the ‘pilus assembly’ complex comprising pilus components and associated factors (Krall et al., 2002; Yuan et al., 2005). In addition to transporting the T-complex and effector proteins from bacteria into plant cells, the VirB/D4 T4SS can translocate an incompatibility group Q (IncQ) plasmid RSF1010 from A. tumefaciens into plant cells (Buchanan-Wollaston et al., 1987) or between agrobacteria (Beijersbergen et al., 1992). Hitherto, little has been known about the contribution(s) of non-VirB proteins to the function of the T4SS; the work presented here suggests a role for the small heat-shock protein HspL.
We have previously used proteomics approaches to identify acetosyringone (AS)-induced proteins and discovered AS induction of HspL (Lai et al., 2006). HspL is an alpha-crystallin-type small heat-shock protein (α-Hsp) that contains a characteristic α-crystallin domain (Narberhaus, 2002). α-Hsps are a diverse protein family of low-molecular-mass chaperones that exist universally in most organisms, including animals, plants, bacteria and archaea. Rhizobiaceae contain a large number of α-Hsp genes, but little is known about their function except for the heat shock induction and chaperone-like activities of some of them; that is, they prevent model substrates from heat-induced aggregation (Munchbach et al., 1999a, b; Rosen et al., 2002; Studer & Narberhaus, 2000). In A. tumefaciens, there are at least four α-Hsp genes: hspC (atu0375) encoded on the circular chromosome, hspL (atu3887) encoded on the linear chromosome and hspAT1 (atu5052) and hspAT2 (atu5449), both encoded on the pAT plasmid (Balsiger et al., 2004). The latter three are induced by heat shock, and heat induction of hspL is regulated by rpoH, which encodes an alternative σ32-like transcription factor (Rosen et al., 2002; Balsiger et al., 2004)
In this study, we characterized the regulation of HspL and its function in the virulence of A. tumefaciens. The results indicate that AS-induced HspL protein accumulation is regulated in a VirB-dependent manner. Further molecular and functional analyses suggest that HspL protein is required for optimal VirB protein accumulation, which may be important for efficient VirB/D4-mediated DNA transfer and virulence.
METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. For vir gene induction, A. tumefaciens cells grown overnight at 25 °C in 523 broth (Kado & Heskett, 1970) with appropriate antibiotics were harvested by centrifugation (8000 g, 10 min) and resuspended in fresh I-medium (AB-MES, pH 5.5) (Lai & Kado, 1998) without antibiotics, at OD600 ∼0.1. After growth at 25 °C to OD600 ∼0.2, the cells were further cultured at 25 °C for different times in the presence of 200 μM acetosyringone (AS) (Sigma-Aldrich) (0.1 %, v/v, of 200 mM stock dissolved in DMSO) until harvesting. The controls were grown in the same conditions without any treatment or treated with DMSO, the solvent used to dissolve AS. The concentrations of antibiotics used were: 100 μg ampicillin (Ap) ml−1, 20 μg tetracycline (Tc) ml−1 and 10 μg gentamicin (Gm) ml−1 for Escherichia coli; 50 μg erythromycin (Em) ml−1, 50 μg rifampicin (Rm) ml−1, 250 μg spectinomycin (Sp) ml−1, 20 μg Tc ml−1 and 50 μg Gm ml−1 for A. tumefaciens.
Table 1.
Bacterial strains and plasmids
| Strain/plasmid | Relevant characteristics | Reference/source |
|---|---|---|
| A. tumefaciens | ||
| A136 | RmR, strain C58 cured of the pTiC58 plasmid | Watson et al. (1975) |
| A348 | RmR, A136 containing octopine-type Ti plasmid pTiA6 | Garfinkel et al. (1981) |
| PC1001–PC1011 | RmR, A348 derivatives each containing a virB gene deletion from pTiA6 | Berger & Christie (1994) |
| NT1RE | RmR EmR, C58 cured of its pTiC58 | Watson et al. (1975) |
| NT1RE-Sp | RmR EmR SpR, NT1RE containing spectinomycin-resistant gene (aadA) | This study |
| NT1RE(pJK270) | RmR EmR KmR/NmR; pJK270 is pTiC58TraC with Tn5 insertion in the T-DNA region without affecting virulence | Kao et al. (1982) |
| NT1RE(pJK107) | RmR EmR KmR/NmR, virA : : Tn5 in pTiC58TraC, virG mutant | Kao et al. (1982) |
| NT1RE(pJK105) | RmR EmR KmR/NmR, virD1 : : Tn5 in pTiC58TraC, virD1 polar mutant | Rogowsky et al. (1987) |
| NT1RE(pJK502) | RmR EmR KmR/NmR, virB3 : : Tn5 in pTiC58TraC, virB3 polar mutant | Lundquist et al. (1984) |
| NT1RE(pJK505) | RmR EmR KmR/NmR, virE : : Tn5 in pTiC58TraC, virE polar mutant | Rogowsky et al. (1987) |
| NT1RE(pJK702) | RmR EmR KmR/NmR, virC1 : : nptII in pTiC58TraC, virC1 mutant | Rogowsky et al. (1987) |
| NT1RE(pJK710) | RmR EmR KmR/NmR, virG : : nptII in pTiC58TraC, virG mutant | Kao et al. (1982) |
| NT1RE(pEL1000) | RmR EmR KmR/NmR, virB operon deletion in pJK270 | This study |
| EML770 | RmR EmR KmR/NmR GmR, hspL replaced by Gm cassette to generate hspL deletion mutant in NT1RE(pJK270) | This study |
| EML815 | RmR EmR KmR/NmR GmR TcR, pHspL in EML770 for complementation experiment | This study |
| EML1057 | RmR EmR KmR/NmR, markerless hspL deletion mutant in NT1RE(pJK270) | This study |
| EML1280 | RmR EmR KmR/NmR TcR, pHspL in EML1057 for complementation experiment | This study |
| E. coli | ||
| DH10B | Host for DNA cloning | Invitrogen |
| S-17 | Host for conjugation | Simon et al. (1983) |
| Plasmids | ||
| pGEMT-Easy | ApR, TA cloning vector | Promega |
| pJQ200KS | GmR, plasmid containing GmR and sacB gene for selection of double crossover | Quandt & Hynes (1993) |
| pUCG Ω1 | GmR, broad-host-range GmR cassettes for site-specific insertion | Schweizer (1993) |
| pGEMT-Sp | ApR SpR, pGEM-T-easy vector containing spectinomycin-resistance gene (aadA) cassette | Wu et al. (2008) |
| pET-22b(+) | ApR, an E. coli overexpression vector to generate C-terminal His-tagged protein | Novagen |
| pRU1064 | ApR TcR, stable broad-host-range promoter-probe vector containing gfpUV and gusA | Karunakaran et al. (2005) |
| pRU1156 | ApR TcR, stable broad-host-range promoter-probe vector containing gfpmut3.1 and gusA | Karunakaran et al. (2005) |
| pRUhspLp | ApR TcR, expression of PhspL-gfp transcriptional fusion on pRU1156 | This study |
| pRUhspCp | ApR TcR, expression of PhspC-gfp transcriptional fusion on pRU1156 | This study |
| pRUhspAT1p | ApR TcR, expression of PhspAT1-gfp transcriptional fusion on pRU1156 | This study |
| pRUhspAT2p | ApR TcR, expression of PhspAT2-gfp transcriptional fusion on pRU1156 | This study |
| pRUvirBp | ApR TcR, expression of PvirB-gfp transcriptional fusion on pRU1156 | This study |
| pRUhspLt | ApR TcR, expression of PhspL-gfp translational fusion protein HspLΔ4–160-GFP on pRU1156 | This study |
| pJM22 | KmR, vector for M2(Flag) epitope tagging | Janine Maddock |
| pEML649 | KmR, pJM22 containing sacB gene | This study |
| pEML651 | KmR GmR, up- and downstream fragments of hspL and G Ω cassette inserted at PstI/SalI site of pEML649 | This study |
| pEML652 | ApR TcR, pRU1064 digested with PstI to remove reporter gene (gfpUV and gusA) | Liu et al. (2008) |
| pEML776 | GmR, up- and downstream fragments of hspL inserted at PstI/SalI site of pJQ200KS | This study |
| pHspL | ApR TcR, expression of hspL gene containing its promoter and ORF on pEML652 | This study |
| pETHspL | ApR, overexpression of HspL-His in E. coli | This study |
| pML122ΔKm | IncQ plasmid RSF1010 derivative pML122 with removal of nptII gene | Labes et al. (1990)</xref> |
| pE1962 | TcR, plasmid for introducing gene into the pgl/picA locus in the chromosome of A. tumefaciens | Lee et al. (2001) |
| pE1962-Sp | TcR SpR, pE1962 containing SpR gene cassette | This study |
Plasmid construction.
Details of the primers used in this study are given in Supplementary Table S1, available with the online version of this paper. The techniques used for DNA cloning and PCR followed standard protocols (Sambrook & Russell, 2001). Plasmid DNA was isolated using the Plasmid Miniprep Purification kit provided by GeneMark. To construct plasmids for promoter activity assay, DNA fragments of the hspL, hspC, hspAT1, hspAT2 and virB promoter regions were amplified by PCR and digested with SpeI and HindIII prior to ligation into the promoter-probe vector pRU1156 at the same sites. The translational fusion between the first three amino acids of HspL and GFP (HspLΔ4–160-GFP) was constructed by ligating HindIII/SpeI-digested hspLt PCR product and SpeI/XbaI-digested gfp PCR product into the HindIII/XbaI site of pRU1156. To generate the hspL deletion mutant, the plasmids pEML651 and pEML776 were constructed for gene replacement experiments. Plasmid pEML651 was constructed by ligating PstI/EcoRI-digested HspL1 PCR product (upstream of hspL), EcoRI-digested GmR gene cassette and EcoRI/SalI-digested HspL2 PCR product (downstream of hspL), into the PstI/SalI sites of the suicide vector pEML649. Plasmid pEML649 was generated by ligating a BamHI-digested sacB PCR product into pJM22 at the BamHI site. Plasmid pEML776 was constructed by ligating PstI/EcoRI-digested HspL1 PCR product and EcoRI/SalI-digested HspL2 PCR product into the PstI/SalI sites of the suicide vector pJQ200KS. Plasmid pHspL was constructed by ligating a HindIII-digested HspL PCR product (containing promoter and ORF) into the same site of pEML652 for complementation test. To produce His-tagged HspL proteins, the DNA fragment containing the hspL ORF without the stop codon was amplified by PCR with specific primers, digested with NdeI and XhoI, and inserted at the same site of pET-22b(+) to result in plasmid pETHspL. The plasmid constructs obtained were confirmed by restriction mapping and DNA sequencing.
GFP quantification.
To quantify the GFP activities of A. tumefaciens cells expressing a gfp transcriptional or translational fusion, the bacterial cells were collected and normalized to OD600 0.2 with 0.9 % NaCl. A 100 μl cell suspension was loaded into a Nunc F96 MicroWell plate and analysed for GFP fluorescence with a multilabel plate reader (Chameleon; Hidex Ltd) at 535/485 nm for emission and excitation. Promoter activity was expressed as relative fluorescence units (RFU) after subtracting the fluorescence signal detected from a vector control strain and normalized at OD600 0.1.
Real-time RT-PCR.
A. tumefaciens strain NT1RE(pJK270) was grown for 16 h at 25 °C in I-medium with the addition of DMSO or AS. Total RNA was extracted (Zuber & Losick, 1983) and subjected to reverse transcription with SuperScript III RNase H− Reverse Transcriptase (Invitrogen) (Lai et al., 2006) with the appropriate 3′ primers (Supplementary Table S1). All primers were designed with the software primer express 2.0 (Applied Biosystems). PCR was performed in 25 μl SYBR Master Mix with 100 ng template cDNA and use of an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems) according to the methods described previously (Lin et al., 2008). To compare data from different PCR runs or cDNA samples, CT values for all target genes were normalized to the CT value of 16S rRNA, a constitutively expressed gene with approximately equal PCR efficiency in cells treated with AS or DMSO.
Gene replacement by double crossover.
Plasmid pEML651 was used to generate the hspL deletion mutant with replacement of the GmR gene cassette, and plasmid pEML776 was used to generate the markerless hspL deletion mutant in A. tumefaciens strain NT1RE. Plasmid pE1962-Sp was used to generate NT1RE-Sp, the recipient strain for RSF1010 conjugation assay, by transfer of the SpR gene cassette into the pgl/picA locus of A. tumefaciens strain NT1RE. A 5 μl volume of overnight culture (grown in LB broth without antibiotics) of E. coli strain S-17 containing the respective plasmid and A. tumefaciens strain NT1RE were mixed and incubated at 28 °C on LB agar overnight. The bacterial cells were then streaked out on LB agar containing Em, Rm and Km and incubated at 28 °C for 2 days to obtain the first crossover events. Three colonies were randomly selected and streaked out on the same selection medium for further colony purification. Each of three independent colonies was grown in 5 ml I-medium without antibiotics at 20 °C overnight; serial dilutions (up to 10−2) were plated onto 523 agar containing 5 % (w/v) sucrose and incubated at 20 °C for 4–7 days. The colonies were then selected for the respective antibiotic resistance and confirmed by colony PCR. The Ti plasmid pJK270 was transferred into the confirmed mutants by conjugation.
HspL antibody production.
The overexpression of HspL-His followed the instructions of the pET user manual (Novagen, EMD Biosciences). HspL-His was purified with use of Ni-NTA His Bind resins (Novagen), following the manufacturer's instructions. A 1 mg sample of purified HspL-His protein was separated by 15 % (w/v) glycine SDS-PAGE (Sambrook & Russell, 2001), followed by Coomassie brilliant blue R-250 staining (Sambrook & Russell, 2001). The major 19 kDa protein, corresponding to the putative HspL-His, was cut out for polyclonal antibody production in rabbits (GlycoNex, Taipei, Taiwan).
Western blot analysis.
Proteins were resolved by glycine SDS-PAGE (Sambrook & Russell, 2001) or Tricine SDS-PAGE (Schagger & von Jagow, 1987). Western blot analysis was performed as described previously (Lai & Kado, 1998) with use of primary polyclonal antibodies against HspL, VirB (Baron et al., 2001; Shirasu & Kado, 1993), VirD4 (Chen & Kado, 1996), VirE2 (Baron et al., 2001) and neomycin phosphotransferase II (NptII) (Sigma-Aldrich) followed by secondary antibody using horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (chemichem) and detection by the use of the Western Lightning System (Perkin Elmer). Chemiluminescent signals were visualized on X-ray film (Kodak).
Tumour assay on potato tuber discs.
Quantitative tumorigenesis assays with potato tuber discs were as described previously (Shurvinton & Ream, 1991; Wu et al., 2008) except that bacterial cells at OD600 0.4–0.6 were collected and resuspended in PBS at 108 and 107 c.f.u. ml−1 for inoculation. The potato tuber discs were placed on water agar, infected with 10 μl bacterial culture and incubated at 24 °C for 2 days. Discs were then placed on water agar supplemented with 100 μg timentin ml−1 to kill bacteria and incubated at 24 °C for 3 weeks before tumours were scored.
Conjugal transfer analysis of IncQ plasmid RSF1010.
The conjugation assay was as described by Fullner & Nester (1996) with minor modifications. The donor strains were NT1RE(pJK270) and its derivatives and the recipient strain was NT1RE-Sp. Cultures of donor and recipient strains were grown overnight at 25 °C in 523 broth with antibiotics. The cells of donor and recipient strains were collected by centrifugation (8000 g, 5 min) and resuspended in fresh I-medium without antibiotics to OD600 ∼0.1. After growth at 25 °C with shaking for 6 h, 200 μM AS was added to the cultures, which continued to grow at 25 °C for an additional 2 h. Donor and recipient cells were mixed together at a ratio of 10 : 1, and 10 μl of the mating mix was spotted on sterilized 1 cm2 nylon paper placed on I-medium agar in the presence of 200 μM AS. After incubation at 25 °C for 3 days, the cells from the nylon paper were resuspended in 1 ml 0.9 % NaCl. The bacterial suspensions with or without dilution were plated onto 523 agar supplemented with appropriate antibiotics and incubated at 28 °C for 2 days to select the transconjugants (EmR, GmR, SpR), input donors (EmR, GmR), and recipients (EmR, SpR). The number of transconjugants present in the selected input donors was ignored because of their low frequency (∼10−5) in the population.
RESULTS
AS-induced hspL expression
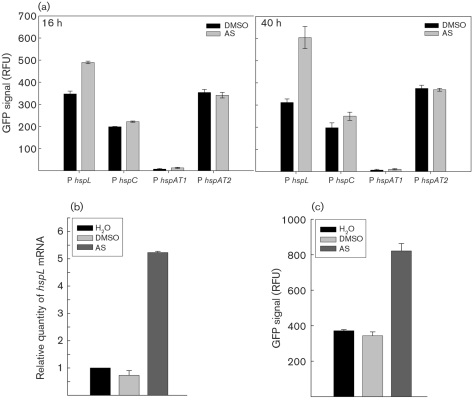
The small heat-shock protein HspL was previously identified as an AS-induced protein in A. tumefaciens (Lai et al., 2006). To determine whether AS also induces the three other α-Hsp genes (hspC, hspAT1, hspAT2), we analysed the promoter activity of each α-Hsp gene transcriptionally fused to gfp. The PhspL-gfp transcriptional fusion was upregulated 1.5- to 2-fold in cells grown in the presence of AS for 16 h or 24 h as compared with the non-induced (DMSO) controls (Fig. 1a). In contrast, the promoter activities of hspC, hspAT1 and hspAT2 were not affected by AS treatment. Quantitative RT-PCR, analysis of translational fusion and Western blot analysis were carried out to monitor the individual steps of hspL expression. We detected about 5- to 6-fold increased hspL mRNA level (Fig. 1b), 2-fold increase of HspLΔ4–160-GFP translational efficiency (Fig. 1c), and 50-fold higher HspL protein level (Fig. 2a) in cells grown in the presence of AS for 16 h in comparison to the DMSO or H2O controls. These data suggest that AS-induced transcription of hspL is a specific response rather than a general effect of AS treatment on heat-shock gene expression.
Fig. 1.
AS-induced hspL expression. (a) Relative GFP signal of A. tumefaciens strain NT1RE(pJK270) containing a gfp transcriptional fusion to the promoter of hspL, hspC, hspAT1 or hspAT2. The bacteria were grown at 25 °C for 16 or 40 h in I-medium with the addition of DMSO or AS. (b) Quantitative RT-PCR analysis of the hspL mRNA level of strain NT1RE(pJK270) and (c) relative GFP signal of strain NT1RE(pJK270) expressing HspLΔ4–160-GFP fusion protein (driven by its native promoter), grown at 25 °C for 16 h in I-medium without (H2O) or with DMSO or AS. The relative GFP signals for promoter activity are shown as the mean±sd of three independent experiments.
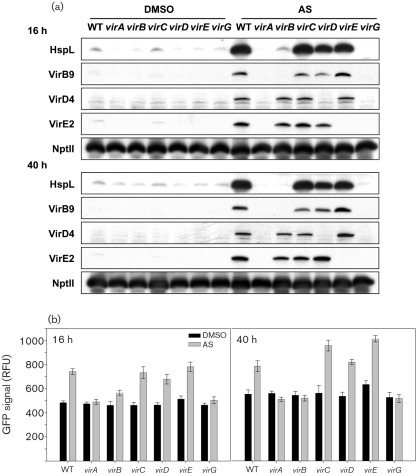
Fig. 2.
AS-induced HspL protein accumulation is regulated in a VirB-dependent manner. (a) Western blot analysis of HspL, VirB9, VirD4, VirE2 and NptII when wild-type and different vir mutants were grown at 25 °C for 16 or 40 h in I-medium with addition of DMSO or AS. (b) Relative GFP signals of A. tumefaciens strains containing plasmid pRUhspLp, expressing the PhspL-gfp transcriptional fusion, grown at 25 °C for 16 or 40 h in I-medium with DMSO or AS. Strains: WT, NT1RE(pJK270); virA, NT1RE(pJK107); virB, NT1RE(pJK502); virC, NT1RE(pJK702); virD, NT1RE(pJK105); virE, NT1RE(pJK505); virG, NT1RE(pJK710). The relative GFP signals for promoter activity are shown as the mean±sd of three independent experiments. Western blotting was performed for at least three independent experiments with similar results. NptII protein levels were determined as the controls.
HspL protein accumulation is induced by AS in a VirB protein-dependent manner
Similar to other known Vir proteins that are regulated by the VirA/VirG two-component system, AS-induced HspL accumulation is also dependent on virA and virG (Lai et al., 2006). Interestingly, in contrast to the presence of VirG-binding sequences (vir box) in the promoters of known vir regulon genes (Cho & Winans, 2005), the apparent absence of a vir box within the putative hspL promoter region suggests that hspL is not directly activated by the VirG response regulator. To address this question, the PhspL-gfp transcriptional fusion construct was transformed into different vir mutants. As expected, hspL promoter activity was upregulated by AS in the wild-type and the virC, virD and virE mutants, but not in the virA or virG mutants, after either 16 h or 40 h AS induction (Fig. 2b). Surprisingly, hspL promoter activity was also compromised in a polar virB3 mutant (Fig. 2b) and the steady-state level of AS-induced hspL mRNA was also diminished in the virB3 mutant (data not shown). Furthermore, HspL protein levels were increased by AS in the wild-type and the virC, virD and virE mutants but not in the polar virB3 mutant (Fig. 2a). The VirA/VirG two-component system is still functional in the virB3 polar mutant because other Vir proteins such as VirD4 and VirE2 were still induced by AS (Fig. 2a). Taken together, these data suggest that AS-induced hspL transcription is not directly activated by the response regulator VirG but rather is linked to the expression of virB genes.
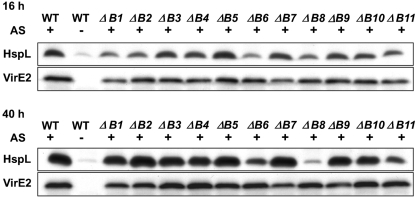
To determine which virB gene(s) are responsible for AS-induced HspL protein accumulation, individual virB nonpolar deletion mutants were analysed, and the HspL protein was detected by Western blot analysis. Reduced levels of HspL protein were observed in the virB1 and virB2 mutants, but only at 16 h after AS induction (Fig. 3). More clearly, after 16 h and 40 h of AS induction, HspL protein levels were reduced in virB6, virB8 and virB11 deletion strains as compared with the wild-type and the other virB mutants. VirE2 levels, as a control, were normal in these strains. These data suggest that HspL protein accumulation is likely induced by the expression of one or a subset of VirB proteins.
Fig. 3.
HspL protein accumulation is compromised in the virB6, virB8 and virB11 nonpolar deletion mutants: Western blot analysis of HspL and VirE2 in A. tumefaciens strains containing octopine-type Ti plasmid pTiA6 and its variants. A348, the wild-type strain, was treated with AS (+) or DMSO (−) and each of the virB non-polar deletion mutants (ΔB1–ΔB11 represent deletions of virB1 to virB11) induced by AS were determined. Western blotting was performed for at least three independent experiments with similar results. VirE2 protein levels were determined as the controls.
The absence of HspL causes reduced VirB protein accumulation at an early stage of AS induction without affecting virB transcription
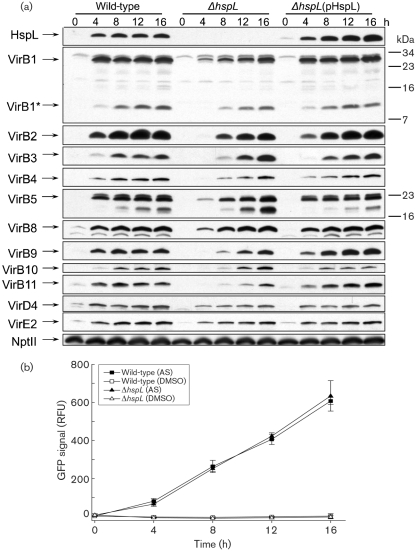
We investigated the physiological role of HspL protein expression and accumulation in response to certain VirB proteins. Small heat-shock proteins are generally thought to bind selectively to non-native proteins to prevent their aggregation and degradation (Narberhaus, 2002; Sun & MacRae, 2005). Thus, HspL might play a role as a chaperone in stabilizing VirB proteins and thereby contribute to efficient T-DNA transfer and tumorigenesis. To test this hypothesis, we determined the effect of deletion of hspL on VirB protein accumulation. The loss of HspL in the deletion mutant and its complementation by expression of hspL (driven by its native promoter) on an IncP plasmid was demonstrated by Western blotting (Fig. 4a). The level of all VirB proteins analysed was lower in the hspL deletion mutant than in the wild-type soon after AS induction (4 h and 8 h). In contrast, levels of other AS-induced proteins such as VirD4 and VirE2 and the internal control protein NptII were not substantially reduced, which suggests that AS-induced HspL acts mainly on VirB proteins. The effects were complemented by hspL expression in trans (Fig. 4a). In contrast, hspL plays no role in AS-induced virB transcription because the virB promoter was induced by AS at similar levels in both the wild-type and the hspL mutant up to 16 h (Fig. 4b). Taken together, the data suggest that HspL plays a role in optimal VirB protein accumulation likely via maintaining their stability during the assembly process.
Fig. 4.
The absence of HspL causes reduced VirB protein accumulation at early stages of AS induction without affecting virB operon transcription. (a) Wild-type NT1RE(pJK270), ΔhspL (hspL deletion mutant, EML770) and ΔhspL(pHspL) (complemented strain, EML815) were grown in I-medium at 25 °C in the presence of AS to induce vir gene expression. The total cell lysates were subjected to Tricine-SDS-PAGE followed by Western blot analysis. Numbers on the right are molecular masses of reference proteins in kDa. NptII served as an internal control. At least three independent experiments were carried out with similar results. (b) Relative GFP signal of A. tumefaciens NT1RE(pJK270) or ΔhspL (hspL deletion mutant, EML770) containing plasmid pRUvirBp expressing the PvirB-gfp transcriptional fusion. The bacteria were grown at 25 °C in I-medium with the addition of DMSO or AS and collected at different times for GFP quantification. The relative GFP signals for hspL promoter activity are shown as the mean±sd of at least three independent experiments.
The absence of HspL causes reduced VirB/D4-mediated DNA transfer and tumorigenesis efficiency
In view of the reduced VirB protein accumulation in the hspL deletion mutant, we were curious to determine whether HspL is involved in the VirB/D4-mediated DNA transfer and virulence of A. tumefaciens. An IncQ plasmid, RSF1010, can be transferred between strains of A. tumefaciens by the Ti plasmid-encoded VirB/D4 machinery in an AS-induced environment (Beijersbergen et al., 1992). We used this alternative functional T4SS assay to determine whether hspL contributes to the conjugal transfer using the RSF1010-derived plasmid pML122ΔKm. The transfer efficiency was consistently reduced, by 30 % on average, in the hspL mutant as compared with the wild-type (Table 2). The mobilization ability of the hspL deletion mutant was restored and even increased by complementation with an hspL-expressing plasmid, which indicates that HspL contributes to efficient mobilization of pML122ΔKm between A. tumefaciens strains. The observed mobilization of pML122ΔKm was indeed mediated by the Ti VirB/D4 T4SS because no transconjugants were detected in the mutant with deletion of the entire virB operon (Table 2) or when the conjugation experiment was carried out in the absence of AS (data not shown). We also determined whether the deletion of hspL affects the function of another Ti plasmid-encoded T4SS, the trb locus, which mediates the conjugal transfer of the Ti plasmid between agrobacteria (Li et al., 1998; von Bodman et al., 1989). The conjugal transfer efficiency of pTiC58-derived pJK270 was not affected in the hspL deletion mutant or in its complemented strain as compared with in the wild-type (data not shown), which suggests that HspL is not a general factor involved in DNA and plasmid transfer. Thus, HspL promotes RSF1010 conjugal transfer, which further substantiates its role in Ti plasmid-encoded VirB/D4 T4SS-dependent function.
Table 2.
Effect of hspL on mobilization of pML122ΔKm in A. tumefaciens
| Donor strain | Relevant genotype | Conjugation frequency (%)* | ||
|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 3 | ||
| NT1RE(pEL1000) | virB operon deletion mutant | <4.35×10−8 (<0.27) | <3.61×10−8 (<0.29) | <4.14×10−8(<0.21) |
| NT1RE(pJK270) | Wild-type | 1.63×10−5 (100) | 1.26×10−5 (100) | 1.96×10−5 (100) |
| EML1057 | ΔhspL | 1.08×10−5 (66.02) | 7.96×10−6 (63.2) | 1.56×10−5 (79.3) |
| EML1280 | ΔhspL(pHspL) | 2.98×10−5 (182.25) | 1.64×10−5 (130.2) | 4.01×10−5 (204.0) |
*The conjugation frequency is expressed as number of transconjugants per input donor.
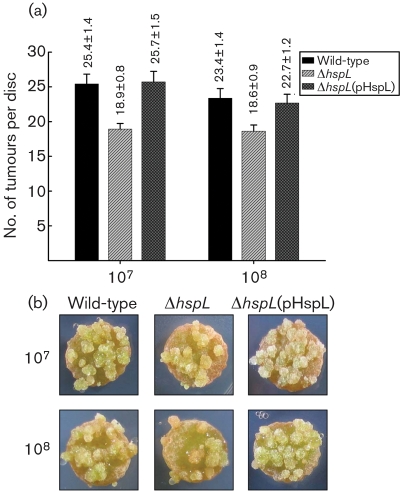
To determine the effects on A. tumefaciens virulence, quantitative tumorigenesis assays on potato tuber discs were used to determine the effect of HspL on tumour formation. The tumorigenesis efficiency was consistently 20–25 % lower in the hspL mutant than in the wild-type with inoculation of 107 and 108 c.f.u. ml−1 of bacterial cells (Fig. 5). Complementation of hspL from a plasmid in the hspL deletion mutant restored tumour formation to wild-type levels, which indicated that the attenuated virulence of the hspL mutant was caused specifically by the loss of hspL (Fig. 5). The reduced tumorigenesis efficiency caused by the deletion of hspL was also observed in infected Arabidopsis thaliana roots (Supplementary Fig. S1). These data strongly suggest that HspL contributes to promote VirB/D4-mediated DNA transfer and disease development.
Fig. 5.
Quantitative tumorigenesis assay on potato tuber discs. (a) Wild-type NT1RE(pJK270), ΔhspL (hspL deletion mutant, EML770) and ΔhspL(pHspL) (complemented strain, EML815) were examined for their tumorigenesis efficiency on potato tuber discs by inoculation at 108 and 107 c.f.u. ml−1. Tumorigenesis efficiency is expressed as the number of tumours per disc (mean±se, calculated from results of 60 potato tuber discs for each analysed strain in each independent experiment). (b) Representative results; at least four independent experiments were carried out with similar results.
DISCUSSION
Most of the components required for conjugal DNA transfer and tumour formation by A. tumefaciens are encoded on the Ti plasmid. In this study, we demonstrated that the chromosomally encoded small heat-shock protein HspL is induced upon expression of certain VirB proteins, the major components of the Ti VirB/D4 T4SS required for gene transfer to plants in A. tumefaciens. Our genetic and functional evidence suggests a role for HspL in promoting VirB protein stability, VirB/D4-mediated DNA transfer and tumorigenesis.
The VirB-induced HspL expression resembles the induction of heat-shock proteins and proteases via the extracytoplasmic (or envelope) stress responses observed in many Gram-negative bacteria (Raivio, 2005; Rowley et al., 2006). The known envelope stress response is regulated via the CpxAR two-component regulatory system or the alternative sigma factor σE (Raivio, 2005). In E. coli, the expression and assembly of functional plasmid R1-determined T4SS pili and of type IV bundle-forming pili were found to elicit the envelope stress responses via the CpxAR two-component regulatory system (Nevesinjac & Raivio, 2005; Zahrl et al., 2006). The discovery of VirB-induced HspL in this study and the identification of Cpx regulation of α-Hsp genes ibpA and ibpB in E. coli (Lau-Wong et al., 2008) suggest the involvement of α-Hsps in envelope stress responses. To our knowledge, however, HspL is the first α-Hsp demonstrated to be involved in the function of a protein secretion system. The mechanism of hspL induction is not clear because CpxAR components could not be identified in the A. tumefaciens C58 genome based on blast analysis. RpoH (σ32) is required for heat-shock-induced HspL protein accumulation (Rosen et al., 2002) and likely regulated at the transcriptional level due to the presence of an RpoH-dependent promoter of hspL (Balsiger et al., 2004). However, RpoH was not essential for AS-induced HspL protein accumulation under non-heat-shock conditions because the HspL protein level accumulated to the wild-type level in the A. tumefaciens rpoH mutant after AS induction at 25 °C (data not shown). Therefore, the expression of certain VirB proteins may trigger an as yet unknown regulator(s) that upregulate(s) hspL expression in A. tumefaciens under non-heat-shock conditions.
Although the assembly of pili by the IncFII plasmid R1 T4SS triggered the envelope stress response and a decreased response was observed in a traA pilin mutant (Zahrl et al., 2006), the exact T4SS component that mediates its induction is unknown. Our data indicate that AS-induced HspL protein accumulates to the wild-type level in most of the single virB deletion mutants after 40 h induction (Fig. 3), which suggests that T4SS-induced HspL protein accumulation requires neither the presence of an intact secretion system nor the formation of the T pilus. The evidence that the HspL protein level was markedly reduced in the virB6, virB8 and virB11 deletion mutants suggests that HspL protein accumulation may be triggered by one or a subset of T4SS components. We noticed that VirB8 and VirB11 protein levels were reduced in the virB6 deletion mutant (data not shown). Since deletion of virB6 had a negative effect on downstream gene expression (virB7–virB11) (Liu & Binns, 2003), it remains to be determined whether the requirement of VirB6 in triggering HspL protein accumulation is direct or indirect. VirB6, VirB8 and VirB11 are inner-membrane components directly involved in the T-DNA/substrate translocation pathway (Cascales & Christie, 2004b); however, T-DNA translocation through this T4SS channel is not required for HspL induction because the VirD4 coupling protein is dispensable for this effect (Fig. 2a, b). Interestingly, the deletion of virB8 caused the greatest decrease in HspL protein level (Fig. 3). Because VirB8 is an assembly factor that may initiate T4SS assembly (Baron, 2006), we speculate that hspL transcription and its protein accumulation may be triggered by the formation of the early subassembly complex.
Although the VirB protein level was clearly reduced, we did not observe effects on virB transcription in the hspL deletion mutant as compared with the wild-type (Fig. 4), which suggests that HspL may function as a VirB chaperone. Interestingly, although hspL seems to be expressed at a basal level and is upregulated only about twofold by AS at the transcriptional level, based on our promoter activity assay (Fig. 1a) and microarray data (Anand et al., 2008), HspL protein is barely detectable in the absence of AS but accumulates markedly – about 50-fold – upon AS induction, in a VirB protein-dependent manner (Fig. 2a). The twofold upregulation of the HspLΔ4–160-GFP translational fusion by AS (Fig. 1c) further suggests a post-translational regulation of AS-induced HspL accumulation. Both a chaperone and its interacting substrate become stabilized when they interact with each other (Narberhaus, 2002; Sun & MacRae, 2005). Thus, we speculate that HspL protein might be stabilized when interacting with its substrates such as VirB proteins and may be rapidly degraded in the absence of its substrate. Likewise, the substrates may be more susceptible to proteolysis in the absence of the chaperone. We are currently investigating whether HspL interacts directly with VirB protein(s) and functions as a VirB chaperone to prevent VirB from aggregation and degradation, thereby maintaining the stability and/or functionality of the individual VirB proteins and/or the assembled T4SS complexes.
The discovery of HspL as a non-VirB factor contributing to T4SS protein stability is novel, but most importantly, the decreased VirB protein accumulation in the absence of HspL also correlates with the reduced tumorigenesis efficiency of the hspL mutant as compared with the wild-type (Fig. 5, Supplementary Fig. S1). Obviously HspL plays a specific role for the Ti VirB/D4 T4SS because VirB/D4-mediated RSF1010 transfer but not Trb-mediated Ti plasmid transfer between agrobacteria was decreased in the absence of HspL (Table 2). This specificity was further supported by evidence that hspL but none of the other three α-Hsp genes (hspC, hspAT1 and hspAT2) was upregulated by AS (Fig. 1a) and no deleterious effects on growth or membrane lipid composition were detected in the absence of hspL (data not shown). However, one may argue that HspL does not contribute an essential function for A. tumefaciens virulence because the reduced VirB protein accumulation and tumorigenesis efficiency was not as drastic in the hspL mutant as the wild-type (Fig. 4a; compare with Fig. 5, Supplementary Fig. S1). Functional redundancy of α-Hsps was found in E. coli, in which the simultaneous presence of α-Hsps IbpA and IbpB enhanced the stabilization of thermally aggregated proteins as compared with the presence of IbpA or IbpB alone (Matuszewska et al., 2005). Thus, it is possible that the basal-level expression of the other three α-Hsps may partially substitute for the function of HspL in VirB protein stability and T4SS function in the absence of HspL. Examining the effect on the stability of VirB proteins/complexes, VirB/D4-mediated DNA transfer and tumorigenesis in single or multiple α-Hsp mutants would be an interesting future study.
In general, the expression of bacterial α-Hsp genes is low or undetectable under normal growth conditions but is induced to high levels under heat shock or other stress conditions (Narberhaus, 2002). The induction of α-Hsp genes was previously reported during bacterial infection with the human pathogens Mycobacterium tuberculosis and Mycobacterium leprae. The α-Hsp genes acr1 and acr2 are induced in M. tuberculosis-infected macrophages and acr2 was further demonstrated to be required for the pathogenicity (Stewart et al., 2005, 2006; Wilkinson et al., 2005). The expression of another α-Hsp gene, shsp18, encoding a surface-exposed antigen of M. leprae (Ilangumaran et al., 1994; Lini et al., 2008), was activated in macrophages (Dellagostin et al., 1995). Our findings that the phytopathogen A. tumefaciens exploits the VirB-induced HspL expression to promote its tumorigenesis add to the list of α-Hsp participation in bacterial virulence. Further study could explore the importance of small heat-shock proteins in the virulence of other bacterial pathogens and elucidate the molecular mechanisms underlying their regulation and involvement in their infection processes under non-heat-shock conditions.
Acknowledgments
We are grateful to Clarence Kado for providing several A. tumefaciens strains and antibodies of VirB2, VirB9 and VirD4. We also thank Peter Christie for providing A348-derived virB mutants. Acknowledgements also go to Shu-Hsing Wu, Kuo-Chen Yeh and the lab members for discussion and technical assistance. This work was supported by the Taiwan National Science Council (NSC) grant NSC 95-2320-B-001-009 to E.-M. L. and an NSC postdoctoral fellowship to Y.-L. T, by grant MOP-84239 from the Canadian Institutes of Health Research (CIHR) to C. B., and a DFG grant (German Research Foundation, SFB 480) to F. N.
Abbreviations
AS, acetosyringone
α-Hsp, α-crystallin-type small heat-shock protein
RFU, relative fluorescence units
T4SS, type IV secretion system
T-DNA, transferred DNA
Ti, tumour-inducing (plasmid)
Footnotes
A supplementary table of primers and a supplementary figure showing the reduced tumorigenesis efficiency of the hspL mutant are available with the online version of this paper.
References
- Anand, A., Uppalapati, S. R., Ryu, C. M., Allen, S. N., Kang, L., Tang, Y. & Mysore, K. S. (2008). Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol 146, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri, K., Cascales, E. & Christie, P. J. (2004). Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol 54, 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsiger, S., Ragaz, C., Baron, C. & Narberhaus, F. (2004). Replicon-specific regulation of small heat shock genes in Agrobacterium tumefaciens. J Bacteriol 186, 6824–6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, C. (2006). VirB8: a conserved type IV secretion system assembly factor and drug target. Biochem Cell Biol 84, 890–899. [DOI] [PubMed] [Google Scholar]
- Baron, C., Domke, N., Beinhofer, M. & Hapfelmeier, S. (2001). Elevated temperature differentially affects virulence, VirB protein accumulation, and T-pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J Bacteriol 183, 6852–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijersbergen, A., Dulk-Ras, A. D., Schilperoort, R. A. & Hooykaas, P. J. (1992). Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256, 1324–1327. [DOI] [PubMed] [Google Scholar]
- Berger, B. R. & Christie, P. J. (1994). Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol 176, 3646–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V., Passiatore, J. E. & Cannon, F. (1987). The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature 328, 172–175. [Google Scholar]
- Cascales, E. & Christie, P. J. (2004a). Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci U S A 101, 17228–17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales, E. & Christie, P. J. (2004b). Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304, 1170–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. Y. & Kado, C. I. (1996). Osa protein encoded by plasmid pSa is located at the inner membrane but does not inhibit membrane association of VirB and VirD virulence proteins in Agrobacterium tumefaciens. FEMS Microbiol Lett 135, 85–92. [DOI] [PubMed] [Google Scholar]
- Cho, H. & Winans, S. C. (2005). VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc Natl Acad Sci U S A 102, 14843–14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, P. J. (2001). Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol Microbiol 40, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, P. J., Atmakuri, K., Krishnamoorthy, V., Jakubowski, S. & Cascales, E. (2005). Biogenesis, architecture, and function of bacterial type iv secretion systems. Annu Rev Microbiol 59, 451–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellagostin, O. A., Esposito, G., Eales, L. J., Dale, J. W. & McFadden, J. (1995). Activity of mycobacterial promoters during intracellular and extracellular growth. Microbiology 141, 1785–1792. [DOI] [PubMed] [Google Scholar]
- Fullner, K. J. & Nester, E. W. (1996). Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J Bacteriol 178, 1498–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel, D. J., Simpson, R. B., Ream, L. W., White, F. F., Gordon, M. P. & Nester, E. W. (1981). Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27, 143–153. [DOI] [PubMed] [Google Scholar]
- Ilangumaran, S., Shankernarayan, N., Ramu, G. & Muthukkaruppan, V. (1994). Antibody response to recombinant 65-kDa, 70-kDa and 18-kDa mycobacterial antigens in leprosy patients and healthy contacts in a leprosy-endemic population. Int J Lepr Other Mycobact Dis 62, 245–255. [PubMed] [Google Scholar]
- Judd, P. K., Kumar, R. B. & Das, A. (2005). Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc Natl Acad Sci U S A 102, 11498–11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado, C. I. & Heskett, M. G. (1970). Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopathology 60, 969–976. [DOI] [PubMed] [Google Scholar]
- Kao, J. C., Perry, K. L. & Kado, C. I. (1982). Indoleacetic acid complementation and its relation to host range specifying genes on the Ti plasmid of Agrobacterium tumefaciens. Mol Gen Genet 188, 425–432. [DOI] [PubMed] [Google Scholar]
- Karunakaran, R., Mauchline, T. H., Hosie, A. H. & Poole, P. S. (2005). A family of promoter probe vectors incorporating autofluorescent and chromogenic reporter proteins for studying gene expression in Gram-negative bacteria. Microbiology 151, 3249–3256. [DOI] [PubMed] [Google Scholar]
- Krall, L., Wiedemann, U., Unsin, G., Weiss, S., Domke, N. & Baron, C. (2002). Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 99, 11405–11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labes, M., Puhler, A. & Simon, R. (1990). A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene 89, 37–46. [DOI] [PubMed] [Google Scholar]
- Lai, E. M. & Kado, C. I. (1998). Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol 180, 2711–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E. M. & Kado, C. I. (2000). The T-pilus of Agrobacterium tumefaciens. Trends Microbiol 8, 361–369. [DOI] [PubMed] [Google Scholar]
- Lai, E. M., Shih, H. W., Wen, S. R., Cheng, M. W., Hwang, H. H. & Chiu, S. H. (2006). Proteomic analysis of Agrobacterium tumefaciens response to the vir gene inducer acetosyringone. Proteomics 6, 4130–4136. [DOI] [PubMed] [Google Scholar]
- Lau-Wong, I. C., Locke, T., Ellison, M. J., Raivio, T. L. & Frost, L. S. (2008). Activation of the Cpx regulon destabilizes the F plasmid transfer activator, TraJ, via the HslVU protease in Escherichia coli. Mol Microbiol 67, 516–527. [DOI] [PubMed] [Google Scholar]
- Lee, L. Y., Humara, J. M. & Gelvin, S. B. (2001). Novel constructions to enable the integration of genes into the Agrobacterium tumefaciens C58 genome. Mol Plant-Microbe Interact 14, 577–579. [DOI] [PubMed] [Google Scholar]
- Li, P. L., Everhart, D. M. & Farrand, S. K. (1998). Genetic and sequence analysis of the pTiC58 trb locus, encoding a mating-pair formation system related to members of the type IV secretion family. J Bacteriol 180, 6164–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. Z., Chen, H. Y., Kao, R., Chang, S. P., Chang, S. J. & Lai, E. M. (2008). Proteomic analysis of rice defense response induced by probenazole. Phytochemistry 69, 715–728. [DOI] [PubMed] [Google Scholar]
- Lini, N., Rehna, E. A., Shiburaj, S., Maheshwari, J. J., Shankernarayan, N. P. & Dharmalingam, K. (2008). Functional characterization of a small heat shock protein from Mycobacterium leprae. BMC Microbiol 8, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. & Binns, A. N. (2003). Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J Bacteriol 185, 3259–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, A. C., Shih, H. W., Hsu, T. & Lai, E. M. (2008). A citrate-inducible gene, encoding a putative tricarboxylate transporter, is downregulated by the organic solvent DMSO in Agrobacterium tumefaciens. J Appl Microbiol 105, 1372–1383. [DOI] [PubMed] [Google Scholar]
- Lundquist, R. C., Close, T. J. & Kado, C. I. (1984). Genetic complementation of Agrobacterium tumefaciens Ti plasmid mutants in the virulence region. Mol Gen Genet 193, 1–7. [DOI] [PubMed] [Google Scholar]
- Matuszewska, M., Kuczynska-Wisnik, D., Laskowska, E. & Liberek, K. (2005). The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J Biol Chem 280, 12292–12298. [DOI] [PubMed] [Google Scholar]
- McCullen, C. A. & Binns, A. N. (2006). Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu Rev Cell Dev Biol 22, 101–127. [DOI] [PubMed] [Google Scholar]
- Munchbach, M., Dainese, P., Staudenmann, W., Narberhaus, F. & James, P. (1999a). Proteome analysis of heat shock protein expression in Bradyrhizobium japonicum. Eur J Biochem 264, 39–48. [DOI] [PubMed] [Google Scholar]
- Munchbach, M., Nocker, A. & Narberhaus, F. (1999b). Multiple small heat shock proteins in rhizobia. J Bacteriol 181, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus, F. (2002). Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev 66, 64–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevesinjac, A. Z. & Raivio, T. L. (2005). The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J Bacteriol 187, 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt, J. & Hynes, M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21. [DOI] [PubMed] [Google Scholar]
- Raivio, T. L. (2005). Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol 56, 1119–1128. [DOI] [PubMed] [Google Scholar]
- Rashkova, S., Zhou, X. R., Chen, J. & Christie, P. J. (2000). Self-assembly of the Agrobacterium tumefaciens VirB11 traffic ATPase. J Bacteriol 182, 4137–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowsky, P. M., Close, T. J., Chimera, J. A., Shaw, J. J. & Kado, C. I. (1987). Regulation of the vir genes of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol 169, 5101–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, R., Buttner, K., Becher, D., Nakahigashi, K., Yura, T., Hecker, M. & Ron, E. Z. (2002). Heat shock proteome of Agrobacterium tumefaciens: evidence for new control systems. J Bacteriol 184, 1772–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley, G., Spector, M., Kormanec, J. & Roberts, M. (2006). Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol 4, 383–394. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. (2001). Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Schagger, H. & von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Schweizer, H. D. (1993). Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15, 831–834. [PubMed] [Google Scholar]
- Shirasu, K. & Kado, C. I. (1993). Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol Lett 111, 287–294. [DOI] [PubMed] [Google Scholar]
- Shurvinton, C. E. & Ream, W. (1991). Stimulation of Agrobacterium tumefaciens T-DNA transfer by overdrive depends on a flanking sequence but not on helical position with respect to the border repeat. J Bacteriol 173, 5558–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, R., Priefer, U. & Puhler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1, 784–791. [Google Scholar]
- Stewart, G. R., Newton, S. M., Wilkinson, K. A., Humphreys, I. R., Murphy, H. N., Robertson, B. D., Wilkinson, R. J. & Young, D. B. (2005). The stress-responsive chaperone alpha-crystallin 2 is required for pathogenesis of Mycobacterium tuberculosis. Mol Microbiol 55, 1127–1137. [DOI] [PubMed] [Google Scholar]
- Stewart, J. N., Rivera, H. N., Karls, R., Quinn, F. D., Roman, J. & Rivera-Marrero, C. A. (2006). Increased pathology in lungs of mice after infection with an alpha-crystallin mutant of Mycobacterium tuberculosis: changes in cathepsin proteases and certain cytokines. Microbiology 152, 233–244. [DOI] [PubMed] [Google Scholar]
- Studer, S. & Narberhaus, F. (2000). Chaperone activity and homo- and hetero-oligomer formation of bacterial small heat shock proteins. J Biol Chem 275, 37212–37218. [DOI] [PubMed] [Google Scholar]
- Sun, Y. & MacRae, T. H. (2005). Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci 62, 2460–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bodman, S. B., McCutchan, J. E. & Farrand, S. K. (1989). Characterization of conjugal transfer functions of Agrobacterium tumefaciens Ti plasmid pTiC58. J Bacteriol 171, 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, D. V., Draper, O., Zupan, J. R. & Zambryski, P. C. (2002). Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci U S A 99, 11493–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, B., Currier, T. C., Gordon, M. P., Chilton, M. D. & Nester, E. W. (1975). Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol 123, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, K. A., Stewart, G. R., Newton, S. M., Vordermeier, H. M., Wain, J. R., Murphy, H. N., Horner, K., Young, D. B. & Wilkinson, R. J. (2005). Infection biology of a novel alpha-crystallin of Mycobacterium tuberculosis: Acr2. J Immunol 174, 4237–4243. [DOI] [PubMed] [Google Scholar]
- Wu, H. Y., Chung, P. C., Shih, H. W., Wen, S. R. & Lai, E. M. (2008). Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J Bacteriol 190, 2841–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Q., Carle, A., Gao, C., Sivanesan, D., Aly, K. A., Hoppner, C., Krall, L., Domke, N. & Baron, C. (2005). Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J Biol Chem 280, 26349–26359. [DOI] [PubMed] [Google Scholar]
- Zahrl, D., Wagner, M., Bischof, K. & Koraimann, G. (2006). Expression and assembly of a functional type IV secretion system elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli. J Bacteriol 188, 6611–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber, P. & Losick, R. (1983). Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell 35, 275–283. [DOI] [PubMed] [Google Scholar]