Abstract
Transcription factors represent one of the largest groups of proteins regulated by SUMO (small ubiquitin-like modifier) modification, and their sumoylation is usually associated with transcriptional repression. To investigate whether sumoylation plays a general role in regulating transcription in yeast, we determined the occupancy of sumoylated proteins at a variety of genes by chromatin immunoprecipitation (ChIP) using an antibody that recognizes the yeast SUMO peptide. Surprisingly, we detected sumoylated proteins at all constitutively transcribed genes tested but not at repressed genes. Ubc9, the SUMO conjugation enzyme, was not present on these genes, but its inactivation reduced SUMO at the constitutive promoters and modestly decreased RNA polymerase II levels. In contrast, activation of the inducible GAL1, STL1, and ARG1 genes caused not only a striking accumulation of SUMO at all three promoter regions, but also recruitment of Ubc9, indicating that gene activation involves sumoylation of promoter-bound factors. However, Ubc9 inactivation, while reducing sumoylation at the induced promoters, paradoxically resulted in increased transcription. Providing an explanation for this, the reduced sumoylation impaired the cell's ability to appropriately shut off transcription of the induced ARG1 gene, indicating that SUMO can facilitate transcriptional silencing. Our findings thus establish unexpected roles for sumoylation in both constitutive and activated transcription, and provide a novel mechanism for regulating gene expression.
Keywords: SUMO, sumoylation, Smt3, transcription, activation, deactivation
Protein sumoylation involves the covalent and reversible attachment of the small ubiquitin-like modifier (SUMO) peptide to lysine side chains in acceptor proteins. Mechanistically similar to ubiquitylation, this involves a cascade of enzymatic activities, including activation by an E1 enzyme (Aos1/Uba2) and conjugation to target proteins by a sole E2 enzyme (Ubc9), guided to appropriate substrates by the SUMO E3 ligases (Johnson 2004; Geiss-Friedlander and Melchior 2007). Budding yeast express a single SUMO isoform, encoded by the essential SMT3 gene, whereas three main isoforms are found in mammalian cells: SUMO1, and the highly similar SUMO2 and SUMO3. At the molecular level, attachment of the SUMO peptide to a substrate can promote its association with interacting proteins through recognition of its SUMO-modified form, or the SUMO moiety can interfere with protein–protein interactions by blocking interaction sites. The consequences of altered protein–protein interactions through sumoylation are diverse, and include changes in subcellular localization, protein activity, and protein stability (Geiss-Friedlander and Melchior 2007).
SUMO orthologs have been identified in all eukaryotic species, and sumoylation modifies proteins involved in a wide range of cellular processes, indicating that regulation by sumoylation is widespread (Zhao 2007; Makhnevych et al. 2009). Gene expression, however, appears to be particularly regulated by sumoylation, because a large number of known SUMO conjugates in yeast and mammals are transcription factors (Gill 2005; Zhao 2007; Makhnevych et al. 2009). Blocking sumoylation of gene-specific transcription factors C/EBP (Kim et al. 2002), c-Jun (Muller et al. 2000), ELK-1 (Yang et al. 2003), and many others (Girdwood et al. 2004; Gill 2005) through mutation of SUMO acceptor sites results in increased transcription of target genes. Consequently, sumoylation is generally associated with transcriptional repression. This is supported by other findings, including observations that transcriptional corepressors, including histone deacetylase complexes (HDACs), preferentially associate with sumoylated forms of transcription factors (Garcia-Dominguez and Reyes 2009; Ouyang and Gill 2009). For example, the coactivator p300 associates with HDAC6 in a SUMO-dependent manner (Girdwood et al. 2003). Additionally, human histone H4 is sumoylated (Shiio and Eisenman 2003), as are all four core histones in yeast (Nathan et al. 2006). Through mutations that attenuate sumoylation, or by the use of histone-SUMO fusion proteins, it was shown that histone sumoylation represses transcription (Nathan et al. 2006). This is thought to occur through the recruitment of HDACs by SUMO-modified histones, or by interference with transcription-promoting histone modifications such as acetylation or ubiquitylation (Shiio and Eisenman 2003; Nathan et al. 2006). However, it is not known whether histone sumoylation is a general mechanism of repression at transcriptionally silent genes. Despite the large number of studies linking SUMO with repression, in a few cases, sumoylation of gene-specific transcription factors is associated with activating transcription (Lyst and Stancheva 2007; Guo and Sharrocks 2009 and references therein), indicating that SUMO does not have a solely repressive role in transcription.
In addition to gene-specific transcription factors, large-scale proteomics screens identified components of the general transcription machinery as SUMO targets in both yeast (Panse et al. 2004; Wohlschlegel et al. 2004; Zhou et al. 2004; Denison et al. 2005; Hannich et al. 2005; Wykoff and O'Shea 2005; Makhnevych et al. 2009) and mammalian (Zhao et al. 2004; Rosas-Acosta et al. 2005) cells. These include multiple subunits of the general transcription factors (GTFs) TFIIA, TFIIF, and TFIID (TBP, as well as several TAFs); Mediator; and subunits of RNA polymerase II (RNAP II) itself. Of the multiple yeast subunits of RNAP II found to be sumoylated, Rpb1 sumoylation was characterized and appears to occur as part of the UV response, but blocking Rpb1 sumoylation does not affect transcription elongation or cell growth (Chen et al. 2009). Sumoylation of the human TFIID subunit TAF5 was shown to impair TFIID binding to promoter DNA (Boyer-Guittaut et al. 2005), suggesting that sumoylation might be a negative regulator of TFIID as well.
The finding that multiple components of the general transcription machinery are modified by SUMO suggests that, in addition to regulating several gene-specific transcription factors, sumoylation may help control transcription at many genes. Here, we provide evidence that this is indeed the case. We first asked, using chromatin immunoprecipitation (ChIP) assays, whether SUMO is associated with a panel of genes in Saccharomyces cerevisiae, in normally growing cells or under conditions where transcription of certain genes is induced. In apparent contrast to the fact that SUMO is strongly associated with transcriptional repression, we found that SUMO is present at all transcriptionally active genes tested, including both constitutive and inducible promoters, but not at repressed or uninduced genes. We did not detect Ubc9 at constitutive genes, implying that factors are sumoylated prior to association with these genes. In contrast, we show that Ubc9 is recruited to promoters of inducible genes when they are activated. Interestingly, however, impairing Ubc9 function results in increased transcription of these genes. Providing an explanation for this, we found that reducing sumoylation at the induced ARG1 promoter impairs the ability of the cell to shut off its transcription when the activating signal disappears. Our results indicate that sumoylation occurs at promoters of active genes during induced transcription, and functions to regulate transcription levels by facilitating shutting off transcription.
Results
SUMO associates with transcriptionally active genes
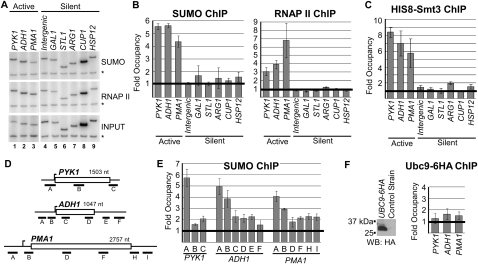
Considering the preponderance of studies linking sumoylation of transcription factors with transcriptional repression, we wished to establish whether sumoylation has a general role in repressing transcription at promoters of silenced genes. To this end, we determined whether sumoylated proteins could be detected at a panel of repressed promoters in yeast cells by ChIP using an antibody that recognizes the yeast SUMO peptide Smt3. Chromatin was prepared from cells grown in standard conditions, and samples were immunoprecipitated with the Smt3 antibody (SUMO) or an antibody recognizing the largest subunit of RNAP II as a control (Fig. 1A). Immunoprecipitated samples were analyzed by PCR for the presence of promoter DNA relative to a nontranscribed region of Chromosome V (Fold Occupancy in Fig. 1B; see the Materials and Methods). Five inducible genes that are repressed during normal growth but activated under different conditions were analyzed: GAL1, STL1, ARG1, CUP1, and HSP12. Presumably as controls, we also tested three constitutively transcribed genes and an intergenic region of Chromosome VII, representing a transcriptionally silent region. Under normal growth conditions, RNAP II ChIP confirmed that RNAP II was associated with only the transcriptionally active genes (constitutively transcribed PYK1, ADH1, and PMA1), and not the repressed genes or the intergenic region (RNAP II) (Fig. 1A,B). Surprisingly, however, SUMO ChIP showed that significant levels of SUMO were present on only the three constitutively transcribed gene promoters (Fig. 1B). We did not detect significant levels of SUMO at the intergenic region, or most notably at any of the five repressed gene promoters (Fig. 1B).
Figure 1.
SUMO associates with constitutively active genes. (A) Representative ChIP analysis of indicated transcriptionally active and silent gene promoters, and a nontranscribed region of Chromosome VII (Intergenic). Occupancy of SUMO and RNAP II was determined by ChIP performed with an Smt3 antibody (SUMO) or an antibody recognizing RNAP II (antibody 8WG16; RNAP II). Control primers recognizing sequences in an untranscribed region of Chromosome V were included in the PCR reactions (*), and input chromatin was also analyzed by PCR for comparison and quantification (INPUT). (B) Quantification of ChIP analyses as shown in A. Quantification (described in the Materials and Methods) is fold over background; a value of 1 indicates no signal detected above background, as indicated with a heavy bar. (C) Occupancy of 8xHIS-tagged Smt3 at indicated genes or intergenic region in the HIS8-SMT3 strain, determined by ChIP as in A and B. (D) Gene diagrams for PYK1, ADH1, and PMA1 indicating gene length, approximate position of transcriptional start site (bent arrow), and regions amplified by indicated ChIP primers. Lengths of genes and amplification products are to scale with respect to each other. (E) Quantification of detailed ChIP analysis of SUMO occupancy at indicated positions across indicated constitutive genes. (F, left) Cell lysates from UBC9-6HA and a control strain were analyzed by Western blot with anti-HA antibody to confirm tagging of Ubc9. (Right) Occupancy of 6xHA-tagged Ubc9 at promoters of indicated genes in the UBC9-6HA strain, determined by ChIP.
The Smt3 antibody used in our ChIP experiments was described previously (Montpetit et al. 2006), and, when used in a Western blot using yeast whole-cell extract, gave the expected pattern of multiple sumoylated proteins (see Fig. 2A; cf. Wohlschlegel et al. 2004). However, to confirm that the signal detected by ChIP was indeed due to the presence of sumoylated proteins, we repeated the ChIP experiments using a strain expressing an 8xHIS epitope-tagged version of Smt3 (Wohlschlegel et al. 2004). As shown in Figure 1C, ChIP using an anti-HIS antibody in the HIS8-SMT3 strain resulted in the same pattern observed with the Smt3 antibody, confirming that the ChIP signal was in fact due to the presence of SUMO at transcriptionally active genes. Low levels of HIS8-Smt3 at the ARG1 promoter likely reflect weak activation of this gene detected in uninduced cells (Govind et al. 2005). These observations are in contrast to what was expected based on models postulating that SUMO is inherently repressive to transcription (Chupreta et al. 2005), and instead indicate that, in normally growing yeast, SUMO is present at the promoter regions of active genes.
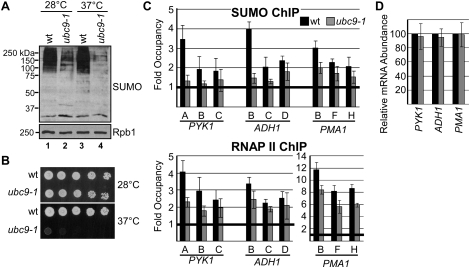
Figure 2.
Reduced sumoylation in ubc9-1 cells correlates with reduced RNAP II occupancy at constitutive genes. (A) Whole-cell yeast extracts were prepared from ubc9-1 cells and isogenic wild-type cells, and were analyzed by Western blot with Smt3 antibody (SUMO) and an antibody recognizing the large subunit of RNAP II (Rpb1; antibody 8WG16) as a loading control. Cells were grown at 28°C, or shifted to 37°C for 30 min prior to analysis as indicated. (B) Spot assay comparing growth of ubc9-1 cells and an isogenic wild-type strain on complete minimal medium at the indicated temperature. Approximately 1.5 × 104 cells were plated in the first spot on the left, with fivefold dilutions in each subsequent spot toward the right. Growth was for 2 d. All ChIP experiments using ubc9-1 and the isogenic control were performed using cells grown at 28°C. (C) Occupancy of SUMO and RNAP II was determined at the indicated positions (see Fig. 1D) across PYK1, ADH1, and PMA1 in ubc9-1 cells (gray bars) and the isogenic wild-type strain (black bars). Statistical analysis (Studetnt's t-test) indicates that SUMO levels and RNAP II levels are significantly different in the wild-type and ubc9-1 data sets (P = 0.004 for SUMO, and P = 0.003 for RNAP II). (D) Semiquantitative RT–PCR analysis of steady-state RNA levels of indicated genes in ubc9-1 cells (gray bars) and in isogenic wild-type cells (black bars). RNA levels are not significantly different in the wild-type and ubc9-1 data sets (P > 0.1).
To determine whether SUMO detected at constitutively transcribed genes was restricted to promoter regions, we performed more detailed ChIP analyses. Primer pairs representing positions across the PYK1, ADH1, and PMA1 genes were used to determine the level of SUMO cross-linking at promoter, internal, and terminator regions, as indicated in the gene diagrams shown in Figure 1D. Low levels of sumoylated proteins were detected at all positions across the genes, but higher levels were detected around promoters (about twofold to threefold higher than at other regions). This indicates that proteins associated with transcriptionally active genes, particularly those associated with promoters, are sumoylated.
The presence of SUMO at transcriptionally active genes indicates that either “presumoylated” proteins assemble specifically at active genes, or sumoylation of assembled factors takes place at active genes. To distinguish between these possibilities, we determined whether Ubc9 could also be detected at the active genes tested. We constructed a yeast strain expressing 6xHA C-terminal-tagged Ubc9 (Fig. 1F, left) and performed ChIP analysis. As shown in Figure 1F (right), significant levels of Ubc9-6HA were not detected at the PYK1, ADH1, or PMA1 promoter regions, even though Ubc9-6HA was detected at other genes using this strain (see Fig. 3A). Although it is possible that low levels of Ubc9 transiently associate with these genes to catalyze the sumoylation that we detect, our data are consistent with recruitment of proteins that are sumoylated before they associate with the gene.
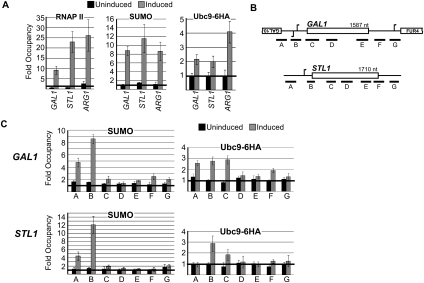
Figure 3.
Activation of inducible genes involves sumoylation of promoter-bound factors. (A) Occupancy of RNAP II, SUMO, and Ubc9-6HA at the inducible GAL1, STL1, and ARG1 gene promoters in normal growth conditions, or in their respective inducing conditions (see the Materials and Methods). (B) Gene diagrams for GAL1 and STL1 as in Figure 1D. (C) Occupancy of RNAP II, SUMO, and Ubc9-6HA at indicated positions across the GAL1 and STL1 genes in uninduced (black bars) or induced conditions (gray bars).
Lowering SUMO levels at constitutive genes results in reduced RNAP II levels
The presence of sumoylated factors specifically on transcriptionally active genes suggests that SUMO plays a positive role in transcription by RNAP II. To determine whether SUMO affects transcription of these genes, we analyzed the effect of reduced sumoylation on levels of RNAP II on the genes. Both SMT3 and UBC9 are essential, preventing the use of SMT3- or UBC9-null strains. Instead, we used a mutant yeast strain, ubc9-1, that expresses a less stable form of the enzyme (Seufert et al. 1995; Betting and Seufert 1996). At normal growth temperatures (28°C–30°C), ubc9-1 cells grew essentially as well as isogenic wild-type cells, but growth was severely impaired at elevated temperatures (37°C) (Fig. 2B). However, examination of whole-cell extract by Western blot showed significantly reduced levels of total sumoylation in ubc9-1 cells compared with wild-type cells at 28°C, while growth at 37°C further exacerbated this effect (Fig. 2A).
To assay the effects of reduced sumoylation on transcription, we compared wild-type and ubc9-1 cells grown at 28°C, avoiding the possibility of growth-related indirect effects. First, we measured levels of SUMO at positions across the three constitutive genes in wild-type and ubc9-1 cells. Overall, somewhat less SUMO was detected on the constitutive genes in this strain background (DF5) than in the strain used in our initial analysis (YPH499), but the pattern of SUMO occupancy across the genes was the same (cf. Figs. 1B,E and 2C, top). As expected, the reduction in global sumoylation in ubc9-1 cells resulted in reduced SUMO levels at the tested genes (Fig. 2C, top). RNAP II ChIP analysis showed an overall reduction in RNAP II levels across the constitutive genes in ubc9-1 cells (Fig. 2C, bottom). Approximately 30% less RNAP II was detected at all positions across the PMA1 gene in ubc9-1 cells compared with wild-type cells, whereas most of the reduction in the PYK1 and ADH1 genes occurred at promoter-proximal regions, with only a modest reduction detected near the 3′ end. An analysis performed after switching wild-type and ubc9-1 cells to 37°C for 30 min resulted in similar reductions in RNAP II densities in ubc9-1 cells (data not shown). These results indicate that sumoylation is required for efficient recruitment of RNAP II to the promoters of the constitutive genes tested. However, this did not significantly affect steady-state transcript levels for these genes (Fig. 2D). This observation is not surprising, considering that ubc9-1 cells grow normally at the permissive temperature, and suggests that the primary function of SUMO at the promoters of constitutive genes likely is not in regulating overall expression levels (see the Discussion).
SUMO and Ubc9 are both recruited to promoters of induced genes during activation
To explore further the function of SUMO in transcription, we next examined whether SUMO associates with activated genes. Three of the inducible genes that did not associate with SUMO when repressed (Fig. 1A,B) were examined in their induced states. GAL1, induced when galactose is the carbon source in growth medium, STL1, an osmoresponse gene induced by the addition of NaCl to the medium, and ARG1, an amino acid biosynthesis gene induced by amino acid starvation conditions, were examined by ChIP. RNAP II ChIP at the GAL1, STL1, and ARG1 promoters showed robust recruitment of RNAP II to the genes under their respective activating conditions (Fig. 3A, left). Strikingly, analysis of SUMO levels showed the appearance of sumoylated proteins coincident with induction of the genes (Fig. 3A, middle), at levels approximately twice as great as at the constitutive genes tested (cf. Figs. 1B and 3A; data not shown). The appearance of SUMO at the three inducible genes, under different respective induction conditions, strongly reinforces our assertion that SUMO associates with active sites of transcription in yeast.
We next wished to investigate whether the high SUMO levels detected at the induced genes might reflect active sumoylation at the promoters. To this end, we examined association of Ubc9 with these promoters under induced and uninduced conditions. As the above experiments were conducted in the UBC9-6HA strain, we performed ChIP analysis using an HA antibody. In contrast to the constitutive genes analyzed above (Fig. 1F), significant levels of Ubc9-6HA were present on each of the induced genes during activation (Fig. 3A, right). Detailed ChIP analysis at positions across GAL1, STL1, and ARG1 demonstrated that the strong SUMO signal detected upon induction was restricted to promoter-proximal regions of the genes (Figs. 3C, 4A,B). Similar analysis of Ubc9-6HA recruitment along the GAL1 and STL1 genes indicated that Ubc9-6HA was localized primarily to promoter-proximal regions as well (Fig. 3C, right; note that GAL1 promoter elements are shared with the upstream coregulated GAL10 gene). The concurrent appearance of SUMO and Ubc9 at induced promoters strongly suggests that promoter-bound factors are sumoylated during gene activation. Thus, while SUMO associates with constitutively expressed genes, likely due to recruitment of sumoylated factors, Ubc9-mediated SUMO modification of promoter-bound factors occurs during activation of inducible genes. This represents a previously unappreciated aspect of gene activation in yeast.
Figure 4.
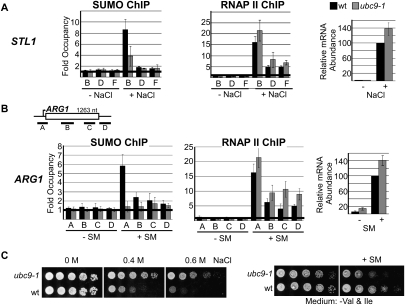
Sumoylation has a negative effect on transcription of inducible genes. (A, B) Occupancy of SUMO (left) and RNAP II (middle) was determined in uninduced and induced conditions (indicated; as in Fig. 3A) for STL1 (A) and ARG1 (B) in ubc9-1 cells (black bars) and isogenic wild-type cells (gray bars). (Right) Steady-state STL1 (A) and ARG1 (B) mRNA abundance in wild-type and ubc9-1 cells was determined by RT–PCR in untreated or induced cells, as indicated. Values are shown relative to transcript abundance in wild-type cells after induction, which is set to 100. Statistical analysis indicates that RNAP II levels are significantly different in the wild-type and ubc9-1 data sets (P < 0.03 for STL1 data, and P < 0.008 for ARG1 data). RNA levels were also significantly different in wild-type and ubc9-1 cells, with P-values of 0.02 and 0.03 for STL1 and ARG1, respectively. (C) Growth comparison of wild-type and ubc9-1 cells in 0, 0.4, and 0.6 M NaCl in rich medium (left) or minimal medium lacking Val and Ile, and the same medium supplemented with 0.5 μg/mL SM (right). Spot assays were performed as in Figure 2B. Growth was for 2 d at 28°C.
Reduced sumoylation at induced promoters correlates with increased transcription
We next wished to determine the effect of impaired sumoylation on activation of inducible genes. To this end, we compared RNAP II recruitment to the STL1 and ARG1 promoters under their respective induction conditions in wild-type and ubc9-1 cells at 28°C. SUMO ChIP indicated that reduced global sumoylation (see Fig. 2A) resulted in less SUMO associated with the promoters of the induced STL1 and ARG1 genes (Fig. 4A,B, left). Unexpectedly, and in contrast to what we observed at constitutive genes, for both STL1 and ARG1, reduced SUMO at promoters correlated with small but consistent increases of RNAP II levels at the promoters and across the genes (Fig. 4A,B, middle). These higher levels of RNAP II resulted in increased steady-state transcript levels (by ∼40%) for both genes, as shown by RT–PCR (Figs. 4A,B, right). These results suggest that, although sumoylation is detected at inducible promoters only after activation, it has a negative effect on RNAP II recruitment and, consequently, transcription.
Next, we asked whether growth of ubc9-1 cells (which is normal at 28°C) (see Fig. 2B), is affected under conditions where gene induction is necessary. Induction of osmoresponse genes such as STL1 is essential for growth in the presence of NaCl (Rep et al. 2000). Whereas isogenic wild-type cells showed growth defects at 0.4 and 0.6 M NaCl, ubc9-1 cells grew relatively well across this range of NaCl concentrations (Fig. 4C, left). Under amino acid starvation conditions (for example, in the presence of the drug sulfometuron [SM], which inhibits expression of Val and Ile) (Falco and Dumas 1985), induction of amino acid biosynthesis genes such as ARG1 is critical. In this case, reduced sumoylation in the ubc9-1 strain resulted in reduced growth compared with isogenic wild-type cells (Fig. 4C, right). Although reduced sumoylation likely affects multiple systems involved in the response to changes in growth conditions, including exposure to NaCl and amino acid starvation, it is reasonable to speculate that deregulated, increased transcription of response genes can affect cell growth. For example, increased expression of osmoresponse genes can facilitate sustained growth in the presence of NaCl, whereas extended expression of amino acid biosynthesis genes beyond their need can be energetically costly and result in reduced growth.
Sumoylation is required for complete deactivation of induced ARG1
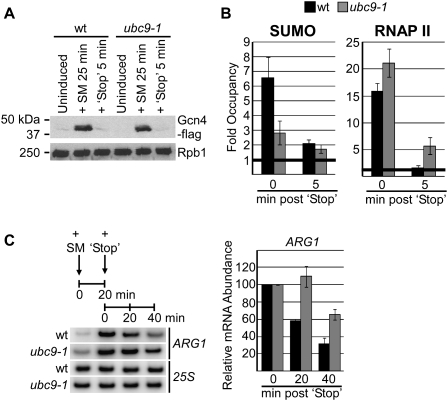
The increased RNAP II densities and elevated transcription of induced genes that we observed in ubc9-1 cells could result in two distinct ways: from increased recruitment of the polymerase to activated promoters, or from prolonged activation of induced genes after the inducing signal is no longer present. In light of our finding that sumoylation has a positive effect on RNAP II recruitment to constitutive genes, we hypothesized that the negative effect of sumoylation on inducible transcription is due to a role in terminating activation of induced promoters, a process that we refer to as deactivation. To investigate whether sumoylation indeed functions in deactivation, we devised a method by which activation of the ARG1 gene could be very rapidly stopped. Upon exposure to SM, depletion of Val and Ile leads to efficient translation and stabilization of the Gcn4 activator protein, which binds the ARG1 promoter and activates its transcription (Qiu et al. 2004, 2005; Govind et al. 2005). The short half-life of Gcn4 (∼5 min under normal growth conditions) (Kornitzer et al. 1994) permits rapid response to changes in amino acid levels. We found that adding a high concentration of Val and Ile (five times their normal concentration, referred to as “stop mix”) to SM-treated ubc9-1 or isogenic wild-type cells resulted in the very rapid disappearance of Gcn4 (no signal in a Western blot 5 min after adding the stop mix) (Fig. 5A), and, as described below, in the complete disappearance of RNAP II from the ARG1 promoter in wild-type cells.
Figure 5.
Sumoylation is required for deactivation of the induced ARG1 gene. (A) ubc9-1 and isogenic wild-type cells transformed with pGcn4-Flag plasmid were grown at 28°C, and cell lysates were prepared from aliquots removed prior to induction (Uninduced), 25 min after adding SM, then 5 min after adding stop mix consisting of fivefold concentrated Val and Ile. Western blot is shown of Gcn4-Flag expression using a Flag antibody and 8WG16 antibody for loading control (Rpb1). (B) Occupancy of SUMO and RNAP II at the ARG1 promoter (position A in Fig. 4B) was determined 25 min after induction with SM, and 5 min after adding the stop mix to wild-type (black bars) and ubc9-1 (gray bars) cells. Statistical analysis of RNAP II levels associated with ARG1 after addition of the stop mix indicates that significant levels were detected in ubc9-1 cells, and no significant levels were detected in wild-type cells (P < 0.04 and P > 0.1, respectively, compared with null hypothesis of fold occupancy equal to 1). (C, left) Transcript abundance of ARG1 was determined by RT–PCR on total RNA isolated from the indicated samples at 0 and 20 min post-addition of SM, and 20 and 40 min post-addition of the stop mix. RNAP I-transcribed 25S RNA analysis is shown as a control. Quantification of three RT–PCR analyses, showing values for 0, 20 and 40 min post-addition of stop mix, is shown at right, normalized to abundance of ARG1 mRNA 20 min post-addition of SM in wild-type and ubc9-1 cells (0 min post-stop).
We used the strategy just described to determine whether reduced sumoylation in ubc9-1 cells affects deactivation of ARG1. To this end, wild-type and ubc9-1 cells were treated with SM for 25 min, then treated with the stop mix for 5 min, or simply treated with SM for 25 min with no further treatment. Analysis of RNAP II occupancy at the ARG1 promoter in wild-type chromatin samples showed that the stop mix was highly effective in deactivating ARG1; RNAP II was no longer detectable on the gene 5 min after its addition (Fig. 5B, RNAP II ChIP). SUMO ChIP with wild-type chromatin indicated that SUMO levels were also greatly reduced at the ARG1 promoter upon its deactivation (Fig. 5B, SUMO ChIP). Thus, SUMO associates with ARG1 only while it is transcriptionally active. In fact, examination of the ARG1 promoter upon activation and deactivation showed that Ubc9, SUMO, and RNAP II show similar patterns of recruitment to the promoter during activation, and all were efficiently cleared from the promoter upon deactivation (Supplemental Fig. S1). Strikingly, analysis of RNAP II occupancy before and after addition of the stop mix shows that clearance of RNAP II from the ARG1 promoter was impaired in ubc9-1 cells compared with wild-type cells. Significant levels of RNAP II remained at the ARG1 promoter in ubc9-1 cells 5 min after addition of the stop mix, whereas deactivation was complete at this point in wild-type cells (Fig. 5B, right). We also examined whether clearance of Gcn4 from the ARG1 promoter was affected by reduced global sumoylation in cells expressing Flag-tagged Gcn4. Indeed, as shown in Supplemental Figure S2, significant levels of Gcn4-Flag remained associated with the ARG1 promoter in ubc9-1 cells compared with wild-type cells 5 min after addition of the stop mix. This is a strong indication that sumoylation at inducible gene promoters plays an important role in deactivation of transcription by facilitating clearance of promoter-bound factors.
Prolonged activation of genes in ubc9-1 cells after elimination of the activating signal would be expected to result in prolonged transcription and delayed clearance of transcripts. To determine if transcription was prolonged in ubc9-1 cells compared with wild-type cells, we measured steady-state levels of ARG1 (which has an approximate half-life of 18–21 min) (Crabeel et al. 1990) by RT–PCR at 20-min intervals post-addition of the stop mix (Fig. 5C, quantification of three experiments is shown at right). Decay of the ARG1 mRNA signal in wild-type cells was as expected, with approximately half as much signal appearing after each 20-min period. In sharp contrast, in ubc9-1 cells, no reduction in ARG1 mRNA levels was observed within the first 20 min after adding the stop mix (Fig. 5C). In fact, a slight increase was detected, possibly reflecting accumulation of the RNA while transcription continued for a short time before deactivation. In the second 20-min interval, after which deactivation was likely completed, decay was normal, with approximately half as much RNA being detected, indicating that the ARG1 mRNA half-life was not affected by the ubc9-1 mutation. Taken together with the analysis of RNAP II occupancy, these results indicate that deactivation of ARG1 transcription is impaired in ubc9-1 cells, reflecting reduced sumoylation at the promoter.
Discussion
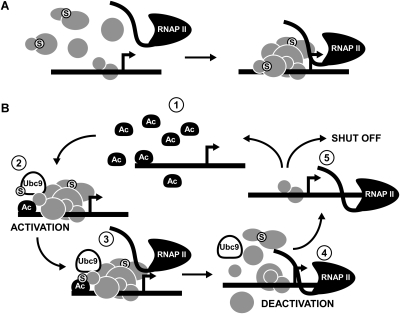
Previous studies have supported the notion that SUMO plays an inherently repressive role in transcription. For example, targeting SUMO isoforms or Ubc9 in human cells to a luciferase reporter gene (through fusions to the Gal4 DNA-binding domain) caused potent reduction of luciferase activity (Holmstrom et al. 2003; Shiio and Eisenman 2003; Chupreta et al. 2005). In fact, a specific surface on the SUMO moiety, present in all human SUMO isoforms as well as in yeast Smt3, was identified that confers repressive effects in the luciferase reporter assay (Chupreta et al. 2005). It is surprising, then, that our data shows that SUMO marks transcriptionally active genes in yeast, both constitutive genes and inducible genes, specifically, when they are activated. Furthermore, since reducing levels of SUMO at the constitutive genes resulted in somewhat lower RNAP II density, efficient sumoylation is important for optimal transcription. Thus, while SUMO is able to repress transcription in some contexts, in the natural context of constitutive transcription, other properties of SUMO must come into play, perhaps in the assembly or organization of the transcription machinery (Fig. 6A). For example, sumoylation plays a role in the appropriate assembly of the human polyadenylation complex on an RNA substrate (Vethantham et al. 2007). Additionally, several nuclear proteins depend on sumoylation for efficient nuclear localization (Pichler and Melchior 2002). Detection of sumoylated proteins at constitutively transcribed genes may thus be a consequence of earlier modification.
Figure 6.
Model for the role of SUMO in active transcription in yeast. (A) At constitutive genes, sumoylation (shown as encircled S) of transcription-related factors (gray) prior to recruitment to the promoter facilitates promoter complex assembly or recruitment of RNAP II (Pol II). (B) For inducible genes, when an activator (Ac) is present at sufficient concentration, it binds to the promoter (step 1), and recruits general transcription factors (gray) as well as Ubc9 (step 2). Note that, as for constitutive genes, some transcription-related factors may be sumoylated prior to promoter recruitment. (Step 3) However, the presence of Ubc9 and detection of SUMO at inducible genes during activation implies that sumoylation of promoter-bound factors is part of the activation process. Although targets of sumoylation have not yet been identified at induced promoters, it is possible that the activator itself is targeted, which facilitates its removal from the promoter, perhaps through SUMO-targeted ubiquitin-mediated degradation by the 26S proteasome (not shown). (Step 4) While RNAP II becomes engaged in transcription, SUMO-mediated loss of the activator (or other transcription factors), leads to deactivation of the promoter. (Step 5) The cleared promoter can now become induced again if sufficient activator is present; otherwise, transcription is shut off. Inability to deactivate the promoter by impairing sumoylation leads to prolonged activation of the induced gene, and reduced ability to shut off transcription.
Our analyses showing the appearance of SUMO and recruitment of Ubc9 indicate that promoter-associated factors at inducible genes become sumoylated coincident with activation, and the SUMO signal disappears when the activating signal is no longer present (at least for ARG1). We did not detect association of either of the two yeast SUMO proteases, Ulp1 and Ulp2, at the ARG1 promoter during its activation or deactivation (E Rosonina and JL Manley, unpubl.), indicating that sumoylated promoter-bound factors are cleared from the deactivated promoter, and not simply desumoylated in that process. Activation-coincident sumoylation was observed for genes that are induced under very different conditions: addition of NaCl (STL1), presence of galactose (GAL1), amino acid starvation (ARG1), and addition of Cu (CUP1) (E Rosonina and JL Manley, unpubl.). Thus, we showed that sumoylation at promoters occurs only while a gene is active, and is likely a general, previously unappreciated aspect of the gene activation process. Because we found that different types of inducible genes all show the same pattern of sumoylation, it is reasonable to assume that components of the general transcription machinery, present at all activated genes, are subject to sumoylation. However, as discussed below, it is also possible that gene-specific transcription factors, a large number of which are known to be sumoylated, are regulated at induced promoters through a common mechanism involving sumoylation.
Our analysis supports a model in which SUMO has multiple roles at actively transcribed genes in yeast. Previously sumoylated proteins assemble at active genes and have a positive effect on RNAP II recruitment (Fig. 6A). These factors are detected at constitutively transcribed genes, but likely contribute to the SUMO signal detected at activated inducible genes as well (Fig. 6B). At inducible genes, sumoylation of promoter-bound factors takes place, which has two consequences. First, we found that sumoylation at induced promoters has a repressive effect on transcription. Recruitment of transcriptional repressors to activated genes has been demonstrated before (e.g., Mot1 and NC2) (Geisberg et al. 2001, 2002; Zanton and Pugh 2004; van Werven et al. 2008), indicating that, in some cases, repressive factors have a role during active transcription. Second, sumoylation functions in shutting off transcription of induced genes at the appropriate time. Both effects of sumoylation on induced transcription can be explained if sumoylation functions in deactivating the promoter, perhaps after every round of transcription initiation. That is, our model posits that sumoylation of some factor(s) is necessary for efficiently clearing the promoter after transcription has initiated, thereby allowing it to respond to another round of activation if a sufficient activation signal (activator) is present, or, if not, to shut down (Fig. 6B). Failure to clear the promoter efficiently is expected to cause elevated activation and prolonged transcription when the activator is no longer available, as we observed in ubc9-1 cells. Immobilized template experiments, performed in yeast nuclear extract, showed that, once RNAP II has cleared the promoter and initiated transcription, a scaffold of factors remains at the promoter, including a subset of GTFs and Mediator (Yudkovsky et al. 2000). We propose that sumoylation is a general mechanism by which promoters are cleared of these factors and/or the activator, thereby regulating transcription levels and facilitating shut off of inducible genes.
The idea that promoters are cleared after every round of transcription, allowing the cell to respond to activator abundance in the nucleus, has some support, particularly for activators with acidic activation domains (Chi et al. 2001; Tansey 2001; Bhaumik and Malik 2008). Ubiquitin-mediated proteolysis of these activators once they are “spent” is thought to allow activator turnover at induced promoters. This is supported by studies that detect components of the 26S proteasome occupying most genes in yeast (Auld et al. 2006; Sikder et al. 2006). Interestingly, the strongest evidence for this comes from studies on Gcn4, which is targeted for ubiquitylation by a phosphorylation mark left by a kinase component of the RNAP II Mediator complex Srb10, implying that degradation of Gcn4 is linked to transcriptional activation (Chi et al. 2001). Promoter-associated factors, such as the repressor Mot1 in yeast, and PARP-1, which associates with the human HSP70.1 promoter (see below), are targets for ubiquitylation via prior sumoylation through the function of SUMO-targeted ubiquitin E3 ligases (STUbLs) (Martin et al. 2009; Wang and Prelich 2009). Thus, ubiquitin-mediated proteolysis of promoter-associated factors, including activators, is a plausible mechanism by which SUMO facilitates promoter clearance/deactivation at induced promoters (Fig. 6B).
Is the connection between active transcription and the presence of sumoylated proteins that we detected in yeast evolutionarily conserved among eukaryotes? In human cells, heat shock results in recruitment of Ubc9 and the SUMO2 isoform to the heat-shock-induced HSP70.1 promoter, where a number of promoter-associated factors are thought to become sumoylated (Martin et al. 2009), suggesting that our findings may be extrapolated to higher eukaryotes. However, in mouse retina cells, ChIP analysis demonstrated that SUMO is present at both activated and repressed photoreceptor-specific genes (Onishi et al. 2009). Although this phenomenon might be specific to this class of genes in the retina, it contrasts with our findings in yeast, where we found no evidence that SUMO is associated with repressed genes. Thus, although the role of sumoylation at activated genes may be evolutionarily conserved, it appears that, in higher eukaryotes, SUMO plays an additional role in maintaining transcriptional repression. Through evolution, higher eukaryotes may have taken advantage of the ability of SUMO to inhibit transcription, which we detect at activated genes, in an additional role in keeping repressed genes silent.
Our findings have several interesting implications. For example, our analysis suggests that one reason so many transcription factors are SUMO substrates is because sumoylation is a general aspect of gene activation that regulates transcription at induced promoters. Furthermore, our results suggest that sumoylation of transcription factors is usually found to impart a negative effect on transcription because sumoylation may be a common mechanism of clearing transcription factors from promoters, such that blocking sumoylation results in apparent increased transcription of target genes. Finally, although the coordinated recruitment of factors to activated promoters has been studied extensively, it is not known how assembled factors are removed from the promoter when the activating signal disappears and the gene product is no longer needed—effectively, how genes are shut off. Our model posits that sumoylation of promoter-bound factors is a general mechanism of deactivating transcription, and suggests that shutting off transcription of many genes, or of individual genes, is a regulated process. In any event, and in contrast to the view that SUMO functions principally at transcriptionally repressed genes, the data presented here indicates that sumoylation is an important and general mechanism by which gene expression is regulated at actively transcribed genes.
Materials and methods
Plasmids and yeast strains
Yeast strains used in this study are listed in Supplemental Table S1. For strain ERYM434B, UBC9 was tagged at its genomic locus by homologous recombination as described previously (Knop et al. 1999). To generate plasmid pGCN4-Flag, GCN4, with 1 kb each of flanking upstream and downstream genomic sequence, was cloned into the URA3-marked CEN vector pRS316. Site-directed mutagenesis was used to create a unique EcoRI site at the 3′ end of the GCN4 ORF, upstream of the stop codon, that was used to introduce a Flag epitope tag sequence. Further cloning details for this plasmid and yeast strains used are available on request.
Yeast growth conditions
For galactose induction (induction of GAL1), strains were grown in rich medium containing 2% raffinose until the O.D. at 595 nm of the sample reached 0.9. Cells were then collected by centrifugation and washed in rich medium containing either 2% glucose (uninduced sample) or 2% galactose (induced sample), then resuspended in the same medium used for washing, and incubated for 90 min. For induction of STL1, cells were grown in complete minimal medium to an O.D. of 0.6–0.8, then NaCl was added to a final concentration of 0.4 M (induced sample) or cells were mock-treated (uninduced) and incubated for an additional 10 min. For induction of ARG1, cells were grown in minimal medium lacking Val and Ile to O.D. 0.6–0.8, then SM was added to a final concentration of 0.5 μg/mL (induced) or cells were untreated (uninduced) for 30 min or as indicated. For analysis of constitutive genes, cells were grown in complete minimal medium, then prepared for ChIP or RNA analysis as indicated below.
ChIP
Two-hundred milliliters of cells was grown at 28°C as per the conditions above, and ChIP was performed as described previously (Rosonina et al. 2009). For SUMO ChIP, 0.8 μL of previously described Smt3 antibody (Montpetit et al. 2006) was used per immunoprecipitation; for RNAP II ChIP, 0.5 μg 8WG16 (Covance) was used; and 1 μg of HA (Sigma) or HIS (Santa Cruz Biotechnology) antibodies was used for HA- and HIS-tagged protein ChIP, respectively. Experiments were performed at least three times for each quantification, which was performed as described previously (Komarnitsky et al. 2000) by determining the ratio of the gene-specific amplification signal to the background amplification signal from the untranscribed region performed in the same reaction, for the immunoprecipitated samples. This ratio was then divided by the same ratio determined for the input material, resulting in a normalized, “fold over background” value and referred to as fold occupancy. Primer sequences used for PCR amplification are listed in Supplemental Table S2. Where statistical analysis was performed to compare data sets, a Student's t-test was applied, with P-values indicated in the figure legends.
RT–PCR analysis
For preparation of total RNA samples, 50-mL cultures were grown as described above, and RNA was isolated and analyzed by RT–PCR as described previously (Rosonina et al. 2009). Primer sequences are listed in Supplemental Table S2. Analyses were performed at least three times, and the result of semiquantitative analyses is shown. Where statistical analysis was performed to compare data sets, a Student's t-test was applied, with P-values indicated in the figure legends.
Acknowledgments
We thank Ben Montpetit and Philip Hieter (University of British Columbia) for generously sharing the Smt3 antibody, and James Wohlschlegel (The Scripps Research Institute) and Stefan Jentsch (Max Planck Institute) for yeast strains. We also thank Patricia Richard for insightful discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health to J.L.M.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1917910.
Supplemental material is available at http://www.genesdev.org.
References
- Auld KL, Brown CR, Casolari JM, Komili S, Silver PA 2006. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol Cell 21: 861–871 [DOI] [PubMed] [Google Scholar]
- Betting J, Seufert W 1996. A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin- and proteasome-dependent pathway. J Biol Chem 271: 25790–25796 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Malik S 2008. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit Rev Biochem Mol Biol 43: 419–433 [DOI] [PubMed] [Google Scholar]
- Boyer-Guittaut M, Birsoy K, Potel C, Elliott G, Jaffray E, Desterro JM, Hay RT, Oelgeschlager T 2005. SUMO-1 modification of human transcription factor (TF) IID complex subunits: Inhibition of TFIID promoter-binding activity through SUMO-1 modification of hsTAF5. J Biol Chem 280: 9937–9945 [DOI] [PubMed] [Google Scholar]
- Chen X, Ding B, LeJeune D, Ruggiero C, Li S 2009. Rpb1 sumoylation in response to UV radiation or transcriptional impairment in yeast. PLoS One 4: e5267 doi: 10.1371/journal.pone.0005267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev 15: 1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupreta S, Holmstrom S, Subramanian L, Iniguez-Lluhi JA 2005. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol 25: 4272–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M, Lavalle R, Glansdorff N 1990. Arginine-specific repression in Saccharomyces cerevisiae: Kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol Cell Biol 10: 1226–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP 2005. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics 4: 246–254 [DOI] [PubMed] [Google Scholar]
- Falco SC, Dumas KS 1985. Genetic analysis of mutants of Saccharomyces cerevisiae resistant to the herbicide sulfometuron methyl. Genetics 109: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dominguez M, Reyes JC 2009. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta 1789: 451–459 [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Holstege FC, Young RA, Struhl K 2001. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol Cell Biol 21: 2736–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Kuras L, Struhl K 2002. Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae. Mol Cell Biol 22: 8122–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F 2007. Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Gill G 2005. Something about SUMO inhibits transcription. Curr Opin Genet Dev 15: 536–541 [DOI] [PubMed] [Google Scholar]
- Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT 2003. P300 transcriptional repression is mediated by SUMO modification. Mol Cell 11: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Girdwood DW, Tatham MH, Hay RT 2004. SUMO and transcriptional regulation. Semin Cell Dev Biol 15: 201–210 [DOI] [PubMed] [Google Scholar]
- Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG 2005. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol Cell Biol 25: 5626–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Sharrocks AD 2009. Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between sumoylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Mol Cell Biol 29: 3204–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 280: 4102–4110 [DOI] [PubMed] [Google Scholar]
- Holmstrom S, Van Antwerp ME, Iniguez-Lluhi JA 2003. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc Natl Acad Sci 100: 15758–15763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES 2004. Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Kim J, Cantwell CA, Johnson PF, Pfarr CM, Williams SC 2002. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J Biol Chem 277: 38037–38044 [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E 1999. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer D, Raboy B, Kulka RG, Fink GR 1994. Regulated degradation of the transcription factor Gcn4. EMBO J 13: 6021–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, Stancheva I 2007. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans 35: 1389–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T, Sydorskyy Y, Xin X, Srikumar T, Vizeacoumar FJ, Jeram SM, Li Z, Bahr S, Andrews BJ, Boone C, et al. 2009. Global map of SUMO function revealed by protein–protein interaction and genetic networks. Mol Cell 33: 124–135 [DOI] [PubMed] [Google Scholar]
- Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, Zhang XD, Bischof O, Seeler JS, Dejean A 2009. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J 28: 3534–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B, Hazbun TR, Fields S, Hieter P 2006. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol 174: 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem 275: 13321–13329 [DOI] [PubMed] [Google Scholar]
- Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, et al. 2006. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev 20: 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi A, Peng GH, Hsu C, Alexis U, Chen S, Blackshaw S 2009. Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron 61: 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Gill G 2009. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics 4: 440–444 [DOI] [PubMed] [Google Scholar]
- Panse VG, Hardeland U, Werner T, Kuster B, Hurt E 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 279: 41346–41351 [DOI] [PubMed] [Google Scholar]
- Pichler A, Melchior F 2002. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3: 381–387 [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Yoon S, Natarajan K, Swanson MJ, Hinnebusch AG 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol Cell Biol 24: 4104–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol 25: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Krantz M, Thevelein JM, Hohmann S 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem 275: 8290–8300 [DOI] [PubMed] [Google Scholar]
- Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG 2005. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics 4: 56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Willis IM, Manley JL 2009. Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III. Mol Cell Biol 29: 2308–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373: 78–81 [DOI] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN 2003. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci 100: 13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikder D, Johnston SA, Kodadek T 2006. Widespread, but non-identical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J Biol Chem 281: 27346–27355 [DOI] [PubMed] [Google Scholar]
- Tansey WP 2001. Transcriptional activation: Risky business. Genes Dev 15: 1045–1050 [DOI] [PubMed] [Google Scholar]
- van Werven FJ, van Bakel H, van Teeffelen HA, Altelaar AF, Koerkamp MG, Heck AJ, Holstege FC, Timmers HT 2008. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev 22: 2359–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley JL 2007. Sumoylation modulates the assembly and activity of the pre-mRNA 3′ processing complex. Mol Cell Biol 27: 8848–8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Prelich G 2009. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol 29: 1694–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR 3rd 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem 279: 45662–45668 [DOI] [PubMed] [Google Scholar]
- Wykoff DD, O'Shea EK 2005. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics 4: 73–83 [DOI] [PubMed] [Google Scholar]
- Yang SH, Jaffray E, Hay RT, Sharrocks AD 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol Cell 12: 63–74 [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408: 225–229 [DOI] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF 2004. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc Natl Acad Sci 101: 16843–16848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J 2007. Sumoylation regulates diverse biological processes. Cell Mol Life Sci 64: 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Kwon SW, Anselmo A, Kaur K, White MA 2004. Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J Biol Chem 279: 20999–21002 [DOI] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, Zhou H 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem 279: 32262–32268 [DOI] [PMC free article] [PubMed] [Google Scholar]