Abstract
Meiotic recombination is initiated by the programmed induction of DNA double-strand breaks (DSBs) catalyzed by the evolutionarily conserved Spo11 protein. Studies in yeast have shown that DSB formation requires several other proteins, the role and conservation of which remain unknown. Here we show that two of these Saccharomyces cerevisiae proteins, Mei4 and Rec114, are evolutionarily conserved in most eukaryotes. Mei4−/− mice are deficient in meiotic DSB formation, thus showing the functional conservation of Mei4 in mice. Cytological analyses reveal that, in mice, MEI4 is localized in discrete foci on the axes of meiotic chromosomes that do not overlap with DMC1 and RPA foci. We thus propose that MEI4 acts as a structural component of the DSB machinery that ensures meiotic DSB formation on chromosome axes. We show that mouse MEI4 and REC114 proteins interact directly, and we identify conserved motifs as required for this interaction. Finally, the unexpected, concomitant absence of Mei4 and Rec114, as well as of Mnd1, Hop2, and Dmc1, in some eukaryotic species (particularly Neurospora crassa, Drosophila melanogaster, and Caenorhabditis elegans) suggests the existence of Mei4–Rec114-dependent and Mei4–Rec114-independent mechanisms for DSB formation, and a functional relationship between the chromosome axis and DSB formation.
Keywords: Meiosis, recombination, double-strand break
In sexually reproducing organisms, the chromosome content of diploid germ cells must be halved during gamete formation. This is achieved by the specialized meiotic cycle composed of one replication phase followed by two divisions, thereby leading to the formation of four haploid gametes from a diploid cell. During the first division (the reductional division), homologous chromosomes segregate from each other, whereas during the second division, sister chromatids segregate. The reductional division is unique to meiotic cells and requires specialized mechanisms to connect homologous chromosomes for their proper orientation at metaphase I and faithful separation by the segregation machinery. In most eukaryotes, these connections are established by crossovers (COs), which are reciprocal recombination events between homologous chromosomes, visualized in the cell as chiasmata (Petronczki et al. 2003). The mechanical role of COs implies a precise regulation of the frequency of these events, at least one per chromosome. These recombination events, in addition to ensuring the mechanism of reductional segregation, also have evolutionary consequences as they increase genome diversity (Coop and Przeworski 2007).
The molecular mechanism of CO formation has been analyzed in details in Saccharomyces cerevisiae and Schizosaccharomyces pombe (Hunter 2007), and several of the main properties are conserved in mammals (Handel and Schimenti 2010) and other eukaryotes. Meiotic recombination is initiated by the formation of DNA double-strand breaks (DSBs). DSBs are repaired by homologous recombination using the homologous chromosome as a template, leading to a gene conversion either without CO (NCO) or with CO (Baudat and de Massy 2007). A fraction of DSBs might be repaired by recombination with the sister chromatid. The formation of the right number of COs depends therefore on the regulation of both DSB formation and repair. How this regulation precisely works remains to be understood (Lynn et al. 2007). One important feature of the mechanism and control of DSB formation and repair is the coordination between the activities that generate DSBs and the structural components of the chromosome axis, as shown by the comparative mapping of DSBs and the axis-associated protein (Blat et al. 2002) and as discussed by Kleckner (2006).
DSB formation is catalyzed by the evolutionarily conserved Spo11 protein, which shares similarity with the catalytic subunit of type II DNA topoisomerases (Bergerat et al. 1997; Keeney et al. 1997). Several other proteins have been shown to be required for DSB formation in different species (Keeney 2008). For instance, in S. cerevisiae, Ski8, Rec102, Rec104, Rec114, Mei4, Mer2, Rad50, Xrs2, and Mre11 are also involved. Spo11 and Ski8 form a subcomplex that interacts with the Rec102–Rec104 subcomplex, and they are mutually dependent for their chromatin binding (Jiao et al. 2003; Kee et al. 2004; Prieler et al. 2005; Maleki et al. 2007). Rec114, Mei4, and Mer2 form another subcomplex (Li et al. 2006; Maleki et al. 2007) that is thought to interact with Rec102–Rec104 through direct binding of Rec114 to the Rec102–Rec104 complex (Maleki et al. 2007).
One striking observation, however, is that, apart from Spo11, the evolutionary conservation of several of these meiotic-specific proteins (Rec102, Rec104, Rec114, Mer2, and Mei4) is uncertain, given that only Rec114 has been found to have a functional ortholog (i.e., S. pombe Rec7) (Molnar et al. 2001). (S. pombe Mei4 is a transcription factor, and is not related by sequence or function to S. cerevisiae Mei4.) Rapid evolutionary divergence could partially explain the lack of identified orthologs in many species, with the exception of ascomycetes (Richard et al. 2005). Furthermore, some of the functional requirements for DSB formation observed in S. cerevisiae also appear not to be conserved. For instance, in Arabidopsis thaliana, the Ski8 ortholog is not required for meiotic recombination (Jolivet et al. 2006). Likewise, Rad50 and Mre11 are not needed for DSB formation in S. pombe (Young et al. 2004), A. thaliana (Bleuyard et al. 2004; Puizina et al. 2004), and Coprinopsis cinerea (Gerecke and Zolan 2000; Merino et al. 2000).
To gain insight into genes involved in DSB formation, we searched for orthologs of several of the S. cerevisiae proteins that are apparently poorly conserved, and that show a high degree of divergence even among Saccharomycotinae. Using a strategy that identifies small blocks of conservation, and thus orthologs, among increasingly distant species, we could find conserved motifs in Mei4 and Rec114, and identify their orthologs among filamentous ascomycetes, basidiomycetes, plants, and higher eukaryotes. We then analyzed Mus musculus MEI4 and REC114, and found that the role of MEI4 in DSB formation, based on the analysis of Mei4−/− mutant mice, and its interaction with REC114 are evolutionarily conserved. We determined the immunolocalization of MEI4 protein in mouse spermatocytes and oocytes, which reveals unique properties providing insight into its function in the process of DSB formation.
Results
Mei4 and Rec114 are conserved among eukaryotes
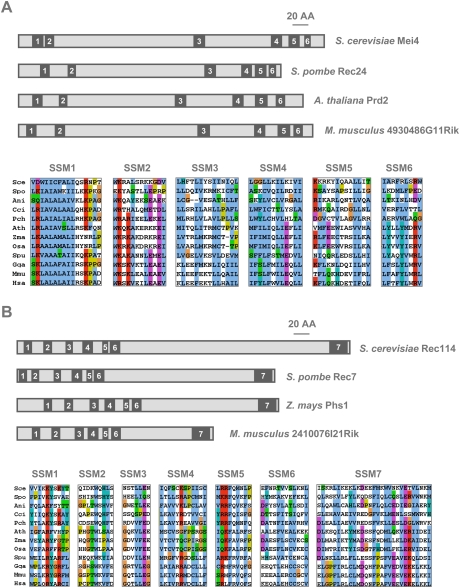
To investigate the evolutionary conservation of the mechanism of DSB formation at meiosis, we searched for orthologs of S. cerevisiae Mei4 and Rec114. To this end, as a great number of completely sequenced fungal and other eukaryotic genomes (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=eukaryote) are currently available, a series of phylogenomic-oriented PSI-BLAST homology searches coupled to MAFFT alignment and secondary structure prediction analyses were performed, as described previously for the analysis of the Mediator complex subunits (Bourbon 2008). Mei4 and Rec114 orthologs could be found in most of the examined Saccharomycetales, including Candida albicans and other CTG clade yeasts, as well as the three schizosaccharomycetes (Fig. 1; Supplemental Fig. S1). The only exception was the CTG clade yeast Candida lusitaniae, which has lost several components of the synaptonemal and synapsis initiation complexes (Butler et al. 2009; Reedy et al. 2009). In fission yeast, a previous report suggested that the Rec114 ortholog was Rec7 (Malone et al. 1997), and here we show that the Mei4 ortholog is predicted to be Rec24, a protein known to be essential for initiation of meiotic recombination (Martin-Castellanos et al. 2005). The overall identity and similarity between S. cerevisiae Mei4 and S. pombe Rec24 was quite low, with only 7% identity and 21% similarity over their entire length (data not shown), although both proteins have roughly similar sizes (407 amino acids vs. 350 amino acids).
Figure 1.
Evolutionary conservation of Mei4 and Rec114 in eukaryotes. The relative localization of the conserved SSMs within the primary structures of fungal, plant, and mouse Mei4 (A) or Rec114 (B) orthologs are shown in the top panels. Alignments of the corresponding SSMs from the same and other representative eukaryotes are shown in the bottom panels. Conserved sequence blocks were extracted from multiple protein sequence alignments generated by MAFFT and were colored with Jalview using default ClustalX color schemes (using the MPI bioinformatics Web resources; see the Materials and Methods). Except for glycines and prolines, colors (i.e., WLVIMFAC in blue, KR in red, TSNQ in green, DE in magenta, G in orange, HY in cyan, and P in yellow) were assigned to residues if the amino acid profile at the given position was conserved. (Sce) Saccharomyces cerevisiae; (Spo) Schizosaccharomyces pombe; (Ani) Aspergillus nidulans; (Cci) Coprinopsis cinerea; (Pch) Phanerochaete chrysosporium; (Ath) Arabidopsis thaliana; (Zma) Zea mays; (Osa) Oryza sativa; (Spu) Strongylocentrotus purpuratus; (Gga) Gallus gallus; (Mmu) Mus musculus; (Hsa) Homo sapiens sapiens.
Primary structure alignments of the identified Mei4 and Rec114 hemiascomycetal orthologs revealed several evolutionarily conserved short signature sequence motifs (SSMs), located mainly at their N-terminal and C-terminal parts (Fig. 1; Supplemental Fig. S1). Altogether, in both Mei4 and Rec114, these motifs represented about one-fifth of the whole primary sequence, providing a rationale for the low overall amino acid identity. The six SSMs of the Mei4 orthologs, including the basic alanine-rich signature motif (SSM1) found at the N terminus of all presumptive orthologs, were predicted to adopt α-helical or coiled-coil structures, which are prone to be involved in protein–protein interactions. In Rec114 orthologs, an amphiphilic α-helical motif (SSM7) was predicted at the C terminus of all orthologs, whereas all other SSMs (1–6) were clustered in the N-terminal moiety and were predicted to adopt a β-sheet structure, most likely part of a structural domain of unknown biochemical function.
To identify putative orthologs in filamentous Ascomycetes, Basidiomycetes (including the mushroom C. cinerea), green plants such as A. thaliana and Zea mays), and animals (particularly mammals), a second series of PSI-BLAST analyses were undertaken using as query sequences the overall alignments of previously identified hemiascomycetal Mei4 and Rec114 orthologs (see the Materials and Methods). Among the PSI-BLAST protein hits, candidate Mei4 and Rec114 orthologs were identified by focusing not only on the highest E-value scores, but also on the conservation of most, if not all, SSMs (including their relative spacing within the primary sequences). The candidates were then validated as likely orthologs based on the conservation of key signature residues (for example, a tryptophan in the middle of the Rec114 SSM2 motif) as well as on the secondary structure predictions. This approach allowed us to identify putative Mei4 and Rec114 orthologs in all examined Eurotiomycetes (such as Aspergillus nidulans), in most Basidiomycetes and Microsporidians (such as Encephalitozoon cuniculi, for which only Rec114 was detected), and in all investigated green plants, as well as in most animals (including mammals) (Supplemental Figs. S1–S3), but not in Sordariomycetes (such as Neurospora crassa), Caenorhabditis elegans, and Drosophila melanogaster, nor in any closely related Nematodes or Dipterans (data not shown).
Plants carrying mutations in the presumptive Mei4 and Rec114 orthologs identified here have been described previously and shown to have meiotic phenotypes: mutants in the genes AtPRD2 in A. thaliana (De Muyt et al. 2009) and Phs1 in Z. mays and A. thaliana (Pawlowski et al. 2004; Ronceret et al. 2009), respectively. AtPRD2 and Phs1 displayed sequence motifs with significant homologies (primary sequence similarities and secondary structure predictions) to the SSMs identified in fungal Mei4/Rec24 (Fig. 1A) and Rec114/Rec7 (Fig. 1B), respectively. Furthermore, as a further support for their key structural importance, equivalent motifs were also identified in the putative mammalian Mei4 and Rec114 orthologs (Fig. 1; Supplemental Fig. S2). S. cerevisiae and M. musculus Mei4 orthologs showed 8% identity and 18% similarity, and the Rec114 orthologs showed 6% identity and 15% similarity (Supplemental Fig. S2). Altogether, our comparative genomics approach provides strong evidence for the conservation of Mei4 and Rec114 in most eukaryotes, from fungi to plants and metazoans.
Mei4 and Rec114 are expressed in testis and embryonic ovary
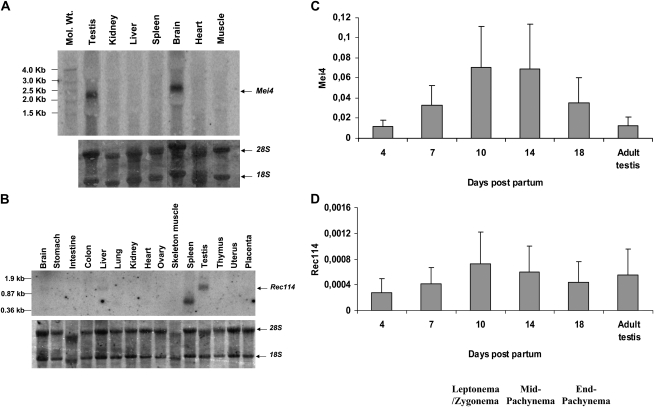
If the function of Mei4 and Rec114 at meiosis is evolutionarily conserved in eukaryotes, mouse Mei4 and Rec114 should be expressed in gametocytes during meiosis at the beginning of the first meiotic prophase, when DSBs are formed (Mahadevaiah et al. 2001). In male mice, the first wave of entry of B-type spermatogonia into meiosis occurs at day 8 post-partum (dpp), and cells enter into meiotic prophase at 9 dpp. Additional waves of meiotic differentiation will follow during their entire life. In female mice, a single wave of entry into meiosis is initiated at embryonic day 12.5 (E12.5), and meiotic prophase extends from E13 to E18. We thus analyzed Mei4 and Rec114 expression in various tissues by Northern blot, and determined the expression levels by RT–qPCR amplification during the first wave of entry into meiosis in prepubertal mice.
Both Mei4 and Rec114 were expressed in adult testes and in embryonic ovaries (Fig. 2A,B; Supplemental Fig. S4). The size of the Mei4 transcript, ∼2.5 kb, was compatible with mRNAs of 2.7 kb (BC096613) and 3.1 kb (AK043929) that code for the full-length protein (389 amino acids), as documented in databases. These reference mRNAs are not from testis, but from eye and neonate cortex, respectively, suggesting that Mei4 might be expressed at low levels in some somatic cells. We also noted a significant expression of Mei4 in the brain. Rec114 mRNA was ∼1 kb long, a size compatible with the reference mRNA of 883 bases (NM_028598.1). A very low expression of Rec114 was detected in the liver, and a shorter transcript was observed in the spleen. To determine the mRNA steady-state levels before and during the first wave of entry into meiosis, Mei4 and Rec114 expression were analyzed by qPCR in 4-, 7-, 10-, 14-, and 18-dpp testes. Expression of both genes was highest at 10 and 14 dpp, when spermatocytes are in the first half of meiotic prophase, from leptonema to mid-pachynema, and when DSBs are formed and engaged into repair (Fig. 2C,D). The amount of Mei4 mRNA was at least 100-fold higher than that of Rec114, indicating a higher level of expression and/or a greater mRNA stability.
Figure 2.
Mei4 and Rec114 are expressed in testis and during the first wave of spermatogenesis. (A,B) Northern blot hybridization of RNA extracted from various tissues of adult mice with the Mei4 (A) or Rec114 (B) probe. Bottom panels show rRNAs (18S and 28S) on the membranes stained with methylene blue. (C,D) RT-qPCR amplification of RNA extracted from testes of juvenile and adult mice. For each gene, qPCR was performed with two different primer pairs specific for Mei4 (C) or Rec114 (D). Plotted values are the expression levels normalized to β-Actin (means of five qPCR assays with 95% confidence interval).
MEI4 and REC114 interact
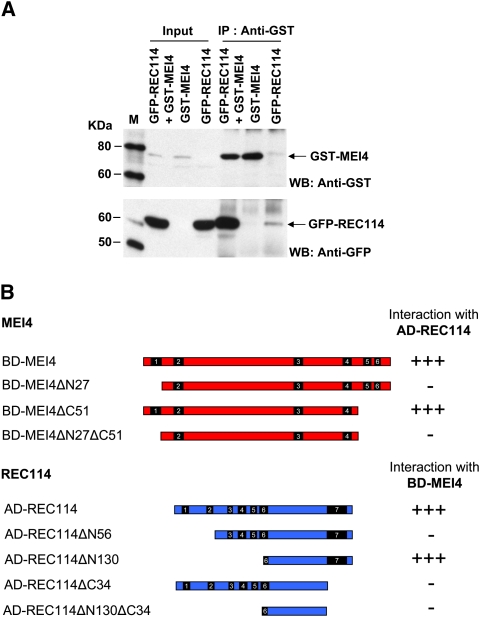
Since S. cerevisiae Mei4 and Rec114 associate in a subcomplex (Arora et al. 2004; Li et al. 2006), we tested whether this interaction was conserved in the mouse using three different approaches.
First, both MEI4 and REC114 were expressed in HeLa cells as fusions proteins with GFP or GST. In those cells, a robust interaction between GST-MEI4 and GFP-REC114 was detected by immunoprecipitation with either anti-GST or anti-GFP antibodies (Fig. 3A; Supplemental Fig. S5A). This interaction was confirmed by immunoprecipitation experiments performed using in vitro synthesized GST-MEI4 and His-REC114 (Supplemental Fig. S5B). We then validated these results and mapped the interaction domains by yeast two-hybrid assay (Fig. 3B; Supplemental Fig. S6). Based on the mapping of their interaction domains in S. cerevisiae (Maleki et al. 2007), and on the positions of the SSMs in the different orthologs (Fig. 1), we generated MEI4 variants with an N-terminal deletion (ΔN27, lacking SSM1), a C-terminal deletion (ΔC51, lacking SSM5 and SSM6) or both deletions, and REC114 variants with an N-terminal deletion (ΔN56, lacking SSM1 and SSM2; ΔN130, lacking SSM1–5), a C-terminal deletion (ΔC34, lacking SSM7), or both. Analysis of the deletion mutants showed that the N-terminal region, but not the C-terminal region, of MEI4 was required for the interaction, and that both N-terminal and C-terminal regions of REC114 were needed. However, in the REC114 ΔN130 mutant, the C-terminal region appeared to be sufficient to allow interaction with MEI4, whereas this was not the case for REC114ΔN56 (Fig. 3B; Supplemental Fig. S6B). This discrepancy might indicate distinct folding behaviors of the two deletion mutants, leading to an inaccessible or accessible SSM7-containing region. The two mutants were expressed at similar levels (Supplemental Fig. S6C), but we cannot exclude that these results might be an artifact of the yeast two-hybrid assay, for instance, due to a defective nuclear localization of REC114ΔN56.
Figure 3.
MEI4 interacts with REC114. (A) Coimmunoprecipitation of MEI4 and REC114. HeLa cells were transfected with plasmids expressing mouse GST-MEI4 and/or GFP-REC114. Protein extracts were immunoprecipitated with anti-GST or anti-GFP (Supplemental Fig. S5A) antibodies. Inputs (1% of total) and immunoprecipitated fractions (40% of total) were detected with anti-GST or anti-GFP antibodies. Protein standards were used as molecular weight markers (M). (B) Mapping the MEI4 and REC114 interaction domains by yeast two-hybrid assay. The ability to interact of full-length and N-terminal or C-terminal deletion mutants of MEI4 and REC114 was assessed in a yeast two-hybrid assay. Either strong interaction (+++) or no interaction (−) was observed, based on growth on selective media (see also Supplemental Fig. S6).
MEI4 is localized on the lateral elements of the synaptonemal complex
The immunolocalization of proteins involved in meiotic recombination has been a powerful tool to analyze various aspects of this process, in particular for studies on proteins located on chromosome cores and proteins involved at various steps of DSB formation (at leptonema) and repair (from zygonema to pachynema). Since DSBs are formed in sequences that are part of chromatin loops and are located on the axes upon DSB repair (Blat et al. 2002; Storlazzi et al. 2003), we wanted to determine whether MEI4 and REC114, which are proteins involved in DSB formation, interacted with the chromatin loops, or the chromosome axes or had a distinct nuclear localization.
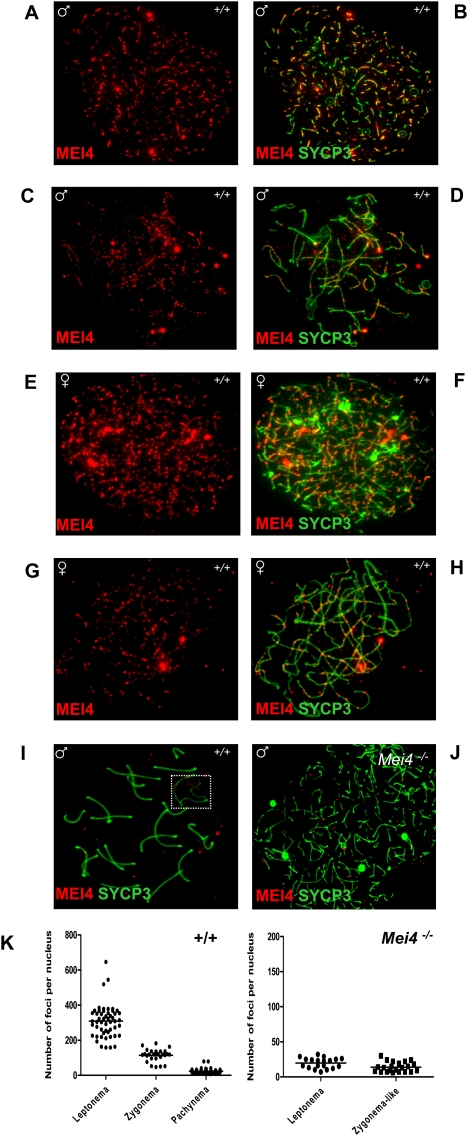
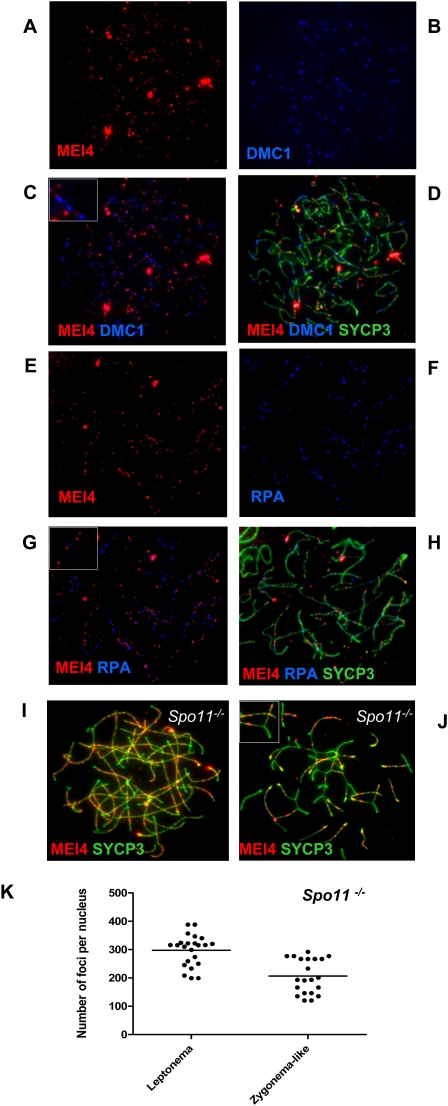
We thus generated rabbit polyclonal antibodies against MEI4 that allowed us to localize the protein on spreads of mouse spermatocytes and oocytes as multiple foci on chromosome axes at the leptotene and zygotene stages (Fig. 4A–H). The absence of signal on chromosome axes after preabsorption of the antibody with recombinant MEI4 (Supplemental Fig. S7) and when it was tested in spermatocytes from Mei4−/− mice (Fig. 4J; Supplemental Fig. S7) indicates that the axes-associated foci correspond to MEI4. Nonspecific interaction of the antibody detected foci in the chromatin, in both wild-type and Mei4−/− spermatocytes, and some bright signals near chromosome ends observed with only one of the two rabbit antisera (data not shown). We cannot exclude, however, that some nonaxis foci correspond to MEI4. The number of MEI4 foci was highest at leptonema (on average, 309 foci per nucleus), decreased at zygonema, and was strongly reduced at pachynema and subsequent stages (Fig. 4I,K). We could also observe multiple MEI4 foci in some early leptotene cells in which DSB formation may not have occurred yet, based on the relatively low γH2AX expression (data not shown). At zygonema, when the number of MEI4 foci started to decrease, most foci were located on unsynapsed axes. At pachynema, MEI4 foci were absent from synapsed axes on autosomes, and from the unsynapsed axes of sex chromosomes (Fig. 4I). We then asked whether MEI4 colocalized with foci from DSB repair proteins (RPA, DMC1, RAD51, etc.), as they are also detected on chromosome axes at leptonema and zygonema (Moens et al. 2002). Colabeling of DMC1 and MEI4, and of RPA and MEI4 showed that these two DSB repair proteins do not colocalize with MEI4 (Fig. 5A–H). Quantification of DMC1 foci showed that only 12% ± 6% colocalized with MEI4, a frequency lower than expected if MEI4 and DSB repair sites were distributed randomly along axes (see the Materials and Methods). Finally, given the potential function for MEI4 in initiation of meiotic recombination, we tested if MEI4 localization required SPO11. On spermatocyte spreads from Spo11−/− mice, MEI4 localization on unsynapsed regions of chromosomes at leptotene and zygotene stages and the number of foci (on average, 298 foci per nucleus at leptonema) were similar to those observed in wild-type mice (Fig. 5I–K). Conversely, in Spo11−/− mice, MEI4 was absent from synapsed regions that involve interactions between nonhomologous chromosomes (Baudat et al. 2000; Romanienko and Camerini-Otero 2000).
Figure 4.
Discrete MEI4 foci on chromosome axes of spermatocytes at leptonema and zygonema. MEI4 was detected in leptotene or zygotene spermatocytes (A–D) and oocytes at E16 (E–H). MEI4 was not observed on the axes of autosomes and sex chromosomes (white rectangle) at pachynema (I), and in spermatocytes from Mei4−/− mice (J). (K) Foci revealed by the anti-MEI4 antibody were quantified in wild-type at leptonema (mean = 309 foci per nucleus, n = 51), zygonema (mean = 114, n = 28), and pachynema (mean = 24, n = 36). Counts included all nuclear foci, of which 67%, 49%, and 14% were on axes at leptonema, zygonema, and pachynema, respectively. On average, 20 and 14 foci per nucleus were detected in the control experiment using Mei4−/− leptotene and zygotene-like nuclei, of which 20% and 23% were axis-associated, respectively. The anti-SYCP3 antibody was used to detect axial elements. (A,C,E,G:) Anti-MEI4 antibody alone. (B,D,F,H,I,J) Anti-MEI4 and anti-SYCP3 antibodies.
Figure 5.
MEI4 foci do not colocalize with DMC1 and RPA, and do not require SPO11. MEI4 was detected in parallel with DMC1 (A–D) or RPA (E–H). Insets in C and G show enlarged views of MEI4, DMC1, and RPA foci. MEI4 foci were detected and quantified in Spo11/ spermatocytes. On average, 298 and 207 foci per nucleus were detected at leptonema (n = 23) and zygonema-like (n = 21), respectively (I–K). Inset in J shows the absence of MEI4 in a synapsed region.
Developmental defects during spermatogenesis and oogenesis in Mei4−/− mice
To test the in vivo function of MEI4, we generated by gene targeting Mei4−/− mice in which the second exon of Mei4, which contains the translation initiation codon and the conserved motifs SSM1 and SSM2, was deleted (Supplemental Fig. S8).
Mei4−/− homozygote mice grew normally without any detectable developmental defect, indicating that MEI4 is not essential for viability. Intercrosses between heterozygous males and females resulted in offspring with a Mendelian ratio of 19 Mei4+/+:22 Mei4+/−:15 Mei4−/− (χ2, P = 0.2077). To investigate the function of MEI4 during spermatogenesis, we examined the presence of spermatozoa and the size of male gonads in 8-wk-old Mei4−/− and wild-type mice. The epididymis of Mei4−/− mice was devoid of spermatozoa, and the weight of mutant testes was 70%–50% lower than in wild-type animals (n = 3). Moreover, whereas wild-type testes showed a full spectrum of spermatogenic cells, including primary spermatocytes, round spermatids, and spermatozoa, in Mei4−/− seminiferous tubules, although primary spermatocytes were observed, no post-meiotic cells were found, suggesting an arrest of spermatogenesis during the early stages meiotic prophase (Fig. 6A,B). Specifically, some seminiferous tubules contained only spermatogonia and Sertoli cells; others were characterized by the presence of one or two layers of primary spermatocytes, sometimes including apoptotic cells, as judged from the condensed nuclear staining. TUNEL assay in testes from 4-wk-old Mei4−/− and wild-type mice (Supplemental Fig. S9) confirmed the presence of a higher number of apoptotic cells in Mei4−/− testes (27% of Mei4−/− seminiferous tubules contained >30 TUNEL-positive nuclei in comparison with only 2% in wild-type animals), suggesting that, in Mei4−/− mice, apoptosis is initiated to eliminate meiotic-arrested spermatocytes.
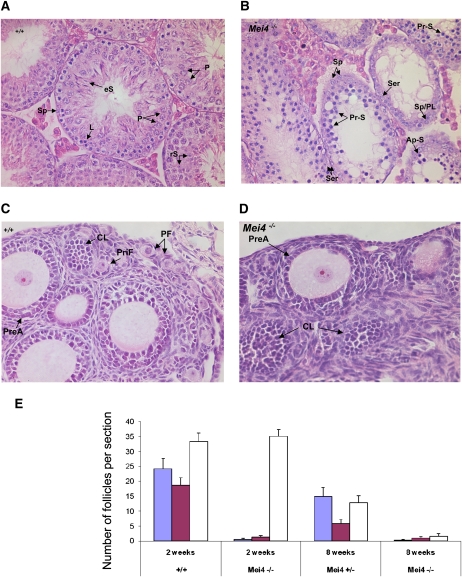
Figure 6.
Meiotic defects in testis and ovary from Mei4−/− mice. (A,B) Hematoxylin-eosin staining of testis from adult wild-type (A) and Mei4−/− (B) mice (n = 2 for each genotype). Among primary spermatocytes, some stages observed in wild-type testis were not present in Mei4−/− (i.e., pachynema, diplonema), whereas spermatocytes with apoptotic nuclei were frequent. Haploid cells (spermatids and spermatozoa) were never observed in Mei4−/−. (Sp) Spermatogonia; (eS) elongated spermatids; (rS) round spermatids; (P) pachynema; (Z) zygonema; (L) leptonema; (Ser) Sertoli; (Pr-S) primary spermatocyte at leptonema or zygonema; (PL) preleptonema; (Ap-S) apoptotic nuclei. (C,D) Hematoxylin-eosin staining of ovaries from 2-wk-old wild-type (C) or Mei4−/− (D) mice shows alteration in oogenesis. (PF) Primordial follicle; (CL) corpus luteum; (PreA) preantral oocyte; (PriF) primary follicle. (E) Quantification of primordial follicles (blue), primary follicles (purple), and growing follicles (white) in 2-wk-old (n = 3 for each genotype) and 8-wk-old (n = 1 for each genotype) wild-type, Mei4+/−, and Mei4−/− ovaries (error bars indicate 95% confidence intervals).
In females, the pool of oocytes is determined during fetal development, when oocytes complete meiotic prophase I and arrest at the dictyate stage. At birth, oocytes are surrounded by somatic cells to form primordial follicles. A cohort of these primordial follicles is stimulated shortly after birth to develop into primary, secondary, and more advanced follicular stages. To investigate oocyte and follicular development in Mei4−/− mice, we analyzed ovaries from 2-wk-old and 8-wk-old mice (Fig. 6C–E). In 2-wk-old wild-type and Mei4−/− animals, no obvious differences were observed in the presence of growing follicles at the preantral and antral stages. Conversely, Mei4−/− ovaries were nearly devoid of primordial and primary follicles, suggesting abnormal oogenesis in the absence of MEI4, as already reported in Spo11−/− mice. In 8-wk-old animals, many follicles at advanced stages of development were present in wild-type ovaries, but few follicles were detected in the mutant ovaries (Fig. 6E).
These histological properties of male and female gonads in Mei4−/− mice are highly similar to what has been described in the two already known mouse DSB-deficient mutants: Spo11−/− and Mei1−/− (Baudat et al. 2000; Romanienko and Camerini-Otero 2000; Libby et al. 2002; Barchi et al. 2005; Di Giacomo et al. 2005).
Mei4−/− mice are defective in DSB formation and homologous synapsis
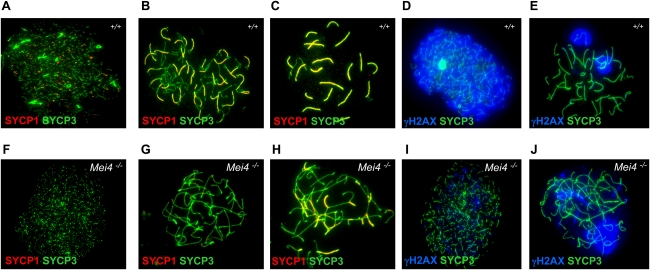
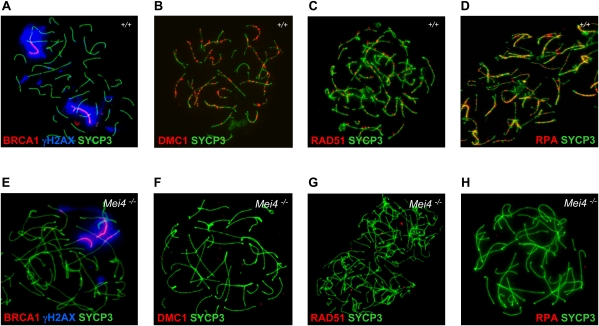
To define the meiotic defects, we analyzed spreads of spermatocytes from Mei4−/− and wild-type mice for chromosome axis formation, synapsis, recombination, and prophase progression by immunostaining. At leptonema, spreads of spermatocytes from Mei4−/− mice showed short or long chromosome axes with apparently normal morphology, based on SYCP3 staining (Fig. 7A,F). Conversely, zygotene-like nuclei with fully elongated chromosome axes were strikingly abnormal when compared with wild-type nuclei. In some nuclei, although the two lateral elements from distinct chromosomes were overlapping, no SYCP1 staining, which monitors synapsis, could be detected (Fig. 7B,G). Nuclei with SYCP1 staining and partner switches of chromosomes axes were also observed, suggesting heterologous synapsis. No nuclei with fully synapsed chromosomes could be detected (Fig. 7C,H). Similar defects in chromosomes synapsis have been observed in mutant mice with defects in DSB formation or repair (Handel and Schimenti 2010). To distinguish between these two possibilities, we monitored the expression of γH2AX, a marker of DSB formation, and DMC1, RAD51, and RPA, markers of DSB repair activity. Greatly reduced levels of γH2AX staining were observed at leptotene stage in Mei4−/− mice compared with wild-type animals (Fig. 7D,I), suggesting a defect in DSB formation. Low residual level of γH2AX has also been reported in Spo11−/− (Supplemental Fig. S10C,D) and Mei1−/− mutants (Libby et al. 2003; Barchi et al. 2005; Chicheportiche et al. 2007), and this could indicate the presence of DSBs formed through a different pathway and/or other DNA lesions. Moreover, in more advanced zygotene-like Mei4−/− nuclei, based on the chromosome axis morphology, γH2AX signal tended to cluster, a feature reminiscent of the pseudosex bodies observed in Spo11−/− spermatocytes (Fig. 7E,J; Barchi et al. 2005; Bellani et al. 2005). Indeed, like in Spo11−/− spermatocytes, BRCA1 was accumulated on chromosome cores surrounded by γH2AX (Fig. 8A,E). The defect in DSB formation at meiosis in Mei4−/− nuclei should lead to absence of the DSB repair proteins RAD51, DMC1, and RPA. All three proteins were indeed undetectable in leptotene and zygotene-like nuclei in Mei4−/− spermatocytes (Fig. 8; Supplemental Fig. S10). Although, based on chromosome morphology, the most advanced nuclei of Mei4−/− spermatocytes appeared to be similar to wild-type zygotene nuclei, this cannot be considered as a proof that meiotic prophase was arrested at zygonema in Mei4−/− cells. To address this issue, we assessed whether the testis-specific histone H1 variant H1t, which is normally detected from the middle of pachytene stage in wild-type spermatocytes (Drabent et al. 1996), was expressed in Mei4−/− spermatocytes. A significant fraction (18%, n = 84) of zygotene-like nuclei of Mei4−/− spermatocytes was enriched in H1t (Supplemental Fig. S11), indicating that they can progress to a stage that presents mid-pachynema features, as also reported in some other mouse lines with defects in meiotic DSB formation or repair (Barchi et al. 2005).
Figure 7.
Mei4 is required for DSB and homologous synapsis formation. Spreads of wild-type (A–E) and Mei4−/− (F–J) spermatocytes (n = 3 for each genotype) were analyzed with antibodies directed against SYCP1 (which is used to monitor synapsis formation) and SYCP3 (A,B,C,F,G,H), and γH2AX (a marker of DSB formation) and SYCP3 (D,E,I,J).
Figure 8.
Mei4 is required for sex body formation and loading of the DSB repair proteins DMC1, RAD51, and RPA. Spreads of wild-type (A–D) and Mei4−/− (E–H) spermatocytes (n = 3 for each genotype) were analyzed with antibodies directed against BRCA1/γH2AX/SYCP3 (A,E), DMC1/SYCP3 (B,F), RAD51/SYCP3 (C,G), and RPA/SYCP3 (D,H).
Discussion
Using a phylogenomic approach to investigate the evolutionary conservation of factors that regulate meiotic DSB formation, we report the identification of presumptive orthologs of S. cerevisiae Mei4 and Rec114 in most of the sexually reproducing eukaryotes. We then show that mouse MEI4 is localized at discrete sites on chromosome axes before or at the stage of DSB formation, independently from SPO11, suggesting that DSB formation might be activated when the SPO11 complex is located on chromosome axes. Moreover, the defect in DSB formation observed in Mei4−/− mice indicates that MEI4 function is conserved. The interaction between MEI4 and REC114 suggests that the function of mouse REC114 may also be conserved. Finally, based on the apparent absence of Mei4 and Rec114, correlated with that of Mnd1, Hop2, and Dmc1, in N. crassa, C. elegans, and D. melanogaster, we propose a functional relationship between chromosome axis and DSB formation.
The evolutionary conservation of MEI4 and REC114
In S. cerevisiae, Mei4, Rec114, and Mer2 are required for DSB formation and associate in a protein complex (Keeney 2008). Using phylogenomic-directed BLAST searches, we identified orthologs of Mei4 and Rec114 in most fungi, plants, and animals. The phylogenic trees of these two proteins indicate a rapid divergence during evolution, such that, even among yeasts, their homology cannot be recognized easily by standard BLAST analysis. This idea is supported by the fact that our phylogenomic approach could not identify nonfungal orthologs of Mer2, suggesting a more pronounced divergence and/or an intrinsic difficulty to identify orthologs due to the presence of coil-coiled domains throughout the primary sequences (Rockmill et al. 1995). In the case of Mei4 and Rec114, the divergence was not homogenously distributed along the proteins, and low but significant conservation was found in only a few specific regions that we called SSMs and that are predicted to play an important role in the function of these proteins. Most SSMs have a predicted α-helical structure, except for the N-terminal region of Rec114 that contains evolutionarily conserved charged residues within SSMs that might adopt a β-sheet conformation to form a compact structural domain of unknown biochemical function. Our analysis of the interaction between mouse MEI4 and REC114 proteins (see below) shows that some of these conserved motifs play a role in protein–protein interactions.
We could not identified Mei4 and Rec114 orthologs in three widely used model species: N. crassa, D. melanogaster, and C. elegans. Obviously, we cannot exclude that these species possess more divergent orthologs that have escaped our detection. However, this possibility seems highly unlikely because no orthologs could be detected in any of the examined Sordariales (20 species), Nematodes (10 species), and Dipterans (15 species). On the other hand, Rec114 orthologs were detected in the Filarian Nematode Brugia malayi and in the Lepidopteran Bombyx mori (data not shown). Thus, we propose that Mei4 and Rec114 have been lost in some species, and that DSB formation can occur in the absence of Mei4 and Rec114 as discussed below.
Several of the Mei4 and Rec114 predicted orthologs we identified in the current study have already been reported to be essential for meiotic DSB formation, and thus have indeed been functionally conserved. In S. pombe, Rec24 (Mei4) and Rec7 (already proposed as the Rec114 homolog) have been shown to be needed for meiotic DSB formation (Cervantes et al. 2000; Martin-Castellanos et al. 2005) and code for proteins that interact (Steiner et al. 2010). The proposed A. thaliana Mei4 homolog AtPrd2 is also functionally conserved, given the DSB defect deduced from the analysis of the AtPrd2 mutant (De Muyt et al. 2009). In A. thaliana and Z. mays, the Phs1 genes for which mutations have been described (Pawlowski et al. 2004; Ronceret et al. 2009) are Rec114 orthologs. In Z. mays, the Phs1 mutant fails to form Rad51 foci and is deficient in homologous synapsis, but DSBs could be detected (Pawlowski et al. 2004). Therefore, it cannot be excluded that meiotic DSB formation is Rec114-independent in Z. mays. However, these DSBs might also be due to a background signal in the cells assayed, and/or to some spontaneous damages independent from Spo11, as seen in mouse spermatocytes, for instance. These data, together with our finding that Mei4−/− mice also present a defect in DSB formation, indicate that MEI4 (and possibly its interaction with REC114) are functionally conserved.
Mei4 is required for meiotic DSB formation in mice
Based on histological and immunocytological analysis of oocytes and spermatocytes, in Mei4−/− mice, meiosis entry occurs normally, but its progression is altered. The first meiotic defect observed is the failure to induce DSB formation at the leptotene stage of meiotic prophase, as shown by the strong reduction of γHAX staining and the absence of DMC1, RAD51, and RPA foci. At the zygotene stage, a failure of homologous synapsis is observed, with the presence of univalent chromosomes and synapsis involving several chromosomes with partner switches, suggestive of formation of heterologous synapsis. Mutant spermatocytes do not progress further than a zygotene-like stage, although detection of H1t in some nuclei suggests that they can reach a stage with characteristics of mid-pachynema, and undergo apoptosis. During oogenesis, Mei4−/− mice show strong reduction of primordial and primary follicles and reduced number of growing follicles, particularly in 8-wk-old, adult mice. All of these phenotypes are similar, if not identical, to those described in Spo11−/− or Mei1−/− mice, which is consistent with the idea that these proteins act at the same step of meiotic recombination (Baudat et al. 2000; Romanienko and Camerini-Otero 2000; Libby et al. 2003).
The localization of MEI4 on meiotic chromosomes
MEI4 localizes in discrete foci on oocyte and spermatocyte chromosome axes, and this process is SPO11-independent. How MEI4 is recruited to specific sites on the chromosome axis remains to be understood, and no other mouse protein has been described to have such a localization pattern. SPO11 was detected as foci on chromatin of leptotene spermatocytes (Romanienko and Camerini-Otero 2000). Among the proteins found on unsynapsed axes of mouse meiotic chromosomes, either structural components (SYCP2, SYCP3, HORMAD1, and HORMAD2) or components of the cohesin complex (REC8, SMC1β, STAG3, and SMC3) could contribute directly or indirectly to MEI4 localization (Revenkova and Jessberger 2005; Wojtasz et al. 2009). However, all of these proteins seem to have a more continuous pattern of localization as compared with MEI4, and thus some additional factors, or a combination of factors, might specify the discrete chromosomal axis localization of MEI4. S. pombe Rec7 (Rec114) localization shares striking similarities with mouse MEI4. Indeed, localization of Rec7 on chromosome axes at early prophase is independent of Rec12 (Spo11) and is dependent on Rec10, a structural component of the chromosome axis (Lorenz et al. 2006). In S. cerevisiae, although Mei4 foci formation is also Spo11-independent, Mei4 appears to be associated, at least in part, with chromatin loops (Maleki et al. 2007). The Mei4/Rec114/Mer2 complex was suggested to have a dynamic and transient interaction with the Spo11 complex and DSB sites, based on the distinct localization of Mer2 and Rec102 or Mre11 (Li et al. 2006). In S. cerevisiae, DSBs take place within DNA sequences on chromatin loops (Blat et al. 2002), whereas repair occurs on chromosome axes, suggesting a loop-to-axis movement of DSB sites (Kleckner 2006). However, it is unclear whether DSB formation takes place before or upon axis association. MEI4 localization at discrete sites on chromosome axes before or at the stage of DSB formation, independently from SPO11, suggests that DSB formation occurs when DSB sites are located on chromosome axes with the SPO11 complex interacting (directly or indirectly) with MEI4.
MEI4 is absent from DSB repair foci
Two interesting features of mouse MEI4 foci are the lack of colocalization with DMC1 and RPA and the depletion from synapsed regions even in the absence of DSBs. The absence of colocalization of MEI4 with DMC1 and RPA could be interpreted in at least two ways: Either MEI4 binds to sites distinct from those where DSB repair takes place, or MEI4, DMC1, and RPA bind to the same sites but at different times. Given that MEI4 (with its partners) is predicted to interact with the SPO11 complex, the first hypothesis (different binding sites), would require two rounds of interactions for the SPO11 complex: a first one with MEI4 (and partners), and a second one with some other axis components that will tether the SPO11 complex to different sites on the chromosome core. In addition, dissociation of MEI4 from chromosome axes would then have to be coordinated indirectly with DSB repair; for instance, during progression into meiotic prophase. In the second hypothesis, MEI4 would be removed from the axis upon DSB repair. The kinetics of foci formation is compatible with this interpretation, as the highest number of MEI4 foci is observed at early leptonema and then declines progressively until the end of zygonema, whereas the number of DMC1 and RPA foci increases from early leptonema to the end of leptonema or zygonema (Moens et al. 2002). Such coordination between DSB repair and MEI4 localization would also explain the absence of MEI4 on unsynapsed sex chromosomes. Similarly, S. pombe Rec7 foci, which also do not colocalize with Rad51, were interpreted as being present at a different time than Rad51 on the axis (Lorenz et al. 2006). In addition to the displacement of MEI4 upon DSB repair, MEI4 also appears to be displaced by the synaptonemal complex, as shown by the lack of MEI4 foci on heterologous synapses in Spo11−/− spermatocytes and their persistence on unsynapsed axes in Spo11−/− zygotene-like nuclei. This persistence argues against the hypothesis that the displacement of MEI4 in wild-type mice is due to meiotic progression into zygonema. It is interesting to note that HORMAD1 and HORMAD2, two axis proteins, are also known to be displaced from synapsed chromosome axes (Wojtasz et al. 2009).
DSB formation in the presence or absence of MEI4: a connection between chromosome axis and DSB formation
One important complementary finding from our phylogenomic analysis is the absence of Mei4 and Rec114 in all or most of the examined Nematodes, Drosophilidae, and Sordariales. All of these species also lack Mnd1, Hop2, and Dmc1 (Malik et al. 2007), three proteins that are known to be involved in DSB repair. Although it is not possible to draw mechanistic conclusions from the presence or absence of genes in different species, these correlations are intriguing, and may suggest a specific functional link between MEI4/REC114 and DMC1/MND1/HOP2. The recombinase Dmc1 is expressed only during meiosis, and although its function is partially overlapping to that of Rad51, it appears to be specialized for interhomolog recombination (Hunter 2007). This role, and therefore the requirement for Dmc1 activity, varies in different species, and may depend on the structure and/or composition of chromosome axis (Sheridan and Bishop 2006). If so, MEI4 being associated with chromosome axes, the correlation we observe, and the functional link we suggest between MEI4 and DMC1 may be indirect, and rather reflect an interaction between MEI4 and some components of the chromosome axis. Distinct properties of chromosome axes in Nematodes, Drosophilidae, and Sordariales—illustrated by the absence of Hop1 in D. melanogaster and N. crassa (Malik et al. 2007), and by the presence of three Hop1 paralogs in C. elegans, with one, HTP-3, required for DSB formation (Goodyer et al. 2008)—may thus provide alternative contexts, allowing DSB formation to occur in the absence of MEI4 and REC114.
The mechanism of meiotic DSB formation thus appears to be much more conserved than originally thought. According to our data, MEI4 and, presumably, REC114 play a structural role in activating DSB formation, but the precise function of these proteins is not yet known. However, they appear to be at the interface of several key components of the regulation of meiotic recombination, and one goal obviously will be to identify the connections between these proteins and other structural components of chromosomes. Indeed, it has been shown recently in C. elegans that a condensin complex regulates DSB formation and distribution (Mets and Meyer 2009). How these components are actually connected to each other will be a very exciting challenge for the future.
Materials and methods
Search for Mei4 and Rec114 orthologs
PSI-BLAST and TBLASTN searches were undertaken using the MPI (http://toolkit.tuebingen.mpg.de/psi_BLAST) and NCBI (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=eukaryote) databases. BLAST analyses were performed mostly using the BLOSUM62 “substitution matrix,” and with an inclusion threshold of 0.001. A “phylogenomic” approach was applied to increase the probability of identifying orthologs, as described previously (Bourbon 2008). SSMs characteristic of the Mei4 or Rec114 identified in 23 yeast species (Supplemental Fig. S1) were inferred from alignments obtained using MAFFT (http://align.bmr.kyushu-u.ac.jp/mafft/online/server). SSMs are evolutionarily conserved motifs comprising at least seven amino acid residues that should be identical or similar (according to Dayhoff's rules) in at least half of the aligned sequences. To detect orthologs among nonyeast fungi and in nonfungal eukaryotes, short sequences encompassing SSMs were used as inputs in BLAST analyses, in which the “expect” (E) threshold was generally 10 (default) and the “low complexity filter” was mostly omitted. Candidate proteins were assigned as putative Mei4 and Rec114 orthologs by PSI-BLAST analyses undertaken using the MPI database (http://toolkit.tuebingen.mpg.de/psi_blast). Whole-sequence alignments generated by MAFFT analyses (above) were used as inputs to derive position-specific scoring matrixes (PSSMs). Only sequences assigned previously as likely orthologs by PSI-BLAST analyses (E-values > 0.001) and secondary structure predictions (below) were included in the alignments. Indeed, only “jump-starting” PSI-BLAST analyses, which were used as inputs PSSMs generated from MAFFT alignments that included validated proteins, allowed the detection of remote homologs. The Smith-Waterman algorithm was implemented during the searches. Secondary structure predictions were done using the Network Protein Sequence Analysis program of the “Pôle Bio-Informatique Lyonnais” bioinformatics facility (PBIL, http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html).
Plasmids
Full-length Mei4 and Rec114 cDNAs were obtained from Geneservice (Rec114, clone ID 2410076121; Mei4, clone ID 6469435), and were subcloned into different vectors using the Gateway technology (Invitrogen). The vectors used either were from Invitrogen (pDEST15 and pDEST27) or were donated by E. Bertrand and N. Bonneaud (pSpo for GFP fusion, pcDNAMyc for Myc fusion, pAS2ΔΔ for Gal4BD fusion, pGAD for Gal4AD fusions, and pET15bder for His fusion).
Northern blotting, RT–PCR, and qPCR assays
Total RNA from various mouse tissues was prepared with the Gene Elute Mammalian Total RNA purification kit (Sigma) according to manufacturer's instructions. After electrophoresis of 10 μg of total RNA of each tissue and transfer to nitrocellulose membranes (GE Healthcare), Northern blot analysis was performed with an α-32P-dCTP-labeled probe specific for Mei4 (obtained using the Mei4-specific oligonucleotides Mei4-T7U56 and Mei4-PolTL50). To detect Rec114 expression, a membrane (MN-MT-1, Zyagen) that contains total RNA from various mouse tissues was hybridized with an α-32P-dCTP-labeled Rec114 probe, amplified using the oligonucleotides Rec114-1U56 and Rec114-777L52.
For RT–PCR amplification, first strand DNA was synthesized by using oligo d(T)18, SuperScript III (Invitrogen), and total RNA (1–2 μg) extracted from testes of juvenile mice and from embryonic and postnatal ovaries. ORFs of Rec114 and Mei4 were amplified under standard PCR conditions with the primer pairs Rec114orfU/Rec114orfL and Mei4orfU/Mei4orfL, respectively. PCR cycling conditions were 3 min at 94°C, and 35 cycles of 15 sec at 94°C, 30 sec at 48°C or 52°C, 90 sec at 72°C, and 5 min at 72°C. Primers to amplify Spo11 as control were Spo11:116U22 and Spo11:655L22.
qPCR assays in 10-μL volume with duplicates were performed using 1× LC480 SYBR Green mix (Roche) together with primer pairs specific for either Rec114 or Mei4, and 2.0 μL of reverse-transcribed products in an LC480 LightCycler (Roche). Amplification conditions were 5 min at 95°C, and 42 cycles of 10 sec at 95°C, 15 sec at 54°C–56°C, and 15 sec at 72°C, with 4.3°C, 2°C, and 4.3°C per second ramping rates, respectively, and fluorescence acquisition during the extension step. The standard curve was calculated from the amplification of a set of serially diluted (from 10,000 to 16 copies) plasmid DNAs containing either Rec114 or Mei4 ORF under similar cycling conditions. β-Actin amplification was used as normalizing control. Amplifications were carried out with primer pairs as follows: Rec114 (qRec114-200MrhU20/qRec114-320MrhL21 and qRec114-586MrhU19/qRec114-725MrhL20), Mei4 (qMei4-1100U19/qMei4-1198L19 and qMei4-431U20/qMei4-558L19), β-Actin (β-Actin-F/β-Actin-R), and Spo11 (qSpo545U19/qSpo698L18). Oligonucleotides are listed in Supplemental Table S1.
Protein interaction assays
Interaction of REC114 with MEI4 was analyzed by coimmunoprecipitation and yeast two-hybrid assays. HeLa cells were transfected to express either GST-MEI4 or GFP-REC114, or to coexpress both proteins. Cell lysates prepared in HNTG buffer (20 mM HEPES at pH 7.9, 150 mM NaCl, 1% Triton, 10% glycerol, 1 mM MgCl2, 1 mM EDTA, complete protease inhibitor [Roche]) were incubated with anti-GST antibodies (Sigma) overnight at 4°C. Protein–antibody complexes were recovered with Protein G-Sepharose followed by washing with HNTG buffer four times. Protein complexes were eluted in Laemmli buffer, and were identified by Western blot analysis with anti-GST (Sigma) or anti-GFP (Torrey Pines) antibodies.
Full-length or truncated Mei4 and Rec114 ORFs were amplified with appropriate primers (full-length ORFs: Mei4: mMei4U53/mMei4L49 and Rec114: Rec114-Mrh1U56/Rec114-Mrh777L52; truncated ORFs: Mei4ΔN27: MeiΔN27U56/mMei4L49, Mei4ΔC51: mMei4U53/MeiΔC51L51, Rec114ΔN56: MrhΔN56U56/Rec114-Mrh777L52, Rec114ΔN130: MrhΔN130U55/Rec114-Mrh777L52, and Rec114ΔC34: Rec114-Mrh1U56/MrhΔC34L51), and were cloned in Gateway-compatible vectors (pAS2 and pGAD derivatives; gift from N. Bonneaud) to generate fusion proteins with the Gal4 DNA-binding domain (Gal4BD) or activation domain (Gal4AD). AH109 and Y187 (Clontech) yeast haploid strains were transformed with constructs encoding Gal4BD and Gal4AD fusion proteins. After mating in YPD plates, diploid cells expressing Gal4BD and Gal4AD fusion proteins were selected in SD/−LW, a drop-out medium without leucine and tryptophan. Protein interactions were assayed by growing diploids cells for 3 d at 30°C on selective media lacking leucine, tryptophan, histidine, and adenine (SD/−LWH and SD/−LWHA). The presence of fusion proteins in diploid strains was verified by Western blot analysis with antibodies against Gal4BD (Clontech, reference no. 630403) or Gal4AD (Millipore, reference no. 06-283).
Production and purification of anti-MEI4 antibodies
Antibodies against MEI4 were raised by immunizing two rabbits with recombinant MEI4 protein fused to 6× HIS tag at the N-terminal part using the standard protocol of the animal facility. Antibodies were then affinity-purified by incubation with PVDF membranes containing recombinant MEI4 protein.
In vitro translation
In vitro translation of Rec114 and Mei4 ORFs cloned into either pDEST15 (Invitrogen) or pET15 (Novagen) to produce GST- or His-tagged fusion proteins was carried out in the presence of 35S-methionine (GE Healthcare) using the TnT T7 Quick Coupled Transcription Translation System (Promega) according to the manufacturer's instructions. For coimmunoprecipitation assays, 4 μL of each in vitro translated protein was incubated in 30 μL of HNTG buffer for 30 min at 30°C. Then, the reaction volume was increased to 300 μL and GST fusion proteins were immunoprecipitated with anti-GST antibodies. Protein–antibody complexes were recovered with Protein G-Sepharose (GE Healthcare) after several washes with HNTG buffer, followed by SDS-PAGE and Western blot analysis.
Histological analysis
Testes and ovaries were fixed in Bouin's fixative and 4% paraformaldehyde, respectively, for 20 h at 4°C. After dehydration and embedding in paraffin, 5-μm sections were prepared and stained with hematoxylin and eosin as described (Mark et al. 2007). Apoptotic cell detection was performed in testes fixed with 4% paraformaldehyde using the Deadend Fluorometric TUNEL system according to the supplier's instructions (Promega).
Preparation of meiotic spreads
Meiotic nuclear spreads from mouse testes were prepared according to the dry-down technique, as described by Peters et al. (1997).
Immunofluorescence assays
Immunofluorescence assays on meiotic spreads were performed in blocking buffer (5% milk, 5% donkey serum in phosphate buffer saline [PBS]), as described previously (Moens et al. 1997). All primary antibodies used in this study were incubated overnight at room temperature at the following dilutions: affinity-purified rabbit anti-MEI4, 1:50; guinea pig anti-SYCP3, 1:500; mouse anti-SYCP1 (gift from C. Heyting), 1:50; rabbit anti-DMC1 (Santa Cruz Biotechnology), 1:200; rabbit anti-RPA (gift from P. de Boer), 1:1000; rabbit anti-BRCA1 (gift from J. Turner), 1:100; rabbit anti-RAD51 (gift from W. Baarends), 1:500; guinea pig anti-H1t (gift from M.A. Handel), 1:1000; and mouse monoclonal anti-phospho H2AX (Upstate Biotechnologies), 1:20,000. Secondary antibodies (goat anti-guinea pig Alexa Fluor 488 [Molecular Probes], donkey Cy3-conjugated anti-rabbit, and donkey Cy5-conjugated anti-mouse antibodies [Jackson Immunoresearch Laboratories]) were incubated for 1 h at 37°C. For colabeling of DMC1 (or RPA) and MEI4 using rabbit antibodies, the procedure was as follows. Rabbit anti-MEI4 antibody was incubated for 2 h at 37°C, Cy3-conjugated anti-rabbit IgGs were incubated for 45 min at 37°C, rabbit serum (5%) was incubated for 1 h at 37°C, donkey anti-rabbit Fab fragment at 20 μg/mL (Jackson ImmunoResearch Laboratories) was incubated for 1 h at 37°C, rabbit anti-DMC1 (or RPA) antibody and guinea pig anti-SYCP3 were incubated for 2 h at 37°C, and Alexa Fluor 488 anti-rabbit and Cy5-conjugated anti-guinea pig IgGs were incubated for 45 min at 37°C. Nuclei were visualized by staining with DAPI (4-9-6-Diamidino-2-phenylindole) (2 μg/mL). For preabsorption assays, 1 μL of affinity-purified anti-MEI4 antibodies (0.09 mg/mL) was incubated with or without 7.5 μg of recombinant His-MEI4 protein in 50 μL of blocking buffer for 2 h at 4°C. Antibodies bound to recombinant His-MEI4 protein were removed by incubation with 50 μL of Ni-NTA beads (Qiagen) for 2 h at 4°C, followed by centrifugation. The supernatant was then used for immunostaining. Digital images were captured using a cooled charge-coupled device (CCD) camera (Coolsnap HQ; Photometrics) attached to a Leica DM 6000B microscope, and were analyzed using the Metamorph imaging software. After data acquisition, all images were processed with Adobe Photoshop CS2. Foci were counted using the “Integrated Morphometry Analysis” tool of Metamorph (Molecular Devices). For colocalization analysis of MEI4 and DMC1, the background signal calculated from a control experiment without the second primary antibody showed that 2% of MEI4 foci gave false-positive DMC1 foci. This background value was substracted from the observed DMC1/MEI4 colocalization in each nucleus (n = 17). Assuming random distributions of foci on the axes, the expected frequency for a DMC1 focus to colocalize with MEI4 by chance was r = n * d/L, where n is the number of MEI4 foci, d is the average diameter of MEI4 foci, and L is total axis length. For n = 182 (average number of MEI4 foci per leptotene and zygotene nuclei in the colocalization analysis), d = 0.5 μm and L = 465 μm (highest value based on end leptotene measurements; range is from 465 μm to 150 μm at pachytene), and r = 0.20.
Generation of Mei4−/− mice
The Mei4 mutant mouse line was established at the MCI/ICS (Mouse Clinical Institute, Institut Clinique de la Souris, Illkirch, France; http://www-mci.u-strasbg.fr). The targeting vector was constructed as follows. A 2.5-kb fragment encompassing part of Mei4 intron 2 was PCR-amplified and subcloned into an MCI proprietary vector, which contains a floxed Neomycin resistance cassette, resulting in step 1 plasmid. A 1.9-kb fragment encompassing part of intron 3 of Mei4 was then PCR-amplified and subcloned in the step 1 plasmid to generate the final targeting construct. The linearized construct was electroporated in 129S2/SvPas mouse embryonic stem (ES) cells. After selection, targeted clones were identified by PCR amplification using external primers (Ef/Nr and Nf/Er) (Supplemental Table S1), and were further confirmed by Southern blot with a Neo probe (Supplemental Fig. S8). Two positive ES clones were injected into C57BL/6J blastocysts, and the derived male chimaeras showed efficient germline transmission. Mice derived from clone “104” were used for this study. Progeny were genotyped using primers Ef3854/Er3855 (amplification of the deleted allele), Ef3854/Mwt2811L22, and Mwt4477U22/Er3855 (amplification of the wild-type allele). All experiments were carried out according to the CNRS guidelines, and were approved by the regional ethics committee on live animals experimentation (project CE-LR-0812).
Acknowledgments
We thank E. Bertrand and N. Bonneau for Gateway-compatible vectors; K. Bouton, B. Moniot, and F. Poulat for technical advice; M. Mark for help with the histological analysis; Y. Boublick and the Recombinant Protein Facility at CRBM for producing recombinant MEI4; P. de Boer for anti-RPA antibody; W. Baarends for anti-RAD51 antibody; J. Turner for anti-BRCA1 antibody; M.A. Handel for anti-H1t antibody; J. Cau from the Montpellier RIO Imaging Facility for advice on image analysis; F. Arnal and members of the Animal Facility for breeding mice and generation of the rabbit antiserum; D. Zickler for comments on the manuscript; anonymous reviewers for constructive comments; F. Baudat for constant insights during the progress of this project; and laboratory members for discussions and suggestions on the manuscript. The mouse mutant line was established at the Targeted Mutagenesis and Transgenesis Department of the Mouse Clinical Institute (Institut Clinique de la Souris, MCI/ICS). We thank ANR-06-BLAN-0160-01 and EDF for grants to R.K.; an ARC grant to H.M.B.; and CNRS, ANR-06-BLAN-0160-01, ANR-09-BLAN-0269-01, and ARC3939 grants to B.d.M.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.571710.
Supplemental material is available at http://www.genesdev.org.
References
- Arora C, Kee K, Maleki S, Keeney S 2004. Antiviral protein ski8 is a direct partner of spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell 13: 549–559 [DOI] [PubMed] [Google Scholar]
- Barchi M, Mahadevaiah S, Di Giacomo M, Baudat F, de Rooij DG, Burgoyne PS, Jasin M, Keeney S 2005. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol 25: 7203–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, de Massy B 2007. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15: 565–577 [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking spo11. Mol Cell 6: 989–998 [DOI] [PubMed] [Google Scholar]
- Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD 2005. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci 118: 3233–3245 [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417 [DOI] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI 2004. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma 113: 197–203 [DOI] [PubMed] [Google Scholar]
- Bourbon HM 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res 36: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell 5: 883–888 [DOI] [PubMed] [Google Scholar]
- Chicheportiche A, Bernardino-Sgherri J, de Massy B, Dutrillaux B 2007. Characterization of Spo11-dependent and independent phospho-H2AX foci during meiotic prophase I in the male mouse. J Cell Sci 120: 1733–1742 [DOI] [PubMed] [Google Scholar]
- Coop G, Przeworski M 2007. An evolutionary view of human recombination. Nat Rev Genet 8: 23–34 [DOI] [PubMed] [Google Scholar]
- De Muyt A, Pereira L, Vezon D, Chelysheva L, Gendrot G, Chambon A, Laine-Choinard S, Pelletier G, Mercier R, Nogue F, et al. 2009. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet 5: e1000654 doi: 10.1371/journal.pgen.10000654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacomo M, Barchi M, Baudat F, Edelmann W, Keeney S, Jasin M 2005. Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc Natl Acad Sci 102: 737–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabent B, Bode C, Bramlage B, Doenecke D 1996. Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem Cell Biol 106: 247–251 [DOI] [PubMed] [Google Scholar]
- Gerecke EE, Zolan ME 2000. An mre11 mutant of Coprinus cinereus has defects in meiotic chromosome pairing, condensation and synapsis. Genetics 154: 1125–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell 14: 263–274 [DOI] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC 2010. Genetics of mammalian meiosis: Regulation, dynamics and impact on fertility. Nat Rev Genet 11: 124–136 [DOI] [PubMed] [Google Scholar]
- Hunter N 2007. Meiotic recombination. In Molecular genetics of recombination (ed. Aguilera A, Rothstein R), pp. 381–442 Springer-Verlag, Berlin [Google Scholar]
- Jiao K, Salem L, Malone R 2003. Support for a meiotic recombination initiation complex: Interactions among Rec102p, Rec104p, and Spo11p. Mol Cell Biol 23: 5928–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet S, Vezon D, Froger N, Mercier R 2006. Non conservation of the meiotic function of the Ski8/Rec103 homolog in Arabidopsis. Genes Cells 11: 615–622 [DOI] [PubMed] [Google Scholar]
- Kee K, Protacio RU, Arora C, Keeney S 2004. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J 23: 1815–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S 2008. Spo11 and the formation of DNA double-strand breaks in meiosis. In Genome dynamics and stability (ed. Lankenau D-H, Egel R), pp. 81–124 Springer, Heidelberg, Germany: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kleckner N 2006. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194 [DOI] [PubMed] [Google Scholar]
- Li J, Hooker GW, Roeder GS 2006. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 173: 1969–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby BJ, De La Fuente R, O'Brien MJ, Wigglesworth K, Cobb J, Inselman A, Eaker S, Handel MA, Eppig JJ, Schimenti JC 2002. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev Biol 242: 174–187 [DOI] [PubMed] [Google Scholar]
- Libby BJ, Reinholdt LG, Schimenti JC 2003. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci 100: 15706–15711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Estreicher A, Kohli J, Loidl J 2006. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma 115: 330–340 [DOI] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Borner GV 2007. ZMM proteins during meiosis: Crossover artists at work. Chromosome Res 15: 591–605 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27: 271–276 [DOI] [PubMed] [Google Scholar]
- Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S 2007. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma 116: 471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM Jr 2007. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One 3: e2879 doi: 10.1371/journal.pone.0002879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RE, Pittman DL, Nau JJ 1997. Examination of the intron in the meiosis-specific recombination gene REC114 in Saccharomyces. Mol Gen Genet 255: 410–419 [DOI] [PubMed] [Google Scholar]
- Mark M, Teletin M, Antal C, Wendling O, Auwerx J, Heikkinen S, Khetchoumian K, Argmann CA, Dgheem M 2007. Histopathology in mouse metabolic investigations. Curr Protoc Mol Biol 78: 29B.4.1–29B.4.32 doi: 10.1002/0471142727.mb29b04s78 [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, et al. 2005. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol 15: 2056–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino ST, Cummings WJ, Acharya SN, Zolan ME 2000. Replication-dependent early meiotic requirement for Spo11 and Rad50. Proc Natl Acad Sci 97: 10477–10482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mets DG, Meyer BJ 2009. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Chen DJ, Shen ZY, Kolas N, Tarsounas M, Heng HHQ, Spyropoulos B 1997. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106: 207–215 [DOI] [PubMed] [Google Scholar]
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J Cell Sci 115: 1611–1622 [DOI] [PubMed] [Google Scholar]
- Molnar M, Parisi S, Kakihara Y, Nojima H, Yamamoto A, Hiraoka Y, Bozsik A, Sipiczki M, Kohli J 2001. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157: 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski WP, Golubovskaya IN, Timofejeva L, Meeley RB, Sheridan WF, Cande WZ 2004. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303: 89–92 [DOI] [PubMed] [Google Scholar]
- Peters A, Plug AW, vanVugt MJ, deBoer P 1997. Drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5: 66–68 [DOI] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K 2003. Un menage a quatre. The molecular biology of chromosome segregation in meiosis. Cell 112: 423–440 [DOI] [PubMed] [Google Scholar]
- Prieler S, Penkner A, Borde V, Klein F 2005. The control of Spo11’s interaction with meiotic recombination hotspots. Genes Dev 19: 255–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J, Siroky J, Mokros P, Schweizer D, Riha K 2004. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16: 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedy JL, Floyd AM, Heitman J 2009. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol 19: 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Jessberger R 2005. Keeping sister chromatids together: Cohesins in meiosis. Reproduction 130: 783–790 [DOI] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Lafontaine I, Dujon B 2005. Comparative genomics of hemiascomycete yeasts: Genes involved in DNA replication, repair, and recombination. Mol Biol Evol 22: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Rockmill B, Engebrecht JA, Scherthan H, Loidl J, Roeder GS 1995. The yeast MER2 gene is required for chromosome synapsis and the initiation of meiotic recombination. Genetics 141: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD 2000. The mouse spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6: 975–987 [DOI] [PubMed] [Google Scholar]
- Ronceret A, Doutriaux MP, Golubovskaya IN, Pawlowski WP 2009. PHS1 regulates meiotic recombination and homologous chromosome pairing by controlling the transport of RAD50 to the nucleus. Proc Natl Acad Sci 106: 20121–20126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan S, Bishop DK 2006. Red-Hed regulation: Recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev 20: 1685–1691 [DOI] [PubMed] [Google Scholar]
- Steiner S, Kohli J, Ludin K 2010. Functional interactions among members of the meiotic initiation complex in fission yeast. Curr Genet Epub. doi: 10.1007/s00294-010-0296-0 [DOI] [PubMed] [Google Scholar]
- Storlazzi A, Tesse S, Gargano S, James F, Kleckner N, Zickler D 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev 17: 2675–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, et al. 2009. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA–ATPase. PLoS Genet 5: e1000702 doi: 10.1371/ journal.pgen.1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Hyppa RW, Smith GR 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]