Abstract
Neisseria meningitidis, a causative agent of meningitis and septicaemia, expresses type IV pili, a feature correlating with the uptake of exogenous DNA from the environment by natural transformation. The outer membrane complex PilQ, through which pili are extruded and retracted, has previously been shown to bind DNA in its pore region. In order to further elucidate how DNA is transported across the membranes, we searched for DNA binding proteins within the meningococcal inner membrane. Inner membrane fractions from a panel of neisserial strains were subjected to a solid-phase overlay assay with DNA substrates, and MS was subsequently employed to identify proteins that bind DNA. A number of DNA binding components were detected, including the pilus biogenesis component PilG, the competence protein ComL, and the cell division ATP-binding protein FtsE, as well as two hypothetical proteins. The DNA binding activity of these components was not dependent on the presence of the neisserial DNA uptake sequence. Null mutants, corresponding to each of the proteins identified, were constructed to assess their phenotypes. Only mutants defective in pilus biogenesis were non-competent and non-piliated. The DNA binding activity of the pilus biogenesis components PilQ and PilG and the phenotypes of their respective null mutants suggest that these proteins are directly involved as players in natural transformation, and not only indirectly, through pilus biogenesis.
INTRODUCTION
Neisseria meningitidis, the meningococcus, is one of the leading causative agents of meningitis and septicaemia worldwide (Davidsen et al., 2007a). Horizontal gene transfer mediates the exchange of genetic information between bacterial strains and is driven by three processes: conjugation, transformation and transduction. Among these, transformation, which is the uptake and incorporation of exogenous DNA into the bacterial chromosome, is the main source of new DNA introduced into neisserial genomes (Koomey, 1998). The transformation process is dependent on type IV pilus expression (Swanson et al., 1971), the presence of the neisserial DNA uptake sequence (DUS) (5′ ATGCCGTCTGAA 3′) in the incoming DNA (Ambur et al., 2007; Goodman & Scocca, 1988; Mathis & Scocca, 1984) and RecA-dependent homologous recombination (Koomey & Falkow, 1987).
Unlike other Gram-negative bacteria, N. meningitidis is naturally competent for transformation throughout its entire life cycle (Jyssum & Lie, 1965), provided that it expresses type IV pili, which are filamentous, hair-like appendages emanating from the bacterial surface (Frøholm et al., 1973). This association has also been observed in other bacteria, such as Neisseria gonorrhoeae (Biswas et al., 1977; Mathis & Scocca, 1984; Swanson et al., 1971), Eikenella corrodens (Tønjum et al., 1993), Legionella pneumophila (Stone & Kwaik, 1999), Pseudomonas stutzeri (Lorenz & Wackernagel, 1990), Thermus thermophilus (Friedrich et al., 2002) and Moraxella nonliquefaciens (Bøvre, 1964; Bøvre et al., 1970). Type IV pilus expression also has a role in adherence (Swanson et al., 1971; Swanson, 1973), twitching motility (Henrichsen et al., 1972; Mattick, 2002), biofilm formation (O'Toole & Kolter, 1998), bacteriophage infection (Bradley, 1974) and virulence (Bieber et al., 1998; Comolli et al., 1999; Merz et al., 1999; Pujol et al., 1999). Twitching motility is caused by type IV pilus retraction, which is dependent on the ATPase PilT (Merz et al., 2000; Wolfgang et al., 1998).
The biogenesis of type IV pili is dependent on a complex machinery of proteins. A number of proteins required for neisserial transformation have been described, including the secretin PilQ, through which pili are extruded and retracted (Collins et al., 2005; Frye et al., 2006), as well as the competence factors ComA (Facius & Meyer, 1993), ComE (Chen & Gotschlich, 2001) and ComL (Fussenegger et al., 1996). In addition to PilQ, the inner membrane proteins PilG and PilP have previously been found to be essential for pilus biogenesis and, thus, for competence (Balasingham et al., 2007; Tønjum et al., 1995). However, PilQ is the only pilus biogenesis component that has previously been shown to bind DNA (Assalkhou et al., 2007).
The uptake of DNA into the meningococcal cell can be dissected into several steps that encompass crossing of the outer and inner membranes and genome incorporation. Although the neisserial transformation pathway has been described to some extent, little is known about how the transforming DNA is taken up. It is still a conundrum whether the effect of pilus biogenesis components on transformation is of a direct or only of an indirect nature. We suggest that meningococcal transformation is coupled to pilus retraction and that exogenous DNA is taken up through non-specific attachment to retracting pili, while other DNA binding components, such as the outer membrane protein PilQ, promote further entry of DNA into the meningococcal cell.
Thus, pili, pilus biogenesis components and DNA binding proteins act together in the uptake of exogenous DNA into the meningococcal cell, enforced by pilus retraction. In order to pursue this hypothesis, we searched for DNA binding proteins that co-purify with the N. meningitidis inner membrane fraction. DNA binding proteins were detected by solid-phase overlay assays using DNA substrates with or without a DUS. Subsequently, the proteins that bound DNA were identified by MS. The corresponding null mutants were tested to see if they had defects in transformation. The identification of novel DNA binding components might contribute to further elucidation of the transformation process, and of other aspects of DNA metabolism.
METHODS
Bacterial strains and growth conditions.
Bacterial strains employed in the study are listed in Table 1. All neisserial strains were grown on blood agar or GC plates at 37 °C in 5 % CO2, while Escherichia coli strains were cultivated on LB plates at 37 °C.
Table 1.
Bacterial strains and plasmids employed in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| N. meningitidis strains | ||
| H44/76 wild-type | Serogroup B, isolated in Norway, 1976 | Holten (1979) |
| H44/76 ΔPilG | pilG : : mTnErm transposon insertion | Tønjum et al. (1995) |
| MC58 wild-type | Serogroup B, isolated in the UK, 1983 | McGuinness et al. (1991) |
| Z2491 wild-type | Serogroup A, isolated in Gambia, 1983 | Achtman et al. (1991); Crowe et al. (1989) |
| 8013 wild-type | Serogroup C | Caugant et al. (1986) |
| M1080 wild-type | Serogroup B, isolated in the USA, 1969 | Frasch & Chapman (1972) |
| M1080 ΔPilQ | M1080 strain with mTnCm#21 in pilQ | Tønjum et al. (1998) |
| M400 (M1080-A) | Derivative of M1080 with recA6 (tetM)* | Tønjum et al. (1998) |
| N. gonorrhoeae strain | ||
| N400 wild-type | recA6 (tetM) | Tønjum et al. (1995) |
| E. coli strains | ||
| ER2566 | Expression strain with a chromosomal copy of the T7 RNA polymerase gene | New England Biolabs |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10Tetr] | Stratagene |
| Vectors | ||
| p1080mutY-ermr | Plasmid containing an erythromycin-resistance gene inserted into mutY | Davidsen et al. (2007b) |
| pBSK+ | General cloning vector, Ampr | Stratagene |
*recA6 is an IPTG-inducible allele of recA.
Selective antibiotics were added when required.
Cellular fractionation and total membrane enrichment.
Neisserial membrane fractionation was performed according to the method of Pannekoek et al. (1992) with some modifications as follows. Meningococcal cells were resuspended in 180 ml phosphate-buffered saline (PBS), pH 7.5, and collected by centrifugation at 4000 g for 20 min at 4 °C. The cells were resuspended in 30 ml 50 mM Tris/HCl, pH 8.0, and subjected to one round of freeze–thawing at −20 °C and room temperature. Additional cell lysis was conducted by passing the suspension twice through a French press (103 500 kPa), Thermo Electron. Undisrupted cells were removed by centrifugation at 4000 g for 25 min at 4 °C. Remaining cell fragments were discarded by centrifugation twice at 10 000 g for 15 min at 4 °C. The membrane fraction was collected by ultracentrifugation at 215 000 g for 2 h at 4 °C. The membrane pellet was resuspended in 13 ml 50 mM Tris/HCl, pH 8.0. After an additional hour of ultracentrifugation at 215 000 g and 4 °C, the final pellet was resuspended in ∼200 μl distilled water.
Inner membrane isolation.
For solubilization and separation of inner and outer membranes from the membrane fraction, we employed a method based on the ability of N-lauroylsarcosine (Sarkosyl) to selectively solubilize inner membrane proteins (Brossay et al., 1994; Filip et al., 1973; Mietzner et al., 1984). Total membrane fractions (corresponding to 500 μg protein) were washed in 4 ml HEPES buffer, pH 7.4. After 1 h ultracentrifugation at 100 000 g and 4 °C, the pellet was resuspended in 250 μl 10 mM HEPES buffer using a 25 gauge needle. Two-hundred and fifty microlitres of 0.4 % Sarkosyl in 10 mM HEPES buffer was added, giving a final concentration of 0.2 % Sarkosyl, equivalent to 1 μg detergent (μl protein)−1. Incubation at room temperature for 10 min was followed by 1 h ultracentrifugation at 100 000 g and 4 °C. The supernatant, consisting of solubilized inner membrane proteins, was collected, and the pellet was incubated in 0.2 % Sarkosyl and centrifuged once more to remove residual inner membrane proteins. The remaining pellet, constituting the outer membrane fraction, was resuspended in 500 μl 10 mM HEPES buffer, and after 1 h ultracentrifugation at 100 000 g and 4 °C, the final pellet was resuspended in ∼200 μl 10 mM HEPES buffer. Both inner membrane and outer membrane fractions were stored at −70 °C.
Isolation of outer membrane vesicles (OMVs).
Isolation of OMVs from N. meningitidis has previously been described (Balasingham et al., 2007; Frasch & Mocca, 1978). In short, bacteria were grown on blood agar plates, harvested in serotype antigen buffer (0.2 M LiCl, 0.1 M sodium acetate, pH 5.8) and inactivated at 60 °C for 30 min. A bacterial suspension of OD600 20 was prepared and shaken with 2 mm glass beads at 230 r.p.m. for 15 min at room temperature. Cellular debris was removed by two rounds of centrifugation at 4000 g for 20 min and 18 000 g for 15 min, respectively. The supernatant was subjected to ultracentrifugation at 140 000 g for 90 min, the pellet was resuspended in distilled water, and another round of ultracentrifugation at 140 000 g for 90 min was performed. The resulting pellet was resuspended in distilled water and stored at −70 °C.
Determination of protein concentration in membrane fractions.
The protein content in neisserial membrane fractions was determined using a detergent-compatible (DC) protein assay (Bio-Rad Laboratories), which is a modification of the Lowry assay.
Immunoblotting of meningococcal membrane fractions and purified proteins.
A 1 μg sample of purified meningococcal inner and outer membrane proteins, as well as OMV proteins and recombinant PilG protein (E. Lång and others, unpublished results), were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Hybond-C Extra, Amersham GE Healthcare) in Towbin transfer buffer (25 mM Tris/HCl, 192 mM glycine, 20 % methanol, 0.1 % SDS, pH 8.3). Rabbit polyclonal antibodies directed against PilG (E. Lång and others, unpublished results) and PilQ (Tønjum et al., 1998) were used in immunoblotting at dilutions of 1 : 1000 and 1 : 2500, respectively. Procedures for SDS-PAGE and immunoblotting have been previously described (Collins et al., 2005; Tønjum et al., 1998).
Solid-phase overlay assay for protein–DNA interaction.
Protein–DNA interactions were assessed by a solid-phase overlay assay (South-western analysis) (Assalkhou et al., 2007). In short, 3–5 μg of isolated inner membrane proteins and 0.2 μg recombinant PilG protein were separated by SDS-PAGE and transferred to membranes as described above. The membranes were pre-incubated in renaturation buffer (0.25 % gelatin, 0.5 % BSA, 0.2 % Triton X-100, 10 mM Tris/HCl, 5 mM β-mercaptoethanol, 100 mM NaCl, pH 7.5) at room temperature for 1 h prior to the addition of oligonucleotides. Approximately 400 pmol biotinylated DNA substrate (Table 2) was applied to each membrane, and incubation in renaturation buffer was conducted with shaking at 4 °C overnight. The next day, the membranes were incubated for an extra 2 h in renaturation buffer with oligonucleotides at room temperature. The membranes were washed in washing buffer (10 mM Tris/HCl, 100 mM NaCl, pH 7.5). Incubation with alkaline phosphatase (AP)-conjugated streptavidin (1 : 5000) was performed with shaking for 1 h at room temperature. Additional washing with final washing buffer (100 mM Tris/HCl, 0.9 % NaCl, pH 7.5) was then performed. Biotinylated DNA was detected using 5-bromo-4-chloro-3′-indolyl phosphate p-toluidine salt (BCIP) and nitro-blue tetrazolium chloride (NBT) as substrates for the AP. The DNA glycosylase Fpg was used as positive control for the detection of DNA binding and BSA as a negative control. Proteins migrating corresponding to DNA binding bands were excised from a Coomassie blue-stained SDS-PAGE gel, run in parallel with those used in South-western analysis. The excised proteins were pre-treated for MS analysis. Due to the close migration of some proteins in the inner membrane fractions, it was sometimes complicated to excise a single protein. Therefore, proteins were selectively excised from the gel based on the presence of distinct bands both on the Coomassie gel and on the South-western blot(s). The South-western experiments, with parallel SDS-PAGE, and subsequent MS analysis were repeated at least three times.
Table 2.
DNA substrates employed in the study
DUS sequences are underlined.
| Substrate | Sequence | DUS length |
|---|---|---|
| ssDNA | ||
| T1 | 5′ CAACAACAACAACAGCCGTCTGAACCAAATTCAGACGGCAACAACAACAACA 3′ | +, 10 bp |
| T3 | 5′ CAACAACAACAACAGGCCTGTCATCCAAACTGACAGGCCAACAACAACAACA 3′ | − |
| HH8 | 5′ GTTGTTGTTGTTTTCAGACGGCATGTTGGTTCAGACGGCATTTGTTGTTGTT 3′ | +, 12 bp |
| dsDNA | ||
| T1T2 | 5′ CAACAACAACAACAGCCGTCTGAACCAAATTCAGACGGCAACAACAACAACA 3′ | +, 10 bp |
| 5′ TGTTGTTGTTGTTGCCGTCTGAATTTGGTTCAGACGGCTGTTGTTGTTGTTG 3′ | ||
| T3T4 | 5′ CAACAACAACAACAGGCCTGTCATCCAAACTGACAGGCCAACAACAACAACA 3′ | − |
| 5′ TGTTGTTGTTGTTGGCCTGTCATTTTGGATGACAGGCCTGTTGTTGTTGTTG 3′ | ||
| HH7,HH8 | 5′ AACAACAACAAATGCCGTCTGAACCAACATGCCGTCTGAAAACAACAACAAC 3′ | +, 12 bp |
| 5′ GTTGTTGTTGTTTTCAGACGGCATGTTGGTTCAGACGGCATTTGTTGTTGTT 3′ |
Protein identification by peptide mass fingerprinting MALDI-TOF MS.
Proteins, in sections excised from the SDS-PAGE, were subjected to in-gel digestion with trypsin as previously described (Fleckenstein et al., 2004). Tryptic peptide mixtures were desalted and concentrated by running the peptides through a GELoader tip (Eppendorf) column filled with minute amounts of Poros 20 R2 reversed-phase packing sorbent (Applied Biosystems). Finally, a solution containing 70 % acetonitrile (ACN), 0.1 % trifluoroacetic acid (TFA) and 10 mg α-cyano-4-hydroxycinnamic acid ml−1 was used to elute the peptides directly onto a stainless steel target plate (Bruker Daltonics). The samples were allowed to crystallize on the plate and analysed on an Ultraflex II MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) operated in the positive reflector mode. MS lists obtained were searched by MASCOT (Matrix Science) against the NCBI, MSDB and Swiss-Prot databases with N. meningitidis as the selected taxon.
Bioinformatics analyses and screening for DNA binding motifs.
Proteins identified by MS analysis were characterized by using different protein databases and bioinformatics tools. The PROSITE database (Hulo et al., 2008) was used to screen for protein domains and functional sites, whereas EMBOSS (Rice et al., 2000) was used to predict helix–turn–helix structures and define the charge predictions of the deduced amino acids of the proteins, by using the EMBOSS helix–turn–helix and charge package. Uniprot (Boeckmann et al., 2003) provided information about properties of the proteins identified, and MicrobesOnline (Alm et al., 2005) was used to study gene sequence, gene homologues and domain structures.
N. meningitidis mutant construction.
Null mutants corresponding to each of the DNA binding proteins identified were constructed. DNA with homology 400–600 bp upstream and downstream of the target gene was PCR-amplified and ligated with a kanamycin-resistance gene cassette between the two PCR fragments (Menard et al., 1993) into the plasmid pBluescript II SK(+) (pBSK+) through four-point ligation into relevant restriction sites, and plasmid DNA was propagated in E. coli ER2566 or XL-1 Blue (Table 1). The oligonucleotide primers employed are listed in Table 3. N. meningitidis strain M400 was transformed with plasmid DNA carrying the cloned DNA fragments, which recombined and integrated into the host chromosome, allowing each gene to be interrupted by the kanamycin-resistance gene. N. meningitidis transformants were selected by growth on plates containing 100 μg kanamycin ml−1 (Sigma), and the gene disruption(s) were confirmed by PCR and DNA sequence analysis.
Table 3.
Primers employed in construction of mutants
Restriction sites are underlined.
| Primer | Orientation | Sequence (5′→3′) | Restriction site |
|---|---|---|---|
| FtsE (NMB0007) | |||
| KH83 | Forward | GCGCTAGCGTTTCGAACAAGTTTCCAAA | NheI |
| KH10 | Reverse | GCGGATCCCATCAGGGTTTCGTCATGTG | BamHI |
| KH11 | Reverse | GCGAATTCGGATCATAGGAGGTCCTGTAA | EcoRI |
| KH12 | Forward | GCAAGCTTCAAATGGTACAAAGCGCAGA | HindIII |
| ComL (NMB0703) | |||
| KH45 | Forward | GCGCTAGCTCTTGGGTAATCTGGGCATC | NheI |
| KH46 | Reverse | GCGGATCCGTTTGTTGACGACGATGACG | BamHI |
| KH47 | Reverse | GCGAATTCACGAGCTGAACAGCAGCAAT | EcoRI |
| KH48 | Forward | GCAAGCTTCAATATGGCGAGCGATTCTT | HindIII |
| Hypothetical protein (NMB0478) | |||
| KH82 | Forward | GCGCTAGCGAATCAACACTTTCACTACAAGCAA | NheI |
| KH6 | Reverse | GCGGATCCTTCCATCTGTGCGACTTCTG | BamHI |
| KH7 | Reverse | GCAAGCTTTGAAAGTGTTGATTCCATATTAAAC | HindIII |
| KH8 | Forward | GCGGTACCCTTCCGCATCCGATATGACT | KpnI |
| Hypothetical protein (NMB0086) | |||
| KH13 | Forward | GCGAATTCCTCATTGCGCTGCCGTTT | EcoRI |
| KH14 | Reverse | GCGGATCCCTTGCCGTCTATCATCACGA | BamHI |
| KH15 | Reverse | GCGAATTCCGATACATGATGTTCCTTCCAAA | EcoRI |
| KH16 | Forward | GCAAGCTTCCAAATTCGTGTGGTGTCTG | HindIII |
| H.8 (NMB1533) | |||
| KH81 | Reverse | GCGCTAGCGTATCTGGCTCTGATTTCTG | NheI |
| KH2 | Forward | GCAAGCTTCTTCGCCGCCGCCGATCAGTT | HindIII |
| KH3 | Forward | GCGAATTCGCTTTCATAACAAATCTCCAAT | EcoRI |
| KH4 | Reverse | GCGGATCCCATTTTCAGACGGCATGTAT | BamHI |
| Omp85 (NMB0182) | |||
| KH84 | Reverse | GCGCTAGCGCATATCGCCTTTGGCACTT | NheI |
| KH18 | Forward | GCAAGCTTTTCAAATTCGATGTCGGTGA | HindIII |
| KH19 | Forward | GCGAATTCAGTTCCTTGTGGTGCGGAA | EcoRI |
| KH20 | Reverse | GCGGATCCAGCTACCGTCCGTCTGTTGT | BamHI |
Phenotypic analysis of N. meningitidis null mutants.
N. meningitidis null mutants were compared with wild-type strains in phenotypic analyses.
(i) Colony morphology. N. meningitidis strains cultured on clear GC plates were assessed by stereo microscopy to define whether they had an agglutinating (agg+) or non-agglutinating (agg−) colonial morphology (Blake et al., 1989).
(ii) Purification of type IV pilus fibres. Type IV pili were purified from the meningococcal cell surface using ammonium sulfate precipitation of a shearing fraction (Brinton et al., 1978). Meningococcal cells from half a heavily streaked GC plate were resuspended in 1 ml 0.15 M ethanolamine buffer, pH 10.5, and vortexed for 1 min. Cellular debris was removed by centrifugation at 16 000 g for 30 min twice. Pili were precipitated from the supernatant by adding one-tenth of the total volume of saturated ammonium sulfate in 0.15 M ethanolamine buffer and left at room temperature overnight. Precipitated pili were collected by centrifugation for 15 min at 16 000 g, and the pellet was washed twice in 1 ml 50 mM Tris/HCl, 150 mM NaCl, pH 8.0, before being dissolved in distilled water. Total protein was determined in the residual cells with a Bio-Rad DC protein assay. The pilus preparations were analysed on SDS-PAGE gels stained with Coomassie blue, using the total protein concentration from the residual cells to normalize the loading. Each null mutant was analysed in triplicate and the experiment was repeated at least three times.
(iii) Competence screening. Competence for transformation of wild-type and mutant strains was performed using the plasmid p1080 mutY-ermr as donor DNA (Table 1) (Davidsen et al., 2007b). Wild-type and mutant strains were harvested in CO2-saturated liquid GC medium containing 7 mM MgCl2 and 1× IsoVitalex (Becton Dickinson Diagnostic Systems). The bacteria were exposed to either plasmid p1080 mutY-ermr or distilled water (negative control). Addition of 0.1 mg DNase I ml−1 (Sigma) mediated degradation of extracellular DNA before 10 volumes of liquid GC medium were added. The bacterial solutions were incubated with tumbling at 37 °C for 4.5 h and subsequently plated on both plain GC medium and GC medium containing 300 μg erythromycin (Erm) ml−1. The transformation rate was estimated by dividing the number of Erm-resistant c.f.u. by the total number of c.f.u. The assay was repeated at least three times for each null mutant.
RESULTS
Cellular fractionation and inner membrane isolation
Inner membrane fractions from a panel of N. meningitidis strains from three important serogroups (A, B and C) as well as N. gonorrhoeae strain N400 were enriched, using antibody-mediated PilG detection as a marker for inner membrane protein content (Fig. 1). Although Sarkosyl preferentially solubilizes inner membranes, complete separation of neisserial inner and outer membrane proteins is a challenging, if not impossible, task (Masson & Holbein, 1983). In addition to the inner membrane proteins enriched, some outer membrane proteins were also present in the neisserial inner membrane fractions, including PilQ, H.8 and Omp85. These proteins are normally expressed at high levels. The presence of outer membrane proteins within the inner membrane fraction was quantified by using PilQ as a marker (data not shown). In addition, some PilG protein could be detected in the outer membrane fraction (Fig. 1, lane 5), once again indicating that the separation of inner and outer membrane proteins was incomplete, due to the membrane composition and the limitations of the separation methods available. It must however be noted that the vast majority of PilG and other inner membrane proteins were found in the inner membrane fraction and vice versa for PilQ in the outer membrane fraction. The aim of this part of the study was to enrich inner membrane proteins into one fraction in our search for DNA binding proteins. For this purpose, using the detergent Sarkosyl was clearly more efficient than using Triton X-100 for neisserial inner membrane enrichment (data not shown).
Fig. 1.
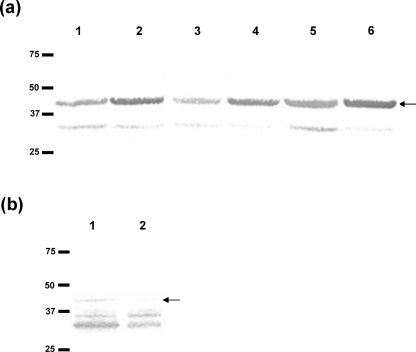
Identification of PilG in the inner membrane (IM). Immunoblotting of meningococcal membrane fractions localizes the PilG protein in the IM of N. meningitidis. Lanes: 1, H44/76 wild-type (wt) IM; 2, H44/76 ΔPilG IM; 3, Z2491 wt IM; 4, M1080 wt IM; 5, H44/76 wt outer membrane. The positions of the 37 and 50 kDa size standards are shown on the left. The arrow on the right indicates the position of the PilG protein.
Detecting DNA binding proteins by a solid-phase overlay assay
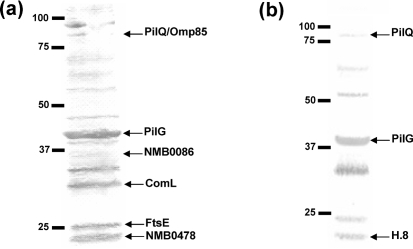
Inner membrane fractions, representing lower complexity than using the entire cellular lysate as a target, from a panel of neisserial strains were searched for DNA binding proteins by solid-phase overlay assay in the form of a South-western analysis. All inner membrane fractions were tested with all the DNA substrates with or without a DUS listed in Table 2. Inner membrane fractions from all neisserial strains included in the study exhibited multiple DNA binding proteins. A selection of the proteins identified by MS is shown in Fig. 2. However, no apparent DUS specificity or preference for single-stranded or double-stranded DNA was detected among the candidate proteins identified. Protein bands to be characterized by subsequent MS analysis were selected on the basis of the reproducibility of their identification as a DNA binding component as well as the gel resolution of proteins of similar size. The complexity in the protein expression profile, differential expression levels and the presence of co-migrating bands make it possible that not all proteins with a putative DNA binding activity would be detected. Thus, the procedure employed might yield only a selection of all DNA binding proteins co-purifying with the neisserial inner membrane (see Discussion).
Fig. 2.
Detection of DNA binding components. Solid-phase overlay assay (South-western analysis) of neisserial inner membrane fractions with biotin-labelled DNA substrates was performed. (a) Identification of PilQ, Omp85, PilG, NMB0086, ComL, FtsE and NMB0478 in the inner membrane fraction of N. meningitidis strain H44/76. (b) Identification of PilQ, Omp85, PilG and H.8 in the inner membrane fraction of N. meningitidis strain 8013. The DNA substrates depicted were ssDNA substrate T1 (a) and T3 (b) with and without the 10 bp DUS, respectively. The positions of the size standards (kDa) are shown on the left.
Identification of DNA binding proteins
DNA binding proteins excised from gels were trypsin-treated and identified by MS (Table 4). Among the inner membrane proteins identified that bound DNA were the competence protein ComL, the cell division ATP-binding protein FtsE, as well as two hypothetical proteins NMB0478 and NMB0086 (Fig. 2). Reports have suggested that the 29 kDa lipoprotein ComL (Fig. 2a) contributes to cleavage of the peptidoglycan layer, providing access for incoming DNA to enter the bacterial cell. However, the main function of ComL is difficult to assess, since most mutants of this essential component are lethal (Fussenegger et al., 1996). The cell division ATP-binding protein FtsE (Corbin et al., 2007) was also identified as an inner membrane protein that binds DNA (Fig. 2). FtsE has been shown to be associated with the inner membrane in E. coli (Gill & Salmond, 1987), and is believed to be involved in cell division due to its interactions with cell division proteins FtsZ and FtsX. The sequences of FtsE and FtsX show homology to ABC transporters; FtsE makes up the ATP-binding cassette (ABC) component, while FtsX anchors the FtsEX complex to the membrane. FtsZ polymerizes into the Z ring and recruits the FtsEX complex to the division site (Corbin et al., 2007; de Leeuw et al., 1999; Schmidt et al., 2004). Even though FtsE is essential in E. coli, studies of FtsE in N. gonorrhoeae indicate that the protein is non-essential in this organism (Bernatchez et al., 2000). Furthermore, the two hypothetical proteins NMB0478 and NMB0086 identified are conserved among the neisserial species, but show no significant homology to other proteins of known function (Fig. 2a).
Table 4.
Phenotypic traits of null mutants corresponding to the DNA binding proteins identified by solid-phase overlay assay and MS analysis
| Protein identified* | Putative function | Colony morphology† | Extracellular pilus expression | Competence for transformation |
|---|---|---|---|---|
| NMB0007 FtsE DUS− | ATPase activity, cell division | agg+ | wt‡ | Yes |
| NMB0703 ComL DUS− | Competence, peptidoglycan function | Not viable | Not viable | Not viable |
| NMB0478 hypothetical protein DUS+ | Unknown | agg+ | wt | Yes |
| NMB0086 hypothetical protein DUS+ | Unknown | agg+ | wt | Yes |
| NMB0333 PilG DUS− | Pilus biogenesis? | agg− | Absent | Non-competent (Tønjum et al., 1995) |
| NMB1812 PilQ DUS− | Outer membrane pore, pilus extrusion transformation | agg− | Absent | Non-competent (Frye et al., 2006) |
| NMB1533 H.8 DUS− | Iron–sulfur binding? | agg+, smaller than wt | wt | Yes |
| NMB0182 Omp85 DUS− | Cell envelope biogenesis | Not viable | Not viable | Not viable |
*The presence/absence of the DUS is given as DUS+/DUS−.
†Colony morphology is described as agglutinating (agg+) or non-agglutinating (agg−).
‡wt, Wild-type levels.
In addition, the pilus-biogenesis protein PilG was found to exhibit DNA binding activity (Fig. 2). Again, no DUS specificity or preference for binding to single-stranded versus double-stranded DNA substrates was observed (Figs 2 and 3; and data not shown). PilG is a polytopic inner membrane protein; however, its exact function in pilus biogenesis is yet to be elucidated.
Fig. 3.
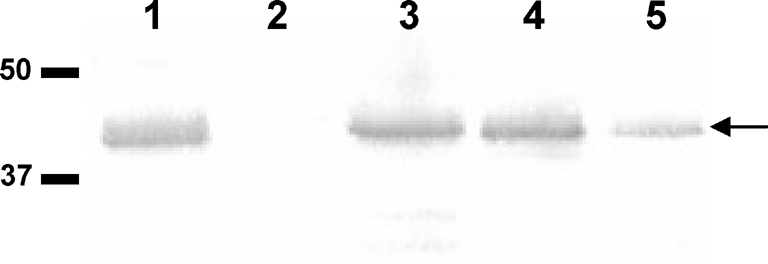
PilG binds DNA without DUS specificity. Solid-phase overlay assay (South-western analysis) shows DNA binding activity of recombinant PilG protein and the PilG protein from the meningococcal inner membrane fraction. No DUS specificity was observed and the position of the PilG protein is indicated by the arrow. (a) DNA binding activity of full-length recombinant PilG. Lanes: 1, ssDNA substrate (T1) with 10 bp DUS; 2, dsDNA substrate (T1T2) with 10 bp DUS; 3, ssDNA substrate (T3) without DUS; 4, dsDNA substrate (T3T4) without DUS; 5, ssDNA substrate (HH8) with 12 bp DUS; 6, dsDNA substrate (HH7,HH8) with 12 bp DUS. (b) Identification of PilG DNA binding activity in the meningococcal inner membrane fraction using ssDNA substrate (T1). Lane 1, H44/76 wild-type; lane 2, H44/76 ΔPilG. The positions of the size standards (kDa) are shown on the left.
Among the DNA binding outer membrane proteins detected that co-purified with neisserial inner membranes were PilQ, H.8, Omp85 and opacity proteins, all of which are outer membrane proteins expressed at high levels in Neisseria species. Contamination of outer membrane proteins in the inner membrane fractions was assessed by monitoring the presence of the secretin PilQ (Collins et al., 2001; Tønjum et al., 1998). PilQ binds DNA without DUS specificity (Assalkhou et al., 2007) and also directly interacts with the inner membrane lipoprotein PilP (Balasingham et al., 2007), promoting its presence in inner membrane fractions. Omp85 is an outer membrane protein, highly conserved among Gram-negative bacteria. This protein has been shown to be essential for survival in N. meningitidis and plays a role in outer membrane assembly (Voulhoux et al., 2003). H.8, also known as Lip in N. meningitidis, is a neisserial-specific outer membrane lipoprotein, primarily expressed in pathogenic Neisseria species and not so frequently expressed in commensal species (Cannon et al., 1984; Hitchcock et al., 1985; Hitchcock, 1989). One study has shown that the H.8 lipoprotein of N. gonorrhoeae stimulates the immune system, indicating a possible role in neisserial pathogenesis (Fisette et al., 2003). Finally, opacity proteins are phase-variable outer membrane proteins that mediate adherence of pathogenic Neisseria spp. to host cells (Dehio et al., 1998).
Bioinformatics analysis of DNA binding components
The deduced amino acid sequences of the DNA binding proteins identified were screened for recognized DNA binding motifs such as helix–turn–helix, helix–hairpin–helix, helix–loop–helix, leucine zipper and zinc finger. No recognized DNA binding motifs were found in any of the candidate proteins. Furthermore, the genes encoding the two hypothetical proteins NMB0478 and NMB0086 contain DUS sequences, which might indicate that they have a role in DNA metabolism (Davidsen et al., 2004; Treangen et al., 2008).
N. meningitidis mutant phenotypes
In order to assess the significance of the DNA binding activity of the proteins identified, the corresponding null mutants of candidates yielding highly reproducible results were constructed. The mutants were assessed with regard to colony morphology (which is dependent on the piliation state), expression of extracellular pili, and competence for transformation as compared with the wild-type strain (Table 4). Only the pilus biogenesis mutants ΔpilG and ΔpilQ were defective in competence, as well as in pilus expression, and showed a non-agglutinating colony morphology (Table 4).
DISCUSSION
The full complement of DNA binding proteins involved in neisserial transformation is yet to be unravelled. The outer membrane complex PilQ has previously been shown to bind DNA. In order to define inner membrane proteins engaged in transformation, we searched for DNA binding components in inner membrane fractions of N. meningitidis by a solid-phase overlay assay and tested the corresponding null mutants with regard to competence for transformation. A number of proteins exhibited DNA binding activity in this in vitro assay, and the predominant eight of these were identified by MS. However, only pilus biogenesis components exerted an effect on transformation. The finding that pilus biogenesis proteins directly bind DNA further supports the already established link between pilus biogenesis and uptake of DNA by natural transformation in N. meningitidis.
Searching for DNA binding proteins by a solid-phase overlay assay presents both opportunities and limitations. The main advantage of the method is that by coupling it to SDS-PAGE, it is possible to separate complex mixtures of proteins, assess the DNA binding abilities of proteins separately and identify DNA binding proteins by MS. During the SDS-PAGE step, proteins are, however, denatured, and if their DNA binding activity is dependent on correct folding, the proteins have to be able to renature under the conditions employed in order to be detected. In addition, the proteins have to be expressed in high enough amounts to give distinct and detectable bands in Coomassie blue-stained gels, enabling excision and identification of proteins by MS. However, some proteins might not renature under the conditions employed and/or are expressed at too low levels to yield a distinct band in SDS-PAGE. In addition, the accuracy of protein excision might vary due to co-migration in the PAGE system. Thus, only a predominant and selected subset of all potential DNA binding proteins in a sample will be detected by this method.
As for any method measuring in vitro protein–DNA interactions, the detection of an interaction in the current assay would only be indicative with respect to the relevance to the biological system under study. Furthermore, due to positive electrostatic charge, DNA binding activity detected in vitro might be non-specific; however, recognized DNA binding motifs also exhibit positive electrostatic charge. Despite these limitations, the strategy employed is a useful approach for screening complex mixtures of proteins and identifying candidates with DNA binding activity. These represent a basis for further testing. In this context, we focused on proteins involved in transformation, which is the major source of the abundance of exogenous DNA introduced into the meningococcal chromosome.
Null mutants corresponding to the DNA binding proteins identified in neisserial inner membranes were constructed and tested for competence for transformation. Only the PilG and PilQ mutants, defective in pilus biogenesis, were non-competent for transformation. For these two components, the interpretation of the biological significance of their DNA binding capabilities is complicated by the fact that they participate in type IV pilus biogenesis, which is required for competence. Thus, it is a conundrum whether the lack of competence in these mutants is due to a defect in their direct binding of DNA, or whether it is indirect, through pilus biogenesis. The DNA binding activity of PilQ in a solid-phase overlay assay has previously been documented, and the PilQ–DNA interaction has also been verified by electromobility shift analysis (EMSA) and mapped to the pore region of the PilQ complex by electron microscopy (Assalkhou et al., 2007). Those findings represent validation of the solid-phase overlay approach for the identification of DNA binding proteins in general. Nevertheless, the biological significance and nature of PilQ DNA binding activity remains to be fully elucidated. The DNA binding activity of PilG has, however, not been previously described.
The null mutants for the cell division ATP-binding protein FtsE, the outer membrane protein H.8 and the two hypothetical proteins NMB0478 and NMB0086 were all competent for transformation. Interestingly, the meningococcal ΔftsE mutant was viable. Even if no effect on transformation was observed, a potential role for their observed DNA binding activity in other cellular events such as conjugation, replication and other DNA metabolism cannot be excluded. However, defining the context of their DNA binding activity was beyond the scope of this study.
For two of the DNA binding proteins identified, ComL and Omp85, viable mutants could not be made, suggesting that they are essential. We were therefore unable to test whether their observed DNA binding activity contributes to transformation. However, ComL has been suggested to play a role in the peptidoglycan-related phase of transformation (Fussenegger et al., 1996). Thus, further studies are needed to determine whether the observed DNA binding has any biological significance in transformation. Whether the DNA binding noted for the proteins Omp85 and H.8 is significant or not must also be assessed in light of the relative abundance of these proteins in the outer membrane.
The ability to take up exogenous DNA is of great importance when it comes to bacterial fitness, survival and evolution (Ochman et al., 2000). The uptake of DNA in N. meningitidis is dependent on DUS sequences, which are present in large numbers throughout the neisserial genome. Interestingly, it has been shown that DUS sequences inside ORFs are preferentially located in genome maintenance and repair genes (Davidsen et al., 2004), indicating that transforming DNA has a function in DNA repair (Treangen et al., 2008). The preferential uptake of DNA containing DUS sequences makes the bacterium more prone to take up DNA from itself and/or closely related species. One central question is how N. meningitidis manages to bind, take up and incorporate new DNA into the genome. The discovery of a DUS-specific receptor would be a great milestone in the elucidation of neisserial natural transformation. A DNA binding protein that exerts DUS specificity in DNA binding was not detected in this study of inner membrane components. It is still an open question whether the discrimination between DUS-positive and DUS-negative DNA occurs before or after the transforming DNA reaches the inner membrane. Since the secretin PilQ appears to function as a non-selective gateway for DNA through the outer membrane, DUS specificity might be exerted in the periplasm/peptidoglycan layer, the inner membrane, or during recombination. In this context, the continued search for neisserial DNA binding components is highly relevant.
Acknowledgments
This work was funded by a grant from the Research Council of Norway. We thank Kristian Alfsnes for the assistance in transformation experiments. We are grateful to Marit Jørgensen for protein identification at the Rikshospitalet-Radiumhospitalet/Universitetet i Oslo (RR/UiO) Core Facility for Proteomics/Mass Spectrometry at Gaustad.
Abbreviations
DUS, DNA uptake sequence
OMV, outer membrane vesicle
References
- Achtman, M., Wall, R. A., Bopp, M., Kusecek, B., Morelli, G., Saken, E. & Hassan-King, M. (1991). Variation in class 5 protein expression by serogroup A meningococci during a meningitis epidemic. J Infect Dis 164, 375–382. [DOI] [PubMed] [Google Scholar]
- Alm, E. J., Huang, K. H., Price, M. N., Koche, R. P., Keller, K., Dubchak, I. L. & Arkin, A. P. (2005). The MicrobesOnline Web site for comparative genomics. Genome Res 15, 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambur, O. H., Frye, S. A. & Tønjum, T. (2007). New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J Bacteriol 189, 2077–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assalkhou, R., Balasingham, S., Collins, R. F., Frye, S. A., Davidsen, T., Benam, A. V., Bjørås, M., Derrick, J. P. & Tønjum, T. (2007). The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology 153, 1593–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasingham, S. V., Collins, R. F., Assalkhou, R., Homberset, H., Frye, S. A., Derrick, J. P. & Tønjum, T. (2007). Interactions between the lipoprotein PilP and the secretin PilQ in Neisseria meningitidis. J Bacteriol 189, 5716–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatchez, S., Francis, F. M., Salimnia, H., Beveridge, T. J., Li, H. & Dillon, J. A. (2000). Genomic, transcriptional and phenotypic analysis of ftsE and ftsX of Neisseria gonorrhoeae. DNA Res 7, 75–81. [DOI] [PubMed] [Google Scholar]
- Bieber, D., Ramer, S. W., Wu, C. Y., Murray, W. J., Tobe, T., Fernandez, R. & Schoolnik, G. K. (1998). Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280, 2114–2118. [DOI] [PubMed] [Google Scholar]
- Biswas, G. D., Sox, T., Blackman, E. & Sparling, P. F. (1977). Factors affecting genetic transformation of Neisseria gonorrhoeae. J Bacteriol 129, 983–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, M. S., MacDonald, C. M. & Klugman, K. P. (1989). Colony morphology of piliated Neisseria meningitidis. J Exp Med 170, 1727–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., Martin, M. J., Michoud, K., O'Donovan, C. & other authors (2003). The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bøvre, K. (1964). Studies on transformation in Moraxella and organisms assumed to be related to Moraxella. 1. A method for quantitative transformation in Moraxella and Neisseria, with streptomycin resistance as the genetic marker. Acta Pathol Microbiol Scand 61, 457–473. [DOI] [PubMed] [Google Scholar]
- Bøvre, K., Bergan, T. & Frøholm, L. O. (1970). Electron microscopical and serological characteristics associated with colony type in Moraxella nonliquefaciens. Acta Pathol Microbiol Scand [B] Microbiol Immunol 78, 765–779. [DOI] [PubMed] [Google Scholar]
- Bradley, D. E. (1974). The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology 58, 149–163. [DOI] [PubMed] [Google Scholar]
- Brinton, C. C., Bryan, J., Dillon, J. A., Guerina, N., Jacobson, L. J., Labik, A., Lee, S., Levine, A., Lim, S. & other authors (1978). Uses of pili in gonorrhoea control: role of bacterial pili in disease, purification and properties of gonococcal pili, and progress in the development of a gonococcal pilus vaccine for gonorrhoeae. In Immunobiology of Neisseria gonorrhoeae, pp. 155–178. Edited by G. E. Brooks, E. C. Gotschlich, K. H. Homes, W. D. Sawyer & F. E. Young. Washington, DC: American Society for Microbiology.
- Brossay, L., Paradis, G., Fox, R., Koomey, M. & Hebert, J. (1994). Identification, localization, and distribution of the PilT protein in Neisseria gonorrhoeae. Infect Immun 62, 2302–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, J. G., Black, W. J., Nachamkin, I. & Stewart, P. W. (1984). Monoclonal antibody that recognizes an outer membrane antigen common to the pathogenic Neisseria species but not to most nonpathogenic Neisseria species. Infect Immun 43, 994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant, D. A., Frøholm, L. O., Bøvre, K., Holten, E., Frasch, C. E., Mocca, L. F., Zollinger, W. D. & Selander, R. K. (1986). Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc Natl Acad Sci U S A 83, 4927–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, I. & Gotschlich, E. C. (2001). ComE, a competence protein from Neisseria gonorrhoeae with DNA binding activity. J Bacteriol 183, 3160–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R. F., Davidsen, L., Derrick, J. P., Ford, R. C. & Tønjum, T. (2001). Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J Bacteriol 183, 3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R. F., Frye, S. A., Balasingham, S., Ford, R. C., Tønjum, T. & Derrick, J. P. (2005). Interaction with type IV pili induces structural changes in the bacterial outer membrane secretin PilQ. J Biol Chem 280, 18923–18930. [DOI] [PubMed] [Google Scholar]
- Comolli, J. C., Hauser, A. R., Waite, L., Whitchurch, C. B., Mattick, J. S. & Engel, J. N. (1999). Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect Immun 67, 3625–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin, B. D., Wang, Y., Beuria, T. K. & Margolin, W. (2007). Interaction between cell division proteins FtsE and FtsZ. J Bacteriol 189, 3026–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe, B. A., Wall, R. A., Kusecek, B., Neumann, B., Olyhoek, T., Abdillahi, H., Hassan-King, M., Greenwood, B. M., Poolman, J. T. & Achtman, M. (1989). Clonal and variable properties of Neisseria meningitidis isolated from cases and carriers during and after an epidemic in The Gambia, West Africa. J Infect Dis 159, 686–700. [DOI] [PubMed] [Google Scholar]
- Davidsen, T., Rødland, E. A., Lagesen, K., Seeberg, E., Rognes, T. & Tønjum, T. (2004). Biased distribution of DNA uptake sequences towards genome maintenance genes. Nucleic Acids Res 32, 1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen, T., Koomey, M. & Tønjum, T. (2007a). Microbial genome dynamics in CNS pathogenesis. Neuroscience 145, 1375–1387. [DOI] [PubMed] [Google Scholar]
- Davidsen, T., Tuven, H. K., Bjørås, M., Rødland, E. A. & Tønjum, T. (2007b). Genetic interactions of DNA repair pathways in the pathogen Neisseria meningitidis. J Bacteriol 189, 5728–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio, C., Gray-Owen, S. D. & Meyer, T. F. (1998). The role of neisserial Opa proteins in interactions with host cells. Trends Microbiol 6, 489–495. [DOI] [PubMed] [Google Scholar]
- de Leeuw, E., Graham, B., Phillips, G. J., ten Hagen-Jongman, C. M., Oudega, B. & Luirink, J. (1999). Molecular characterization of Escherichia coli FtsE and FtsX. Mol Microbiol 31, 983–993. [DOI] [PubMed] [Google Scholar]
- Facius, D. & Meyer, T. F. (1993). A novel determinant (comA) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of a comA defect on pilin variation. Mol Microbiol 10, 699–712. [DOI] [PubMed] [Google Scholar]
- Filip, C., Fletcher, G., Wulff, J. L. & Earhart, C. F. (1973). Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol 115, 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisette, P. L., Ram, S., Andersen, J. M., Guo, W. & Ingalls, R. R. (2003). The Lip lipoprotein from Neisseria gonorrhoeae stimulates cytokine release and NF-κB activation in epithelial cells in a Toll-like receptor 2-dependent manner. J Biol Chem 278, 46252–46260. [DOI] [PubMed] [Google Scholar]
- Fleckenstein, B., Qiao, S. W., Larsen, M. R., Jung, G., Roepstorff, P. & Sollid, L. M. (2004). Molecular characterization of covalent complexes between tissue transglutaminase and gliadin peptides. J Biol Chem 279, 17607–17616. [DOI] [PubMed] [Google Scholar]
- Frasch, C. E. & Chapman, S. S. (1972). Classification of Neisseria meningitidis group B into distinct serotypes. I. Serological typing by a microbactericidal method. Infect Immun 5, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch, C. E. & Mocca, L. F. (1978). Heat-modifiable outer membrane proteins of Neisseria meningitidis and their organization within the membrane. J Bacteriol 136, 1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, A., Prust, C., Hartsch, T., Henne, A. & Averhoff, B. (2002). Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl Environ Microbiol 68, 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøholm, L. O., Jyssum, K. & Bøvre, K. (1973). Electron microscopical and cultural features of Neisseria meningitidis competence variants. Acta Pathol Microbiol Scand [B] Microbiol Immunol 81, 525–537. [DOI] [PubMed] [Google Scholar]
- Frye, S. A., Assalkhou, R., Collins, R. F., Ford, R. C., Petersson, C., Derrick, J. P. & Tonjum, T. (2006). Topology of the outer membrane secretin PilQ from Neisseria meningitidis. Microbiology 152, 3751–3764. [DOI] [PubMed] [Google Scholar]
- Fussenegger, M., Facius, D., Meier, J. & Meyer, T. F. (1996). A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol Microbiol 19, 1095–1105. [DOI] [PubMed] [Google Scholar]
- Gill, D. R. & Salmond, G. P. (1987). The Escherichia coli cell division proteins FtsY, FtsE and FtsX are inner membrane-associated. Mol Gen Genet 210, 504–508. [DOI] [PubMed] [Google Scholar]
- Goodman, S. D. & Scocca, J. J. (1988). Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 85, 6982–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen, J., Frøholm, L. O. & Bøvre, K. (1972). Studies on bacterial surface translocation. 2. Correlation of twitching motility and fimbriation in colony variants of Moraxella nonliquefaciens, M. bovis, and M. kingii. Acta Pathol Microbiol Scand [B] Microbiol Immunol 80, 445–452. [PubMed] [Google Scholar]
- Hitchcock, P. J. (1989). Unified nomenclature for pathogenic Neisseria species. Clin Microbiol Rev 2 (Suppl.), S64–S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock, P. J., Hayes, S. F., Mayer, L. W., Shafer, W. M. & Tessier, S. L. (1985). Analyses of gonococcal H8 antigen. Surface location, inter- and intrastrain electrophoretic heterogeneity, and unusual two-dimensional electrophoretic characteristics. J Exp Med 162, 2017–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holten, E. (1979). Serotypes of Neisseria meningitidis isolated from patients in Norway during the first six months of 1978. J Clin Microbiol 9, 186–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., Cuche, B. A., de Castro, E., Lachaize, C., Langendijk-Genevaux, P. S. & Sigrist, C. J. (2008). The 20 years of PROSITE. Nucleic Acids Res 36, D245–D249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyssum, K. & Lie, S. (1965). Genetic factors determining competence in transformation of Neisseria meningitidis. 1. A permanent loss of competence. Acta Pathol Microbiol Scand 63, 306–316. [DOI] [PubMed] [Google Scholar]
- Koomey, M. (1998). Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl 84, 56–61. [DOI] [PubMed] [Google Scholar]
- Koomey, J. M. & Falkow, S. (1987). Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J Bacteriol 169, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. G. & Wackernagel, W. (1990). Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA. Arch Microbiol 154, 380–385. [DOI] [PubMed] [Google Scholar]
- Masson, L. & Holbein, B. E. (1983). Physiology of sialic acid capsular polysaccharide synthesis in serogroup B Neisseria meningitidis. J Bacteriol 154, 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis, L. S. & Scocca, J. J. (1984). On the role of pili in transformation of Neisseria gonorrhoeae. J Gen Microbiol 130, 3165–3173. [DOI] [PubMed] [Google Scholar]
- Mattick, J. S. (2002). Type IV pili and twitching motility. Annu Rev Microbiol 56, 289–314. [DOI] [PubMed] [Google Scholar]
- McGuinness, B. T., Clarke, I. N., Lambden, P. R., Barlow, A. K., Poolman, J. T., Jones, D. M. & Heckels, J. E. (1991). Point mutation in meningococcal por A gene associated with increased endemic disease. Lancet 337, 514–517. [DOI] [PubMed] [Google Scholar]
- Menard, R., Sansonetti, P. J. & Parsot, C. (1993). Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol 175, 5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz, A. J., Enns, C. A. & So, M. (1999). Type IV pili of pathogenic Neisseriae elicit cortical plaque formation in epithelial cells. Mol Microbiol 32, 1316–1332. [DOI] [PubMed] [Google Scholar]
- Merz, A. J., So, M. & Sheetz, M. P. (2000). Pilus retraction powers bacterial twitching motility. Nature 407, 98–102. [DOI] [PubMed] [Google Scholar]
- Mietzner, T. A., Luginbuhl, G. H., Sandstrom, E. & Morse, S. A. (1984). Identification of an iron-regulated 37,000-Dalton protein in the cell envelope of Neisseria gonorrhoeae. Infect Immun 45, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H., Lawrence, J. G. & Groisman, E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304. [DOI] [PubMed] [Google Scholar]
- O'Toole, G. A. & Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30, 295–304. [DOI] [PubMed] [Google Scholar]
- Pannekoek, Y., van Putten, J. P. & Dankert, J. (1992). Identification and molecular analysis of a 63-kilodalton stress protein from Neisseria gonorrhoeae. J Bacteriol 174, 6928–6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol, C., Eugene, E., Marceau, M. & Nassif, X. (1999). The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc Natl Acad Sci U S A 96, 4017–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, P., Longden, I. & Bleasby, A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Schmidt, K. L., Peterson, N. D., Kustusch, R. J., Wissel, M. C., Graham, B., Phillips, G. J. & Weiss, D. S. (2004). A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol 186, 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, B. J. & Kwaik, Y. A. (1999). Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol 181, 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, J. (1973). Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J Exp Med 137, 571–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, J., Kraus, S. J. & Gotschlich, E. C. (1971). Studies on gonococcus infection. I. Pili and zones of adhesion: their relation to gonococcal growth patterns. J Exp Med 134, 886–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønjum, T., Weir, S., Bøvre, K., Progulske-Fox, A. & Marrs, C. F. (1993). Sequence divergence in two tandemly located pilin genes of Eikenella corrodens. Infect Immun 61, 1909–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønjum, T., Freitag, N. E., Namork, E. & Koomey, M. (1995). Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol 16, 451–464. [DOI] [PubMed] [Google Scholar]
- Tønjum, T., Caugant, D. A., Dunham, S. A. & Koomey, M. (1998). Structure and function of repetitive sequence elements associated with a highly polymorphic domain of the Neisseria meningitidis PilQ protein. Mol Microbiol 29, 111–124. [DOI] [PubMed] [Google Scholar]
- Treangen, T. J., Ambur, O. H., Tonjum, T. & Rocha, E. P. (2008). The impact of the neisserial DNA uptake sequences on genome evolution and stability. Genome Biol 9, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M. & Tommassen, J. (2003). Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265. [DOI] [PubMed] [Google Scholar]
- Wolfgang, M., Lauer, P., Park, H. S., Brossay, L., Hebert, J. & Koomey, M. (1998). PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol 29, 321–330. [DOI] [PubMed] [Google Scholar]