Abstract
The Escherichia coli guaB promoter (PguaB) regulates transcription of two genes, guaB and guaA, that are required for the synthesis of guanosine 5′-monophosphate (GMP), a precursor for the synthesis of guanine nucleoside triphosphates. Transcription from PguaB increases as a function of increasing cellular growth rate, and this is referred to as growth rate-dependent control (GRDC). Here we investigated the role of the factor for inversion stimulation (FIS) in the regulation of this promoter. The results showed that there are three binding sites for FIS centred near positions −11, +8 and +29 relative to the guaB transcription start site. Binding of FIS to these sites results in repression of PguaB in vitro but not in vivo. Deletion of the fis gene results in increased PguaB activity in vivo, but GRDC of PguaB is maintained.
INTRODUCTION
The Escherichia coli guaB promoter (PguaB) regulates transcription of two genes, guaB and guaA, which together constitute the guaBA operon. The guaB and guaA genes encode inosine 5′-monophosphate (IMP) dehydrogenase and guanosine 5′-monophosphate (GMP) synthetase, respectively, and are required for the biosynthesis of GMP from the common purine nucleotide precursor, IMP (Mehra & Drabble, 1981; Tiedeman & Smith, 1984). PguaB is regulated by the cAMP receptor protein (CRP) and a putative CRP binding site is centred near position −117.5 relative to the guaB transcription start site (Hutchings & Drabble, 2000). Furthermore, transcription from PguaB is strongly enhanced by an UP element (Husnain & Thomas, 2008). PguaB also contains a putative binding site for PurR that overlaps the core promoter region, and PurR downregulates expression of guaB (Meng et al., 1990; Davies & Drabble, 1996). DnaA binds to a sequence overlapping the core promoter region and also downregulates transcription from PguaB (Tesfa-Selase & Drabble, 1996). It has also been shown that the rate of transcription from PguaB per unit cell mass increases as a function of increasing cellular growth rate (Davies & Drabble, 1996; Husnain & Thomas, 2008). This phenomenon is commonly referred to as growth rate-dependent control (GRDC) (Gourse et al., 1996; Dennis et al., 2004). GRDC of PguaB requires the UP element and sequences located upstream of the UP element (Husnain & Thomas, 2008).
The factor for inversion stimulation (FIS) regulates transcription by binding to highly degenerate 15 bp DNA sequences (Finkel & Johnson, 1992; Ross et al., 1999). At some E. coli promoters, FIS activates transcription by contacting the C-terminal domain of the RNA polymerase (RNAP) α subunit (αCTD) (Aiyar et al., 2002; McLeod et al., 2002). FIS can also promote transcription by decreasing the negative superhelicity of DNA (Travers et al., 2001). At other promoters, FIS downregulates promoter activity by binding to a site that overlaps or is located downstream from the RNAP binding site, or by forming a complex assembly with other nucleoid proteins (Gonzalez-Gil et al., 1998; Browning et al., 2000, 2004; Jackson et al., 2004). A previous study identified four putative binding sites for FIS that are located upstream of the PguaB core promoter region (Hutchings & Drabble, 2000). The putative FIS binding sites are centred near positions −77, −92, −109 and −126 relative to the guaB transcription start, and were referred to as FIS sites I–IV, respectively (Fig. 1a). FIS contributes to GRDC of the thrU and pdxA promoters, and is required for growth rate-dependent synthesis of 4.5S RNA,  and
and  (Emilsson & Nilsson, 1995; Dong et al., 1996). Moreover, transcription from the fis promoter is coupled to cellular growth rate (Mallik et al., 2006). Cellular levels of FIS and FIS mRNA also change with the growth phase, and they increase dramatically upon entry into the mid-exponential growth phase (Appleman et al., 1998; Ali Azam et al., 1999; Mallik et al., 2006).
(Emilsson & Nilsson, 1995; Dong et al., 1996). Moreover, transcription from the fis promoter is coupled to cellular growth rate (Mallik et al., 2006). Cellular levels of FIS and FIS mRNA also change with the growth phase, and they increase dramatically upon entry into the mid-exponential growth phase (Appleman et al., 1998; Ali Azam et al., 1999; Mallik et al., 2006).
Fig. 1.
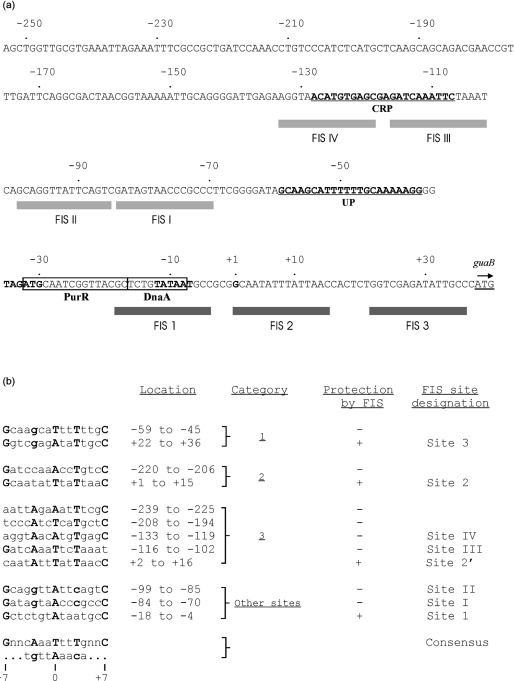
Identification of putative FIS sites at PguaB. (a) Sequence of the guaB promoter from positions −253 to +40 relative to the guaB transcription start site. Putative FIS sites I–IV, FIS sites 1–3, a putative CRP binding site centred at position −117.5 and binding sites for PurR and DnaA are indicated (Hutchings & Drabble, 2000). The core promoter elements (−35 and −10 regions), UP element, and the initiating nucleotide are also shown, in bold. Apart from FIS sites I–IV, putative FIS sites that have been shown not to bind FIS in this work are not indicated. For clarity, FIS site 2′ is not shown. (b) Candidate FIS binding sites were identified by comparing the consensus sequence for FIS (Ross et al., 1999; Shultzaberger et al., 2007) with PguaB sequences located from positions −253 to +36. The consensus sequence employed is indicated accordingly; alternative bases at each position of the consensus sequence are shown in the second line. Conserved bases at the five positions considered to be critical for FIS binding (positions −7, −3, 0, +3 and +7) are emboldened, and the most frequently occurring base is shown in upper case. ‘n' signifies any base. Candidate FIS sites are categorized according to their similarity to the consensus. Category 1 and 2 sites exhibit a 4/5 match at the critical positions, including positions +7 and −7, which are most strongly conserved among FIS sites. However, at category 2 sites, an infrequently occurring base is present at position −3, 0 or +3 (in the case of PguaB, such mismatches only occur at position −3). Category 3 sites also exhibit a 4/5 match at the critical positions but the mismatched base occurs at position +7 or −7. Other pertinent sites that do not fulfil the criteria for inclusion in categories 1–3 are shown as ‘other sites'. FIS sites that were protected by FIS in DNase I footprinting are as indicated.
In this work, we investigated whether FIS plays a role in the regulation of PguaB. We show that the putative FIS binding sites located upstream of the PguaB core elements do not recruit FIS (Hutchings & Drabble, 2000). Moreover, we demonstrate that FIS is recruited to three sites centred near −11, +8 and +29 relative to the guaB transcription start site, and all three sites are necessary for full FIS-mediated repression. We also show that FIS is not required for GRDC of PguaB.
METHODS
Strains and plasmids.
All strains were derivatives of the E. coli K-12 strain VH1000. Each strain contained a chromosomally integrated transcriptional fusion of lacZ to one of three PguaB derivatives: the full-length guaB promoter (i.e. strain VH1000G-253), extending from positions −253 to +36 relative to the guaB transcription start site [PguaB (−253 to +36)] (Husnain & Thomas, 2008), the PguaB (−253 to +10) promoter (i.e. strain VH1000G-25310, this work), which contains the same upstream end point as the full-length promoter but has a downstream end point at +10, and the PguaB (−69 to +36) promoter (i.e. strain VH1000G-69, this work), which has the same downstream end point as the full-length promoter but has an upstream end point at position −69 relative to the guaB transcription start site. Fusions were carried on λ prophage and were constructed using a system based on λimm21 (Simons et al., 1987; Rao et al., 1994). Strain VH1000G-253Δfis was made by introducing the fis : : aadA allele from strain JCB38841 (Ball et al., 1992; Wu et al., 1998) into VH1000G-253 by P1 transduction; strain VH1000G-69Δfis was made by introducing the same allele into VH1000G-69. All plasmids contain PguaB derivatives that were inserted as EcoRI–HindIII fragments. Plasmids pBSG-253 and pBSG-133 are derivatives of pBluescript II KS containing the promoters PguaB (−253 to +36) and PguaB (−133 to +36), respectively. pUCG-253 is a derivative of pUC19 containing PguaB (−253 to +36). The plasmid pRLG770 has been described previously (Ross et al., 1990). pRLG770 derivatives containing promoters PguaB (−253 to +36), PguaB (−133 to +36), PguaB (−59 to +36) and PguaB (−37 to +36) were constructed previously (Husnain & Thomas, 2008). pRLG770 derivatives pRLG-13321, pRLG-1331 and pRLG-25310 contain promoters PguaB (−133 to +21), PguaB (−133 to +1) and PguaB (−253 to +10), respectively.
DNase I footprinting.
The EcoRI–XhoI DNA fragment in pBSG-253 was purified following electrophoresis in a 6 % acrylamide gel (Meng et al., 2000), labelled at the downstream (XhoI) end with [γ-32P]ATP [>7000 Ci (2.59×1014 Bq) mmol−1, MP Biomedicals] and subsequently purified according to a published procedure (Husnain & Thomas, 2008). The EcoRI–XhoI fragment in pBSG-133 was labelled similarly at the upstream end. Labelled DNA fragment (4 nM) was incubated at room temperature for 30 min in a volume of 20 μl containing 20 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM potassium glutamate, 1 mM DTT, 20 μg ml−1 sonicated calf thymus DNA (GE Healthcare) and 5 % (v/v) glycerol, in the absence or presence of purified FIS protein. Purified FIS was a generous gift from T. Gaal and R. L. Gourse (University of Wisconsin–Madison). DNase I footprinting and DNA fragment separation were performed exactly as described previously (Husnain & Thomas, 2008). Footprints were visualized using a FujiFilm FLA3000 phosphorimager.
Electromobility shift assay (EMSA).
A DNA fragment containing PguaB (−253 to +36) was amplified by PCR from pUCG-253, using primers pUC19(for) (5′-ACGTTGTAAAACGACGGCCAG-3′) and pUC19(rev) (5′-GCGCGGATCCATGACCATGATTACGCCAAGCT-3′). A DNA fragment containing the rrnB P1 promoter with FIS site I (positions −87 to +50 relative to the rrnB P1 transcription start site) and an rrnB P1 promoter derivative that did not contain a FIS site (positions −37 to +52 relative to the rrnB P1 transcription start site, and containing non-rrnB P1 sequences upstream to position −92) were PCR amplified from plasmids pRLG1616 and pRLG4720, respectively, using a forward primer with the sequence 5′-GTATCACGAGGCCCT-3′ and reverse primer RLG1620 (5′-GCGCTACGGCGTTTCACTTC-3′), both of which are vector-specific (Newlands et al., 1991; Ross et al., 1998; Meng et al., 2001). PCR products were digested with HindIII, and purified following electrophoresis in a 6 % acrylamide gel (Meng et al., 2000). Fragments were labelled at the HindIII end using [α-32P]dATP [3000 Ci (1.11×1014 Bq) mmol−1, MP Biomedicals] and DNA polymerase I Klenow fragment. Labelled DNA (final concentration 0.4 nM) was incubated at room temperature for 30 min in a volume of 10 μl containing 20 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM potassium glutamate, 1 mM DTT, 10 % (v/v) glycerol and 20 μg ml−1 sonicated calf thymus DNA (GE Healthcare), in the absence or presence of different concentrations of FIS. Samples were loaded (under tension) onto a 6 % acrylamide gel (37.5 : 1 acrylamide : bis acrylamide) containing 7.5 % (v/v) glycerol while running at ∼15 V cm−1, and gels were run for ∼1 h at 4 °C. Radiolabelled DNA was visualized using a FujiFilm FLA3000 phosphorimager.
Measurement of transcription in vitro.
Multiple-round transcription reactions were performed as described previously, using supercoiled pRLG770 derivatives containing PguaB fragments (Husnain & Thomas, 2008). As a control, transcription was also measured from an rrnB P1 promoter derivative that did not contain any FIS sites (plasmid pRLG4238; Estrem et al., 1998). FIS (250 nM) was incubated with DNA in reaction buffer at room temperature for 30 min. Transcription was initiated at 30 °C with 10 nM E. coli RNAP holoenzyme (Epicentre), and reactions were allowed to proceed for 20 min.
Measurement of transcription in vivo.
Strains containing a chromosomally integrated PguaB-lacZ transcriptional fusion were employed in the measurement of promoter activity in vivo. Cells were inoculated from dense starter cultures into media that supported different cellular growth rates, as described previously (Husnain & Thomas, 2008). The β-galactosidase activity was determined following disruption of cells by sonication (Miller, 1972). To measure promoter activity at different stages of the growth cycle, cells were grown overnight in M9 minimal medium with 0.4 % (w/v) glucose, 0.8 % (w/v) Casamino acids and 5 μg thiamine ml−1, and inoculated into fresh medium to an OD600 of ∼0.01. Growth was monitored at OD600, and the β-galactosidase activity was measured at different points on the growth curve after cells were permeabilized with chloroform-SDS (Miller, 1972).
RESULTS
Identification of putative FIS sites at PguaB
The 15 bp consensus sequence for FIS [5′-Gnn(c/t)(A/g)(a/t)(a/t)(T/A)(t/a)(t/a)(T/c)(g/a)nnC-3′] contains five highly conserved positions (underlined) that are the most significant for the recruitment of FIS (Finkel & Johnson, 1992; Hengen et al., 1997; Ross et al., 1999; Shultzaberger et al., 2007; Shao et al., 2008). At each highly conserved position (hereafter referred to as a ‘critical’ position), an upper-case letter indicates the most strongly conserved base at that position, and a lower-case letter indicates a conserved base that is non-consensus. We employed the MatInspector program (Genomatix) to identify putative FIS sites at PguaB that matched the consensus in at least four out of the five critical positions (Quandt et al., 1995). This was achieved by performing a sequence alignment of a modified consensus sequence (5′-GnnnRnnWnnYnnnC-3′, where R=A or G; W=A or T; Y=T or C; n=any base) with PguaB DNA sequences located between positions −253 and +36, and also with the reverse complement of this DNA sequence.
Using this approach, no sites were identified that contained the most highly conserved base at all five critical positions. However, we identified nine candidate FIS binding sites that contained the most highly conserved base at 4/5 critical positions (Fig. 1b). All of these sites contained the consensus A or T base at the central position (position 0) and the conserved T residue at position +3. These sequences were subdivided into three categories. Category 1 sites contain bases that match the consensus at 4/5 critical positions, including the outer bases (positions −7 and +7) that are most strongly conserved among FIS sites and which are presumed to be bound by the D helices of FIS (Shultzaberger et al., 2007). The remaining critical position (position −3) contained the alternative purine base G that occurred less frequently at that position. Two candidate FIS sites were identified that fell into this category (Fig. 1b). Category 2 sites differ from category 1 sites in having a less frequently occurring C or T residue at position −3. Two additional sites fell into this category. Category 3 sites also contain bases that match the consensus at 4/5 critical positions. However, the mismatches occur at one of the highly conserved bases that are located at the outermost positions. Five sequences were classified as category 3 sites (Fig. 1b). Two of them correspond to the previously identified putative FIS sites III and IV located upstream of PguaB (Hutchings & Drabble, 2000). Putative FIS sites I and II were not identified by this analysis as they harbour bases that match the consensus at only 3/5 of the critical positions, although they do include non-consensus but frequently occurring bases at the remaining two critical positions (positions −3 and +3) (Fig. 1b).
Analysis of FIS binding to PguaB
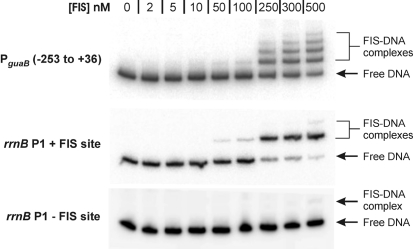
EMSA was employed to determine whether FIS can bind to a DNA fragment containing the guaB promoter. As a comparison, binding of FIS to the rrnB P1 promoter, which is known to bind FIS under physiological conditions, was also analysed. The rrnB P1 promoter fragment employed contained the promoter-proximal FIS site, i.e. FIS site I (Ross et al., 1990; Bokal et al., 1995). The minimum concentration of FIS required to observe FIS–DNA interactions at either PguaB (−253 to +36) or the rrnB P1 fragment by EMSA was 50 nM. Increasing the FIS concentration to 300 nM resulted in the formation of three different complexes between FIS and PguaB (Fig. 2). At this concentration of FIS, a larger fraction of the rrnB P1 promoter fragment was bound by FIS, but there remained only a single FIS–DNA complex, and no FIS–DNA complexes were observed at an rrnB P1 promoter derivative that lacked a FIS site (Fig. 2). At a FIS concentration of 500 nM, an additional FIS–DNA complex was observed at both PguaB and rrnB P1 harbouring FIS site I. As a complex was also observed with the promoter fragment that did not contain a FIS site, it is likely that the additional FIS–DNA interactions observed at 500 nM FIS are non-specific (Fig. 2). These results indicate that FIS binds to at least three sites at or near PguaB under similar conditions to those in which FIS specifically binds to rrnB P1, and thereby suggest that FIS is likely to bind to these sites under physiological conditions.
Fig. 2.
Analysis of FIS binding to PguaB by EMSA. EMSA was employed to compare the relative binding affinity of FIS for a DNA fragment containing PguaB (−253 to +36) with an rrnB P1 promoter derivative containing FIS site I (‘rrnB P1+FIS site’) and rrnB P1 containing no FIS sites (‘rrnB P1–FIS site’). The concentration of FIS in each binding reaction is indicated above the corresponding gel lane.
Mapping the location of FIS binding sites at PguaB by DNase I footprinting
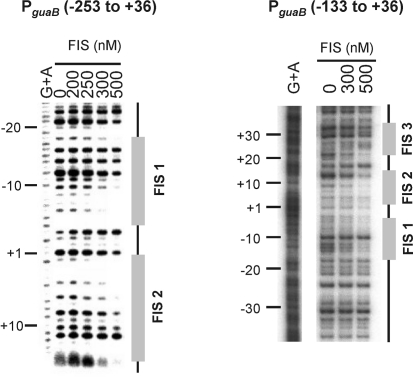
To determine the location of any FIS binding sites at PguaB, a DNA fragment extending from positions −253 to +36 of PguaB [i.e. PguaB (−253 to +36)] was radiolabelled at the downstream end, and DNase I footprinting was performed in the presence or absence of purified FIS. The results show that increasing the concentration of FIS up to 500 nM resulted in increased protection at two sites centred near positions −11 and +8 (FIS site 1 and FIS site 2, respectively) (Fig. 3). DNA fragments corresponding to PguaB sequences downstream of position +16 were not visible on this gel. To determine whether FIS bound to sequences downstream of position +16, a DNA fragment extending from positions −133 to +36 [i.e. PguaB (−133 to +36)] was radiolabelled at the upstream end for DNase I footprinting. The results show that, in addition to the protection observed at FIS sites 1 and 2, a third site (FIS site 3) centred near position +29 was also bound by FIS. FIS sites 2 and 3 were identified by comparison with the consensus as being more likely to recruit FIS (i.e. FIS site 3 is a category 1 site, and FIS site 2 is a category 2 site) (Fig. 1b). However, protection of other category 1 and category 2 FIS sites that were identified by bioinformatic analysis was not observed. Interestingly, FIS site 1 contains mismatches to the consensus at two critical positions, and therefore was not identified by the bioinformatic analysis (Fig. 1b). FIS did not bind to any of the previously predicted FIS sites (FIS sites I–IV). It should be noted that another FIS site (site 2′) is located overlapping site 2, with its centre shifted by one base pair downstream of the centre of site 2 (Fig. 1). As sites that contain both an A at position −4 and a T at position +4 are bound by FIS much less efficiently than are sites that lack a G at position −7 (or a C at +7) (Shao et al., 2008), it is possible that site 2′ may be preferred by FIS over site 2.
Fig. 3.
Mapping the location of FIS sites at PguaB by DNase I footprinting. A DNA fragment containing PguaB (−253 to +36) radiolabelled at the downstream end (relative to the guaB transcription start site), and a DNA fragment containing PguaB (−133 to +36) labelled at the upstream end, were employed in DNase I footprinting in the presence of different concentrations of FIS (as shown). Nucleotide positions are shown relative to the guaB transcription start site, and lanes containing the G+A ladder are indicated accordingly.
Role of FIS in the regulation of transcription from PguaB in vitro
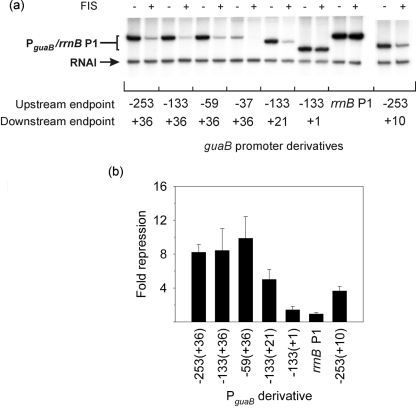
To determine the role of FIS in the regulation of PguaB, multiple-round transcription reactions were performed in the presence or absence of 250 nM FIS. Transcription was measured from PguaB (−253 to +36) and shorter derivatives [PguaB (−133 to +36), PguaB (−59 to +36), PguaB (−37 to +36), PguaB (−133 to +21), PguaB (−133 to +1) and PguaB (−253 to +10)] (end points as indicated). Addition of FIS to the transcription reaction resulted in ∼8–10-fold repression of transcription from PguaB (−253 to +36). Under the same conditions, there was no repressive effect of FIS on transcription from the rrnB P1 promoter (Fig. 4a, b). This indicates that the repression of PguaB afforded by FIS at a concentration of 250 nM was due to a site-specific FIS–DNA interaction. Deletion of sequences upstream of the PguaB UP element [i.e. PguaB (−59 to +36)] did not lead to any significant change in the degree of repression afforded by FIS, confirming that putative FIS sites I–IV do not play a role in the regulation of PguaB by FIS. Deletion of the PguaB UP element [PguaB (−37 to +36)] gave rise to an undetectable level of transcripts from PguaB in the presence of FIS, which meant that the fold repression afforded by FIS could not be determined. Deletion of FIS site 3 [i.e. PguaB (−133 to +21)] led to decreased repression by FIS (approximately sixfold repression), and deletion of both FIS site 2 and FIS site 3 [i.e. PguaB (−133 to +1)] gave rise to an approximately twofold repression by FIS (Fig. 4b). As PguaB (−133 to +1) retains FIS site 1 and is still subject to some degree of repression by FIS, these results indicate that FIS sites 1–3 each contribute to repression of PguaB, and that FIS site 1 can function independently of the other FIS sites. A PguaB derivative harbouring a deletion of FIS site 3 and deletion of the downstream six bases of FIS site 2 [i.e. PguaB (−253 to +10)] was subject to an approximately fourfold repression by FIS, indicating that FIS site 2 had not been completely inactivated (Fig. 4b).
Fig. 4.
Role of FIS in regulation of transcription from PguaB in vitro. (a) Multiple-round in vitro transcription was employed to measure transcription from PguaB derivatives PguaB (-253 to +36), PguaB (-133 to +36), PguaB (-59 to +36), PguaB (-37 to +36), PguaB (-133 to +1), PguaB (-133 to +21) and PguaB (-253 to +10) in the presence (+) and absence (−) of 250 nM FIS. Promoter endpoints are as indicated. PguaB (-133 to +21) lacks FIS site 3, and PguaB (-133 to +1) lacks FIS sites 2 and 3. As a control, transcription was also measured from an rrnB P1 promoter derivative that did not contain any FIS sites. All promoters were cloned in pRLG770 and supercoiled DNA was used for the assays. The vector-encoded replication repressor, RNAI (∼110 nucleotides), is also indicated. (b) The repression afforded by FIS at each promoter is shown. The fold repression by FIS was calculated by dividing the activity in the absence of FIS by the activity in its presence. Values are the mean (with standard deviation) of three independent experiments. The activity of PguaB (-37 to +36) in the presence of FIS was too low to quantitate and hence a value for the fold repression was not obtained.
FIS is not required for growth rate-dependent control of PguaB
GRDC of PguaB (−253 to +36) was measured in a wild-type strain background and in a strain that harboured a deletion in the fis gene. To determine whether FIS site 3 is important for GRDC of PguaB, activity of PguaB (−253 to +10) was also analysed. In exponentially growing wild-type E. coli cells, the activity of PguaB (−253 to +36) increased approximately twofold with every doubling of the growth rate as, shown previously (Fig. 5a, c; Davies & Drabble, 1996; Husnain & Thomas, 2008). In an otherwise isogenic fis strain, the activity of PguaB (−253 to +36) was higher than in the wild-type at all growth rates and was more pronounced at faster growth rates (i.e. an approximately twofold increase in activity was observed at the fastest growth rate) (Fig. 5a). However, the degree of GRDC was similar to that observed in a wild-type strain (i.e. in both cases, a doubling of the growth rate corresponded to an approximately twofold increase in promoter activity) (compare plots of relative activity versus growth rate, Fig. 5c). These results demonstrate that FIS is not required for GRDC of PguaB. Although FIS appears to downregulate transcription from PguaB in vivo, deletion of FIS site 3 and part of FIS site 2 did not lead to a significant change in PguaB activity in exponentially growing wild-type cells in comparison to PguaB containing the full complement of functional FIS sites [i.e. a 1.87-fold increase in PguaB (−253 to +10) activity occurred for every doubling of the growth rate in comparison to a 1.84-fold increase for PguaB (−253 to +36) (Fig. 5a, b)]. These results suggest either that FIS site 1 is able to effect full repression in vivo (contrasting with the results obtained in vitro) or that FIS sites 1–3 may not contribute to repression of PguaB in vivo under the conditions employed, and therefore the observed FIS-dependent repression is indirect.
Fig. 5.
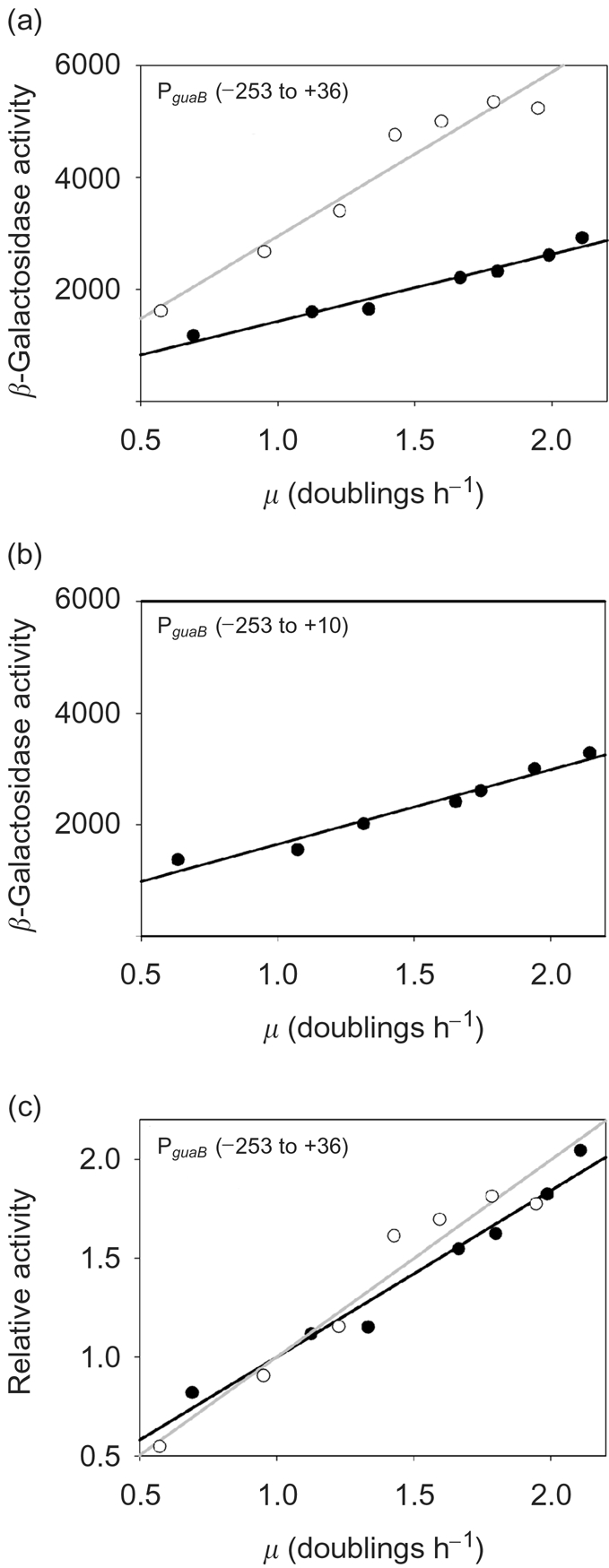
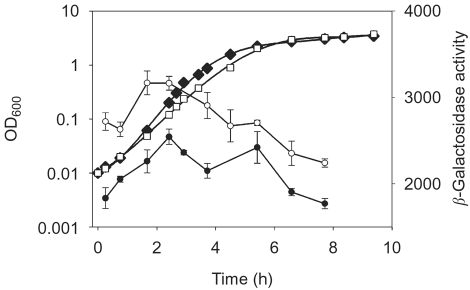
GRDC of PguaB. (a, c) GRDC of a wild-type strain (•) or a strain containing the fis : : aadA allele (○), harbouring a fusion of PguaB (−253 to +36) to lacZ, was analysed. (b) GRDC of a wild-type strain harbouring a PguaB (−253 to +10)-lacZ fusion was also measured. Strains were grown at different cellular growth rates to an OD600 of 0.34–0.45, whereupon the β-galactosidase activity was determined. The promoter activity for PguaB (−253 to +36) in the presence and absence of functional fis is given both in Miller units (β-galactosidase activity) and expressed as a ratio to the activity at 1 doubling per hour (relative activity). The magnitude of the gradient in plots of relative promoter activity versus doublings per hour is proportional to the degree of GRDC. Each data point represents the mean promoter activity or mean growth rate. The mean was calculated using data obtained from three independent experiments.
FIS is not required for growth phase-dependent regulation of PguaB
Previous studies indicate that FIS levels are elevated during the mid-exponential growth phase, and they decrease sharply as cells enter stationary phase (Appleman et al., 1998). This phenomenon is responsible for the known contribution of FIS to growth-phase-dependent regulation of some promoters (Nilsson et al., 1992; Appleman et al., 1998; Mallik et al., 2006; Bradley et al., 2007). To test whether FIS-dependent regulation of PguaB varies with the growth phase, transcription from a PguaB derivative with an upstream end point of −69 that contained all three experimentally determined FIS sites [i.e. PguaB (−69 to +36)] was measured at different stages of growth in a wild-type strain, and in a strain that harboured a deletion in the fis gene. This promoter derivative was chosen as it lacks the putative CRP site centred near position −117.5 (Hutchings & Drabble, 2000). It has previously been shown that FIS represses the crp1 promoter and this may alter cellular levels of CRP (Gonzalez-Gil et al., 1998).
In accordance with the results of the GRDC experiment, the activity of PguaB was higher in the fis background than in the wild-type strain throughout the course of the growth cycle. In the wild-type strain, PguaB activity increased by nearly 40 % as cells entered the mid-exponential growth phase (i.e. the promoter activity at an OD600 of ∼0.15–0.20 was 40 % higher than the activity at an OD600 of ∼0.012). The increase in activity in a fis strain over the corresponding part of the growth curve was less marked (i.e. there was a ∼16 % increase in promoter activity). Upon entry into stationary phase, there was a gradual decrease in the promoter activity in both strain backgrounds (Fig. 6). The results suggest that PguaB is subject to a degree of growth-phase-dependent regulation. However, there was no significant change in the transcription activity profile during the growth cycle when comparing the two strain backgrounds. Furthermore, PguaB activity peaks at the time that FIS levels are expected to be at their highest, and then falls off upon entry into stationary phase when FIS levels fall (Appleman et al., 1998; Ali Azam et al., 1999). These observations suggest that FIS does not significantly influence growth phase-dependent regulation of PguaB under the conditions employed.
Fig. 6.
Growth-phase-dependent regulation of PguaB. A wild-type strain or a strain containing the fis : : aadA allele (Δfis), each harbouring a fusion of PguaB (−69 to +36) to lacZ, were inoculated from a dense culture into fresh growth medium [M9 minimal medium containing 0.4 % (w/v) glucose, 0.8 % (w/v) Casamino acids and 5 μg thiamine ml−1] to an OD600 of ∼0.01. Samples were taken during different stages of growth and the β-galactosidase activity was determined. The promoter activity of the wild-type strain (•) and Δfis strain (○) is given in Miller units (β-galactosidase activity). Values presented are the mean±sd, for three independent experiments. Cell density measurements (OD600) for single cultures that are representative of the growth curve for the wild-type strain (⧫) and the Δfis strain (□), were plotted on a logarithmic axis.
DISCUSSION
A previous study identified four adjacent putative FIS binding sites (FIS sites I–IV) located upstream of the UP element at PguaB. These sites were identified by comparison of the upstream PguaB sequence to a published consensus sequence for FIS (Finkel & Johnson, 1992, Hutchings & Drabble, 2000). By employing sequence alignment to compare PguaB sequences to a more representative consensus for FIS, we have identified four candidate FIS sites that bear a closer resemblance to this consensus than FIS sites I–IV (Ross et al., 1999). Two of these sites, sites 2 (or 2′) and 3 (centred near positions +8 and +29 relative to the guaB transcription start site, respectively), are protected by FIS in DNase I footprinting experiments. One of the other two candidate FIS sites overlaps the UP element, and the remaining site is located at a distance upstream of the core promoter elements (centred at −213). Neither of these sites recruits FIS as judged by DNase I footprinting. Putative FIS sites I–IV also do not bind FIS. Interestingly, we show that a site centred at position −11, which was not identified as a likely FIS site, also recruits FIS. This suggests that although bioinformatic analyses are useful when searching for sites that are bound by FIS, they may not be useful in identifying some FIS binding sites that exhibit a weak match to the consensus.
At the tyrT promoter, FIS binding to sites II and III (centred at positions −91 and −122, respectively) cooperatively affects the binding of FIS to site I centred at −71 bp upstream of the transcription start site (Lazarus & Travers, 1993; Pemberton et al., 2002). As the B-form of DNA has a periodicity of 10.6 bp, this places these three sites on the same face of the DNA helix, with the centres of sites I and II separated by 21 bp. This suggests that cooperative interactions require adjacent FIS dimers to be positioned on the same face of the DNA, two turns of the DNA helix apart. At the rrnB P1 promoter, FIS sites I–III are centred at positions −71, −102 and −143, respectively, also placing them approximately on the same face of the DNA helix. However, FIS does not bind to rrnB P1 cooperatively in the absence of RNAP, and it is noteworthy that the central positions of these sites are separated by 32 or 42 bp (i.e. not 21 bp). Interestingly, at PguaB, FIS sites 2 and 3 are positioned 22 bp apart (centre to centre) which will also place them on the same face of the DNA helix and a similar distance apart as FIS sites I and II at the tyrT promoter. However, the centres of FIS sites 1 and 2 at PguaB are 18 bp apart, making it unlikely that they are located on the same face of the DNA. Therefore, it is possible that FIS binds cooperatively to sites 2 and 3, but it appears less likely that occupancy of sites 2 and/or 3 stimulates binding of FIS to site 1.
Consistent with the location of functional FIS sites, we show that FIS represses transcription from PguaB ∼8–10-fold in vitro. Deletion of FIS site 3 results in partial relief of repression, and deletion of FIS sites 2 and 3 together further relieves repression in vitro. The residual FIS-mediated repression of the guaB promoter fragment containing the +1 downstream end point is likely to occur through interactions with site 1, and would suggest that binding of to site 1 does not require cooperative interactions with FIS dimers bound to adjacent sites. The binding of FIS to site 1 is likely to sterically hinder the recruitment of RNAP to PguaB, as observed at the crp1 promoter (Gonzalez-Gil et al., 1998). The role of FIS sites 2 and 3, which together exert the most influence on transcription from PguaB in vitro, is less clear, although it is likely that the role of FIS site 3 is to stimulate binding of FIS to site 2, which in turn may play more of a direct role in repression. Our results suggest that the presence of FIS should decrease RNAP binding to PguaB. However, DNase I footprinting experiments carried out in the presence of both FIS and RNAP were inconclusive (data not shown).
Although our results demonstrate that FIS represses transcription from PguaB in vitro, evidence for direct repression by FIS in vivo was not obtained (i.e. deletion of FIS sites 2 and 3 did not result in increased PguaB activity in wild-type exponentially growing cells). This is consistent with the results of a chromatin immunoprecipitation (ChIP)-chip analysis carried out under similar conditions, in which FIS binding at PguaB was not detected (see supplementary data in Grainger et al., 2006). However, in a fis strain we observed an increase in the activity of the guaB promoter in derivatives containing all three FIS sites in the presence (PguaB (−253 to +36)) or absence [PguaB (−69 to +36)] of the putative CRP site centred at −117.5. This rules out the possibility that the effect of deleting fis on guaB promoter activity is mediated by changes in CRP abundance [FIS has been shown to modulate transcription of crp (Gonzalez-Gil et al., 1998)]. However, it is possible that the change in transcription activity of PguaB in a fis background occurs as a result of altered regulation of PguaB by a transcription factor other than CRP, for example H-NS or HU (Claret & Rouviere-Yaniv, 1996; Falconi et al., 1996) or by changes in supercoiling (Schneider et al., 1997; Weinstein-Fischer et al., 2000). Another possible explanation is that the potential relief of PguaB repression that occurs upon deleting FIS sites 2 and 3 is masked in vivo through an alternative compensatory regulatory mechanism. A less likely explanation, in view of the poor match to the consensus FIS binding site, is that FIS binding to site 1 mediates full FIS-mediated repression in vivo.
A previous study has shown that the PguaB UP element, and sequences located further upstream, are required for GRDC of PguaB (Husnain & Thomas, 2008). Our results show that FIS does not play a role in GRDC at this promoter, thereby implying that a different cellular factor is required for conferring GRDC on PguaB (Emilsson & Nilsson, 1995; Dennis et al., 2004; Paul et al., 2004). Experiments are under way to uncover the identity of this factor(s). Our results also suggest that PguaB is subject to growth phase-dependent control. However, although levels of FIS protein are also subject to growth phase-dependent control, it does not appear to play an important role in growth phase-dependent control at PguaB. Thus, the physiological role of FIS at the guaB promoter remains to be elucidated.
Acknowledgments
This work was supported by a family PhD sponsorship awarded to S. I. H., kindly provided by S. M. Husnain, and a research project grant awarded to M. S. T. by the Wellcome Trust (grant ref. 073917). We thank W. Ross, T. Gaal, H. Murray and R. L. Gourse (University of Wisconsin–Madison) for strains, plasmids and purified FIS protein. We are grateful to T. Belyaeva (University of Leeds) for advice on EMSA, and also to S. J. W. Busby and D. Browning (University of Birmingham) for strains and advice on DNase I footprinting.
Abbreviations
CRP, cAMP receptor protein
EMSA, electromobility shift assay
FIS, factor for inversion stimulation
GRDC, growth rate-dependent control
RNAP, RNA polymerase
References
- Aiyar, S. E., McLeod, S. M., Ross, W., Hirvonen, C. A., Thomas, M. S., Johnson, R. C. & Gourse, R. L. (2002). Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J Mol Biol 316, 501–516. [DOI] [PubMed] [Google Scholar]
- Ali Azam, T., Iwata, A., Nishimura, A., Ueda, S. & Ishihama, A. (1999). Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol 181, 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleman, J. A., Ross, W., Salomon, J. & Gourse, R. L. (1998). Activation of Escherichia coli rRNA transcription by FIS during a growth cycle. J Bacteriol 180, 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, C. A., Osuna, R., Ferguson, K. C. & Johnson, R. C. (1992). Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol 174, 8043–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokal, A. J., Ross, W. & Gourse, R. L. (1995). The transcriptional activator protein FIS: DNA interactions and cooperative interactions with RNA polymerase at the Escherichia coli rrnB P1 promoter. J Mol Biol 245, 197–207. [DOI] [PubMed] [Google Scholar]
- Bradley, M. D., Beach, M. B., de Koning, A. P., Pratt, T. S. & Osuna, R. (2007). Effects of FIS on Escherichia coli gene expression during different growth stages. Microbiology 153, 2922–2940. [DOI] [PubMed] [Google Scholar]
- Browning, D. F., Cole, J. A. & Busby, S. J. (2000). Suppression of FNR-dependent transcription activation at the Escherichia coli nir promoter by Fis, IHF and H-NS: modulation of transcription initiation by a complex nucleo-protein assembly. Mol Microbiol 37, 1258–1269. [DOI] [PubMed] [Google Scholar]
- Browning, D. F., Beatty, C. M., Sanstad, E. A., Gunn, K. E., Busby, S. J. W. & Wolfe, A. J. (2004). Modulation of CRP-dependent transcription at the Escherichia coli acsP2 promoter by nucleoprotein complexes: anti-activation by the nucleoid proteins FIS and IHF. Mol Microbiol 51, 241–254. [DOI] [PubMed] [Google Scholar]
- Claret, L. & Rouviere-Yaniv, J. (1996). Regulation of HU alpha and HU beta by CRP and FIS in Escherichia coli. J Mol Biol 263, 126–139. [DOI] [PubMed] [Google Scholar]
- Davies, I. J. & Drabble, W. T. (1996). Stringent and growth-rate-dependent control of the gua operon of Escherichia coli K-12. Microbiology 142, 2429–2437. [DOI] [PubMed] [Google Scholar]
- Dennis, P. P., Ehrenberg, M. & Bremer, H. (2004). Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol Mol Biol Rev 68, 639–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H., Kirsebom, L. A. & Nilsson, L. (1996). Growth rate regulation of 4.5 S RNA and M1 RNA the catalytic subunit of Escherichia coli RNase P. J Mol Biol 261, 303–308. [DOI] [PubMed] [Google Scholar]
- Emilsson, V. & Nilsson, L. (1995). Factor for inversion stimulation-dependent growth rate regulation of serine and threonine tRNA species. J Biol Chem 270, 16610–16614. [DOI] [PubMed] [Google Scholar]
- Estrem, S. T., Gaal, T., Ross, W. & Gourse, R. L. (1998). Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci U S A 95, 9761–9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconi, M., Brandi, A., La Teana, A., Gualerzi, C. O. & Pon, C. L. (1996). Antagonistic involvement of FIS and H-NS proteins in the transcriptional control of hns expression. Mol Microbiol 19, 965–975. [DOI] [PubMed] [Google Scholar]
- Finkel, S. E. & Johnson, R. C. (1992). The Fis protein: it's not just for DNA inversion anymore. Mol Microbiol 6, 3257–3265. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gil, G., Kahmann, R. & Muskhelishvili, G. (1998). Regulation of crp transcription by oscillation between distinct nucleoprotein complexes. EMBO J 17, 2877–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse, R. L., Gaal, T., Bartlett, M. S., Appleman, J. A. & Ross, W. (1996). rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol 50, 645–677. [DOI] [PubMed] [Google Scholar]
- Grainger, D. C., Hurd, D., Goldberg, M. D. & Busby, S. J. W. (2006). Association of nucleiod proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res 34, 4642–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengen, P. N., Bartram, S. L., Stewart, L. E. & Schneider, T. D. (1997). Information analysis of Fis binding sites. Nucleic Acids Res 25, 4994–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnain, S. I. & Thomas, M. S. (2008). The UP element is necessary but not sufficient for growth rate-dependent control of the Escherichia coli guaB promoter. J Bacteriol 190, 2450–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings, M. I. & Drabble, W. T. (2000). Regulation of the divergent guaBA and xseA promoters of Escherichia coli by the cyclic AMP receptor protein. FEMS Microbiol Lett 187, 115–122. [DOI] [PubMed] [Google Scholar]
- Jackson, L., Blake, T. & Green, J. (2004). Regulation of ndh expression in Escherichia coli by Fis. Microbiology 150, 407–413. [DOI] [PubMed] [Google Scholar]
- Lazarus, L. R. & Travers, A. A. (1993). The Escherichia coli FIS protein is not required for the activation of tyrT transcription on entry into exponential growth. EMBO J 12, 2483–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik, P., Paul, B. J., Rutherford, S. T., Gourse, R. L. & Osuna, R. (2006). DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J Bacteriol 188, 5775–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod, S. M., Aiyar, S. E., Gourse, R. L. & Johnson, R. C. (2002). The C-terminal domains of the RNA polymerase α subunits: contact site with Fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter. J Mol Biol 316, 517–529. [DOI] [PubMed] [Google Scholar]
- Mehra, R. K. & Drabble, W. T. (1981). Dual control of the gua operon of Escherichia coli K12 by adenine and guanine nucleotides. J Gen Microbiol 123, 27–37. [DOI] [PubMed] [Google Scholar]
- Meng, L. M., Kilstrup, M. & Nygaard, P. (1990). Autoregulation of PurR repressor synthesis and involvement of purR in the regulation of purB, purC, purL, purMN and guaBA expression in Escherichia coli. Eur J Biochem 187, 373–379. [DOI] [PubMed] [Google Scholar]
- Meng, W., Savery, N. J., Busby, S. J. W. & Thomas, M. S. (2000). The Escherichia coli RNA polymerase α subunit linker: length requirements for transcription activation at CRP-dependent promoters. EMBO J 19, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, W., Belyaeva, T., Savery, N. J., Busby, S. J., Ross, W. E., Gaal, T., Gourse, R. L. & Thomas, M. S. (2001). UP element-dependent transcription at the Escherichia coli rrnB P1 promoter: positional requirements and role of the RNA polymerase alpha subunit linker. Nucleic Acids Res 29, 4166–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Newlands, J. T., Ross, W., Gosink, K. K. & Gourse, R. L. (1991). Factor-independent activation of Escherichia coli rRNA transcription. II. characterization of complexes of rrnB P1 promoters containing or lacking the upstream activator region with Escherichia coli RNA polymerase. J Mol Biol 220, 569–583. [DOI] [PubMed] [Google Scholar]
- Nilsson, L., Verbeek, H., Vijgenboom, E., van-Drunen, C., Vanet, A. & Bosch, L. (1992). FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J Bacteriol 174, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, B. J., Ross, W., Gaal, T. & Gourse, R. L. (2004). rRNA transcription in Escherichia coli. Annu Rev Genet 38, 749–770. [DOI] [PubMed] [Google Scholar]
- Pemberton, I. K., Muskhelishvili, G., Travers, A. A. & Buckle, M. (2002). FIS modulates the kinetics of successive interactions of RNA polymerase with the core and upstream regions of the tyrT promoter. J Mol Biol 318, 651–663. [DOI] [PubMed] [Google Scholar]
- Quandt, K., Frech, K., Karas, H., Wingender, E. & Werner, T. (1995). MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, L., Ross, W., Appleman, J. A., Gaal, T., Leirmo, S., Schlax, P. J., Record, M. T., Jr & Gourse, R. L. (1994). Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol 235, 1421–1435. [DOI] [PubMed] [Google Scholar]
- Ross, W., Thompson, J. F., Newlands, J. T. & Gourse, R. L. (1990). Escherichia coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J 9, 3733–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, W., Aiyar, S. E., Salomon, J. & Gourse, R. L. (1998). Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol 180, 5375–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, W., Salomon, J., Holmes, W. M. & Gourse, R. L. (1999). Activation of Escherichia coli leuV transcription by FIS. J Bacteriol 181, 3864–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, R., Travers, A. & Muskhelishvili, G. (1997). FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol Microbiol 26, 519–530. [DOI] [PubMed] [Google Scholar]
- Shao, Y., Feldman-Cohen, L. S. & Osuna, R. (2008). Functional characterisation of the Escherichia coli Fis-DNA binding sequence. J Mol Biol 376, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultzaberger, R. K., Roberts, L. R., Lyakhov, I. G., Sidorov, I. A., Stephen, A. G., Fisher, R. J. & Schneider, T. D. (2007). Correlation between binding rate constants and individual information of E. coli Fis binding sites. Nucleic Acids Res 35, 5275–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, R. W., Houman, F. & Kleckner, N. (1987). Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53, 85–96. [DOI] [PubMed] [Google Scholar]
- Tesfa-Selase, F. & Drabble, W. T. (1996). Specific binding of DnaA protein to a DnaA box in the guaB gene of Escherichia coli K12. Eur J Biochem 241, 411–416. [DOI] [PubMed] [Google Scholar]
- Tiedeman, A. A. & Smith, J. M. (1984). Isolation and characterization of regulatory mutations affecting the expression of the guaBA operon of Escherichia coli K-12. Mol Gen Genet 195, 77–82. [DOI] [PubMed] [Google Scholar]
- Travers, A., Schneider, R. & Muskhelishvili, G. (2001). DNA supercoiling and transcription in Escherichia coli: the FIS connection. Biochimie 83, 213–217. [DOI] [PubMed] [Google Scholar]
- Weinstein-Fischer, D., Elgrably-Weiss, M. & Altuvia, S. (2000). Escherichia coli response to hydrogen peroxide: a role for DNA supercoiling, topoisomerase I and Fis. Mol Microbiol 35, 1413–1420. [DOI] [PubMed] [Google Scholar]
- Wu, H., Tyson, K. L., Cole, J. A. & Busby, S. J. W. (1998). Regulation of transcription initiation at the Escherichia coli nir operon promoter: a new mechanism to account for co-dependence on two transcription factors. Mol Microbiol 27, 493–505. [DOI] [PubMed] [Google Scholar]