Abstract
Azotobacter vinelandii is a nitrogen-fixing soil bacterium that undergoes differentiation to form cysts resistant to desiccation. Upon encystment, this bacterium becomes non-motile. As in enteric bacteria, motility in A. vinelandii occurs through the use of peritrichous flagella. Pseudomonas aeruginosa, a phylogenetically close relative of A. vinelandii, possesses a single polar flagellum. The FlhDC proteins are the master regulators of flagella and motility in enterobacteria, whereas FleQ is the master regulator in P. aeruginosa, and it is under AlgU (sigmaE) negative control. At present, nothing is known about the organization and expression of flagella genes in A. vinelandii. Here, we identified the flagella gene cluster of this bacterium. Homologues of the master regulatory genes flhDC and fleQ are present in A. vinelandii. Inactivation of flhDC, but not fleQ, impaired flagella biogenesis and motility. We present evidence indicating that a negative effect of the AlgU sigma factor on flhDC expression causes loss of motility in A. vinelandii, and that CydR (a homologue of Fnr) is under AlgU control and has a negative effect on flhDC expression. Taken together, these results suggest the existence of a cascade consisting of AlgU and CydR that negatively controls expression of flhDC; the results also suggest that the block in flagella synthesis under encystment conditions centres on flhDC repression by the AlgU–CydR cascade.
INTRODUCTION
Azotobacter vinelandii, a Gram-negative bacterium belonging to the Gammaproteobacteria, is closely related to bacteria of the genus Pseudomonas (Rediers et al., 2004), and undergoes a differentiation process to form cysts resistant to desiccation. In A. vinelandii, motility occurs through the use of peritrichous flagella. When induced for encystment, this bacterium becomes non-motile (Sadoff, 1975). To the best of our knowledge, studies on the genes involved in motility in this organism have not been carried out. Genes involved in the biogenesis and function of flagella have been extensively studied in Escherichia coli and Salmonella, where motility also occurs by peritrichous flagella. In those bacteria, flagella and motility genes comprise a large and complex regulon, with more than 50 genes organized in at least 17 operons (Macnab, 1996). The flagella operons are classified into three hierarchical transcriptional classes, where the class 1 flhDC operon is the master regulator of flagella and motility (for a review see Aldridge & Hughes, 2002). FlhDC proteins are activators of the class 2 genes, which include those encoding proteins involved in the formation of the hook basal body, the flagella sigma factor FliA, and its anti-sigma factor FlgM. FliA is necessary to activate transcription of the class 3 genes, and these include fliC, which encodes the structural component of the filament, the flagellin. Transcription of flhDC is initiated at six start sites, and its regulation is very complex (Clarke & Sperandio, 2005; Francez-Charlot et al., 2003; Kutsukake, 1997; Soutourina et al., 1999; Wei et al., 2001; Yanagihara et al., 1999).
In Pseudomonas aeruginosa, motility occurs through the use of a single polar flagellum. Flagella genes are clustered in three regions of the chromosome. Transcription of the flagella genes requires a number of regulatory proteins, including FleQ (Arora et al., 1997) and the two-component system FleSR (Ritchings et al., 1995), acting in a four-tiered transcriptional regulatory circuit (Dasgupta et al., 2003). The master regulator FleQ, an NtrC-like transcriptional activator, belongs to the top tier of the flagella hierarchy, and is required to activate all other flagella genes, with the exception of fliA (Arora et al., 1997; Dasgupta et al., 2003; Jyot et al., 2002). The anti-activator protein FleN negatively controls the activity of FleQ (Dasgupta & Ramphal, 2001).
In P. aeruginosa and A. vinelandii, the alternative sigma factor AlgU (also named AlgT) controls alginate biosynthesis. The mechanism by which AlgU exerts this control is well understood (Martínez-Salazar et al., 1996; Núñez et al., 2000; Ramsey & Wosniak, 2005). The anti-sigma proteins MucA and MucB negatively control AlgU activity (Mathee et al., 1997; Schurr et al., 1996; Xie et al., 1996). External stresses affecting the folding of periplasmic proteins initiate the cleavage of MucA, and the release of AlgU (Qiu et al., 2007). In P. aeruginosa, this sigma factor is required to activate the alginate biosynthesis operon (Martin et al., 1993; Wozniak & Ohman, 1994). In A. vinelandii, AlgU is required for activation of the alginate biosynthesis genes algD and algC (Campos et al., 1996; Gaona et al., 2004). Thus, in both P. aeruginosa and A. vinelandii, mutations inactivating algU abrogate alginate synthesis (Moreno et al., 1998), whereas mutations in mucA produce an alginate-overproducing phenotype (Martin et al., 1993; Núñez et al., 2000).
In P. aeruginosa, AlgU activity has a negative effect on flagellum synthesis (Garrett et al., 1999). Tart et al. (2005) showed that the flagella regulon is significantly downregulated in the presence of AlgU, and that increased expression of fleQ reverses the AlgU-mediated inhibition, and thus they concluded that AlgU inhibits expression of FleQ. The mechanism of this inhibition has been shown to be indirect, and it acts by promoting the expression of the transcriptional regulator AmrZ (AlgZ), which interacts directly with the fleQ promoter as a repressor (Tart et al., 2006).
When vegetative motile cells of A. vinelandii are induced for encystment, they lose motility (Sadoff, 1975). We show here that this loss is caused by the activity of the sigma factor AlgU. Thus, similar to the findings for P. aeruginosa, we found a negative effect of AlgU on motility and flagella synthesis.
The organization and expression of flagella genes in A. vinelandii are unknown. A search of the A. vinelandii genome for the flagella regulon was carried out in this study, and revealed the presence of homologues of flhDC and fleQ, which are the master regulators in E. coli and Pseudomonas, respectively. Inactivation of these genes indicated that flhDC, but not fleQ, is required for flagella biogenesis and motility in A. vinelandii. We also show that AlgU and CydR have a negative effect on flhDC expression.
METHODS
Microbiological procedures.
The bacterial strains and plasmids used in this work are shown in Table 1. A. vinelandii was grown at 30 °C in Burk's nitrogen-free salts (Kennedy et al., 1986), supplemented with either sucrose at 2 % (BS medium), or 0.2 % n-butanol (BB encystment medium). E. coli DH5α was grown on Luria–Bertani (LB) medium (Miller, 1972) at 37 °C. Antibiotic concentrations used for A. vinelandii and E. coli, respectively, were as follows: ampicillin, not used and 200 μg ml−1; gentamicin, 1.5 and 10 μg ml−1; kanamycin 6 μg ml−1 and not used; tetracycline, 15 and 20 μg ml−1. Triparental matings were carried out as reported by Kennedy et al. (1986). A. vinelandii transformation was carried out as described by Page & von Tigerstrom (1978), as modified by Bali et al. (1992).
Table 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| A. vinelandii | ||
| ATCC 9046 | Wild-type | ATCC |
| SMU88 | ATCC 9046 with an algU : : Km mutation | Moreno et al. (1998) |
| JRA4 | ATCC 9046 with mucA : : Gm mutation | Núñez et al. (2000) |
| SRA4 | ATCC 9046 with a mucA : : Km mutation | This work |
| AQ 20 | ATCC 9046 with a fleQ : : Gm mutation | This work |
| AC 30 | ATCC 9046 with a flhC : : Tc mutation | This work |
| ATCR | ATCC 9046 with a cydR : : Gm mutation | This work |
| E. coli | ||
| DH5α | supE44lacU169hsdR17recA1endA1gyrA96thi1relA1 | Gibco-BRL |
| Plasmids | ||
| pMOSBlue | Plasmid used for cloning PCR products | Amersham |
| pLRQ | pMOSBlue derivative carrying a 2.1 kb DNA fragment containing A. vinelandii fleQ gene amplified by PCR | This work |
| pLRDC | pMOSBlue derivative carrying a 1.6 kb DNA fragment containing A. vinelandii flhCD genes amplified by PCR | This work |
| pLRQ30 | pLRQ derivative containing a fleQ : : Gm mutation | This work |
| pLRDC50 | pLRDC derivative containing a flhC : : Tc mutation | This work |
| pMUC | pMOSBlue derivative carrying a 0.91 kb DNA fragment containing A. vinelandii mucA gene amplified by PCR | This work |
| pSRA4 | pMUC derivative containing a mucA : : Km mutation | This work |
| pMCYDR | pMOSBlue derivative carrying a 1.17 kb DNA fragment containing A. vinelandii cydR gene amplified by PCR | This work |
| pMCYDR-Gm | pCYDR derivative containing a cydR : : Gm mutation | This work |
| pJB3Km1 | Blatny et al. (1997) | |
| pRK2013 | Hedges & Baumberg (1973) | |
| pHP4 Ω-Tc | Source of the Tcr cassette | Fellay et al. (1987) |
| pBSL141 | Source of the Gmr cassette | Alexeyev et al. (1995) |
| pBSL99 | Source of the Kmr cassette | Alexeyev et al. (1995) |
| pSM40 | Source of the Gmr cassette | Peralta-Gíl et al. (2002) |
| pLRGm-DC | pJB3Km1 derivative carrying a 1.6 kb DNA fragment containing A. vinelandii flhCD genes and the Gmrh gene from pBSL14 | This work |
DNA techniques.
DNA isolation, cloning, Southern blotting, and nick translation procedures were carried out as described by Sambrook et al. (1989).
Cloning of flhDC, fleQ, mucA and cydR genes.
The A. vinelandii fleQ and flhDC genes were amplified by PCR using ATCC 9046 chromosomal DNA as a template and the following oligonucleotides: fleQ, upper primer 5′-TTATGCCTTGCTGGGGTTGC-3; fleQ, lower primer 5′-TTCACCCGTTCGTAGGCATC-3′; flhDC, upper primer 5′-AATGCTTCCCAGGCGAGATC-3′; and flhDC, lower primer 5′-GACAACGATGAGACC CAGAG-3′. For mucA and cydR, oligonucleotides mucA-1U 5′-GAAATCGAGGCCACTGTG-3′, mucA-1L 5′-CAACCAATTCTGCGCATC-3′, cydRf 5′-GTTCGTTCGATCTGCATGC-3′ and cydRr 5′-TTACTGGAAGCGGACATGCG-3′ were used. Primers were designed on the basis of the OP strain draft genome sequence available at http://img.jgi.doe.gov/cgi-bin/pub/main.cgi. The resulting 2157 (fleQ), 1667 (flhDC), 910 (mucA) and 1173 (cydR) bp PCR products were cloned in pMOSBlue, producing plasmids pLRQ, pLRDC, pMUC and pMCYDR, respectively (Table 1). Restriction mapping and partial sequencing confirmed the identity of the inserts (data not shown).
Construction of fleQ : : Gm, flhC : : Tc, mucA : : Km and cydR : : Gm mutations.
Plasmid pLRQ was digested with XhoI to eliminate a 230 bp XhoI internal fleQ fragment. The 0.8 kb XhoI fragment containing a gentamicin-resistance cassette isolated from plasmid pBSL141 (Alexeyev et al., 1995) was ligated into the pLRQ XhoI-digested plasmid. Plasmid pLRQ30, containing the fleQ : : Gm mutation was isolated. For inactivation of the flhC gene, a 2.0 kb SmaI fragment, containing a tetracycline-resistance cassette isolated from plasmid pHP4 Ω-Tc (Fellay et al., 1987), was inserted into the StuI site within gene flhC in plasmid pLRDC. Plasmid pLRDC50 containing the flhC : : Tc insertion was isolated. A kanamycin-resistance cassette from plasmid pBSL99 (Alexeyev et al., 1995) was introduced into the XhoI site of mucA in plasmid pMUC, producing plasmid pSRA4. For inactivation of cydR, a 3.0 kb ClaI fragment, containing a gentamicin-resistance cassette from plasmid pMS40 (Peralta-Gíl et al., 2002), was introduced into the ClaI site present within cydR in pMCYDR, producing plasmid pMCYDR-Gm
Plasmids pLRQ30 and pLRDC50, which are unable to replicate in A. vinelandii, were used to introduce the fleQ : : Tc and flhC : : Gm mutations into strain ATCC 9046. Transformants were selected using the corresponding antibiotic, and confirmed by Southern blot analysis to carry the desired mutations (data not shown). Plasmid pSRA4 was transformed into strain ATCC 9046 to generate strain SRA4. The presence of the mucA : : Km mutation in strain SRA4 was confirmed by PCR analysis. Plasmid pMCYDR-Gm was introduced into strains ATCC 9046 and SRA4. A gentamicin-resistant transformant derived from ATCC 9046 was isolated, and named ATCR. No gentamicin transformants derived from strain SRA4 were obtained in three different experiments. The cydR : : Gm gene replacement in ATCR was confirmed by PCR analysis (data not shown).
Construction of plasmid pLRGm-DC.
Plasmid pJB3Km1 (Blatny et al., 1997), which was able to replicate in A. vinelandii, was digested with HindIII and BamHI restriction enzymes to remove a 1 kb fragment that included the kanamycin-resistance gene. This fragment was replaced by a 1.6 kb BamHI–HindIII fragment containing the flhDC genes, including their promoter sequences. A 0.8 kb BamHI fragment encoding the gentamicin-resistance gene was introduced into the plasmid to produce plasmid pLRGm-DC, which was transferred by conjugation into strain AC30 in a triparental mating using plasmid pRK2013.
Motility assay.
To test the motility of A. vinelandii, bacterial strains were grown on BS medium at 30 °C until exponential phase. Samples of the cells were then transferred to BS or BB encystment medium, containing 0.3 % agar. These plates were incubated at 30 °C for 36 or 48 h.
Electron microscopy.
Transmission electron microscopy to visualize flagella was carried out as previously reported (Gaona et al., 2004)
Quantitative RT-PCR (qRT-PCR).
qRT-PCR was performed as reported (Noguez et al., 2008). For RNA extraction, the cultures were grown in BS liquid medium. Cells were collected at the exponential phase of growth for flhC, and at 37 h for cydR. The sequences of the primers used for the qRT-PCR assays were: for cydR, 5′-GGC TGTCGAGACCGTATCC-3′ and 5′-ATTCGACGGGATTGAGAATG-3′; for flhDC, 5′-GAACATCCATTCCTCGCTGT-3′ and 5′-ATAGAGCCGGAAAGCCTTGA-3′.
RESULTS
AlgU activity causes loss of flagella in A.vinelandii
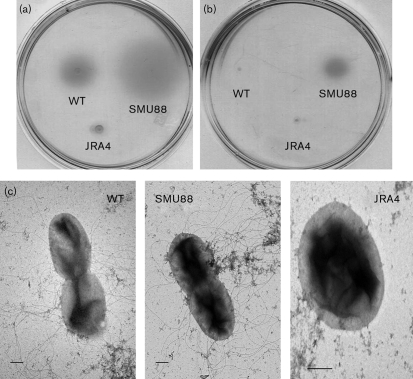
In P. aeruginosa, AlgU has a negative effect on transcription of the flagella regulon by downregulating fleQ. To determine whether AlgU affected flagella synthesis in A. vinelandii, we performed motility assays in BS medium for strains ATCC 9046, SMU88 (algU mutant derivative of ATCC 9046) and JRA4, a derivative carrying a mucA mutation that results in high AlgU activity (Núñez et al., 2000). As shown in Fig. 1(a), the mucA mutant strain was non-motile, as indicated by the absence of a motility zone. In contrast, the algU mutant produced a motility zone larger than that produced by the wild-type strain. When observed under the light microscope, cells of the wild-type strain ATCC 9046 growing on BB encystment medium were non-motile. Therefore, swimming assays were also performed in BB encystment medium for the wild-type, and the mucA and algU mutants (Fig. 1b). No swimming was observed for the wild-type and mucA strains, whereas the algU mutant produced a swimming zone. Using transmission electron microscopy, we examined the mucA and algU strains harvested from cultures growing exponentially on BS medium to determine the presence of flagella. In agreement with the swimming phenotype, no flagella were present for the mucA strain, whereas the algU mutant, similar to the wild-type, produced numerous flagella (Fig. 1c).
Fig. 1.
Motility phenotype of algU and mucA mutants. Swimming assays of A. vinelandii wild-type strain ATCC 9046 (WT), algU mutant SMU88, and mucA mutant JRA4, carried out on motility agar plates with BS medium (a) and BB encystment medium (b). (c) Electron micrographs of negatively stained preparations of strains ATCC 9046, SMU88 and JRA4. Bars, 1.0 μm. Cells for transmission electron microscopy were harvested from cultures growing exponentially on BS medium.
Flagella and motility genes found in the A. vinelandii genome
The flagella gene system of E. coli is one of the best studied, and is composed of over 50 genes for flagella assembly and function (Kutsukake & Nambu, 2000; Macnab, 1996). In order to identify A. vinelandii genes involved in flagella biogenesis and motility, and the possible targets for the AlgU-negative effect, we carried out an in silico analysis to search the draft genome sequence of A. vinelandii OP for genes homologous to bacterial flagella and motility genes. The A. vinelandii genome sequence data were obtained from http://img.jgi.doe.gov/cgi-bin/pub/main.cgi. Putative flagella and motility genes were identified by tblastn searches using E. coli genes. Because of the close phylogenetic relationship between Azotobacter and Pseudomonas species (Rediers et al., 2004), we also used P. aeruginosa genes in the analysis.
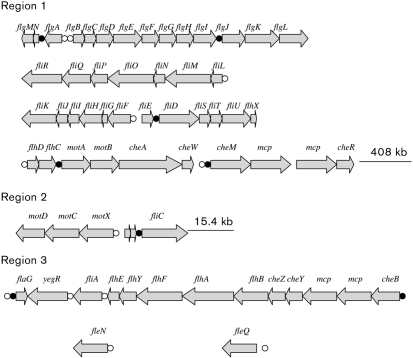
Three regions containing putative flagella and motility genes were identified in the A. vinelandii genome. These genes and the putative proteins they encode are listed in supplementary Table S1 (available with the online version of this paper). The genes include homologues of the flhDC master regulators in E. coli, and the Pseudomonas master regulator fleQ and its anti-activator fleN. In contrast to Pseudomonas spp., where these genes are present in the neighbourhoods of other flagella genes, the A. vinelandii fleQ and fleN genes are not linked to flagella genes. A representation of the regions, as well as the position and orientation of these genes in the A. vinelandii genome, is presented in Fig. 2. Region 1 consists of 39 717 bp, and contains 42 genes. Region 2 harbours four genes, including fliC, and region 3 consists of 12 genes.
Fig. 2.
Schematic representation of the A. vinelandii flagellar and motility regulon. Filled and open circles represent the presence of FliA- and RpoD-recognized consensus sequences, respectively. Genes in region 1 are contiguous. In region 2, motD is located 408 kb downstream of cheR. The fliC gene in region 2 is separated from flaG by 15.3 kb. fleQ and fleN are not linked to flagella genes.
A blast search of A. vinelandii FlhDC proteins revealed the absence of homologues in Pseudomonas spp. A summary of flagella genes present or absent in A. vinelandii, P. aeruginosa and E. coli is presented in supplementary Table S2 (available with the online version of this paper). Most A. vinelandii flagella genes showed the highest identity to genes from Chromohalobacter salexigens and Cupriavidus necator (Table S1). C. salexigens is a gamma proteobacterium that is closely related to Pseudomonas spp. and E. coli. Motility in Chromohalobacter occurs by means of peritrichous flagella (Holt et al., 1994).
We also carried out a search for AlgU-, RpoD- and FliA-recognized consensus sequences within intergenic sequences of flagella genes larger than 80 nt. For putative RpoD (sigma 70)-recognized promoters, we used bprom (http://www.softberry.com/berry.phtml), which is a program for the prediction of bacterial RpoD promoters that has an accuracy of about 80 %. The search for putative FliA and AlgU promoters was carried out by ocular inspection. The results are presented in supplementary Table S3 (available with the online version of this paper) and Fig. 2. The AlgU GAACTT-16/17 bp-TCTgA-recognized sequence (Gaona et al., 2004) was not identified. The FliA-recognized sequence CTAA-15 bp-GCCGATAG was found upstream of eight putative operons. Twelve genes, including the master operon flhDC, were found to possess putative RpoD promoters The putative cheM–mcp–mcp–cheR operon, and the flaG gene, were found to possess both FliA and RpoD consensus sequences.
Inactivation of the flhDC genes, but not fleQ, impairs motility
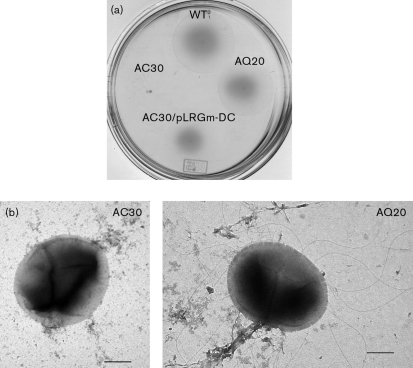
In contrast to E. coli and Pseudomonas spp., A. vinelandii was found to possess flhDC and fleQ regulatory genes. To determine the functionality of the flhDC and fleQ homologues, which are located in the top hierarchy of flagella gene regulation in E. coli and P. aeruginosa, respectively, we constructed, as described in Methods, strain AC30 carrying a flhC : : Tc mutation, and strain AQ20 carrying a fleQ : : Gm mutation. The AC30 and AQ20 mutants were tested for their swimming phenotype. As shown in Fig. 3(a), inactivation of flhC completely inhibited motility. In contrast, the mutant carrying the fleQ mutation showed a motility phenotype similar to the wild-type strain. Electron microscopy revealed the absence of flagella in the flhC mutant, but not in the fleQ mutant (Fig. 3b). These results indicate that flhDC positively controls flagella synthesis and motility in A. vinelandii.
Fig. 3.
Motility phenotype of flhC and fleQ mutants. (a) Swimming plate assay carried out in BS medium for: ATCC 9046 (WT), flhC mutant AC30, fleQ mutant AQ20, and AC30/pLRGm-DC. (b) Electron micrographs of AC30 and AQ20 mutants. Bars, 1.0 μm.
Motility is restored by complementation with the flhDC genes
The flhD and flhC genes overlap by 1 nt, and they are separated from the downstream motAB genes by an intergenic region containing a consensus FliA-recognized sequence (Fig. 2, Supplementary Table S3). Thus, the flhDC genes appear to constitute a bicistronic operon, and the flhC mutation was not expected to affect motAB transcription. To confirm that the swimming defect in strain AC30 was caused by the lack of the FlhC protein, and not by polar effects on downstream genes, and to confirm functionality of the flhDC genes, plasmid pLRGm-DC, containing only the flhDC genes including the promoter sequences, was introduced into strain AC30 by conjugation. The resultant strain AC30/pLRGm-DC showed a swimming phenotype similar to that of the wild-type strain (Fig. 3a).
Effect of AlgU on expression of flhDC
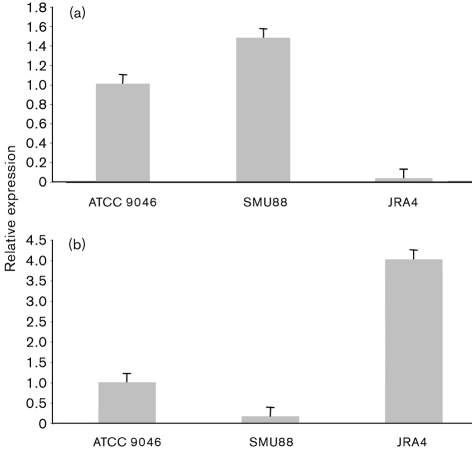
To determine whether AlgU affected flagella synthesis by downregulation of the master operon flhDC, we carried out qRT-PCR analysis to quantify the levels of flhDC mRNA in cells of mutant SMU88 lacking AlgU activity, and in the mucA mutant JRA4, in which the absence of the anti-AlgU protein MucA results in high AlgU activity (Núñez et al., 2000) (Fig. 4). RNA was isolated from cultures of the A. vinelandii strains grown exponentially on BS medium. In the SMU88 algU mutant strain, flhDC mRNA was 40 % higher than in the wild-type. In contrast, flhDC mRNA levels were very low in the non-motile mucA strain JRA4. Based on these results, we conclude that the expression of the master flhDC operon is under the negative control of AlgU, although this control might not be direct.
Fig. 4.
Effect of AlgU on expression of flhDC and cydR. qRT-PCR analysis of flhDC (a), and cydR (b) gene expression from BS cultures of the wild-type strain ATCC 9046, the algU mutant SMU88 and the mucA mutant JRA4. The level of flhDC and cydR transcripts was normalized according to the levels of gyrA mRNA, and data are presented as -fold changes of mRNA levels of SMU88 and JRA4 mutant strains relative to those of the wild-type ATCC 9046.
CydR is under AlgU control, and is likely to be a repressor of flhDC expression
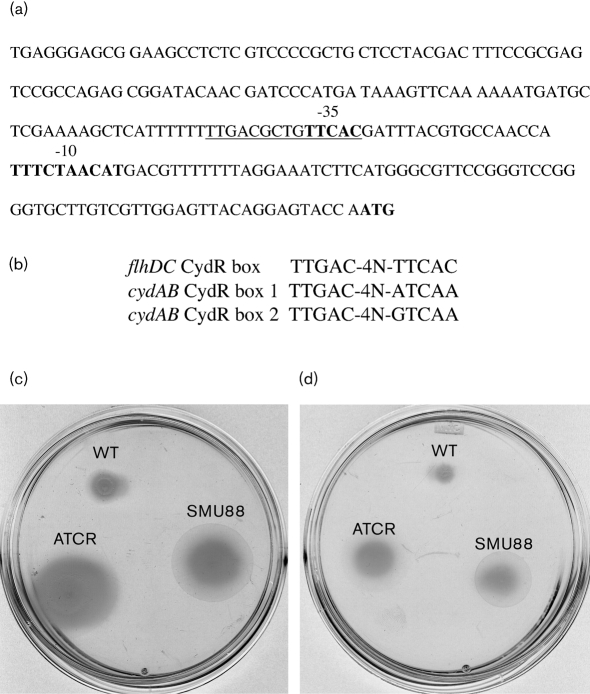
We inspected the 228 nt flhDC promoter region for the presence of putative binding sites for known regulators. A. vinelandii CydR is an Fnr homologue that represses transcription of the oxidase genes cydAB by binding at the CydR boxes located in the cydAB promoter region (Wu et al., 2000). A sequence highly similar to the CydR boxes is present in the flhDC promoter region, and it overlaps the putative –35 sequence (Fig. 5). The presence of a putative CydR box within the flhDC putative promoter led us to hypothesize that CydR might mediate repression of flhDC by AlgU. In order to determine whether cydR transcription was dependent on AlgU, we carried out qRT-PCR analysis to determine the levels of cydR mRNA in cells of the wild-type, the algU mutant SMU88, and the mucA mutant JR4. RNA was isolated from cultures of the A. vinelandii strains grown on BS medium. As shown in Fig. 4(b), in the algU mutant strain, the cydR mRNA level is significantly reduced compared to the wild-type, whereas the levels in the mucA mutant are threefold higher, indicating that AlgU is indeed required for CydR expression. Based on this result, and on the presence of CydR boxes in the flhD promoter, inactivation of cydR was expected to produce a hyper-motility phenotype similar to that of the algU mutant, and to restore the motility phenotype in the mucA mutant. We constructed, as described in Methods, strain ATCR, which is an ATCC 9046 derivative carrying a cydR mutation. Strain ATCR grew very poorly on Burk's medium (data not shown), but produced a motility zone larger than that produced by the algU mutant (Fig. 5c). The swimming of ATCR on BB encystment medium was similar to that of the algU mutant (Fig. 5d). Efforts to construct a mucA–cydR double mutant strain were unsuccessful, probably because of the detrimental effects on growth caused by the cydR mutation, and also because of the effects of the mucA mutation, which reduces the growth rate due to alginate overproduction (Núñez et al., 2000). Taken together, these results indicate that AlgU exerts a positive control on CydR, and that this in turn is a repressor of flhD expression.
Fig. 5.
Motility phenotype of cydR mutant. (a) DNA sequence of the 5′ region of flhDC. The putative CydR-binding site is underlined. The –10 and –35 regions are shown in bold. (b) Sequence alignment of the CydR boxes. (c, d) Swimming plate assay carried out in BS medium (c), and BB encystment induction medium (d), for ATCC 9046 (WT), and the cydR (ATCR), and algU (SMU88) mutants.
DISCUSSION
A. vinelandii undergoes differentiation to form a metabolically dormant cyst resistant to desiccation. A mature cyst consists of a contracted cell known as the central body, which is surrounded by a capsule containing a high proportion of alginate (Page & Sadoff, 1975). Encystment can be induced in laboratory conditions by transferring vegetative motile cells grown in liquid BS medium to Burk's medium supplemented with 0.2 % n-butanol or β-hydroxybutyrate as the sole carbon source. This induction results in loss of flagella (Sadoff, 1975).
The alternative sigma factor AlgU is required for expression of the alginate biosynthesis genes in A. vinelandii (Gaona et al., 2004; Moreno et al., 1998). Alginate is essential for the formation of mature cysts, and mutations in the alginate biosynthesis genes, or in algU, impair alginate synthesis and encystment (Campos et al., 1996; Mejía-Ruíz et al., 1997; Moreno et al., 1998). A link between alginate synthesis and flagellum expression, which are inversely regulated by the alternative sigma factor AlgU, has been shown in P. aeruginosa (Tart et al., 2005, 2006), which is a close relative of A. vinelandii.
We have shown here that loss of motility upon encystment induction in A. vinelandii is caused by AlgU activity. Thus, as in the case of P. aeruginosa, alginate synthesis and flagella biogenesis, are inversely controlled by AlgU. In order to identify possible targets for AlgU among the flagella genes, we identified the A. vinelandii gene homologues of bacterial flagella and motility genes. Most of these genes share the highest identity with the genes of C. salexigens; this bacterium is phylogenetically closely related to A. vinelandii, since their 16S rRNA shares 90.86 % similarity, and this level of identity is second only to Pseudomonas spp., for which the similarity is around 95–96 %.
An important finding of this study was the presence in A. vinelandii of fleQ and flhDC, which are the master regulators of flagella biogenesis. flhDC, but not fleQ, is located in the context of other flagella genes. Inactivation of the flhDC and fleQ genes indicated that the FlhDC proteins are the master regulators of flagella biogenesis in A. vinelandii.
The lack of FleQ involvement in flagella biogenesis is in agreement with the absence of the fleRS genes (Table S2), which are the targets of FleQ regulation in P. aeruginosa (Dasgupta et al., 2003). It is possible that, in A. vinelandii, FleQ participates in regulating the expression of other genes that are not involved in flagella biogenesis.
This study showed that in A. vinelandii, AlgU activity inhibits flagella synthesis in vegetative cells and under encysting conditions. A negative effect of AlgU on transcription of the flhDC operon was also shown here. In P. aeruginosa, negative regulation of flagella synthesis by AlgU is carried out by activating expression of the transcriptional regulator AmrZ, which in turn represses fleQ transcription (Tart et al., 2005, 2006). Interestingly, binding sites for CydR (CydR boxes) within the flhDC promoter region were identified, suggesting that CydR is a repressor of flhDC expression. In agreement with this proposal, inactivation of cydR conferred a hyper-swimming capacity to the wild-type strain. In addition, we showed that the levels of cydR mRNA are significantly reduced in the algU mutant, indicating that AlgU is required for cydR expression, and that CydR mediates the negative effect of AlgU. Inactivation of cydR was therefore expected to restore motility in the mucA mutant. Unfortunately, our efforts to isolate a mucA–cydR double mutant were unsuccessful.
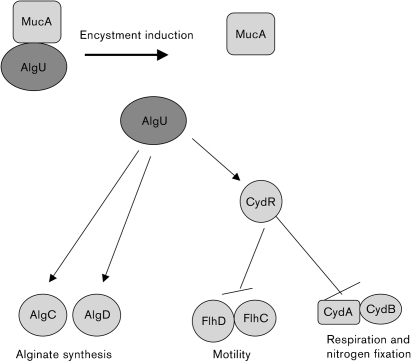
CydR is a homologue of Fnr. In A. vinelandii, CydR has been shown to act as a repressor of the cydAB genes encoding cytochrome bd, which is required for aerotolerant nitrogen fixation (Wu et al., 2000). The loss of flagella and nitrogen fixation activity observed upon induction of encystment (Sadoff, 1975; Hitchins & Sadoff, 1973) can now be explained by the negative effect of CydR on expression of the cydAB and flhDC genes. Taken together, the results presented in this study indicate the existence of this regulatory cascade consisting of AlgU upstream of CydR, which in turn acts as a repressor of flhCD expression. A model for the control of alginate synthesis, motility, respiration and nitrogen fixation, upon encystment induction by the AlgU–CydR regulatory cascade in A. vinelandii, is shown in Fig. 6.
Fig. 6.
Model for the control by AlgU of: motility, alginate synthesis, respiration, and nitrogen fixation upon encystment induction in A. vinelandii. Induction of encystment, which is likely to be caused by a stress condition, results in the release of AlgU by MucA. AlgU in turn activates transcription of alginate biosynthesis genes and CydR, which is the repressor of FlhDC and CydAB.
This study also showed that the common feature in the regulation of motility in both P. aeruginosa and A. vinelandii is the negative effect of AlgU, but that there is a difference in the repressor controlled by AlgU (AmrZ for Pseudomonas, and CydR for Azotobacter), and the targets of these repressors (fleQ for Pseudomonas, and flhDC for A. vinelandii). These differences may be explained by the need of A. vinelandii to coordinate loss of functions such as motility, high respiration rate and nitrogen fixation that occur upon encystment induction, which is a process not carried out by Pseudomonas species.
Acknowledgments
This research was founded in part by grants PAPIIT IN208805 and CONACyT U47781-Q. R. L. was the recipient of a CONACyT Scholarship. We thank Roxana Sanchez and Guadalupe Zavala for support with the electron microscopy studies, Josefina Guzmán and Soledad Moreno for technical support, and Dr Gloria Soberón for critical reading of the manuscript.
Abbreviations
qRT-PCR, quantitative RT-PCR.
Footnotes
Tables of A. vinelandii flagellar and motility genes, and consensus sequences recognized by FliA and RpoD sigma factors present upstream of A. vinelandii flagellar genes are available with the online version of this paper.
References
- Aldridge, P. & Hughes, K. T. (2002). Regulation of flagellar assembly. Curr Opin Microbiol 5, 160–165. [DOI] [PubMed] [Google Scholar]
- Alexeyev, M. F., Shokolenko, I. & Croughan, T. P. (1995). Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160, 63–67. [DOI] [PubMed] [Google Scholar]
- Arora, S. K., Ritchings, B. W., Almira, E. C., Lory, S. & Ramphal, R. (1997). A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol 179, 5574–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali, A., Blanco, G., Hill, S. & Kennedy, C. (1992). Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol 58, 1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatny, J. M., Brautaset, T., Winther-Larsen, H. C., Haugan, K. & Valla, S. (1997). Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl Environ Microbiol 63, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, M.-E., Martínez-Salazar, J. M., Lloret, L., Moreno, S., Núñez, C., Espín, G. & Soberón-Chávez, G. (1996). Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol 178, 1793–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, M. B. & Sperandio, V. (2005). Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorragic Escherichia coli. Mol Microbiol 57, 1734–1749. [DOI] [PubMed] [Google Scholar]
- Dasgupta, N. & Ramphal, R. (2001). Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 183, 6636–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta, N., Wolfgang, M. C., Goodman, A. L., Arora, S. K., Jyot, J., Lory, S. & Ramphal, R. (2003). A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50, 809–824. [DOI] [PubMed] [Google Scholar]
- Fellay, R., Frey, J. & Krisch, H. (1987). Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis. Gene 52, 147–154. [DOI] [PubMed] [Google Scholar]
- Francez-Charlot, A., Laugel, B., Van Gemert, A., Dubarry, N., Wiorowski, F., Castanie-Cornet, M. P., Gutierrez, C. & Cam, K. (2003). RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol 49, 823–832. [DOI] [PubMed] [Google Scholar]
- Gaona, G., Núñez, C., Goldberg, J. B., Linford, A. N., Nájera, R., Castañeda, M., Guzmán, J., Espín, G. & Soberón-Chávez, G. (2004). Characterization of the Azotobacter vinelandii algC gene involved in alginate and lipopolysaccharide production. FEMS Microbiol Lett 238, 199–206. [DOI] [PubMed] [Google Scholar]
- Garrett, E. S., Perlegas, D. & Wozniak, D. J. (1999). Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J Bacteriol 181, 7401–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, R. W. & Baumberg, S. (1973). Resistance to arsenic compounds conferred by a plasmid transmissible between strains of Escherichia coli. J Bacteriol 115, 459–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins, V. M. & Sadoff, H. L. (1973). Sequential metabolic events during encystment in Azotobacter vinelandii. J Bacteriol 113, 1273–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, J. G., Krieg, N. R., Sneath, P. H. A., Staley, J. T. & Williams, S. T. (1994). Bergey's Manual of Determinative Bacteriology, 9th edn, p. 79. Baltimore: Williams & Wilkins.
- Jyot, J., Dasgupta, N. & Ramphal, R. (2002). FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol 184, 5251–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, C., Gamal, R., Hummprey, R., Ramos, J., Brigle, K. & Dean, D. (1986). The nifH, nifM, and nifN genes of Azotobacter vinelandii: characterization by Tn5 mutagenesis and isolation from pLARF1 gene banks. Mol Gen Genet 205, 318–325. [Google Scholar]
- Kutsukake, K. (1997). Autogenous and global control of the flagellar master operon, flhDC, in Salmonella typhimurium. Mol Gen Genet 254, 440–448. [DOI] [PubMed] [Google Scholar]
- Kutsukake, K. & Nambu, T. (2000). Bacterial flagellum: a paradigm for biogenesis of transenvelope supramolecular structures. Recent Res Dev Microbiol 4, 607–615. [Google Scholar]
- Macnab, R. M. (1996). Flagella and motility. In Escherichia coli and Salmonella: Cellular and Molecular Biology, pp. 123–145. Edited by F. C. Neidhardt and others. Washington, DC: American Society for Microbiology.
- Martin, D. W., Schurr, M. J., Mudd, M. H., Govan, J. R., Holloway, B. W. & Deretic, V. (1993). Mechanism of conversion to mucoidy in P. aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A 90, 8377–8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salazar, J. M., Moreno, S., Nájera, R., Boucher, J. C., Espín, G., Soberón-Chávez, G. & Deretic, V. (1996). Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J Bacteriol 178, 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee, K., McPherson, C. J. & Ohman, D. E. (1997). Posttranslational control of the AlgT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol 179, 3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía-Ruíz, H., Moreno, S., Guzmán, J., Nájera, R., León, R., Soberón-Chávez, G. & Espín, G. (1997). Isolation and characterization of an Azotobacter vinelandii algK mutant. FEMS Microbiol Lett 156, 101–106. [DOI] [PubMed] [Google Scholar]
- Miller, J. H. (1972). Experiments in Molecular Genetics, pp. 431–435. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Moreno, S., Nájera, R., Guzmán, J., Soberón-Chávez, G. & Espín, G. (1998). Role of alternative factor AlgU in encystment of Azotobacter vinelandii. J Bacteriol 180, 2766–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguez, R., Segura, D., Moreno, S., Hernandez, A., Juárez, K. & Espín, G. (2008). Enzyme INtr, Npr, and IIANtr are involved in regulation of the polyhydroxybutyrate biosynthetic genes in Azotobacter vinelandii. J Mol Microbiol Biotechnol in press [DOI] [PubMed]
- Núñez, C., León, R., Guzmán, J., Espín, G. & Soberón-Chávez, G. (2000). Role of Azotobacter vinelandii mucA and mucC gene products in alginate production. J Bacteriol 182, 6550–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, W. J. & Sadoff, H. L. (1975). Relationship between calcium and uronic acids in the encystment of Azotobacter vinelandii. J Bacteriol 122, 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, W. J. & von Tigerstrom, M. (1978). Induction of transformation competence in Azotobacter vinelandii iron limited cultures. Can J Microbiol 24, 1590–1594. [DOI] [PubMed] [Google Scholar]
- Peralta-Gíl, M., Segura, D., Guzmán, J., Servín-Gonzalez, L. & Espín, G. (2002). Expression of the Azotobacter vinelandii poly-β-hydroxybutyrate biosynthetic phbBAC operon is driven by two overlapping promoters and is dependent on the transcriptional activator PhbR. J Bacteriol 184, 5672–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, D., Eisinger, V. M., Rowen, D. W. & Yu, H. D. (2007). Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 104, 8107–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, D. M. & Wosniak, D. J. (2005). Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol 56, 309–322. [DOI] [PubMed] [Google Scholar]
- Rediers, H., Vanderleyden, J. & De Mot, R. (2004). Azotobacter vinelandii: a Pseudomonas in disguise? Microbiology 150, 1117–1119. [DOI] [PubMed] [Google Scholar]
- Ritchings, B. W., Almira, E. C., Lory, S. & Ramphal, R. (1995). Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun 63, 4868–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff, H. L. (1975). Encystment and germination in Azotobacter vinelandii. Bacteriol Rev 39, 516–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Schurr, M. J., Yu, H., Martinez-Salazar, J. M., Boucher, J. C. & Deretic, V. (1996). Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol 178, 4997–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina, O., Kolb, A., Krin, E., Laurent-Winter, C., Rimsky, S., Danchin, A. & Bertin, P. (1999). Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181, 7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tart, A. H., Wolfgang, M. C. & Wosniak, D. J. (2005). The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol 187, 7955–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tart, A. H., Blanks, M. J. & Wosniak, D. J. (2006). The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J Bacteriol 188, 6483–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, B. L., Brun-Zinkernagel, A. M., Simecka, J. W., Pruss, B. M., Babitzke, P. & Romeo, T. (2001). Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40, 245–256. [DOI] [PubMed] [Google Scholar]
- Wozniak, D. J. & Ohman, D. E. (1994). Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol 176, 6007–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Cruz-Ramos, H., Hill, S., Green, J., Sawers, G. & Poole, R. K. (2000). Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr). Sensitivity to oxygen species, and nitric oxide. J Biol Chem 275, 4679–4686. [DOI] [PubMed] [Google Scholar]
- Xie, Z. D., Hershberger, C. D., Shankar, S., Ye, R. W. & Chakrabarty, A. M. (1996). Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol 178, 4990–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara, S., Iyoda, S., Ohnishi, K., Iino, T. & Kutsukake, K. (1999). Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet Syst 74, 105–111. [DOI] [PubMed] [Google Scholar]