Abstract
Small nucleolar RNAs (snoRNAs) guide nucleotide modifications within ribosomal RNAs or spliceosomal RNAs by base-pairing to complementary regions within their RNA targets. The brain-specific snoRNA MBII-52 lacks such a complementarity to rRNAs or snRNAs, but instead has been reported to target the serotonin receptor 2C pre-mRNA, thereby regulating pre-mRNA editing and/or alternative splicing. To understand how the MBII-52 snoRNA might be involved in these regulatory processes, we isolated the MBII-52 snoRNP from total mouse brain by an antisense RNA affinity purification approach. Surprisingly, by mass spectrometry we identified 17 novel candidates for MBII-52 snoRNA binding proteins, which previously had not been reported to be associated with canonical snoRNAs. Among these, Nucleolin and ELAVL1 proteins were confirmed to independently and directly interact with the MBII-52 snoRNA by coimmunoprecipitation. Our findings suggest that the MBII-52 snoRNA assembles into novel RNA-protein complexes, distinct from canonical snoRNPs.
Keywords: noncoding RNA, small nucleolar RNA, MBII-52, Nucleolin, ELAVL1
INTRODUCTION
Small nucleolar RNAs (snoRNAs) represent one of the most abundant noncoding RNA species in all organisms and are required for maturation of pre-ribosomal RNA (pre-rRNA) within the nucleolus. SnoRNAs are grouped into two classes, box C/D and box H/ACA snoRNAs, which mediate 2′-O-ribose methylation and pseudouridylation on pre-rRNAs or pre-snRNAs, respectively (Hüttenhofer et al. 2002). Both snoRNA classes exert their function by base-pairing to complementary sites within target rRNAs or snRNAs (Kiss-Laszlo et al. 1996, 1998).
In previous studies, high-throughput screening for noncoding RNAs in mouse revealed a set of snoRNAs (MBII-13, MBII-52, MBII-85, and MBI-36) expressed exclusively in rodent brain (Cavaille et al. 2000; Hüttenhofer et al. 2001, 2002). Unlike known snoRNAs, brain-specific snoRNAs lack complementarities to pre-rRNAs or pre-snRNAs, thus being designated as “orphan snoRNAs.” Although both experimental and computational approaches have uncovered an increasing number of orphan snoRNAs, the biological function of orphan snoRNAs still remains elusive (Yang et al. 2006).
In contrast to canonical snoRNAs targeting snRNAs or rRNAs, the orphan snoRNA MBII-52 has been reported to exhibit an 18-nucleotide-long complementarity to the serotonin receptor 2C (5-HT2CR) pre-mRNA, thus regulating mRNA expression (Cavaille et al. 2000). The targeted site within the 5-HT2CR pre-mRNA is subject to A-to-I RNA editing and is also juxtaposed to an alternative splice site (Canton et al. 1996; Burns et al. 1997; Wang et al. 2000b). In the past, several groups have reported the involvement of MBII-52 in regulation of editing or splicing of the 5-HT2CR pre-mRNA (Vitali et al. 2005; Kishore and Stamm 2006; Doe et al. 2009). In these studies it has been noted that MBII-52 snoRNA transcripts are extremely abundant in mouse brain, compared to canonical snoRNAs (Hüttenhofer et al. 2001). This has been explained by the fact that >100 closely related copies of MBII-52 are clustered within the mouse chromosome 7C locus together with several other snoRNAs (Nicholls and Knepper 2001). A cluster of the human ortholog HBII-52 (now assigned as SNORD115@) is conserved in the syntenic region on chromosome 15q11-q13.
Based on conserved sequence motifs, MBII-52 snoRNA has been classified as a box C/D-type snoRNA. In general, C/D snoRNAs contain conserved boxes C (RUGAUGA) and D (CUGA) at their 5′ and 3′ termini, respectively, and internally also exhibit a second set of box elements, namely C′ and D′. An rRNA- or snRNA-targeting sequence, termed “antisense element,” located upstream of the D (also D′) box. Boxes C/D (or C′/D′), which form a specific structural motif termed kink-turn (K-turn), are required for the assembly of four core proteins, namely, 15.5K, Nop56, Nop58, and a methyltransferase, designated as Fibrillarin, to build up a functional ribonucleo-protein particle (C/D snoRNP) (Samarsky et al. 1998; Watkins et al. 2000; Klein et al. 2001; Watkins et al. 2002; Tran et al. 2004). Fibrillarin is necessary and required to localize the mature snoRNP complex to the nucleolus and also catalyzes the formation of the 2′-O-Methylribose within target RNAs (Lafontaine and Tollervey 2000; Wang et al. 2000a; Verheggen et al. 2001).
As shown for canonical snoRNPs, the nucleolar localization of the MBII-52 snoRNA, although lacking complementarity to rRNA or snRNA targets, has been demonstrated by FISH analysis (Vitali et al. 2005). In addition, by employing specific antibodies against the four canonical C/D RNP core proteins, MBII-52 RNA was coimmunoprecipitated, suggesting that MBII-52 forms a C/D snoRNP in vivo (Vitali et al. 2005). Therefore, these reports have implied a function of the MBII-52-RNP complex as a bona fide C/D snoRNP within the nucleolus. These findings, however, are in contrast to its predicted function in regulation of splicing and/or editing, events occurring within the nucleoplasm, outside the nucleolus. In the present study, we aimed to isolate MBII-52 RNP complexes from total mouse brain. Identification of MBII-52 binding proteins within these RNPs was expected to result in a better understanding on its cellular distribution, as well as its biological function. To this end, we employed an RNA affinity purification procedure, and candidate proteins were identified by mass spectrometry. Thereby, 17 novel candidates for MBII-52 snoRNA binding proteins were identified, which previously had not been reported to be associated with canonical snoRNPs.
RESULTS AND DISCUSSION
Antisense RNA-based affinity purification approaches are powerful tools for analyzing protein components of RNPs. Although in principle this method is applicable to any RNP analysis, it has so far and preferentially been employed for the characterization of spliceosomal complexes (Blencowe and Lamond 1999; Will et al. 2004). In this study, we investigated MBII-52 snoRNP with this approach since the MBII-52 is extremely abundant in mouse brain, compared to canonical snoRNAs (Hüttenhofer et al. 2001).
MBII-52 interacts with novel RNA binding proteins distinct from C/D box core proteins
In order to isolate MBII-52 RNP complexes we prepared nuclear extracts directly from total mouse brain. First, optimal lysis conditions for brain-derived nuclei were determined; thereby, the MBII-52 snoRNP was efficiently extracted in buffer conditions containing >300 mM of NaCl (Fig. 1A, left). Interestingly, MBII-52 was more efficiently solubilized than U3 snoRNA in buffer containing increasing salt concentrations. Immunoprecipitation by an anti-Fibrillarin antibody copurified MBII-52 from the extract, indicating that the mature canonical MBII-52 snoRNP remained intact under the employed buffer conditions (Fig. 1A, right). Fractionation of the MBII-52 snoRNP on a glycerol gradient revealed that the complex migrated at about 12S, a similar sedimentation coefficient as observed for a U3 snoRNP monoparticle (Fig. 2A; Granneman et al. 2003). Taken together, we concluded that the MBII-52 snoRNP was likely to be undissociated under the employed conditions and, for further experiments, isolated the MBII-52 snoRNP from the 12S peak fractions of glycerol gradients.
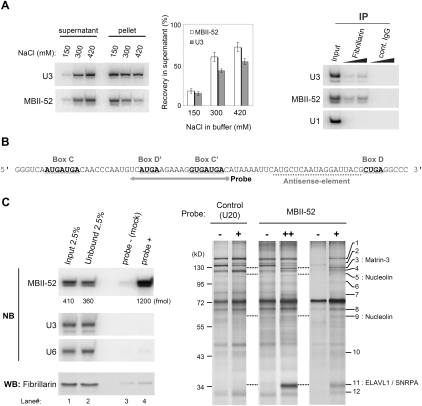
FIGURE 1.
Antisense-RNA affinity purification of MBII-52. (A) Preparation of MBII-52 snoRNP from mouse brain. Solubility of MBII-52 upon lysis of nuclei (left) and immunoprecipitation of Fibrillarin from the lysate containing 300 mM NaCl (right). (B) An antisense RNA probe designed for affinity purification. (C, left) MBII-52 and Fibrillarin recovered from large-scale affinity purification. The quantity of MBII-52 was estimated from a standard curve using in vitro transcribed MBII-52 (not shown). Other abundant RNA species, i.e., U3 snoRNA and U6 snRNA, were not included in the eluate. (C, right) Proteins isolated from affinity-purified MBII-52 snoRNP. Different quantity of MBII-52 probe, indicated as + and ++, recovered ∼1 and 1.2 pmol of MBII-52 snoRNA, respectively. See Table 1 to find the corresponding protein of each band. NB, Northern blotting; WB, Western blotting.
FIGURE 2.
Candidate proteins closely related to MBII-52 RNP. (A) Distribution of candidate proteins in a glycerol gradient of brain nuclear extract. The signal peak of Nucleolin and ELAVL1, as well as Fibrillarin, appeared in lighter fractions than that of MBII-52 Northern blotting, while Matrin-3 and SNRPA appeared in heavier fractions. (B) MBII-52 copurified with an antibody against each candidate from fractions 3–8. Notably, MBII-52 was copurified in association with Fibrillarin from fractions 5 and higher. U6 signal was shown in the right for background control. (C) Immunoprecipitation of Nucleolin and ELAVL1 from individual fractions. Both proteins did not coimmunoprecipitate with each other. NB, Northern blotting; WB, Western blotting.
In order to evaluate the availability of RNA antisense probes for isolation of MBII-52 snoRNP, we performed RNase H digestion assays. Indeed, the antisense element, preceding the D-box motif, as well as an internal loop region of the MBII-52 snoRNA was found to be accessible for hybridization (data not shown). Since RNA probes complementary to the antisense element of MBII-52 likely exhibit an intrinsic competitor, i.e., the 5-HT2CR pre-mRNA, we chose an RNA antisense probe instead that targeted the internal loop of the MBII-52 (Fig. 1B).
We then proceeded to a large-scale isolation of the MBII-52 snoRNP by the RNA antisense affinity purification approach (Fig. 1C). Thereby, we aimed to recover at least 1 pmol of the MBII-52 snoRNP, since it would contain an equimolar amount of Fibrillarin (1 pmol = 34 ng), which would be sufficient to visualize the protein by silver staining. Northern blotting and densitometric analyses revealed that ∼7.3% of the MBII-52 snoRNA were captured by the RNA antisense probe (Fig. 1C, left, lanes 1,2,4). Regardless of significant amounts of the MBII-52 RNP in the purification eluate, the RNA probe did not efficiently recover Fibrillarin as shown by Western blotting (Fig. 1C, lanes 3,4).
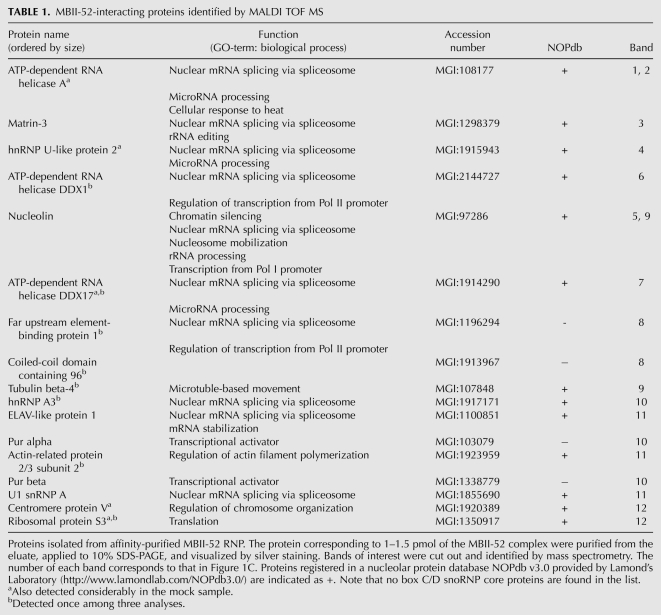
Subsequently, proteins purified from the eluate were separated by SDS-PAGE followed by silver staining. Respective protein bands were cut from the gel, and their sequence was identified by mass spectrometry (Fig. 1C). By comparison to a mock experiment, in which the RNA antisense oligo was omitted (i.e., probe-), Matrin-3, Nucleolin, ELAV-like 1 (ELAVL1), and U1 snRNP A (U1A also known as SNRPA) were identified as likely candidates interacting with MBII-52 (Fig. 1C, right, bands 3,5,9,11). Since all four proteins have previously been reported to bind to nucleic acids, in particular AU-rich RNA sequences (e.g., Nucleolin and ELAVL1), they might directly, and thus unspecifically, bind to the RNA antisense probe (Ma et al. 1996; Sengupta et al. 2004). To rule out this possibility, we employed a different RNA antisense probe, containing a similar AU content, but being complementary to the U20 snoRNA. As a result, none of the identified four proteins were retrieved with this antisense probe, suggesting that the candidate proteins were specific for the MBII-52 snoRNP complex.
Expanding our analysis to retrieved proteins that were less abundant, in total 17 candidate proteins were identified, including hnRNPs, RNA helicases, as well as other proteins, reported to be involved in pre-mRNA splicing or transcriptional regulation (Fig. 1C, right; Table 1). Surprisingly, also among these lesser abundant proteins, Fibrillarin as well as all other C/D snoRNP core proteins (i.e., 15.5K, Nop56, and Nop58) could not be detected by mass spectrometry. However, by employing excess amounts of the MBII-52 antisense probe we were able to detect the presence of Fibrillarin within the MBII-52 snoRNP by Western blotting, but failed to demonstrate its association to the MBII-52 snoRNA by visualization within silver-stained protein gels (data not shown). It is thus likely that the employed RNA antisense probe preferentially binds to MBII-52 RNP complexes different from canonical snoRNPs.
TABLE 1.
MBII-52-interacting proteins identified by MALDI TOF MS
In a recent report, Kishore et al. (2010) employed an affinity-purification approach to purify an MBII-52 snoRNP using an RNA antisense probe against the antisense element of the MBII-52 snoRNA. Thereby, similar to our study, hnRNPs A1/A2/A3/B1/D0 have been identified, as well as TDP43, and Pur-alpha, while the authors were also unable to retrieve Fibrillarin. We speculate that the different conditions (e.g., probe selection, buffer composition, and preparation of cell extract) might be the reason for discrepancy of the remaining proteins identified. Notably, none of the retrieved proteins from this study was verified by coimmunoprecipitation with MBII-52 snoRNA, employing antibodies against respective proteins.
Nucleolin and ELAVL1 are novel components of MBII-52 RNP
We next investigated the distribution of each of the four candidate proteins within the gradient in comparison to the MBII-52 RNP (Fig. 2A). Indeed, we observed that the signal peak of all four candidates, as well as Fibrillarin, coincided or overlapped with the MBII-52-peak fractions (Fig. 2A, fractions 3–8). Notably, Fibrillarin, Nucleolin, and ELAVL1 appeared in fractions 3 and 4, thus being slightly lighter than a 12S particle. In contrast, Matrin-3 and SNRPA were mostly detected in fractions 5 and 6, at a size similar to a 12S particle. To further verify the involvement of these proteins in binding to the MBII-52 snoRNA, we investigated coprecipitation of MBII-52 RNA from individual fractions 3–8 employing antibodies against these proteins (Fig. 2B). Thereby, MBII-52 was copurified in association with Fibrillarin preferentially in fraction 5 and higher. In contrast, the ELAVL1 antibody precipitated MBII-52 predominantly from fractions 3 and 4. The Nucleolin antibody coprecipitated MBII-52 from all investigated fractions (3–8), with a slight increase in fractions 3 and 4. Immunoprecipitation by antibodies against both Matrin-3 and SNRPA did not show unambiguous interaction with MBII-52. These results suggest that Nucleolin and ELAVL1 are associated with the MBII-52 snoRNA. Consistent with the fact that no direct interaction between Nucleolin and ELAVL1 has been reported until now, immunoprecipitation of Nucleolin did not coimmunoprecipitate ELAV1 and vice versa (Fig. 2C), suggesting that each protein might be part of a separate individual MBII-52 RNP particle.
Nucleolin has previously been reported to be involved in numerous cellular events such as ribosomal assembly, nucleo-cytoplasmic transport and chromatin organization (Mongelard and Bouvet 2006). Very recently, Nucleolin has also been shown to be an integral part of the human SSU processome by a transient interaction between 18S pre-rRNA and the U3 snoRNP (Turner et al. 2009). It is thus conceivable, that Nucleolin might also mediate some aspects of MBII-52 RNP function, for example, by guiding binding of the MBII-52 snoRNA to its predicted 5-HT2CR pre-mRNA target. We would like to note, however, that Nucleolin has previously also been copurified in an antisense purification assay of the U11 snRNP, but not in the purification of the U11/U12 particle, and thus was regarded as a contaminant, binding unspecifically to ncRNAs (Will et al. 2004). In contrast, under our experimental conditions, Nucleolin appeared not to unspecifically associate with other abundant ncRNAs, such as U6 snRNA (Fig. 2B, right). Therefore, at this point we are reluctant to exclude Nucleolin from the list of candidates associating with the brain-specific MBII-52 snoRNA.
ELAVL1 was identified as a second candidate binding to the MBII-52 snoRNA. ELAVL1 (also known as HuR in human) belongs to the Hu protein family, which modulates post-transcriptional regulatory events (Hinman and Lou 2008). Hu proteins preferentially bind to AU-rich elements in the 3′ UTR of target mRNAs, resulting in their protection from degradation. In a recent finding, human HuR is shown to regulate alternative splicing of Fas mRNA (Izquierdo 2008). This feature is particularly interesting, since the MBII-52 RNP has been reported to be involved in regulation of alternative splicing of the 5-HT2CR mRNA (Kishore and Stamm 2006).
MBII-52 localizes both nucleoplasm and nucleolus
Both Nucleolin and ELAVL1 are known as RNA-binding proteins that contain four and three RNA-recognition motifs (RRMs), also known as RNA-binding domains (RBDs), respectively. Interestingly, both proteins have been reported to be abundant in the nucleus (Ma et al. 1996; Mongelard and Bouvet 2006), suggesting that novel MBII-52 RNPs might also, at least transiently, locate within the nucleoplasm, consistent with their predicted functions in regulation of splicing and/or editing. This is, however, inconsistent with the reported nucleolar localization of MBII-52, which has previously been demonstrated by FISH analysis (Vitali et al. 2005).
One explanation for this discrepancy might be that by FISH analysis the intense fluorescent signal of the MBII-52 snoRNA within nucleoli may mask a weaker signal of MBII-52 when distributed—to a lesser extent—within the nucleoplasm. Therefore, we re-evaluated the distribution of MBII-52 snoRNA by monitoring its subnuclear distribution in a serial sonication of a nuclear suspension. In brief, nuclei obtained from Neuro-2A cells were sonicated by a 10-sec pulse up to five rounds. As a result, more than 90% of nuclei were broken within 30 sec as demonstrated by microscopic imaging (i.e., differential interference contrast [DIC]) (see Fig. 3A). At each time point, released nucleolar particles were isolated from the nucleoplasmic supernatant by sucrose-density fractionation, followed by RNA/protein purification for Northern and Western blot analyses, respectively. As expected, U3 snoRNA as well as Fibrillarin largely remained in the nucleoli-enriched sediment even after a 50-sec sonication step (Fig. 3B). In contrast, the amount of MBII-52 in this fraction was significantly reduced. Thereby, the subnuclear distribution of MBII-52 snoRNA nearly followed that of molecules, such as U6 snRNA and c-fos protein, residing in the nucleoplasm. We determined the ratio of the distribution of the MBII-52 signal in supernatant versus pellet (i.e., nucleoplasm versus nucleoli) at about 3.5 after 30 sec of sonication (Fig. 3C). This number closely resembles the distribution ratio of the nuclear U6 snRNA, suggesting that the MBII-52 snoRNA might be located, at least partially, within the nucleoplasm, thereby being involved in regulation of splicing and/or editing.
FIGURE 3.
Distribution of MBII-52 in nuclei. (A) Microscopic images (differential interference contrast [DIC]) during nucleoplasmic/nucleolar fractionation by sonication. Nuclei obtained from Neuro-2A cells were broken by five rounds of a 10-sec sonication, each. The RNA-enriched particles visualized by SYBR Green II staining are shown in the inset. Bars = 10 μm. (B) Monitoring the amount of nucleoplasmic/nucleolar components in collected pellets. MBII-52 exhibited a significant decline in its nucleoplasmic/nucleolar distribution close to nucleoplasmic U6 snRNA and c-fos protein. (C) Signal ratio of MBII-52/U3/U6 in supernatant to pellet. After a 30-sec sonication, fractions of supernatant and pellet, i.e., nucleoplasmic and nucleolar fraction, respectively, were purified and analyzed by Northern blotting. MBII-52 was enriched in nucleoplasmic fraction.
In summary, we provide evidence that Nucleolin and ELAVL1 might be novel proteins interacting with MBII-52 snoRNA, thereby assembling into RNPs distinct from canonical C/D snoRNPs. The nucleoplasmic distribution of these candidate proteins thereby sheds some light on the proposed nuclear functions of the MBII-52 snoRNP, in particular the regulation of editing or alternative splicing (Fig. 4). We assume that the size difference in each RNP (Fibrillarin, Nucleolin, or ELAVL1-containing) reflects their functional diversities. For instance, the ELAVL1-containing RNP is relatively small and presumably consists of a few additional proteins. Therefore it might have a function in the regulation of splicing and/or editing. On the other hand, a broad spectrum of Nucleolin-containing RNP fractions suggests that Nucleolin might function as a chaperone or assist shuttling of MBII-52 RNPs between nucleoplasm and nucleoli. At present, we cannot rule out the possibility that also other orphan snoRNAs or even canonical snoRNAs may share such proteins. Further investigations will be needed to reveal the complete protein content of such novel RNP species.
FIGURE 4.
A hypothetical model for MBII-52-protein assembly and function. A small portion of MBII-52 snoRNA may involve ELAVL1 in its complex and exert the function in nucleoplasm, while most MBII-52 assemble canonical C/D core proteins and localize to the nucleolus. Nucleolin may interact with the novel MBII-52 RNP and/or the canonical MBII-52 snoRNP to escort them for transit between nucleoplasm and nucleolus.
MATERIALS AND METHODS
Preparation of mouse brain extract and gradient centrifugation
Preparation of the nuclear pellet was performed as described previously (Jiang et al. 2008), except with minor modifications. Brains were dissected from 5–8-wk-old C57BL/6 mice and quick-frozen in liquid nitrogen. A frozen mouse brain was homogenized in 5 mL of homogenization buffer (20 mM HEPES [pH 7.9], 0.32 M sucrose, 5 mM CaCl2, 3 mM MgCl2, and 0.1% Triton X-100 supplemented with Complete protease inhibitor cocktail [Roche diagnostic]) for 20 gentle strokes by a glass homogenizer. The homogenate was layered onto 7 mL of a sucrose bed (20 mM HEPES [pH 7.9] and 1.8 M sucrose) and spun at 100,000 g for 2.5 h to sediment nuclei. The nuclei pellet was subsequently lysed in LB300 buffer (20 mM HEPES [pH 7.9], 300 mM NaCl, 0.5 mM EDTA, and 0.1% Triton X-100), sonicated, and spun to remove insoluble materials. The extract was immediately used for antisense affinity purification and immunoprecipitation. Alternatively, the lysate was directly layered on 10%–30% (v/v) glycerol gradient, spun at 140,000 g for 17.5 h, and fractionated into 20 aliquots. Upon initial experiments, several salt concentrations, i.e., 150, 300, and 420 mM of NaCl were tested for lysis buffer. As we found that 150 mM NaCl solubilized only a small portion (∼20%) of MBII-52 and that 420 mM NaCl affected downstream experiments, we chose 300 mM NaCl as a salt condition for all experiments.
The expression level of MBII-52 is incomparably higher than that of other snoRNA except for U3 snoRNA. U3 snoRNA is associated with numerous, additional U3-specific proteins and is involved in processing of ribosomal RNA (Granneman et al. 2003; Granneman and Baserga 2004). However, the U3 snoRNP monomer has been shown to contain the four core box C/D proteins as well as hU3-55K (Watkins et al. 2000). Therefore, we chose U3 snoRNA as a control throughout this study.
Immunoprecipitation
Brain nuclear extract or a glycerol fraction was mixed with 0.5–4 μg of a specific antibody (anti-Fibrillarin [ab5821; Abcam], anti-Matrin-3 [sc-55723; Santa Cruz], anti-Nucleolin [ab50279; Abcam], anti-HuR [sc-5483; Santa Cruz], anti-SNRPA [sc-102121; Santa Cruz]) for 2 h at 4°C and further incubated with Protein A/G agarose beads (SC-2003; Santa Cruz) for 2 h. Beads were subsequently washed four times with LB300. The immunocomplex was eluted from the beads by an elution buffer (100 mM Tris [pH 7.5], 12.5 mM EDTA, 150 mM NaCl, and 1% SDS) at 95°C for 5 min. All RNAs, purified from both the precipitant and the supernatant, were analyzed by Northern blotting. For Western blot analysis anti-Fibrillarin antibody H-140 (SC-25397; Santa Cruz) was used at a 1:200 dilution.
Affinity purification of target snoRNPs
Antisense RNA probes for MBII-52 were designed according to the results of RNase H digestion assays (data not shown). An RNA probe for MBII-52 (GUCAUCACCUUUCUUCAUGA) was synthesized with methylated ribonucleotide in combination with a biotin at both 5′ and 3′ ends (Dharmacon). A probe against U20 snoRNA (AUUUUCUUGAGAUUUCAUCA) was used as a control. Nuclear extract was incubated with the biotinylated RNA probe for 2 h. Corresponding amounts of Streptavidin agarose beads (S-1638; Sigma-Aldrich) were added and then incubated for 3 h. After four successive rounds of washing with washing buffer WB250 (20 mM HEPES [pH 7.9], 250 mM NaCl, 1.5 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, and 0.1% Triton X-100), beads were resuspended in elution buffer (1.5 mM sodium citrate, 15 mM NaCl, and 0.1% SDS) and heated at 90°C for 30 min with occasional gentle mixing. After brief centrifugation, the supernatant was recovered as an eluate. Both RNAs and proteins were purified from the eluate by phenol/chloroform treatment and a following ethanol and acetone precipitation, respectively. In addition, to remove excess RNA probes that bind to free proteins, size fractionation of extract/probe mixture was performed. Protein samples obtained from eluate were applied to SDS-PAGE (Tris-Glycine) and visualized by silver staining. Bands were cut out from the gel and proteins were identified by mass spectrometry. For Northern blotting, the following DNA oligonucleotides were end-labeled with 32P by T4 polynucleotide kinase: MBII-52 (CGTAATCCTATTGAGCATGA), U3 snoRNA (GTGGTTTCGGGTGCTC), U1 snRNA (CACCTTCGTGATCATGGTAT), and U6 snRNA (TGGAACGCTTCACGAATTTG).
Mass spectrometry
Affinity purified protein samples were analyzed by SDS-PAGE, and proteins were visualized by silver staining. Relevant protein bands were excised from gels, destained, and in-gel digested with modified trypsin, sequence grade (Promega) as described in Hellman (2000). The in-gel digests were concentrated and desalted using microZipTipC18 (Millipore) by elution of peptides with the acetonitrile solution containing the alfa-cyano-4-hydroxycinnamic acid (Fluka) as a matrix directly onto the target. Mass spectra were acquired using a MALDI-TOF/TOF Ultraflex instrument (Bruker Daltonics). The N2 laser (337 nm wavelength) was used at a 50-Hz frequency. Calibration was done externally using Peptide Calibration Standard II (Bruker Daltonics). Flex Control 2.4 was utilized for data acquisition, and further data processing was carried out using Flex analysis 2.4 and BioTools 2.2 software packages provided by the manufacturer (Bruker Daltonics). Peptide mass fingerprintings (PMF) were interpreted with the MASCOT (http://www.matrixscience.com/) against SwissProt database. A peptide mass tolerance was set to 0.1 Da and one missed cleavage was allowed.
Preparation of nucleoplasmic and nucleolar fractions
Fractionation of nucleoplasm/nucleoli was performed as provided by Lamond Lab (http://www.lamondlab.com/). Exponentially growing Neuro-2A cells in complete media (CM; DMEM supplemented with 10% fetal bovine serum) were harvested by trypsinization and the cell suspension was then centrifuged at 160 g for 5 min. The cell pellet obtained was resuspended and homogenized in 5 mL homogenization buffer (20 mM HEPES [pH 7.9], 0.32 M sucrose supplemented with 1X Complete protease inhibitor cocktail [Roche]) for 40 strokes. The homogenate was then spun at 230 g for 5 min and the pellet was resuspended in buffer 1 (20 mM HEPES [pH 7.9], 0.25 M sucrose, and 10 mM MgCl2). The suspension was layered on buffer 2 (20 mM HEPES [pH 7.9], 0.35 M sucrose, and 0.5 mM MgCl2) and spun at 1450 g for 5 min to sediment nuclei. The nuclear pellet was resuspended in buffer 2, sonicated three times for 10 s with a UPS-200 sonicator at a setting of 60, layered on buffer 3 (20 mM HEPES [pH 7.9], 0.88 M sucrose, and 0.5 mM MgCl2) and centrifuged for 10 min at 2000 g to prepare nucleoplasmic supernatant. The resultant nucleolar pellet was rinsed with buffer 2 and resuspended in a buffer containing 0.6 M sucrose and 0.5 mM MgCl2.
ACKNOWLEDGMENTS
We thank Gerald Brosch, Mathieu Rederstorff, Andreas Ploner, Melanie Lukasser, Roland Hutzinger, and Konstantinia Skreka for technical assistance and discussion, and Günther Bonn for access to mass spectrometry. This work was supported by Austrian Genome Research Grants (D-110420-011-013 and D-110420-011-015) to A.H. and the Austrian Proteomics Platform (APP, GEN-AU) to L.A.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2109710.
REFERENCES
- Blencowe BJ, Lamond AI 1999. Purification and depletion of RNP particles by antisense affinity chromatography. In RNA-protein interaction protocols (ed. Haynes SR), pp. 275–287 Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308 [DOI] [PubMed] [Google Scholar]
- Canton H, Emeson RB, Barker EL, Backstrom JR, Lu JT, Chang MS, Sanders-Bush E 1996. Identification, molecular cloning, and distribution of a short variant of the 5-hydroxytryptamine2C receptor produced by alternative splicing. Mol Pharmacol 50: 799–807 [PubMed] [Google Scholar]
- Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci 97: 14311–14316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CM, Relkovic D, Garfield AS, Dalley JW, Theobald DEH, Humby T, Wilkinson LS, Isles AR 2009. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR mediated behavior. Hum Mol Genet 18: 2140–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ 2004. Ribosome biogenesis: Of knobs and RNA processing. Exp Cell Res 296: 43–50 [DOI] [PubMed] [Google Scholar]
- Granneman S, Gallagher JE, Vogelzangs J, Horstman W, van Venrooij WJ, Baserga SJ, Pruijn GJ 2003. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res 31: 1877–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman U 2000. Sample preparation by SDS/PAGE and in-gel digestion. EXS 88: 43–54 [DOI] [PubMed] [Google Scholar]
- Hinman MN, Lou H 2008. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 65: 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J 2001. RNomics: An experimental approach that identifies 201 candidates for novel, small, nonmessenger RNAs in mouse. EMBO J 20: 2943–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A, Brosius J, Bachellerie JP 2002. RNomics: Identification and function of small, nonmessenger RNAs. Curr Opin Chem Biol 6: 835–843 [DOI] [PubMed] [Google Scholar]
- Izquierdo JM 2008. Hu antigen (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J Biol Chem 283: 19077–19084 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Matevossian A, Huang H, Straubhaar J, Akbarian S 2008. Isolation of neuronal chromatin from brain tissue. BMC Neurosci 9: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Stamm S 2006. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311: 230–232 [DOI] [PubMed] [Google Scholar]
- Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S 2010. The snoRNA MBII-52 (SNORD115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet 19: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Bachellerie JP 1996. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Kiss T 1998. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J 17: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Schmeing T, Moore P, Steitz T 2001. The kink-turn: A new RNA secondary structure motif. EMBO J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Tollervey D 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol Cell Biol 20: 2650–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem 271: 8144–8151 [DOI] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P 2006. Nucleolin: A multiFACeTed protein. Trends Cell Biol 17: 80–86 [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL 2001. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2: 153–175 [DOI] [PubMed] [Google Scholar]
- Samarsky DA, Fournier MJ, Singer RH, Bertrand E 1998. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J 17: 3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta TK, Bandyopadhyay S, Fernandes DJ, Spicer EK 2004. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J Biol Chem 279: 10855–10863 [DOI] [PubMed] [Google Scholar]
- Tran E, Brown J, Maxwell ES 2004. Evolutionary origins of the RNA-guided nucleotide-modification complexes: From the primitive translation apparatus? Trends Biochem Sci 29: 343–350 [DOI] [PubMed] [Google Scholar]
- Turner AJ, Knox AA, Prieto J-L, McStay B, Watkins NJ 2009. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, Nucleolin, RRP5, and DBP4. Mol Cell Biol 29: 3007–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Mouaikel J, Thiry M, Blanchard JM, Tollervey D, Bordonne R, Lafontaine DL, Bertrand E 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors, and a novel nuclear domain. EMBO J 20: 5480–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, Hüttenhofer A 2005. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol 169: 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Boisvert D, Kim KK, Kim R, Kim SH 2000a. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophil, at 1.6 Å resolution. EMBO J 19: 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, O'Brien PJ, Chen CX, Cho DS, Murray JM, Nishikura K 2000b. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem 74: 1290–1300 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Lührmann R 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Dickmanns A, Lührmann R 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol 22: 8342–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, Elbashir S, Tuschl T, Lührmann R 2004. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10: 929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, Chen YQ, Qu LH 2006. snoSeeker: An advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res 34: 5112–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]