Abstract
The CCR4-NOT complex is the main enzyme catalyzing the deadenylation of mRNA. We have investigated the composition of this complex in Drosophila melanogaster by immunoprecipitation with a monoclonal antibody directed against NOT1. The CCR4, CAF1 (=POP2), NOT1, NOT2, NOT3, and CAF40 subunits were associated in a stable complex, but NOT4 was not. Factors known to be involved in mRNA regulation were prominent among the other proteins coprecipitated with the CCR4-NOT complex, as analyzed by mass spectrometry. The complex was localized mostly in the cytoplasm but did not appear to be a major component of P bodies. Of the known CCR4 paralogs, Nocturnin was found associated with the subunits of the CCR4-NOT complex, whereas Angel and 3635 were not. RNAi experiments in Schneider cells showed that CAF1, NOT1, NOT2, and NOT3 are required for bulk poly(A) shortening and hsp70 mRNA deadenylation, but knock-down of CCR4, CAF40, and NOT4 did not affect these processes. Overexpression of catalytically dead CAF1 had a dominant-negative effect on mRNA decay. In contrast, overexpression of inactive CCR4 had no effect. We conclude that CAF1 is the major catalytically important subunit of the CCR4-NOT complex in Drosophila Schneider cells. Nocturnin may also be involved in mRNA deadenylation, whereas there is no evidence for a similar role of Angel and 3635.
Keywords: deadenylation, mRNA decay, mRNA stability, CCR4-NOT complex, post-transcriptional regulation
INTRODUCTION
The CCR4-NOT complex is the main enzyme responsible for the deadenylation of mRNA in yeast (Daugeron et al. 2001; Tucker et al. 2001), flies (Temme et al. 2004), man (Yamashita et al. 2005), and trypanosomes (Schwede et al. 2008). Deadenylation is the first step in the two exonucleolytic mRNA decay pathways, 5′ decay initiated by cap-hydrolysis and 3′ decay (Meyer et al. 2004; Parker and Song 2004; Garneau et al. 2007; Goldstrohm and Wickens 2008; Houseley and Tollervey 2009). Furthermore, deadenylation can contribute to translational repression of mRNAs (Wickens et al. 2000). Therefore, the CCR4-NOT complex plays an important role in the regulation of gene expression. For example, deadenylation by the complex is involved in post-transcriptional control during oogenesis and embryonic development in Drosophila (Morris et al. 2005; Semotok et al. 2005; Zaessinger et al. 2006; Chicoine et al. 2007; Kadyrova et al. 2007). The deadenylase can be directed toward specific mRNAs by association with RNA binding proteins such as the PUF proteins (Goldstrohm et al. 2006) or Drosophila Smaug and its yeast homolog Vts1p (Aviv et al. 2003; Semotok et al. 2005; Zaessinger et al. 2006; Rendl et al. 2008). Alternatively, miRNAs can be responsible for the recruitment of the CCR4-NOT complex to specific mRNAs (Behm-Ansmant et al. 2006; Fabian et al. 2009). ATP dependence of a cell-free deadenylation system derived from Drosophila embryos suggests the involvement of additional factors (Jeske et al. 2006). The CCR4-NOT complex may also play a role in transcriptional regulation (Collart 2003; Denis and Chen 2003; Collart and Timmers 2004). Functions beyond mRNA deadenylation are supported most strongly by the fact that mutations in some genes encoding subunits of the CCR4-NOT cause more severe phenotypes than do mutations inactivating its deadenylase activity (Traven et al. 2009); for example, only NOT1 is an essential gene in yeast (Maillet et al. 2000).
In Saccharomyces cerevisiae, the CCR4-NOT core complex was reported to consist of the Pop2/Caf1, Ccr4, Not1-5, Caf40, and Caf130 proteins (Chen et al. 2001). Not1p is considered the central scaffold of the complex to which the other subunits are attached directly or indirectly. Two subunits of the complex are potential 3′ exonucleases: Ccr4p, a member of the exonuclease III family, carries the main catalytic function of the yeast complex (Chen et al. 2002; Tucker et al. 2002; Viswanathan et al. 2003; Goldstrohm et al. 2007); and Pop2p/Caf1p has similarities to the RNase D family of 3′ exonucleases, but whether or not the S. cerevisiae protein has catalytic activity has been debated (Daugeron et al. 2001; Chen et al. 2002; Tucker et al. 2002; Thore et al. 2003; Goldstrohm et al. 2007). Point mutations in the predicted active site do not cause a detectable phenotype (Viswanathan et al. 2004). Much less is known about functions of the other subunits. Deadenylation is clearly affected by mutations in NOT2 and NOT5, but little or not at all by mutations in NOT3 and NOT4 (Tucker et al. 2002). The Not4 protein is active as a ubiquitin ligase (Albert et al. 2002; Panasenko et al. 2006; Dimitrova et al. 2009; Mersman et al. 2009).
Analyses in other organisms have confirmed much of this picture but modified it in some aspects. Caf130p does not appear to be conserved. Yeasts Not3p and Not5p are similar to each other and are represented by just a single ortholog in flies (Temme et al. 2004), man (Albert et al. 2000), and trypanosomes (Schwede et al. 2008). Not4, while conserved, was not found among the core components of the mammalian CCR4-NOT complex (Morita et al. 2007; Lau et al. 2009). The division of labor among the deadenylase subunits of the complex may also have shifted during evolution: In trypanosomes, Ccr4 seems to be lacking, whereas Caf1 is associated with Not proteins, is required for deadenylation in vivo, and has poly(A) degrading activity in vitro (Schwede et al. 2008). Mammals have two variants each of CAF1 and CCR4 (Albert et al. 2000; Dupressoir et al. 2001), and in addition, a more distant relative of CAF1 (Wagner et al. 2007). The two CAF1 proteins seem to be integrated into the CCR4-NOT complex in a mutually exclusive manner, and the same is true for the two CCR4 proteins (Lau et al. 2009). All four proteins are active as deadenylases in vitro (Viswanathan et al. 2003, 2004; Bianchin et al. 2005; Morita et al. 2007). They play overlapping roles in mRNA deadenylation, and their catalytic activities are essential for their in vivo functions, as shown by dominant-negative effects of mutant variants (Yamashita et al. 2005; Morita et al. 2007; Mauxion et al. 2008; Schwede et al. 2008; Zheng et al. 2008; Aslam et al. 2009). In addition to CCR4, higher eukaryotes, including Drosophila, also have three paralogs, Angel, Nocturnin, and 3635 (CG31759 in flies) (Dupressoir et al. 2001). Whereas all four proteins contain an exonuclease domain, CCR4 is distinguished by a conserved leucine-rich repeat, which is thought to be responsible for its interaction with CAF1 and, through CAF1, with the rest of the complex (Dupressoir et al. 2001; Clark et al. 2004). Xenopus and mouse Nocturnin have been reported to have poly(A) degrading activity (Baggs and Green 2003; Garbarino-Pico et al. 2007).
The Drosophila genome contains orthologs for most of the subunits of the CCR4-NOT complex (Temme et al. 2004): CCR4, CAF1 (=POP2), NOT1, NOT2, NOT3 (homologous to both yeast Not3p and Not5p), NOT4, and CAF40. This article reports that these polypeptides, with the exception of NOT4, associate as a stable complex. CAF1, NOT1, NOT2, and NOT3 are required for mRNA deadenylation. CCR4 appears to be dispensable for deadenylation in Schneider cells, and CAF1 carries the major catalytic activity.
RESULTS
Subunits of the CCR4-NOT complex
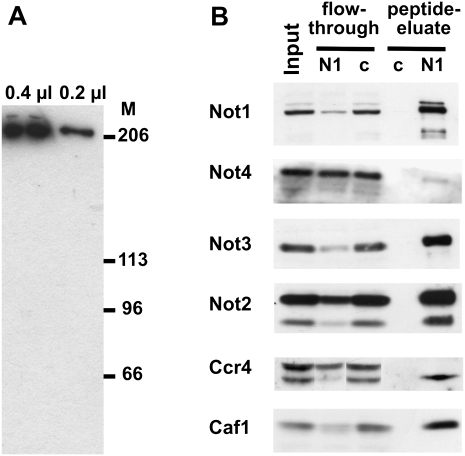
In order to facilitate purification of the CCR4-NOT complex under native conditions, mouse monoclonal antibodies were generated against a peptide from NOT1 (see Materials and Methods). One monoclonal antibody, 2G5, recognized two polypeptides of the anticipated molecular weight of about 200 kDa in Western blots (Fig. 1A). This antibody was used for immunoprecipitation of NOT1 from RNase-treated 0–2 h embryo extracts, and NOT1, together with associated polypeptides, was eluted by the addition of an excess of the peptide antigen. Two rabbit sera raised against the same peptide and a different NOT1 peptide, respectively, both recognized a doublet of about 200 kDa in the eluate and also in extracts of embryos and S2 cells (Fig. 1B; data not shown). The intensity of both bands was reduced upon treatment of S2 cells with double-stranded NOT1 RNA (see Fig. 6 below). These results confirmed the specificity of the antibodies and suggested that the two polypeptides were isoforms of NOT1. Analysis with antibodies directed against the other anticipated subunits of the CCR4-NOT complex (Temme et al. 2004; Jeske et al. 2006) revealed a depletion of CAF1, CCR4, NOT1, NOT2, and NOT3 in the supernatant of the immunoprecipitation. The same polypeptides were enriched in the peptide eluate (Fig. 1B). The association of these subunits was stable when the KCl concentration in the wash buffer was raised to 500 mM (data not shown). These data indicate that at least a significant fraction of these polypeptides is associated in a stable complex. In contrast, the NOT4 polypeptide was not noticeably depleted in the supernatant, and only small amounts were detectable in the precipitate. When immunoprecipitations were carried out under slightly different conditions, NOT4 was not coprecipitated (data not shown). When the NOT1 precipitate was analyzed by mass spectrometry, NOT4 was at the detection limit (see below). Thus, NOT4 is not stably associated with the complex. No appropriate antibody was available for the CAF40 protein, but numerous CAF40 peptides were detected in the mass spectrometric analysis of the NOT1 precipitate (see below). Thus, with the exception of NOT4, all anticipated subunits of the CCR4-NOT complex are in fact stably associated with each other.
FIGURE 1.
Association of the CCR4-NOT polypeptides in a stable complex. (A) Detection of NOT1 with a monoclonal antibody. Indicated amounts of an extract from 0- to 2-h-old Drosophila embryos were separated on a 6% SDS–polyacrylamide gel, blotted to a PVDF membrane, and probed with the monoclonal antibody CISS 2G5. Sizes of marker proteins are indicated. (B) Western blot analysis of NOT1 immunoprecipitation from embryo extract. The monoclonal NOT1 antibody CISS 2G5 and a monoclonal rat antibody against human hnRNP K were used for immunoprecipitation (lanes labeled N1 and c, respectively). Equal amounts of input and flow-through were loaded. Bound proteins were eluted with the peptide used for immunization. A polyclonal antibody (CR#1) directed against the same peptide was used for detection of NOT1. Polyclonal antibodies for detection of the other subunits are indicated on the left. Note that the CCR4 antibody recognizes a doublet in embryo extracts. Only the lower band (arrowhead) is depleted by NOT1 immunoprecipitation and enriched in the peptide eluate. The identity of the upper band is unknown.
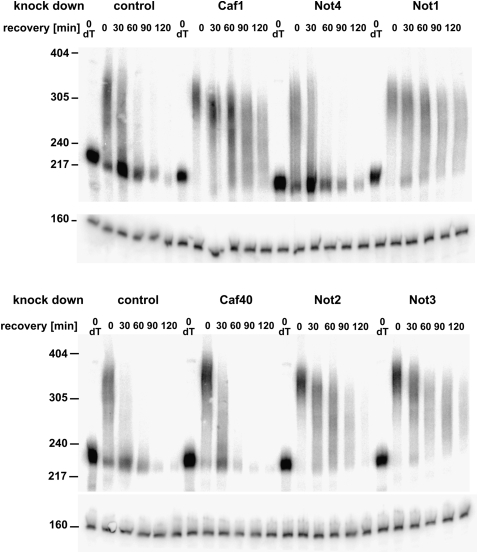
FIGURE 6.
Involvement of individual subunits of the CCR4-NOT complex in hsp70 mRNA deadenylation. Schneider cells treated with the double-stranded RNAs indicated were heat-shocked for 30 min at 35.5°C and then allowed to recover at 25°C. RNA was prepared at the indicated times after recovery and digested with RNase H in the presence of an hsp70-specific oligonucleotide. dT indicates Oligo(dT) was included in the RNase H digestion to mark the fully deadenylated RNA. Products were analyzed by Northern blot with probes against the 3′UTR of hsp70 and against U1 RNA serving as loading control. For control of the knock-down efficiency, see Figure 5; RNA samples were from the same cells.
Localization and expression of the CCR4-NOT complex
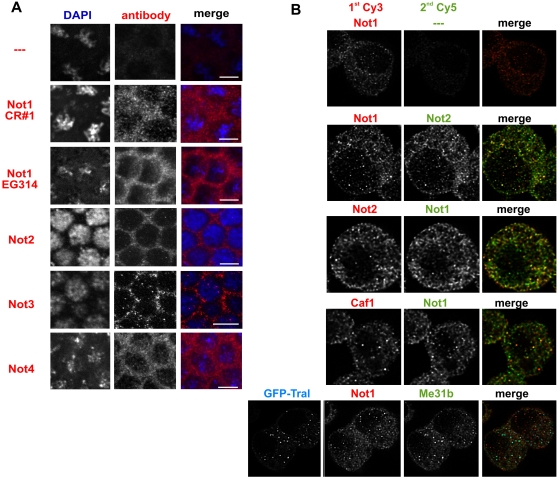
CAF1 and CCR4 are found mostly in the cytoplasm in ovaries and embryos, and show a nonhomogenous, punctate distribution (Temme et al. 2004; Zaessinger et al. 2006). Polyclonal antibodies against NOT1, NOT2, and NOT3 revealed a similar distribution of the respective antigens in embryos and Schneider (S2) cells (Fig. 2A; data not shown) Nuclear staining was much weaker. Unfortunately, the Not1 monoclonal antibody 2G5 was not useful for immunofluorescence experiments. Therefore, sequential staining of different subunits of the CCR4-NOT complex with polyclonal rabbit antibodies directed against a NOT1 peptide, full-length NOT2, and CAF1 was used to investigate colocalization of these polypeptides in S2 cells. The first primary antibody was detected with a Cy3-labeled antibody at saturating concentrations before the second primary antibody was applied and detected with a Cy5-labeled secondary antibody. Controls in which the second primary antibody was omitted gave no Cy5 signal, confirming that the first primary antibody had been saturated with secondary antibody (Fig. 2B, upper panel). Although staining intensity was variable, the pairwise combinations demonstrated that the NOT2 antibody labeled the same structures as the NOT1 and CAF1 sera. Predominant colocalization of the antigens is in agreement with the biochemical evidence for their association (Fig. 2B).
FIGURE 2.
Localization of subunits of the CCR4-NOT complex. (A) Confocal images of immunostained Drosophila embryos. Blastoderm stage embryos were incubated with the antiserum indicated on the left and a Cy3-labeled secondary antibody (middle panel). DAPI was included in the mounting medium to label the nuclei (left panel). The white bar in the merge (right panel) represents 5 μm in size. Colors in the merge are blue for the DAPI staining and red for the immunostaining. (B) Colocalization of different CCR4-NOT complex subunits in S2 cells. S2 cells were sequentially stained for two different subunits of the complex. The primary antibodies used are indicated above each image. The first secondary antibody was always Cy3-labeled (left panel); the second secondary, always Cy5-labeled (middle panel). The top panel shows a control in which the second primary antibody was omitted. No Cy5 signal was seen, confirming that the first secondary antibody had been used at saturating concentrations. The bottom panel shows sequential staining for NOT1 and Me31B in cells transiently expressing GFP-Tral for labeling of P-bodies. In the merged images (right panel), the Cy3 channel is in red, the Cy5-channel in green, and GFP in blue. The overlay of red and green becomes yellow, and the overlay of green and blue becomes turquoise.
mRNA decay by the 5′ pathway can occur in cytoplasmic aggregates termed P-bodies (Eulalio et al. 2007a; Parker and Sheth 2007). To test the relationship of the CCR4-NOT containing cytoplasmic structures with P-bodies, a GFP-Tral expressing plasmid was transiently transformed into S2 cells. Tral is a known component of P bodies (Eulalio et al. 2007b). The GFP-Tral expressing cells were immunostained with antibodies either against NOT1 or Me31B, another known P-body marker (Eulalio et al. 2007b; Lin et al. 2008). Confocal imaging of the cells showed that endogenous Me31B and GFP-Tral colocalized as expected, whereas little colocalization could be seen between GFP-Tral and NOT1 (Fig. 2B, lower panel). This indicates that the CCR4-NOT complex is not a major component of P-bodies, in agreement with data obtained in Drosophila nurse cells and blastoderm embryos (Lin et al. 2008).
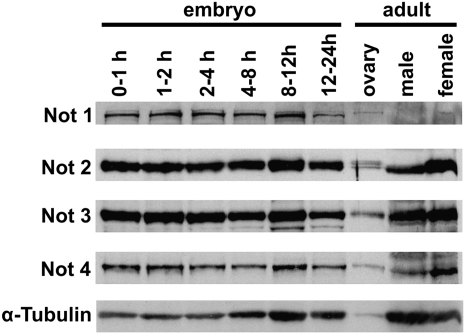
Western blotting of Drosophila extracts with antibodies directed against NOT1, NOT2, NOT3, and NOT4 detected expression of the proteins at all embryonic stages and in adult flies (Fig. 3). This is similar to the developmental profile of CCR4 and CAF1 expression (Temme et al. 2004). The presence of the NOT proteins before the onset of zygotic transcription (0–2 h) demonstrated a maternal contribution.
FIGURE 3.
Developmental expression profile of the subunits of the CCR4-NOT complex. Protein extracts were from 20 embryos, 0.2 ovaries, 0.5 females, and one male. The blot was probed with the monoclonal anti-NOT1 antibody 2G5 and with polyclonal antibodies against NOT2, 3, and 4 as indicated on the left. α-Tubulin served as a loading control. The same blot was stripped and reprobed with the different antibodies.
Association of Nocturnin with the CCR4-NOT complex
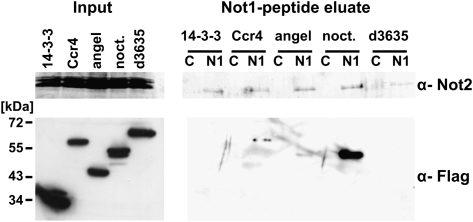
In addition to CCR4, encoded by the gene twin, the Drosophila genome also encodes three paralogs: Angel, Nocturnin (encoded by curled) (Grönke et al. 2009), and 3635. Reverse transcription-polymerase chain reaction (RT-PCR) analysis showed that twin, angel, and 3635 are expressed in S2 cells, but curled is not (data not shown). Stably transformed S2 cell lines were generated, each of which expressed one of the four proteins as a Flag-tagged fusion. Flag-tagged 14-3-3ɛ was expressed as a control. A possible integration of the CCR4 paralogs into the CCR4-NOT complex was tested by immunoprecipitation experiments with the monoclonal NOT1 antibody. Bound proteins were eluted with the NOT1 peptide and analyzed by Western blot for coprecipitated proteins. As expected, the NOT1 antibody but not the control antibody coprecipitated NOT2 from all extracts (Fig. 4). Western blot analysis with an anti-Flag antibody showed that, although all fusion proteins were expressed in comparable amounts, only CCR4 and Nocturnin were coprecipitated with NOT1. The lower amount of coprecipitated CCR4 compared with Nocturnin was possibly due to the competition of CCR4-Flag with internal CCR4. The data were confirmed by precipitation with the Flag antibody: CAF1, NOT2, and NOT3 as representatives of the CCR4-NOT complex were coprecipitated with Nocturnin and, as a positive control, CCR4, but not with 3635 (Supplemental Fig. 1) We conclude that Nocturnin can be stably incorporated into the CCR4-NOT complex but Angel and 3635 cannot. We were unable to determine whether Flag-Nocturnin and endogenous CCR4 can be part of the same complex due to a low abundance of the coprecipitated proteins and a high background of the CCR4 antiserum.
FIGURE 4.
Association of CCR4 and its paralogs with the CCR4-NOT complex. S2 cells were stably transformed with plasmids expressing individual members of the CCR4 family with C-terminal Flag tags under the control of a metallothionein promoter. 14-3-3 ɛ-Flag was used as a control for nonspecific association with the CCR4-NOT complex. Expression was induced for 20 h before the cells were lysed. A Western blot (left panel; 2.5% input) showed that the Flag-tagged CCR4 family members were expressed at similar levels, whereas the 14-3-3 control protein was more strongly expressed. NOT2 served as a loading control. The monoclonal NOT1 antibody CISS 2G5 was used for immunoprecipitation (lanes labeled N1), and an unrelated monoclonal antibody served as a control (lanes labeled C). Proteins were eluted with the immunogenic NOT1 peptide; 2.5% of the input and 25% of the eluate were separated on a 6% gel and a 10% SDS gel, blotted, and probed with antibodies against NOT2 (6% gel) or the Flag epitope (10% gel). Input and eluate of the Flag Western had different exposure times. The Flag-reactive band that appears to be present in the peptide eluate of the Angel expressing cells is not Angel, as the molecular weight does not match that of Angel in the left panel.
Proteomic analysis of the CCR4-NOT complex
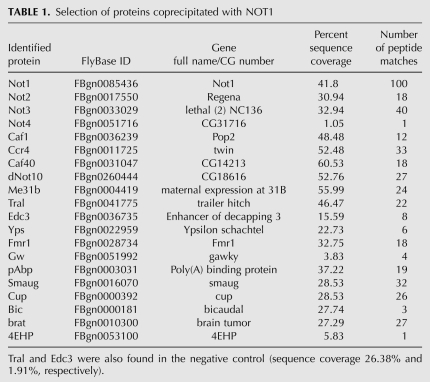
The polypeptides copurified from an RNase-treated Drosophila embryo extract by immunoprecipitation with the monoclonal NOT1 antibody and peptide elution were identified by peptide mass fingerprint analysis using nano-electrospray ionization LTQ-Orbitrap mass spectrometry. The most important results are summarized in Table 1, and complete lists are found in Supplemental Tables 1 and 2. CCR4, CAF1, NOT1, NOT2, NOT3, and CAF40 were identified, each with a sequence coverage of 30%–60%. In contrast, NOT4 was represented only by a single peptide and was thus below the threshold set for protein identification (see Materials and Methods). CNOT10 is considered a subunit of the human CCR4-NOT complex (Gavin et al. 2002; Morita et al. 2007; Lau et al. 2009), and the corresponding Drosophila protein (CG18616) was also present. TAB182, another constituent of the mammalian complex (Morita et al. 2007; Lau et al. 2009), does not seem to have an ortholog in the fly, and consequently, none was identified here. C2ORF29, also present in the mammalian complex (Lau et al. 2009), was not identified either, even though an orthologous protein (CG13567) is encoded in the Drosophila genome. None of the CCR4 paralogs was found, but we do not know if they are expressed at these stages of embryo development. BTG/TOB, which would be expected to associate with the CCR4-NOT complex (Mauxion et al. 2009), was not detected either. Again, expression in embryos has not been tested.
TABLE 1.
Selection of proteins coprecipitated with NOT1
Several proteins involved in mRNA decay and/or translational regulation were copurified with the complex (Table 1). These included Me31B, the orthologs of which associate with the CCR4-NOT complex in yeast (Hata et al. 1998; Maillet and Collart 2002) and in trypanosomes (Schwede et al. 2008). Me31B is known to associate with either Tral or Edc3 in two mutually exclusive complexes, the former of which also contains Cup (Tritschler et al. 2008, 2009). All three proteins were present in the immunoprecipitate, although a lower level of Tral and Edc3 (as judged by sequence coverage) was also present in the negative control. Me31B and Tral also associate with the Drosophila Y box protein, Ypsilon Schachtel (Yps) (Wilhelm et al. 2005), which was found in the NOT1 immunoprecipitate. Related interactions are conserved in Xenopus (Minshall et al. 2007). The fragile X mental retardation protein, Fmr1 in flies, cooperates with Me31B and Tral in translational regulation (Barbee et al. 2006). Gawky, the fly ortholog of GW182 (Schneider et al. 2006) functions in miRNA-mediated silencing, in part by activating CCR4-NOT-dependent deadenylation (Behm-Ansmant et al. 2006; Eulalio et al. 2009). The interaction between Gawky and the CCR4-NOT complex is thought to be mediated by the cytoplasmic poly(A) binding protein, PABP (Fabian et al. 2009; Zekri et al. 2009; Jinek et al. 2010).
mRNA-specific regulatory proteins previously reported to be associated with the CCR4-NOT complex and identified here include Smaug (Semotok et al. 2005; Zaessinger et al. 2006) and its interaction partner Cup (Table 1; Nelson et al. 2004). Brain Tumor has not itself been linked to the CCR4-NOT complex, but its associated proteins Nanos and Pumilio have (Sonoda and Wharton 2001; Kadyrova et al. 2007). Brain Tumor interacts with the regulatory protein 4EHP (Cho et al. 2006), which was also present in the NOT1 precipitate, although its representation with a single peptide was below the threshold set for protein identification. Bicaudal is involved in post-transcriptional regulation of the nanos RNA, which is a substrate for the CCR4-NOT complex (Markesich et al. 2000; Jeske et al. 2006; Zaessinger et al. 2006).
Involvement of subunits of the CCR4-NOT complex in mRNA deadenylation
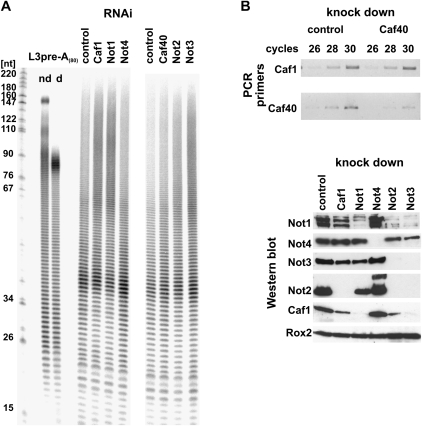
Knock-down of CAF1 in S2 cells causes an increase in steady-state length of bulk poly(A) and strongly retards the deadenylation of hsp70 mRNA (Temme et al. 2004; Bönisch et al. 2007). Knock-down of CCR4 had no effect in S2 cells, but an increase in bulk poly(A) tail length and some retardation of hsp70 mRNA decay was detectable in twin (encoding CCR4) mutant flies (Temme et al. 2004). The NOT2 subunit, encoded by the Regena gene, was also found to be involved in mRNA deadenylation both by knock-down experiments in cell culture and by the analysis of mutant flies. An effect on bulk poly(A) was also reported for knock-down of NOT1 and NOT3 (Temme et al. 2004). We have now examined the role of known or potential subunits of the CCR4-NOT complex in mRNA deadenylation more completely. Bulk poly(A) tail length was measured by 3′-labeling of total RNA isolated from the cells, digestion of the non-poly(A) sequences by a combination of RNases A and T1, and analysis by denaturing gel electrophoresis (Fig. 5A; Supplemental Fig. 2). Knock-down of CAF1 served as a positive control and showed the anticipated increase in bulk poly(A) tail length. Knock-down of NOT1 or NOT3 led to similar phenotypes. Thus, these two subunits are required for mRNA decay, in agreement with their stable association with the rest of the CCR4-NOT complex and earlier data (Temme et al. 2004). Depletion of NOT4 or CAF40 had no detectable effect, even though the knock-down, checked by Western blotting in the case of NOT4 and by RT-PCR in the case of CAF40, was efficient (Fig. 5B). Most of these RNAi treatments reduced the abundance not only of the targeted protein but also of other subunits of the CCR4-NOT complex (Fig. 5B). Thus, the requirement for any individual subunit in deadenylation might be indirect. As depletion of one subunit often reduces the stability of the remaining parts of a protein complex, these data are in agreement with the assembly of the affected polypeptides in a stable complex. Interestingly, NOT4 was not affected by any of the other knock-downs, and the NOT4 knock-down did not change the abundance of any of the other polypeptides examined. This observation supports the notion that NOT4 is not a stable subunit of the CCR4-NOT complex.
FIGURE 5.
Involvement of individual subunits of the CCR4-NOT complex in bulk mRNA deadenylation. (A) Analysis of the length of bulk poly(A) in S2 cells after 5 d of incubation with dsRNA as indicated. DsRNA against luciferase served as a control. Total RNA was isolated from the cells, 3′-labeled, digested with RNases A and T1, and separated on a 9% polyacrylamide-urea gel. The RNA samples were from the 90-min recovery time point of Fig. 6. As a control for the specificity of the RNase digestion, in vitro synthesized L3preA(80) RNA (Kerwitz et al. 2003) was digested like the experimental RNA samples (d, digested; nd, nondigested). Lengths of DNA markers (in nucleotides) are indicated on the left. (B) Control of the knock-downs by RT-PCR (CAF40; upper panel) or Western blot (all others; lower panel). Oligo(dT)-primed reverse transcription was performed on DNase-treated RNA from knock-down and control cells. PCR primers were used to amplify either the CAF40 ORF or the CAF1 ORF as control. Aliquots of the PCR reactions were taken after 26, 28, or 30 cycles of amplification and analyzed on a 1% agarose gel. For the Western blot, control or knock-down cells were lysed in SDS sample buffer after 5 d of incubation with dsRNA and analyzed on 6% or 10% SDS polyacrylamide gels. Western blots were performed against the proteins indicated. PABP2 (Benoit et al. 1999) served as loading control.
The effects of the same subunit depletions on the rate of hsp70 mRNA deadenylation were examined in a pulse–chase protocol: Synthesis of the hsp70 mRNA was induced by a brief heat shock. During heat shock, mRNA decay is inhibited, and the hsp70 mRNA is stable. Synthesis of this RNA ceases upon return to the normal growth temperature, and at the same time, its decay is initiated by deadenylation (Dellavalle et al. 1994; Temme et al. 2004; Bönisch et al. 2007). Poly(A) tail lengths at various time points of recovery were measured by Northern blotting after shortening of the RNA with RNase H and a DNA oligonucleotide hybridizing close to the 3′ end of the coding sequence. The results were in agreement with those of bulk poly(A) analysis: Knock-down of NOT1 and NOT3 and, as positive controls, CAF1 and NOT2 (Temme et al. 2004) strongly retarded deadenylation of the hsp70 message. In contrast, the knock-down of NOT4 and CAF40 had no detectable effect (Fig. 6). While a negative result of a knock-down experiment is not conclusive, the lack of an effect of the NOT4 knock-down on both bulk poly(A) and the rate of hsp70 mRNA deadenylation is consistent with its unstable association with the CCR4-NOT complex and suggests that this polypeptide is dispensable for deadenylation.
CAF1 carries the main catalytic activity for deadenylation in S2 cells
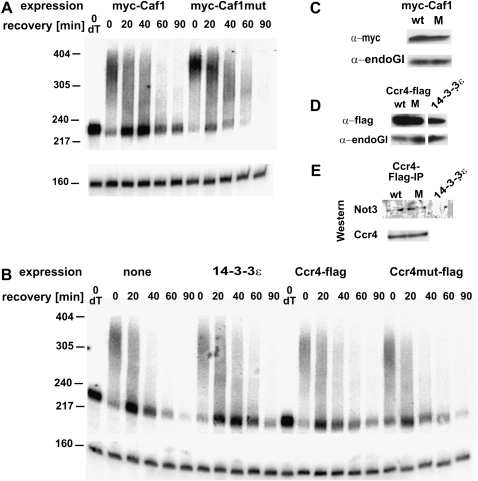
Results presented previously suggested that CAF1 might be more important for mRNA decay in S2 cells than CCR4 (Temme et al. 2004). Overexpression of catalytically inactive mutants of the two proteins was used to find out which of the polypeptides is responsible for the catalysis of deadenylation in S2 cells. For this purpose, cells were stably transformed with constructs expressing CCR4-Flag or myc-Caf1, either wild-type or with inactivating double point mutations in the active sites. Expression was controlled by an inducible promoter. Approximately 20 h after induction of the proteins, cells were heat shocked, and decay of the hsp70 mRNA was analyzed during recovery. Whereas overexpression of wild-type CAF1 did not affect the rate of hsp70 deadenylation, overexpression of the mutant polypeptide had a dominant-negative effect (Fig. 7A). This was most clearly visible at the 0-min time point (end of heat shock): All intermediates of deadenylation, including completely deadenylated RNAs, were visible in the two control samples, expressing the wild-type protein or no additional CAF1. In comparison, the corresponding sample from the cells overexpressing mutant CAF1 had almost exclusively fully polyadenylated RNAs. In these cells, the transient accumulation of the deadenylated decay intermediate during further decay was much reduced, and polyadenylated species remained more highly populated. In contrast, expression of mutant CCR4 did not change the time course of hsp70 mRNA deadenylation compared with expression of wild-type protein or of an irrelevant Flag-tagged control protein (Fig. 7B). Analyses of bulk poly(A) were in agreement with the hsp70 mRNA decay experiments, showing longer poly(A) tails upon overexpression of the CAF1 mutant but not of the CCR4 mutant (Supplemental Fig. 3). Wild-type and mutant versions of myc-CAF1 were expressed to the same extent, and the same was true for CCR4-Flag (Fig. 7C,D). Both versions of CCR4-Flag also coprecipitated similar amounts of NOT3 (Fig. 7E). Thus, the lack of an effect of the CCR4 mutant was presumably not due to its inability to be incorporated into the CCR4-NOT complex. In summary, the results suggest that the catalytic activity of CCR4 has little or no importance for the deadenylation of the hsp70 mRNA in S2 cells, and CAF1 is the relevant catalytic subunit in these cells.
FIGURE 7.
CAF1 carries the catalytic activity for mRNA deadenylation in S2 cells. (A) Expression of myc-CAF1 or a variant containing a double point mutation in the first exonuclease motif was induced for 20 h in stably transformed S2 cells. Cells were heat-shocked for 30 min at 35.5°C and then allowed to recover at 25°C. RNA was prepared at the indicated times after recovery and digested with RNase H in the presence of an hsp70-specific oligonucleotide. dT indicates Oligo(dT) was included in the RNase H digestion to mark the fully deadenylated RNA. Products were analyzed by Northern blot with probes against the 3′UTR of hsp70 and against U1 RNA serving as loading control. (B) An equivalent experiment was carried out with cells expressing flag tagged CCR4, a CCR4 variant containing a double point mutation in the exo III domain, or a control protein. (C, D) Expression of the tagged proteins was monitored by Western blotting with the antibodies indicated. EndoGI (Temme et al. 2009) served as loading control. (E) Wild-type and mutant CCR4-Flag were precipitated with anti-Flag antibodies and eluted with Flag peptide. Equal amounts of the eluates were separated on a 10% gel, blotted, and probed with antibodies against NOT3 or CCR4.
Nocturnin may be involved in mRNA decay, but the other paralogs of CCR4 are not
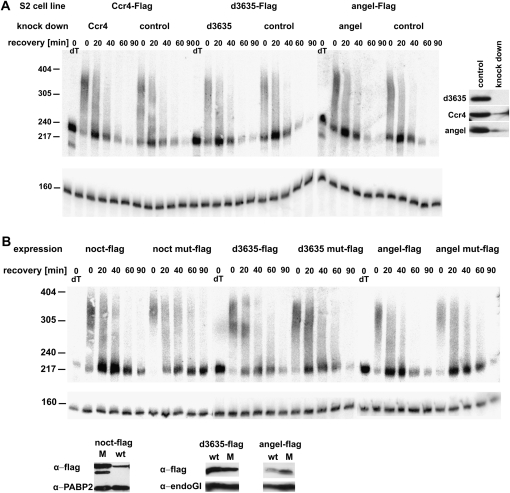
A possible explanation for the apparent irrelevance of CCR4 in deadenylation is its replacement by one or more of its paralogs. Thus, their involvement in mRNA decay was also examined by RNAi. Since we were unable to amplify curled fragments from S2 cell cDNA, the encoded Nocturnin does not seem to be expressed in these cells and was not included in the RNAi experiments. For assays examining a role of the two other paralogs, the stable lines described above, expressing Flag-tagged Angel or 3635 under the control of a regulated promoter, were used. Knock-down of the endogenous proteins was initiated under conditions in which the tagged variants were not expressed. In order to examine the efficiency of RNAi, synthesis of tagged Angel or 3635 was then induced in an aliquot of the knock-down cells 4 d after the addition of double-stranded RNA (dsRNA), and the level of protein was examined 4 h later by Western blotting. Knock-down of the Flag-tagged proteins was efficient; we infer that knock-down of the endogenous proteins (if present) was also efficient. A heat shock was applied to the remaining knock-down cells, in which overproduction of the CCR4 paralog had not been induced, and the rate of hsp70 mRNA deadenylation and steady-state length of bulk poly(A) were examined. Neither the knock-down of Angel nor that of 3635 had any detectable effect (Fig. 8A). The results were supported by analyses of bulk poly(A) (Supplemental Fig. 4). A triple knock-down of CCR4, Angel, and 3635 had no effect either (data not shown). The same knock-down experiments were also carried out with regular S2 cells that had not been transformed with the expression plasmids; no effects on mRNA decay were observed.
FIGURE 8.
Angel and 3635 do not play a detectable role in mRNA deadenylation (A) RNAi experiment: S2 cells stably transformed with expression constructs for Flag-tagged CCR4, Angel, or 3635 were treated with the corresponding dsRNA for 4 d under noninducing conditions. Then a 30-min heat shock at 35.6°C was applied. After return to 25°C, RNA was prepared at the indicated times of recovery and digested with RNase H in the presence of an hsp70-specific oligonucleotide. dT indicates Oligo(dT) was included in the RNase H digestion to mark the fully deadenylated RNA. Products were analyzed by Northern blot with probes against the 3′UTR of hsp70 and against U1 RNA serving as loading control. (Right panel) Expression of the flag-tagged proteins was induced for 4 h in small aliquots of the RNAi cells and in controls cells. Knock-down efficiency was examined by Western blot analysis with an anti-Flag antibody. Equal loading was controlled by Ponceau staining (data not shown). (B) Dominant-negative experiment: S2 cells expressing either Flag-tagged CCR4 paralogs or variants containing double point mutations in their exo III domains were grown under inducing conditions for 20 h, heat-shocked for 30 min at 35.5°C, and then allowed to recover at 25°C. RNA was prepared at the indicated times of recovery, and hsp70 mRNA was analyzed as above. RNA samples from nontransformed cells with and without addition of copper sulfate were analyzed in parallel and were indistinguishable from the samples expressing the wild-type CCR4 paralogs (data not shown). (Bottom panel) Expression of the tagged proteins was monitored by Western blotting with the antibodies indicated. EndoGI (Temme et al. 2009) and PABP2 served as loading controls.
In addition, CCR4 paralogs with point mutations in the predicted active sites were expressed and the cells examined for dominant-negative effects. Overexpression of either Flag-Angel or Flag-3635 with inactivating point mutations had no visible effect on the rate of hsp70 deadenylation or on bulk poly(A), and neither had the expression of the wild-type versions (Fig. 8B; data not shown). We conclude that these two proteins are unlikely to play a role in the degradation of the hsp70 mRNA. Although Nocturnin is not expressed in S2 cells, its potential involvement in mRNA deadenylation was also tested by a dominant-negative approach. In this case, there was a detectable delay of hsp70 mRNA decay, which was most clearly visible in the population of undegraded poly(A) tails at the beginning of recovery (Fig. 8B). The visual impression was confirmed by quantitative analyses of the gel images (data not shown). Thus, in agreement with its ability to associate with the other subunits of the CCR4-NOT complex, Nocturnin might be involved in mRNA decay in those cells where the protein is expressed (see Discussion).
DISCUSSION
The CCR4-NOT complex is the most prominent mRNA deadenylase in eukaryotic cells. This study shows that most of the subunits initially identified in yeast form a stable complex in Drosophila as well: CCR4, CAF1, NOT1-3, and CAF40. The association was shown directly by coprecipitation from extract and supported by the observation that RNAi-mediated depletion of individual subunits also led to a depletion of other subunits, presumably due to reduced stability of partial complex assemblies. The strongest link in this regard was between NOT2 and NOT3, which were codepleted with very high efficiency. Yeast Not2p and Not5p can associate in the absence of the other subunits of the CCR4-NOT complex (Bai et al. 1999). As Drosophila NOT3 is related to both yeast Not3p and Not5p, and these are related to each other, the Drosophila NOT2-NOT3 interaction and the yeast Not2p-Not5p interaction are in good agreement. RNAi-mediated depletion of most individual subunits of the CCR4-NOT complex delayed mRNA deadenylation, confirming the functional significance of the polypeptides' association. However, the fact that RNAi against any individual subunit led to the codepletion of others also means that this type of experiment cannot prove the role of particular subunits in mRNA decay. Still, CAF40 may be dispensable for mRNA decay, as its depletion had no effect on this process. In addition to the subunits known from the analysis of the yeast complex, the fly ortholog of CNOT10, a protein stably associated with the human CCR4-NOT complex (Gavin et al. 2002; Morita et al. 2007; Lau et al. 2009), was also found in the NOT1 precipitate. CNOT10 has sometimes been called an ortholog of yeast CAF130, but we have not detected significant sequence similarity between the two proteins.
Whereas Not4p is considered a bona fide subunit of the yeast CCR4-NOT complex (Chen et al. 2001), NOT4 was not detected in stable association with the mammalian complex (Gavin et al. 2002; Morita et al. 2007; Lau et al. 2009). We found only trace amounts of NOT4 in immunoprecipitates of the Drosophila complex, either by Western blotting or by MS analysis. It is unlikely that this negative result was caused by a disruption of an interaction between NOT4 and the rest of the complex by the precipitating antibody, as similar results were obtained with different precipitating antibodies. Also, NOT4 abundance was not affected by knock-down of any subunit of the CCR4-NOT complex, and vice versa. Knock-down of NOT4 had no detectable effect on bulk poly(A) tail length or the rate of deadenylation of the hsp70 mRNA. In summary, the data suggest that NOT4 is not a stable component of the CCR4-NOT complex in Drosophila and is not involved in mRNA deadenylation. Deletion of yeast NOT4 also had at most a minor effect on deadenylation (Tucker et al. 2002). It remains possible that NOT4 is a regulatory subunit associated with the complex in specific cells or under specific conditions. In Drosophila embryos, cyclin B mRNA is regulated by the CCR4-NOT complex in the primordial germ cells located at the posterior pole of the embryo. In these cells, the CCR4-NOT complex has been proposed to be recruited to cyclin B mRNA by a direct interaction between Nanos and NOT4 (Kadyrova et al. 2007).
Knock-down of CCR4 had no effect on mRNA deadenylation in S2 cells (Temme et al. 2004), and this study shows that overexpression of a catalytically inactive mutant caused no phenotype either, even though the mutant protein associated with the other CCR4-NOT complex components to the same extent as the overexpressed wild-type protein. In addition to bulk poly(A), we have only examined one individual transcript, and negative results in knock-down or overexpression experiments are not entirely conclusive; nevertheless, the data suggest that, at least in S2 cells, the catalytic activity of CCR4 may be of minor importance or even dispensable for mRNA decay. This is supported by the strong effects of CAF1 knock-down and overexpression of an inactive CAF1 mutant. Thus, CAF1 seems to be the major catalyst of deadenylation. We note that an increase in bulk poly(A) tail length and some retardation of hsp70 mRNA decay was detectable in twin (=CCR4) mutant flies (Temme et al. 2004). Moreover, in twin mutants, specific mRNAs important during oogenesis and in early embryos have significantly longer poly(A) tails (Morris et al. 2005; Zaessinger et al. 2006). It remains to be seen whether CCR4 functions as a deadenylase in these cases or whether the protein is required to permit deadenylation by CAF1.
Among the CCR4 orthologs, Angel and 3635 did not associate with NOT1, and neither RNAi nor overexpression of inactive mutants had any effect on mRNA decay. In contrast, Nocturnin was found associated with NOT1, and overexpression of an inactive mutant had a dominant-negative effect on deadenylation. This is in agreement with a report that Xenopus and mouse Nocturnin have poly(A) degrading activity (Baggs and Green 2003; Garbarino-Pico et al. 2007) and suggests a possible involvement in mRNA decay. As CAF1 is the main active subunit of the CCR4-NOT complex in S2 cells and a catalytic function of CCR4 was not detectable, it is not straightforward to explain how the replacement of CCR4 with inactive Nocturnin should reduce the rate of deadenylation. Although a reduced rate of hsp70 deadenylation was seen only with the mutant Nocturnin variant, not with the wild-type, we note that the effect was relatively subtle compared with the dominant-negative effect of mutant CAF1. Thus, it remains possible that the slight inhibition of deadenylation is caused by the replacement of CCR4 by Nocturnin per se. In fact, overexpression of wild-type Nocturnin caused lengthening of steady-state poly(A) (data not shown). In any case, the data are consistent with the association of Nocturnin with subunits of the CCR4-NOT complex detected by immunoprecipitation.
Some extent of association between the CCR4-NOT complex and P-bodies has been found in other studies (Eulalio et al. 2007a; Parker and Sheth 2007). Although we found little association detectable by microscopy, analysis of the NOT1 immunoprecipitation revealed a coprecipitation of several prominent constituents of P-bodies, like Tral, Me31B, EDC3, and Gawky. The interaction between Me31B and the CCR4-NOT complex is in agreement with a physical and functional interaction between the Me31B homolog Dhh1p and the CCR4-NOT complex in other organisms (Hata et al. 1998; Maillet and Collart 2002; Schwede et al. 2008). Me31B, Dhh1p, and the vertebrate ortholog RCK/p54 are known as repressors of translation (Minshall et al. 2001, 2009; Nakamura et al. 2001; Coller and Parker 2005; Weston and Sommerville 2006), and Xenopus p54 has been proposed to function in translational repression in association with RAP55 (ortholog of Tral) and eIF4E-T (ortholog of Cup) (Tanaka et al. 2006; Minshall et al. 2007). Several Drosophila mRNAs like nanos (Jeske et al. 2006; Zaessinger et al. 2006) and Cyclin B (Benoit et al. 2005; Kadyrova et al. 2007; Vardy and Orr-Waver 2007) are regulated both by CCR4-NOT-dependent deadenylation and by a deadenylation-independent mechanism of translational repression. The association of Me31B, Cup, and Tral with the CCR4-NOT complex might indicate the formation of large complexes involved in both deadenylation and repression of translation.
An unexpectedly large number of additional proteins copurified with the CCR4-NOT complex (Supplemental Tables 1, 2). At the moment, it is impossible to say how many of these associations may be nonspecific or reflect the abundance of the copurified proteins, for example, ribosomal proteins and translation factors. Although quite harsh RNase digestion conditions were used, RNA-dependent associations cannot be completely excluded either. However, quite a few of the associated proteins match interactions reported previously. For example, many polypeptides of the proteasome system were present in the NOT1 immunoprecipitation, and association of the yeast CCR4-NOT complex with the proteasome has been reported before (Laribee et al. 2007). The presence of polypeptides involved in transcription and its regulation is in agreement with the proposed role of the CCR4-NOT complex in this process (Collart 2003; Denis and Chen 2003; Collart and Timmers 2004). The recovery of most components of the pre-mRNA cleavage/polyadenylation complex and of Wispy, the cytoplasmic GLD2-type poly(A) polymerase, may reflect the regulation of key mRNAs in early development by competing deadenylation and polyadenylation (Zaessinger et al. 2006; Belloc and Méndez 2008; Benoit et al. 2008). The Pan Gu-Plutonium complex was also present in the NOT1 coprecipitation. The Pan Gu kinase plays an essential role during egg activation, promoting the translation of smaug and cyclin B mRNAs by poly(A)-dependent and poly(A)-independent mechanisms (Tadros et al. 2007; Vardy and Orr-Waver 2007). The cyclin B mRNA is also regulated by the CCR4-NOT complex (Kadyrova et al. 2007). Perhaps the most unexpected components of the NOT1 precipitate were proteins of the endoplasmic reticulum (ER) and protein trafficking. Several proteins associated with the CCR4-NOT complex—Me31B, Cup, Tral, and BicC—have been implicated in protein secretion from the ER, presumably by regulation of mRNAs encoding components of the secretion pathway directly on the surface of the ER (Wilhelm et al. 2005; Thomson et al. 2008; Kugler et al. 2009).
MATERIALS AND METHODS
Plasmids and dsRNAs
For the expression of CCR4, Angel, Nocturnin (splice variant RE), and 3635 in S2 cells, coding sequences were amplified by RT-PCR with 3′ primers replacing the natural stop codon with a Flag-tag followed by a stop codon and a suitable restriction site, and ligated into a pMT/V5-His C vector (Invitrogen). Mutations were introduced through site-directed mutagenesis to replace the conserved amino acids aspartate and asparagine in the predicted active sites of the CCR4 paralogs by alanine (CCR4: amino acids 411 + 413; Angel: amino acids 249 + 251; Nocturnin: amino acids 470 + 472; 3635: amino acids 488 + 490). The corresponding mutations have been shown to inactivate yeast Ccr4p (Tucker et al. 2002). For the expression of myc-Caf1, an oligonucleotide encoding the myc tag sequence was cloned into the KpnI/EcoRI site of the pM T/V5-His C vector (Invitrogen). The CAF1 ORF was then cloned into the EcoRV site of the vector in frame with the N-terminal tag. Site-directed mutagenesis was used to replace the conserved amino acids aspartate 53 and glutamate 55 in the exo I motif by alanine. All constructs were verified by sequencing. The GFP-Tral expressing plasmid (Tritschler et al. 2008) was a gift from Elisa Izaurralde (Max Planck Institute for Developmental Biology).
dsRNAs used for RNAi were generated as described (Bönisch et al. 2007). Primer pairs have either been published (Temme et al. 2004; Bönisch et al. 2007) or are listed in Supplemental Table 3.
Antibodies, Western blots, and immunofluorescence
A peptide comprising amino acids 349–362 (ISSGPGTEPIYRNS) of Drosophila NOT1 was synthesized and coupled to keyhole limpet hemocyanin and ovalbumin. For monoclonal antibody production, mice were immunized subcutaneously and intraperitoneally with a mixture of 50 μg peptide-KLH, 5 nmol CpG oligonucleotide 1668 (TIB MOLBIOL), 250 μL PBS, and 250 μL incomplete Freund's adjuvant. A boost without adjuvant was given 6 wk after the primary injection. Fusion was performed using standard procedures. Supernatants were tested by a differential ELISA with the NOT1 peptide coupled to ovalbumin and irrelevant peptides coupled to the same carrier. MAbs that reacted specifically with the NOT1 peptide were further analyzed in Western blots and immunoprecipitation of Drosophila embryo extract. The mAb CISS 2G5, subclass IgG2b, was used in this study. The polyclonal Not1 peptide antibody CR#1 was generated by immunization of rabbits with the same peptide mentioned above (Charles River Company). The rabbit polyclonal Not1 antibody EG314 was generated against the peptide CDTDDPPGLQEKTEFL (Eurogentec). The antibody against Me31B was a kind gift from Akira Nakamura (RIKEN Center for Developmental Biology). A rat monoclonal antibody against a peptide from human hnRNP K similar to the one described (Naarmann et al. 2008) was obtained from B. Moritz (University of Halle) and A. Ostareck-Lederer (University of Halle). The anti-myc antibody was from Roche (mouse monoclonal 9E10) and the α-tubulin antibody from Sigma-Aldrich (T5168). Other antibodies have been described (Benoit et al. 1999; Temme et al. 2004; Jeske et al. 2006; Temme et al. 2009).
Western blots were performed as described (Temme et al. 2004; Jeske et al. 2006). The polyclonal rabbit sera were diluted 1:500 (CCR4) or 1:1000 (all others). The CAF1 antibody was affinity purified on recombinant protein and diluted 1:500. The monoclonal NOT1 antibody CISS 2G5 was diluted 1:250. Immunofluorescence stains were performed as described (Temme et al. 2009). For sequential staining of two proteins, the cells were washed five times for 10 min in PBS-0.1% Triton (PBST) after application of the first secondary antibody, blocked again in PBST+3% BSA, and then incubated with the second primary antibody or with blocking solution as a control. The cells were mounted in Prolong Gold Mounting medium with DAPI (Invitrogen) and analyzed on a confocal laser scanning microscope (Leica, SP5) using a 100× TIRF lens and the Leica 3D deconvolution software. For immunofluorescence stains the NOT1 polyclonal serum EG 314 and the NOT2 serum were diluted 1:1000, and affinity-purified CAF1 antibody 1:25. Cy5- or Cy3-labeled secondary antibodies were purchased from Dianova and diluted 1:1000.
Immunoprecipitation experiments and MS analysis
For large-scale purification of the CCR4-NOT complex from Drosophila embryo extract, 200 μL Protein G-Agarose (Roche) was equilibrated in 0.1 M Tris-HCl (pH 8) and incubated for 3 h at room temperature on a rotating wheel with 1.9 mL CISS 2G5 culture supernatant adjusted to 0.1 M Tris (pH 8). An antibody directed against human hnRNP K was used as a control. Unbound antibodies were removed by five washes with IP buffer (50 mM Tris at pH 7.4, 150 mM KCl, 1.5 mM MgCl2, 0.1% NP-40). One milliliter of embryo extract was diluted with 2 mL IP-buffer and supplemented with 200 μg RNase A. After a preincubation for 10 min at room temperature, the extract was divided in two, and one-half was incubated with the immobilized 2G5 antibody, and the other half was incubated with the control antibody, both for 90 min in the cold room. The supernatant was removed, and the beads were quickly washed three times with 1 mL buffer and then two more times for 15 min under rotation. Proteins were eluted with 500 μL buffer containing 1 mg/mL of the NOT1 peptide used to generate the antibody and 5 mM DTT. After rotation for 1 h, the slurries were applied to microspin columns (Bio-Rad) to separate the eluates from the beads. Proteins in the eluates were TCA precipitated, acetone washed, and dissolved in 30 μL 2× SDS sample buffer. Equal amounts of the input and the supernatants (7.5 μL each) and 5 μL aliquots of the peptide eluates were separated on an SDS gradient gel (7%–18%) and analyzed by Western blotting. Twenty microliters of the eluates was separated on the same type of gel and stained with colloidal Coomassie blue G250 for mass spectrometric analysis.
For peptide mass fingerprint analysis, lanes containing the peptide eluate of the NOT1 immunoprecipitate and of the control were excised from the gel and cut into 37 and 29 bands, respectively. The proteins were in-gel digested following standard protocols. Briefly, protein disulfides were reduced with DTT, and cysteines were alkylated with iodoacetamide. Trypsin digestion was performed overnight at 37°C as described (Schulz et al. 2004). Peptides were analyzed by LC/MS on an UltiMate Nano-HPLC system (LC Packings) coupled to an LTQ-Orbitrap XL mass spectrometer (ThermoFisher Scientific) equipped with a nano-electrospray ionization source (Proxeon). The samples were loaded onto a trapping column (Acclaim PepMap C18, 300 μm × 5 mm, 5 μm, 100 Å, LC Packings) and washed for 15 min with 0.1% trifluoroacetic acid (TFA) at a flow rate of 30 μL/min. Trapped peptides were eluted with the separation column (Acclaim PepMap C18, 75 μm × 150 mm, 3 μm, 100 Å, LC Packings), which had been equilibrated with 100% A (5% acetonitrile, 0.1% formic acid). Peptides were separated with a linear gradient: 0%–50% B (80% acetonitrile, 0.1% formic acid) in 60 min, 50%–100% B in 2 min, 100% B for 17 min, 100%–0% B in 2 min, 0% B for 17 min. The column was kept at 30°C, and the flow rate was 300 nL/min. During gradient elution, online MS data were acquired in data-dependent MS/MS mode: each high-resolution full scan (m/z 300 to 2000, resolution 60,000) in the Orbitrap analyzer was followed by five collision-induced dissociation product ion scans in the linear trap for the five most intense signals in the full-scan mass spectrum (isolation window, 2.3 Th). Dynamic exclusion (repeat count was 3; exclusion duration, 180 sec) was enabled to allow detection of less abundant ions. Data analysis was performed using the Proteome Discoverer 1.0 (ThermoFisher Scientific). MS/MS data of precursor ions in the m/z range 600–6000 (S/N threshold 2, no spectrum clustering) were searched against the NCBInr Database (version 07/04/25, taxonomy Drosophila, 53,042 entries) using Mascot (version 2.2) and against the uniprot database (version 07/03/07, taxonomy Drosophila, 3115 entries) using the Sequest algorithm. Mass accuracy was set to 2 ppm and 0.8 Da for precursor and fragment ions, respectively; carbamidomethylation of cysteines was set as fixed modification and oxidation of methionine as possible modification; two missed cleavages of trypsin were allowed. Proteins identified by at least two peptides with an expectation value <0.01 (Mascot search) or an Xcorr >1.5, 2.0, 2.5, 3 for 1+, 2 + 3+ or ≥4+ charged precursor ions, respectively (Sequest search), were considered unambiguously identified, but proteins identified by a single peptide are also listed (Supplemental Table 2).
To detect the association of the CCR4-NOT complex with the overexpressed CCR4 paralogs, extracts were prepared from stable S2 cell lines that had been treated with 0.5 mM CuSO4 to induce expression of the Flag-tagged proteins. The procedure was as described (Temme et al. 2009) except that the lysis buffer was 30 mM HEPES (pH 7.4), 10 mM KAc, 1.5 mM MgCl2, 0.5 mM DTT, 2 μg/mL leupeptin and pepstatin, and 0.5 μg/mL Pefabloc (Roche). Thirty microliters of CISS 2G5-coupled beads was incubated with 300 μL extract diluted with 300 μL IP buffer for 2 h. After removal of the supernatant, the beads were quickly washed twice with 600 μL IP buffer and then once more for 15 min under rotation. For elution, the beads were incubated for 2 h with 60 μL Not1 peptide containing buffer in a 0.2-mL tube under rotation. After centrifugation, 50 μL of the supernatant was removed and mixed with SDS sample buffer. We analyzed 7.5 μL of the diluted extracts (input), and 30 μL of the eluates were analyzed by Western blot using the M2-Flag antibody (Sigma). Flag immunoprecipitation was performed using the same extracts mentioned above essentially as described (Temme et al. 2009). Proteins were eluted with 60 μL 0.5 mM Flag peptide.
S2 cells and flies
All procedures for S2 cell culture and RNAi experiments were as described (Bönisch et al. 2007) except that Schneider cell medium from Invitrogen was used. Flies were kept and 0–2 h embryo extracts prepared as des LTQ-Orbitrap cribed (Jeske et al. 2006; Jeske and Wahle 2008).
RNA analysis
All procedures were as described (Bönisch et al. 2007).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Elisa Izaurralde, Akira Nakamura, Bodo Moritz, and Antje Ostareck-Lederer for the gift of materials; Melinda Diver and Jana Peplinski for help with some experiments; Nadine Stöhr for help with confocal microscopy; and Sven-Erik Behrens and Stefan Hüttelmaier for letting us use their microscopes. C.T. was the recipient of an EMBO short-term fellowship. The work was supported by a grant from the Deutsche Forschungsgemeinschaft to E.W. and by CNRS UPR1142, “ANR Blanche” (ANR-06-BLAN-0343) and FRM (Equipe FRM 2007, DEQ20071210560) to M.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2145110.
REFERENCES
- Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart M, Timmers HTM 2000. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res 28: 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TK, Hanzawa H, Legtenberg YIA, de Ruwe MJ, van den Heuwel FAJ, Collart MA, Boelens R, Timmers HTM 2002. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J 21: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam A, Mittal S, Koch F, Andrau J-C, Winkler GS 2009. The Ccr4-Not deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol Biol Cell 20: 3840–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA 2003. The RNA binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol 10: 614–621 [DOI] [PubMed] [Google Scholar]
- Baggs JE, Green CB 2003. Nocturnin, a deadenylase in Xenopus laevis retina: A mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol 13: 189–198 [DOI] [PubMed] [Google Scholar]
- Bai Y, Salvadore C, Chiang Y-C, Collart M, Liu H-Y, Denis CL 1999. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4 and NOT5. Mol Cell Biol 19: 6642–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko A-M, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al. 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52: 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloc E, Méndez R 2008. A deadenylation negative feedback mechanism governs meiotic metaphase arrest. Nature 452: 1017–1021 [DOI] [PubMed] [Google Scholar]
- Benoit B, Nemeth A, Aulner N, Kühn U, Simonelig M, Wahle E, Bourbon H-M 1999. The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic Acids Res 27: 3771–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M 2005. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev Cell 9: 511–522 [DOI] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M 2008. PAP- and GLD 2-like poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development 135: 1969–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchin C, Mauxion F, Sentis S, Seraphin B, Corbo L 2005. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA 11: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönisch C, Temme C, Moritz B, Wahle E 2007. Degradation of hsp70 and other mRNAs in Drosophila via the 5′-3′ pathway and its regulation by heat shock. J Biol Chem 282: 21818–21828 [DOI] [PubMed] [Google Scholar]
- Chen J, Rappsilber J, Chiang Y-C, Russell P, Mann M, Denis CL 2001. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol 314: 683–694 [DOI] [PubMed] [Google Scholar]
- Chen J, Chiang Y, Denis CL 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 21: 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P 2007. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell 13: 691–704 [DOI] [PubMed] [Google Scholar]
- Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N 2006. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol 16: 2035–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LB, Viswanathan P, Quigley G, Chiang Y-C, McMahon JS, Yao G, Chen J, Nelsbach A, Denis CL 2004. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J Biol Chem 279: 13616–13623 [DOI] [PubMed] [Google Scholar]
- Collart M 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313: 1–16 [DOI] [PubMed] [Google Scholar]
- Collart MA, Timmers HTM 2004. The eukaryotic Ccr4-Not complex: A regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol 77: 289–322 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron M-C, Mauxion F, Seraphin B 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res 29: 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellavalle RP, Petersen R, Lindquist S 1994. Preferential deadenylation of Hsp70 mRNA plays a key role in regulating Hsp70 Expression in Drosophila melanogaster. Mol Cell Biol 14: 3646–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis CL, Chen J 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol 73: 221–250 [DOI] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T 2009. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem 284: 10343–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Morel A-P, Barbot W, Loireau M-P, Corbo L, Heidmann T 2001. Identification of four families of yCCR4- and Mg-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E 2007a. P bodies: At the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E 2007b. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Izaurralde E 2009. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. RNA 15: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, Green CB 2007. Immediate early response of the circadian poly(A) ribonuclease nocturnin to two extracellular stimuli. RNA 13: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ 2007. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Wickens M 2008. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 9: 337–344 [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Seay DJ, Hook BA, Wickens M 2007. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem 282: 109–114 [DOI] [PubMed] [Google Scholar]
- Grönke S, Bickmeyer I, Wunderlich R, Jäckle H, Kühnlein RP 2009. curled encodes the Drosophila homolog of the vertebrate circadian deadenylase Nocturnin. Genetics 183: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A 1998. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Jeske M, Wahle E 2008. Cell-free deadenylation assays using Drosophila embryo extracts. Methods Enzymol 448: 107–118 [DOI] [PubMed] [Google Scholar]
- Jeske M, Meyer S, Temme C, Freudenreich D, Wahle E 2006. Rapid ATP-dependent deadenylation of nanos mRNA in a cell-free system from Drosophila embryos. J Biol Chem 281: 25124–25133 [DOI] [PubMed] [Google Scholar]
- Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA 2010. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol 17: 238–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, Wharton RP 2007. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134: 1519–1527 [DOI] [PubMed] [Google Scholar]
- Kerwitz Y, Kühn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E 2003. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J 22: 3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler J-M, Chicoine J, Lasko P 2009. Bicaudal-C associates with a trailer hicht/Me31B complex and is required for efficient Gurken secretion. Dev Biol 328: 160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, Weissman JS, Briggs SD, Krogan NJ, Strahl BD 2007. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc Natl Acad Sci 104: 5836–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N-C, Kolkman A, van Schaik FMA, Mulders KW, Pijnappel WWMP, Heck AJR, Timmers HTM 2009. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J 422: 443–453 [DOI] [PubMed] [Google Scholar]
- Lin M-D, Jiao X, Grima D, Newbury SF, Kiledjian M, Chou T-B 2008. Drosophila processing bodies in oogenesis. Dev Biol 322: 276–288 [DOI] [PubMed] [Google Scholar]
- Maillet L, Collart M 2002. Interaction between Not1p, a component of the Ccr4-Not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J Biol Chem 277: 2835–2842 [DOI] [PubMed] [Google Scholar]
- Maillet L, Tu C, Hong YK, Shuster EO, Collart M 2000. The essential function of Not1 lies within the Ccr4-Not complex. J Mol Biol 303: 131–143 [DOI] [PubMed] [Google Scholar]
- Markesich DC, Gajewski KM, Nazimiec ME, Beckingham K 2000. bicaudal encodes the Drosophila β NAC homolog, a component of the ribosomal translational machinery. Development 127: 559–572 [DOI] [PubMed] [Google Scholar]
- Mauxion F, Faux C, Seraphin B 2008. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J 27: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauxion F, Chen C-AA, Seraphin B, Shyu A-B 2009. BTG/TOB factors impact deadenylases. Trends Biochem Sci 34: 640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersman DP, Du H-N, Fingerman IM 2009. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev 23: 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E 2004. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit Rev Biochem Mol Biol 39: 197–216 [DOI] [PubMed] [Google Scholar]
- Minshall N, Thom G, Standart N 2001. A conserved role of a DEAD box helicase in mRNA masking. RNA 7: 1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Reiter MH, Weil D, Standart N 2007. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem 282: 37389–37401 [DOI] [PubMed] [Google Scholar]
- Minshall N, Kress M, Weil D, Standart N 2009. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol Biol Cell 20: 2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Suzuki T, Nakamura T, Yokoyama K, Miysaka T, Yamamoto T 2007. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA levels and impairs cell growth. Mol Cell Biol 27: 4980–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Hong A, Lilly MA, Lehmann R 2005. twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development 132: 1165–1174 [DOI] [PubMed] [Google Scholar]
- Naarmann IS, Harnisch C, Flach N, Kremmer E, Kühn H, Ostareck DH, Ostareck-Lederer A 2008. mRNA silencing in human erythroid cell maturation. Heterogeneous nuclear ribonucleoprotein K controls the expression of its regulator c-src. J Biol Chem 283: 18461–18472 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S 2001. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128: 3233–3242 [DOI] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA 2004. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J 23: 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko O, Landrieux E, Feuermann M, Finka A, Paquet N, Collart MA 2006. The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J Biol Chem 281: 31389–31398 [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Parker R, Song H 2004. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Rendl LM, Bieman MA, Smibert CA 2008. S. cerevisiae Vts1p induces deadenylation-dependent transcript degradation and interacts with the Ccr4p-Pop2p-Not deadenylase complex. RNA 14: 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MD, Najand N, Chaker S, Pare JM, Haskins J, Hughes SC, Hobman TC, Locke J, Simmonds AJ 2006. Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J Cell Biol 174: 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz DM, Ihling C, Clore GM, Sinz A 2004. Mapping the topology and determination of a low-resolution three-dimensional structure of the calmodulin-melittin complex by chemical cross-linking and high-resolution FTICRMS: Direct demonstration of multiple binding modes. Biochem 43: 4703–4715 [DOI] [PubMed] [Google Scholar]
- Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C 2008. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res 36: 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA 2005. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol 15: 284–294 [DOI] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP 2001. Drosophila Brain Tumor is a translational repressor. Genes Dev 15: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Waver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 12: 143–155 [DOI] [PubMed] [Google Scholar]
- Tanaka KJ, Ogawa K, Takagi M, Imamoto N, Matsumoto K, Tsujimoto M 2006. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem 281: 40096–40106 [DOI] [PubMed] [Google Scholar]
- Temme C, Zaessinger S, Simonelig M, Wahle E 2004. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J 23: 2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Weissbach R, Lilie H, Wilson C, Meinhart A, Meyer S, Golbik R, Schierhorn A, Wahle E 2009. The Drosophila melanogaster gene cg4930 encodes a high affinity inhibitor for endonuclease G. J Biol Chem 284: 8337–8348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Liu N, Arkov A, Lehmann R, Lasko P 2008. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev 125: 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Mauxion F, Seraphin B, Suck D 2003. X-ray structure and activity of the yeast Pop2 protein: A nuclease subunit of the mRNA deadenylase complex. EMBO Rep 4: 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A, Beilharz TH, Lo TL, Lueder F, Preiss T, Heierhorst J 2009. The Ccr4-Pop2-NOT mRNA deadenylase contributes to septin organization in Saccharomyces cerevisiae. Genetics 182: 955–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V 2008. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol 28: 6695–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O 2009. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell 33: 661–668 [DOI] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 21: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L, Orr-Waver TL 2007. The Drosophila PNG kinase complex regulates the translation of Cyclin B. Dev Cell 12: 157–166 [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Chen J, Chiang Y-C, Denis CL 2003. Identification of multiple RNA features that influence CCR4 deadenylation activity. J Biol Chem 278: 14949–14955 [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Ohn T, Chiang Y-C, Chen J, Denis CL 2004. Mouse CAF1 can function as a processive deadenylase/3′-5′ exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem 279: 23988–23995 [DOI] [PubMed] [Google Scholar]
- Wagner E, Clement SL, Lykke-Andersen J 2007. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear Cajal bodies. Mol Cell Biol 27: 1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston A, Sommerville J 2006. Xp54 and related (DDX6-like) RNA helicases: Roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res 34: 3082–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze M 2000. Translational control of developmental decisions. In Translational control of gene expression, (ed. N Sonenberg et al. ). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Wilhelm JE, Buszczak M, Sayles S 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell 9: 675–685 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Chang T-C, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, Shyu A-B 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M 2006. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133: 4573–4583 [DOI] [PubMed] [Google Scholar]
- Zekri L, Huntzinger E, Heimstädt S, Izaurralde E 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol 29: 6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzedine N, Chen C-Y, Zhu W, He X, Shyu A-B 2008. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol 182: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]