Abstract
Diverse steps in gene expression are tightly coupled. Curiously, the La-motif-containing protein Sro9 has been shown to play a role in transcription and translation. Here, we show that Sro9 interacts with nuclear and cytoplasmic protein complexes involved in gene expression. In addition, Sro9 shuttles between nucleus and cytoplasm and is exported from the nucleus in an mRNA export-dependent manner. Importantly, Sro9 is recruited to transcribed genes. However, whole genome expression analysis shows that loss of Sro9 function does not greatly change the level of specific transcripts indicating that Sro9 does not markedly affect their synthesis and/or stability. Taken together, Sro9 might bind to the mRNP already during transcription and accompany the mature mRNP to the cytoplasm where it modulates translation of the mRNA.
Keywords: Sro9, transcription, translation, nucleocytoplasmic shuttling
INTRODUCTION
La-motif-containing proteins constitute a highly conserved family of RNA binding proteins involved in various RNA-related processes. Originally identified in humans as an autoimmune antigen of patients with rheumatic diseases such as systemic lupus erythematosus and Sjogren's syndrome (Mattioli and Reichlin 1974; Alspaugh and Tan 1975), La proteins are highly conserved throughout evolution (for review, see Wolin and Cedervall 2002). Authentic La proteins are nuclear proteins (Hendrick et al. 1981; Yoo and Wolin 1994) that protect the 3′ end of newly synthesized RNA polymerase III transcripts. This binding event not only protects the small RNAs from exonucleases, but also determines the correct subsequent processing of these RNAs (for review, see Wolin and Cedervall 2002). In addition, La proteins are important for the stabilization of newly synthesized U6 RNAs (Pannone et al. 1998) and histone mRNAs (McLaren et al. 1997). Surprisingly for a nuclear protein, La proteins might also enhance the translation of viral and cellular mRNAs (Meerovitch et al. 1993; Svitkin et al. 1994; Holcik and Korneluk 2000; see also Discussion).
More recently, however, sequence analysis revealed a second class of La motif-containing proteins to be present in all sequenced eukaryotic genomes (Sobel and Wolin 1999). These proteins contain a phylogenetically different La motif that is positioned N-terminally, centrally, or C-terminally—in contrast to the predominantly N-terminal localization of the La motif in authentic La proteins—and display no sequence homology except in the La motif itself. The Saccharomyces cerevisiae protein Sro9 belongs to this new class of La motif-containing proteins. Sro9 is a cytoplasmic protein that was originally identified in genetic screens as a suppressor of mutants of the secretory pathway (Tsukada and Gallwitz 1996), a mutant affecting bud formation (Imai et al. 1996), and mutants in actin or tropomyosin (Kagami et al. 1997). Subsequently, Sobel and Wolin (1999) reported that Sro9 binds to RNA and associates with translating ribosomes. As deletion of Sro9 reduces sensitivity toward translation inhibitors, Sro9 might act as a molecular chaperone stabilizing mRNAs in the correct translational conformation or might influence mRNP rearrangements for efficient translation of the mRNA. In addition, Sro9 was shown to function in transcription by RNA polymerase II (Tan et al. 2000). High copy Sro9 suppresses transcription defects caused by deletion of Rpb4, a nonessential subunit of RNA polymerase II. Furthermore, addition of recombinant Sro9 in in vitro transcription reactions restores the transcription defects, pointing to a direct role of Sro9 in transcription. In addition, overexpression of Sro9 increases total mRNA levels in Δrpb4 cells indicating a role for Sro9 in mRNA stability. Thus, Sro9, which localizes to the cytoplasm at steady state (Kagami et al. 1997; Sobel and Wolin 1999), functions in processes as diverse as transcription, translation, and mRNA stability.
Gene expression encompasses multiple steps that are highly interconnected. During transcription the genetic information stored in the protein coding genes is transcribed into mRNA. The mRNA is processed (capped, spliced, and polyadenylated), packaged into a mature mRNP, transported through the nuclear pore complex to the cytoplasm, and finally translated into the encoded protein by the ribosomes (for review, see Erkmann and Kutay 2004; Fasken and Corbett 2005; Olesen et al. 2005; Kohler and Hurt 2007; Iglesias and Stutz 2008; Röther and Sträßer 2009). Here, we show that Sro9 might function in gene expression processes as distant as transcription and translation by shuttling between nucleus and cytoplasm. Consistent with this model, Sro9 associates with protein complexes involved in nuclear and cytoplasmic steps of gene expression. Importantly, Sro9 is recruited to actively transcribed genes. However, Sro9 is most likely not needed for the synthesis or stability of specific mRNAs as revealed by genome-wide expression analysis. According to our model, Sro9 is cotranscriptionally recruited to the nascent transcript and shuttles to the cytoplasm as a component of the exported mRNP, where it is important for modulation of translation of the bound mRNA.
RESULTS AND DISCUSSION
Sro9 associates with multiple protein complexes of the gene expression pathway
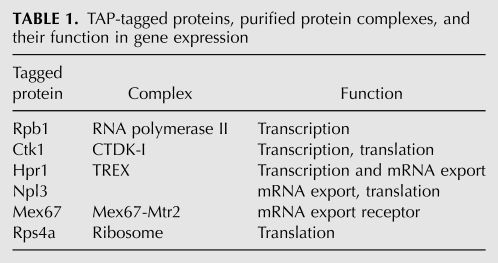
As Sro9 was reported to be involved in nuclear transcription as well as cytoplasmic translation, we chose representative protein complexes along the gene expression pathway (Table 1) to assess at which stages Sro9 is associated. We purified RNA polymerase II (Rpb1), the transcription elongation factor CTDK-I (Ctk1), the TREX complex that couples transcription to mRNA export (Hpr1), the mRNP-bound protein Npl3, the mRNA export receptor Mex67-Mtr2 (Mex67), and ribosomes (Rps4a) by tandem affinity purification (TAP) using the TAP-tagged subunit indicated in brackets and we tested a putative association of Sro9 by Western blotting (Fig. 1). The CTDK-I complex phosphorylates the C-terminal domain of RNA polymerase II during transcription elongation (for review, see Prelich 2002; Svejstrup 2004). In addition, this protein complex has a second function in gene expression by increasing the accuracy of amino acid incorporation during translation elongation (Röther and Sträßer 2007). TREX is a highly conserved complex that is recruited to the nascent transcript during transcription elongation and interacts downstream with the mRNA export receptor Mex67-Mtr2, thereby coupling transcription to nuclear export of the mRNA (Sträßer et al. 2002; Reed and Cheng 2005). Npl3 is a serine-arginine (SR)-rich protein that binds cotranscriptionally to the mRNP, is exported with the mature mRNP, and needs to be dissociated from the mRNA for efficient translation to occur (Gilbert et al. 2001; Gilbert and Guthrie 2004; Windgassen et al. 2004). The heterodimeric mRNA export receptor Mex67-Mtr2 is recruited to the mRNA by the TREX complex and exports the mRNA to the cytoplasm by direct interaction with nuclear pores (Segref et al. 1997; Sträßer and Hurt 2000; Sträßer et al. 2000). In the cytoplasm, ribosomes finally translate the mRNA into the encoded protein (for review, see Preiss and Hentze 2003).
TABLE 1.
TAP-tagged proteins, purified protein complexes, and their function in gene expression
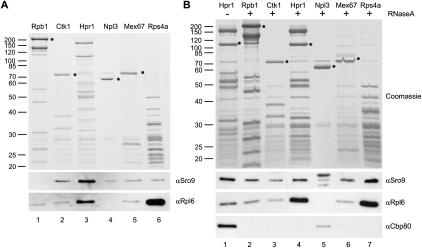
FIGURE 1.
Sro9 interacts with protein complexes involved in gene expression. (A) RNA polymerase II (Rpb1), catalyzing mRNA synthesis, CTDK-I (Ctk1), involved in transcription and translation elongation, TREX (Hpr1), a cotranscriptionally recruited complex coupling transcription and mRNA export, the SR-protein Npl3, which is also involved in translation, the mRNA export receptor Mex67-Mtr2 (Mex67), and ribosomes (Rps4a) were purified by tandem affinity purification. Copurification of Sro9 and the ribosomal protein Rpl6 was assessed by Western blotting. Sro9 does not consistently copurify with RNA polymerase II but with protein complexes downstream in the gene expression process—independently of ribosomes. Circles indicate the TAP-tagged proteins. (B) Same as in A, but lysates were treated with RNase A prior to protein complex purification to assess RNA dependence of the interactions observed in A. To assess the efficiency of RNase treatment the presence of Cbp80, which binds in an RNA-dependent manner to TREX, in the eluted complexes was analyzed.
Sro9 copurified with CTDK-I, TREX, Npl3, Mex67-Mtr2, and ribosomes (Fig. 1A, lanes 2–6, αSro9). The observed interaction of Sro9 with ribosomes is consistent with the comigration of Sro9 with ribosomal proteins (subunits, mono-, and polysomes) in sucrose density gradients (Sobel and Wolin 1999; S Röther and K Sträßer, unpubl.). Importantly, though, copurification of Sro9 with those protein complexes is independent of the level of ribosomal proteins present in each purification (Fig. 1A, lanes 2–5, cf. αRpl6 and αSro9) showing that Sro9 is not a ribosome-associated contaminant. In contrast, copurification of Sro9 with RNA polymerase II, i.e., the transcription machinery, varied between experiments (Fig. 1A, lane 1, αSro9). This might indicate that Sro9 interacts only transiently with RNA polymerase II and might be quickly dissociated from the transcription machinery in vivo.
Since La proteins are known to bind RNA we tested whether the association of Sro9 with the above protein complexes is mediated by mRNA. Degradation of mRNA by RNase A treatment was tested by assessing the presence of the cap binding protein component Cpb80, which binds to TREX in an RNA-dependent manner (Fig. 1B, lanes 1,4, αCpb80). In contrast, RNase A treatment did not disrupt copurification of Sro9 (Fig. 1B, αSro9). Taken together, Sro9 binds to protein complexes involved in different steps of gene expression independently of RNA.
Sro9 shuttles between nucleus and cytoplasm
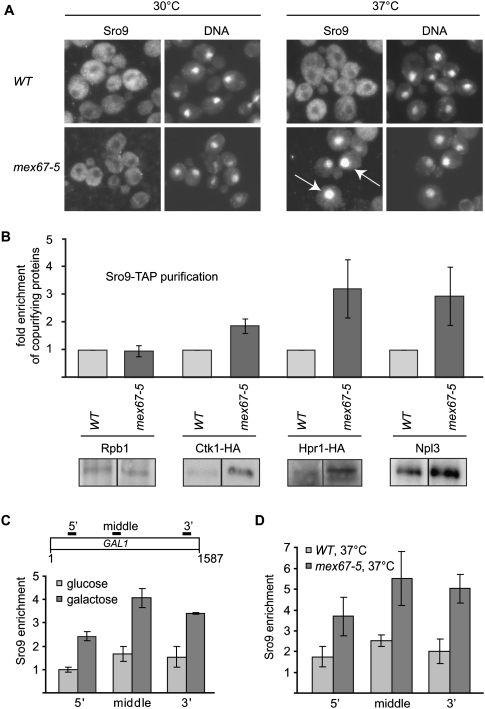
Consistent with a function of Sro9 in transcription (Tan et al. 2000) we found that Sro9 interacts with protein complexes mainly localized to the nucleus, although Sro9 localizes to the cytoplasm at steady state. A function of Sro9 in the nucleus would thus require Sro9 to shuttle between nucleus and cytoplasm. Due to the association of Sro9 with protein complexes of the mRNA export machinery, we analyzed the localization of Sro9 after inhibition of mRNA export. In mex67-5 cells, a temperature-sensitive mutant of the general mRNA export receptor Mex67-Mtr2, mRNA export is blocked at the nonpermissive temperature (37°C) und poly(A)+ mRNA accumulates in the nucleus (Segref et al. 1997). In wild-type (WT) and mutant (mex67-5) cells, Sro9 was exclusively localized to the cytoplasm at the permissive temperature (30°C) (Fig. 2A, left panel). In contrast, Sro9 accumulates in the nucleus of mex67-5 cells at the nonpermissive temperature (37°C), i.e., when mRNA export is blocked (Fig. 2A, right panel). Thus, Sro9 shuttles between nucleus and cytoplasm and is exported from the nucleus in an mRNA export-dependent manner.
FIGURE 2.
Sro9 shuttles between nucleus and cytoplasm and is recruited to an actively transcribed gene. (A) Sro9 is exported to the cytoplasm dependent on mRNA export. At the permissive temperature (30°C) Sro9 localizes to the cytoplasm. However, at the nonpermissive temperature (37°C), when mRNA export is blocked in the mex67-5 mutant, Sro9 accumulates in the nucleus, indicating that Sro9 is part of the exported mRNP. White arrows point to the accumulated nuclear protein. Nuclear DNA was stained with DAPI. (B) Association of Sro9 with nuclear protein complexes of the gene expression pathway occurs in vivo and increases upon inhibition of mRNA export. Sro9 was purified after cross-linking at the nonpermissive temperature (37°C) from wild-type (WT) and mex67-5 cells and the copurification of nuclear proteins was assessed by Western blotting. Sro9 binds to Ctk1, Hpr1, and Npl3 in vivo (WT). Compared to WT, association of Sro9 with Ctk1, Hpr1, and Npl3 increased two- to threefold when mRNA export is blocked (mex67-5). In contrast, the association of Sro9 with Rpb1, which could not be observed consistently, remained unchanged. Representative Western blots are shown. Values represent means of four to five experiments with standard deviation as error bars. (C) Sro9 is recruited to an actively transcribed gene. Chromatin immunoprecipitation experiments show that Sro9 is recruited to the induced GAL1 gene. Wild-type cells were grown in glucose- or galactose-containing medium at 30°C. Primer pairs amplify a 5′, middle, and 3′ region of GAL1 as indicated in the upper panel. Enrichment of Sro9 at GAL1 was calculated relative to the Sro9 occupancy at a nontranscribed region. Columns and error bars represent the mean ± standard deviation from three independent experiments. The values for Sro9 recruitment to the repressed versus activated GAL1 gene differ significantly (P-values are 0.0004 [5′], 0.0012 [middle], and 0.0019 [3′]; t-test). (D) Recruitment of Sro9 to GAL1 increases, when Sro9 accumulates in the nucleus upon inhibition of mRNA export (mex67-5, 37°C). Cells were grown in galactose-containing medium at 37°C, and the experiment was performed as in C. Columns and error bars represent the mean ± standard deviation from four independent experiments; P-values are 0.0103 (5′), 0.0042 (middle), and 0.0006 (3′); t-test.
Based on the interaction of Sro9 with nuclear protein complexes (Fig. 1) and its nucleocytoplasmic shuttling, we asked whether these protein–protein interactions are enhanced when mRNA export is blocked and Sro9 accumulated in the nucleus. To preserve these potentially enhanced interactions and to avoid unspecific association of protein complexes when nucleo- and cytoplasm are mixed during cell lysis, we cross-linked WT and mex67-5 cells after shift to the nonpermissive temperature and purified Sro9-TAP under high salt conditions to dissociate any proteins that were not associated with Sro9 in vivo; i.e., during cross-linking. Purification of Sro9 under these conditions (see Materials and Methods) but without cross-linking did not yield any of the tested proteins (data not shown). Importantly, the above proteins (with the exception of Rpb1, whose copurification was inconsistent) (Figs. 1, 2B) copurified with Sro9 under these conditions, showing that these interactions occur specifically in vivo and are not simply the result of mixing cellular compartments during cell lysis. Furthermore, in comparison with WT cells, association of Sro9 with Ctk1, Hpr1, and Npl3 increased two- to threefold when Sro9 accumulated in the nucleus (Fig. 2B, mex67-5). In summary, even though Sro9 localizes to the cytoplasm at steady state, it shuttles to the nucleus, where it associates with protein complexes involved in processes along the gene expression pathway before it is exported to the cytoplasm in an mRNA export-dependent manner.
Sro9 is recruited to an actively transcribed gene
As Sro9 might be part of the exported mRNP, we wanted to know whether Sro9 is recruited to the nascent transcript or associates with the mRNP at a later time point. Using chromatin immunoprecipitation (ChIP) experiments, we tested whether Sro9 can be found at an actively transcribed gene. To be able to easily regulate transcription, we chose the inducible GAL1 gene. Sro9 is not recruited to the GAL1 locus when cells are grown in glucose-containing medium, and consequently, transcription of GAL1 is repressed (Fig. 2C, light gray). Importantly, when cells are grown in galactose-containing medium and transcription of GAL1 is active, Sro9 is recruited to this gene in WT cells (Fig. 2C, dark gray). Expectedly, association of Sro9 with GAL1 increases when mRNA export is blocked in the mex67-5 mutant and Sro9 accumulates in the nucleus (Fig. 2D, dark gray, mex67-5, 37°C). Taken together, as many other mRNP components (also see below), Sro9 is already present at the transcribed gene and thus most likely loaded cotranscriptionally onto the nascent transcript.
Sro9 is not essential for expression or stability of specific transcripts
Next, we wanted to assess whether Sro9 has a direct function in transcription, mRNA export, or stability. Even though Sro9 is a nonessential protein, its deletion leads to a mild temperature-sensitive phenotype at 37°C (data not shown). Therefore, we tested for a potential mRNA export defect in the absence of Sro9 at this temperature. However, Δsro9 cells do not accumulate bulk poly(A)+ RNA in the nucleus at 37°C (data not shown), indicating that Sro9 is not needed for efficient mRNA export.
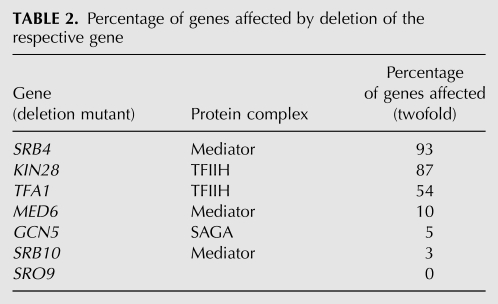
Overexpression of Sro9 alleviates the in vitro transcription defect; suppresses the reduced induction of GAL1, but not PHO5 or INO1 genes; and increases bulk mRNA stability in Δrpb4 cells (Tan et al. 2000). Thus, an influence of Sro9 function on steady-state levels of a specific set of mRNAs was assessed using whole genome expression profiling of Δsro9 in comparison with WT cells. With data processing techniques commonly applied in microarray analysis (see Materials and Methods), only two genes apart from SRO9 itself showed significantly—i.e., more than twofold—altered mRNA levels as compared with the WT strain: YHR087W coding for a protein of unknown function and GPH1 coding for a nonessential glycogen phosphorylase. However, since after multiple test correction (Benjamini and Hochberg 1995) no differentially expressed genes were identified, these two genes most likely represent false positives. For comparison, deletion of bona fide transcription factors (TFs) such as certain subunits of the mediator complex, the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex, or general transcription factors of Pol II, usually alters the expression of several hundred genes but at least 3% of total genes (Table 2; Holstege et al. 1998). Thus, the absence of Sro9 does not lead to changes in the steady-state level of specific transcripts, which might indicate the presence of additional factors with a function redundant to that of Sro9. One likely factor is the Sro9 homolog Slf1. Sro9 and Slf1 share about 30% identity throughout their amino acid sequence and are thought to result from an ancient gene duplication event (Wolfe and Shields 1997). Interestingly, when Slf1 is overexpressed, Sro9 protein levels are decreased (Sobel and Wolin 1999), indicating that Slf1 could take over the function(s) of Sro9. However, SRO9 and SLF1 do not interact genetically (Sobel and Wolin 1999), suggesting that either Sro9 or Slf1 does not function redundantly or that its (partially) redundant function is not essential. Thus, further experiments are needed to dissect the distinct and/or overlapping functions of Sro9 and Slf1. Taken together, whole genome expression profiling revealed that Sro9 is not essential for expression and stability of specific transcripts.
TABLE 2.
Percentage of genes affected by deletion of the respective gene
Concluding remarks
In recent years, it became widely accepted that nuclear gene expression processes are intimately linked (for review, see Erkmann and Kutay 2004; Fasken and Corbett 2005; Olesen et al. 2005; Kohler and Hurt 2007; Iglesias and Stutz 2008; Röther and Sträßer 2009). Many mRNA processing steps depend on preceding steps and determine subsequent ones: already during ongoing transcription, the mRNA is capped, spliced, and (after cleavage) polyadenylated. In addition, proteins package the mRNA into a messenger ribonucleoprotein particle (mRNP). This mRNP is then transported to the cytoplasm, where the ribosomes translate the mRNA into the encoded protein (Preiss and Hentze 2003; Holcik and Sonenberg 2005). In contrast, transcription and mRNA processing on the one hand and translation on the other hand were considered to be independent processes. However, considering the fact that from transcription to translation the mRNA is covered by various proteins and protein complexes, proteins loaded on the mRNA during nuclear maturation of the message can influence the cytoplasmic translation of the mRNA. One example of such a connection is Tap-p15, the human homolog of the yeast mRNA exporter Mex67-Mtr2. Tap-p15 is nuclear at steady state but shuttles between nucleus and cytoplasm and promotes translation of the viral CTE mRNA it exports (Jin et al. 2003). Second, the yeast SR-protein Npl3 binds to the mRNA already cotranscriptionally, but needs to be dissociated for efficient translation to occur (Windgassen et al. 2004). Third, the yeast cyclin-dependent kinase Ctk1 phosphorylates RNA polymerase II during transcription elongation in the nucleus and is also important in the maintenance of accuracy during translation in the cytoplasm (Röther and Sträßer 2007). Fourth, the exon–exon junction complex (EJC) of frogs, plants, and humans, important in nuclear steps of gene expression, travels together with the mRNP to the cytoplasm, where it mediates nonsense-mediated decay and enhances translational efficiency (Wiegand et al. 2003; Nott et al. 2004; Belostotsky and Rose 2005). Fifth, the cotranscriptionally recruited mRNP component Dbp5, an RNA helicase important in the directionality of mRNA export, has recently been shown to be important in translation termination (Gross et al. 2007). Along the same line, Gle1 does not only stimulate the RNA-dependent ATPase activity of Dbp5 (Alcazar-Roman et al. 2006; Weirich et al. 2006) to ensure mRNA transport directionality (Tran and Wente 2006), but it is also essential for translation initiation and termination (Bolger et al. 2008). Finally, as already mentioned in the Introduction, the nuclear human La protein has been implicated in translation initiation of cellular and viral mRNAs (for review, see Wolin and Cedervall 2002).
mRNPs are known to be highly dynamic particles, whose protein content undergoes constant remodeling. To date, the distinct mRNP composition at each stage of gene expression is not known. However, the above-mentioned examples suggest that some mRNP components function in nuclear as well as cytoplasmic steps of gene expression, accompany the mRNA from the site of transcription to the ribosomes, the site of translation, and thereby could couple these two spatially separate processes. An increasing number of reports suggests that these proteins could mediate a new layer of gene expression, where the nuclear protein composition of the mRNP is decisive for the translational fate of the bound mRNA, such as initiation (Gle1, La), efficiency (Tap-p15, Npl3, or EJC components), accuracy (Ctk1), or termination (Dbp5, Gle1) of translation.
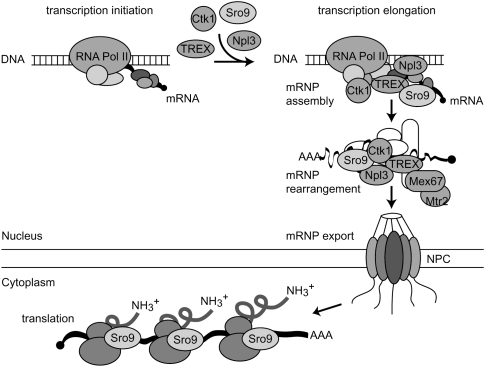
Interestingly, purification of some of the above-mentioned proteins (Ctk1, Npl3, and Mex67) yields Sro9—a protein with a function in transcription, translation, and mRNA stability—as a copurifying protein. In addition, Sro9 is recruited to actively transcribed genes and shuttles between nucleus and cytoplasm. While it is possible that there are separate pools of Sro9 that act independently, we propose that Sro9 is loaded onto the nascent transcript during transcription, where it contributes to robust transcription and might already stabilize the mRNA (Tan et al. 2000), perhaps by binding to the nascent mRNA. Sro9 then travels to the cytoplasm most likely as part of the mRNP, where it modulates translation (Sobel and Wolin 1999). Thus, Sro9 might be a member of the growing group of mRNP binding proteins that link nuclear and cytoplasmic steps of gene expression (Fig. 3).
FIGURE 3.
Model of the functions of Sro9 in transcription and translation. Similar to Ctk1, TREX, and Npl3, Sro9 is recruited to the nascent mRNA already during transcription. After maturation of the messenger ribonucleoprotein particle, Sro9 is exported to the cytoplasm as part of the mRNP, where it reaches the ribosomes. In line with this model, Sro9 is able to function in transcription, mRNA stability (Tan et al. 2000), and translation (Sobel and Wolin 1999).
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains and plasmids are listed in Table 3. TAP-tagged strains were generated by integration of the TAP tag into the genome C-terminally of the respective genes by homologous recombination according to Puig et al. (2001). To delete SRO9, a sro9∷HIS3 construct was cloned by polymerase chain reaction (PCR) of a 500-base-pair (bp) SalI-BamHI promoter and a 500-bp BamHI-NotI terminator fragment of the SRO9 gene and ligation of these fragments into pBluescriptIIKS(+), followed by insertion of the BamHI HIS3 fragment of YDp-H (Berben et al. 1991). The SalI-NotI sro9∷HIS3 fragment was transformed into RS453, HIS+ transformants were selected, and SRO9 deletion checked by colony PCR. Positive colonies were transformed with pRS316-SRO9 (Sobel and Wolin 1999).
TABLE 3.
Strains and plasmids
Protein purification
Rpb1, Ctk1, Hpr1, Npl3, Mex67, and Rps4a, and associated proteins were purified by TAP according to Puig et al. (2001). For RNase treatment the lysis buffer contained 0.1 mg/mL RNase A. Copurifying proteins were analyzed by sodium dodecyl sulfate (SDS)-gel electrophoresis and Western blotting using antibodies directed against Sro9, Rpl6, and Cbp80. For purification of cross-linked protein complexes, SRO9-TAP CTK1-HA/HPR1-HA MEX67 and SRO9-TAP CTK1-HA/HPR1-HA mex67-5 cells were cultivated at 30°C to an OD600 of 0.6. Cultures were divided and further cultivated for 2 h at 30°C and 37°C, respectively. Formaldehyde was added to a final concentration of 1% and cultures incubated further for 10 min at 30°C and 37°C, respectively. To stop cross-linking, glycine was added prior to harvesting. Sro9 and associated proteins were purified by tandem affinity purification as described above with some modifications. Briefly, cross-linked cells were lysed in an equal volume of high salt (1 M NaCl) buffer and glass beads by vortexing at full speed for 6 × 3 min with 3-min breaks on ice. After sonication and removal of cell debris, the supernatant was incubated overnight with 500 μL IgG sepharose beads. After washing with high salt buffer, immunoprecipitated material was cleaved from the beads with TEV protease. Copurifying proteins were analyzed by SDS-gel electrophoresis and Western blotting using antibodies directed against Sro9, Npl3, Rpb1 (8WG16, Convance), and HA (Roche Diagnostics). The Anti-Sro9 antibody was a kind gift from S. Wolin (Yale University), anti-Rpl6 from G. Dieci (University of Parma), anti-Cbp80 from D. Görlich (MPI for Biophysical Chemistry), and anti-Npl3 from C. Guthrie (University of California at San Francisco). Copurification of Rpb1, Ctk1, Hpr1, and Npl3 with Sro9 was calculated from four to five experiments relative to the amount of purified Sro9.
Indirect immunofluorescence
To analyze the localization of Sro9 by indirect immunofluorescence SRO9-TAP MEX67 and SRO9-TAP mex67-5 cells were cultivated at 30°C or shifted to 37°C for 2 h. Formaldehyde was added to a final concentration of 3.3% and cells were fixed for 90 min at 30°C, pelleted, washed, and spheroblasted by Zymolyase treatment. Spheroblasts were applied to polylysine-coated multiwell slides and fixed onto the slides by consecutive immersions of the slide in −80°C methanol (6 min) and −80°C acetone (30 s). After drying and rehydration, anti-proteinA (Sigma-Aldrich) was applied for 2 h, followed by 1 h incubation with Alexa488-goat–anti-rabbit antibody (Invitrogen). Nuclear DNA was stained with DAPI, and slides were analyzed using an Olympus BX60 fluorescence microscope (Olympus).
Chromatin immunoprecipitation
SRO9-TAP MEX67 and SRO9-TAP mex67-5 cells were cultivated in full medium containing either 2% glucose or 2% galactose at 30°C to an OD600 of 0.4. Cultures were divided and 40 mL each further cultivated for 2 h at 30°C or 37°C and cross-linked with formaldehyde for 25 and 20 min, respectively. ChIP was done as described (Sträßer et al. 2002) with some modifications. Briefly, cross-linked cells were lysed with an equal volume of glass beads by vortexing at full speed for 6 × 3 min with 3-min breaks on ice. Chromatin lysate corresponding to 14 A280 was used for immunoprecipitation with 15 μL of IgG-coupled Dynabeads for 3.5 h at 20°C. After washing and elution from the beads according to Kuras and Struhl (1999), the IP eluates as well as 0.08 A280 for input samples were treated with proteinase K for 2 h at 37°C and cross-links were reversed by overnight incubation at 65°C. DNA was purified over spin-columns (Macherey-Nagel). Quantitative PCR with input and IP samples was performed on an Applied Biosystems StepOnePlus cycler, using Applied Biosystems' Power SYBRGreen PCR Master Mix to assess Sro9 binding to the endogenous GAL1 gene. As a control, primers for a nontranscribed region (NTR) of chromosome V were used (sequences available on request). PCR efficiencies (E) were determined with standard curves. Sro9 enrichment over the NTR was calculated according to 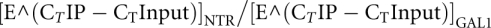 . Means were calculated from four to five independent experiments.
. Means were calculated from four to five independent experiments.
Genome-wide expression profiling
RNA isolation
Wild-type and Δsro9 cells were grown in SDC(-leu) at 30°C. RNA was extracted with phenol, purified with the Qiagen RNeasy MinElute Kit, and subjected to microarray analysis.
Microarray handling
Yeast RNA was hybridized to Affymetrix GeneChipYeast Genome 2.0 Arrays (Affymetrix) essentially as described by Dengl et al. (2009). To minimize errors, samples were processed in parallel and arrays were scanned the same day. Biological duplicate measurements were performed for the WT strain and the Δsro9 strain.
Gene expression data analysis
Raw signal intensities for each probe set in the .CEL files were analyzed using version 6.3 of the Partek Genomics Suite software (Partek, Inc.). Data were filtered by application of an expanded mask file that was based on the s_cerevisiae.msk file of Affymetrix, to mask the Schizosaccharomyces pombe probe sets, unspecific probe sets, and replicate probe sets of S. cerevisiae. The robust multiarray average (RMA) normalization method (Irizarry et al. 2003) was used for RMA background correction, quantile normalization, and median-polish probe set summarization. Expression values were transformed to log2 before statistical analysis. A sample intensity plot was calculated, showing that the data are normally distributed for all samples, as well as the homogeneity of variance (no outlier), which also indicated the great similarity of the biological replicates. Genes that were differentially expressed between WT and mutant strains were detected with one-way ANOVA, a statistical technique used to compare means of two or more samples implemented in the software package. A linear contrast was used to compare mutant samples with baseline WT samples. The recovered P-values of the comparisons were then corrected using a step-up false discovery rate value of 5% (Benjamini and Hochberg 1995). The resulting list of significantly expressed genes was filtered to include only genes that demonstrated twofold or greater up- or down-regulation. Microarray data were deposited in the ArrayExpress database with accession number E-MEXP-2678.
ACKNOWLEDGMENTS
We thank S. Wolin (Yale University) for plasmid pRS316-SRO9 and the Sro9 antibody, G. Dieci (University of Parma) for the Rpl6 antibody, D. Görlich (MPI for Biophysical Chemistry) for the Cbp80 antibody, C. Guthrie (University of California at San Francisco) for the Npl3 antibody, P. Cramer (Gene Center Munich) for help with gene expression analysis, and D. Martin (Gene Center Munich) for critical reading of the manuscript. This work was supported by grants from the European Research Council (ERC), Sonderforschungsbereich SFB646, the EMBO Young Investigator Programme, and the “Fonds der Chemischen Industrie.”
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2089110.
REFERENCES
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol 8: 711–716 [DOI] [PubMed] [Google Scholar]
- Alspaugh MA, Tan EM 1975. Antibodies to cellular antigens in Sjögren's syndrome. J Clin Invest 55: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky DA, Rose AB 2005. Plant gene expression in the age of systems biology: Integrating transcriptional and post-transcriptional events. Trends Plant Sci 10: 347–353 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Berben G, Dumont J, Gilliquet V, Bolle PA, Hilger F 1991. The YDp plasmids: A uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7: 475–477 [DOI] [PubMed] [Google Scholar]
- Bolger TA, Folkmann AW, Tran EJ, Wente SR 2008. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134: 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengl S, Mayer A, Sun M, Cramer P 2009. Structure and in vivo requirement of the yeast Spt6 SH2 domain. J Mol Biol 389: 211–225 [DOI] [PubMed] [Google Scholar]
- Erkmann JA, Kutay U 2004. Nuclear export of mRNA: From the site of transcription to the cytoplasm. Exp Cell Res 296: 12–20 [DOI] [PubMed] [Google Scholar]
- Fasken MB, Corbett AH 2005. Process or perish: Quality control in mRNA biogenesis. Nat Struct Mol Biol 12: 482–488 [DOI] [PubMed] [Google Scholar]
- Gilbert W, Guthrie C 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell 13: 201–212 [DOI] [PubMed] [Google Scholar]
- Gilbert W, Siebel CW, Guthrie C 2001. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA 7: 302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H 2007. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315: 646–649 [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA 1981. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: Further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Biol Cell 1: 1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Korneluk RG 2000. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: Role of La autoantigen in XIAP translation. Mol Cell Biol 20: 4648–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N 2005. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Iglesias N, Stutz F 2008. Regulation of mRNP dynamics along the export pathway. FEBS Lett 582: 1987–1996 [DOI] [PubMed] [Google Scholar]
- Imai J, Toh-e A, Matsui Y 1996. Genetic analysis of the Saccharomyces cerevisiae RHO3 gene, encoding a rho-type small GTPase, provides evidence for a role in bud formation. Genetics 142: 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jin L, Guzik BW, Bor YC, Rekosh D, Hammarskjold ML 2003. Tap and NXT promote translation of unspliced mRNA. Genes Dev 17: 3075–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M, Toh-e A, Matsui Y 1997. SRO9, a multicopy suppressor of the bud growth defect in the Saccharomyces cerevisiae rho3-deficient cells, shows strong genetic interactions with tropomyosin genes, suggesting its role in organization of the actin cytoskeleton. Genetics 147: 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399: 609–613 [DOI] [PubMed] [Google Scholar]
- Mattioli M, Reichlin M 1974. Heterogeneity of RNA protein antigens reactive with sera of patients with systemic lupus erythematosus. Description of a cytoplasmic nonribosomal antigen. Arthritis Rheum 17: 421–429 [DOI] [PubMed] [Google Scholar]
- McLaren RS, Caruccio N, Ross J 1997. Human La protein: A stabilizer of histone mRNA. Mol Biol Cell 17: 3028–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol 67: 3798–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ 2004. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes Dev 18: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen JR, Libri D, Jensen TH 2005. A link between transcription and mRNP quality in Saccharomyces cerevisiae. RNA Biol 2: 45–48 [DOI] [PubMed] [Google Scholar]
- Pannone BK, Xue D, Wolin SL 1998. A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J 17: 7442–7453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Hentze MW 2003. Starting the protein synthesis machine: Eukaryotic translation initiation. Bioessays 25: 1201–1211 [DOI] [PubMed] [Google Scholar]
- Prelich G 2002. RNA polymerase II carboxy-terminal domain kinases: Emerging clues to their function. Eukaryot Cell 1: 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Reed R, Cheng H 2005. TREX, SR proteins, and export of mRNA. Curr Opin Cell Biol 17: 269–273 [DOI] [PubMed] [Google Scholar]
- Röther S, Sträßer K 2007. The RNA polymerase II CTD kinase Ctk1 functions in translation elongation. Genes Dev 21: 1409–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röther S, Sträßer K 2009. mRNA export—an integrative component of gene expression. In Nuclear transport (ed. Kehlenbach R), pp. 1–2 Landes Bioscience, Austin, TX [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Lührmann R, Hurt E 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J 16: 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Wolin SL 1999. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol Biol Cell 10: 3849–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K, Hurt E 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 19: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K, Bassler J, Hurt E 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol 150: 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ 2004. The RNA polymerase II transcription cycle: Cycling through chromatin. Biochim Biophys Acta 1677: 64–73 [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Sonenberg N 1994. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol 68: 7001–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Li X, Sadhale PP, Miyao T, Woychik NA 2000. Multiple mechanisms of suppression circumvent transcription defects in an RNA polymerase mutant. Mol Cell Biol 20: 8124–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Wente SR 2006. Dynamic nuclear pore complexes: Life on the edge. Cell 125: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Tsukada M, Gallwitz D 1996. Isolation and characterization of SYS genes from yeast, multicopy suppressors of the functional loss of the transport GTPase Ypt6p. J Cell Sci 109: 2471–2481 [DOI] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol 8: 668–676 [DOI] [PubMed] [Google Scholar]
- Wiegand HL, Lu S, Cullen BR 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci 100: 11327–11332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H 2004. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol 24: 10479–10491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Cedervall T 2002. The La protein. Annu Rev Biochem 71: 375–403 [DOI] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL 1994. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: A yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol 14: 5412–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]