Abstract
RNA viruses recruit the host translational machinery by different mechanisms that depend partly on the structure of their genomes. In this regard, the plus-strand RNA genomes of several different pathogenic plant viruses do not contain traditional translation-stimulating elements, i.e., a 5′-cap structure and a 3′-poly(A) tail, and instead rely on a 3′-cap-independent translational enhancer (3′CITE) located in their 3′ untranslated regions (UTRs) for efficient synthesis of viral proteins. We investigated the structure and function of the I-shaped class of 3′CITE in tombusviruses—also present in aureusviruses and carmoviruses—using biochemical and molecular approaches and we determined that it adopts a complex higher-order RNA structure that facilitates translation by binding simultaneously to both eukaryotic initiation factor (eIF) 4F and the 5′ UTR of the viral genome. The specificity of 3′CITE binding to eIF4F is mediated, at least in part, through a direct interaction with its eIF4E subunit, whereas its association with the viral 5′ UTR relies on complementary RNA–RNA base-pairing. We show for the first time that this tripartite 5′ UTR/3′CITE/eIF4F complex forms in vitro in a translationally relevant environment and is required for recruitment of ribosomes to the 5′ end of the viral RNA genome by a mechanism that shares some fundamental features with cap-dependent translation. Notably, our results demonstrate that the 3′CITE facilitates the initiation step of translation and validate a molecular model that has been proposed to explain how several different classes of 3′CITE function. Moreover, the virus–host interplay defined in this study provides insights into natural host resistance mechanisms that have been linked to 3′CITE activity.
Keywords: cap-independent translation, 3′-cap-independent translational enhancer, RNA virus, RNA structure, RNA–protein interaction, plant virus
INTRODUCTION
Viruses are dependent upon their hosts for the translation of viral proteins. However, the host translational machinery is geared to function efficiently with cellular mRNAs, most of which contain terminal 5′-m7G cap structures and 3′-poly(A) tails that facilitate the process (Gingras et al. 1999). The 5′-cap is bound by the cap-binding protein, eukaryotic initiation factor (eIF) 4E, which is part of an eIF4F complex that includes eIF4G (Sonenberg 2008). Plants also possess eIF4A, an RNA helicase, but, in contrast to mammals (Hernández and Vazquez-Pianzola 2005), it is not present in isolated eIF4F complexes (Browning 1996). Additionally, plants contain a second isoform of eIF4F, eIFiso4F, which contains its own eIFiso4E and eIFiso4G subunits (Browning et al. 1992). Binding of the plant eIF4F or eIFiso4F to the 5′-cap structure mediates recruitment of the 43S ribosomal subunit to the 5′ untranslated region (UTR) via an eIF3-bridged interaction with the 4G subunits (Browning 1996); however, depending on structural properties of the message, the two eIF4F isoforms exhibit different levels of activity (Gallie and Browning 2001). The lured 43S subunit enters at the 5′ end of the message and then scans in a 5′-to-3′ direction until it encounters an initiation codon, at which point the large 60S subunit joins it to form an 80S initiation complex (Browning 1996; Gingras et al. 1999). The 4G subunits of the 5′-cap-bound eIF4Fs also interact with poly(A)-binding protein (PABP) associated with the 3′-poly(A) tail (Imataka et al. 1998), thereby circularizing the message (Wells et al. 1998) and leading to synergistic enhancement of translation (Gallie 1998).
The central role for the 5′-cap and 3′-poly(A) tail in conventional translation means that plus-strand RNA viruses that lack one or both of these terminal structures must use alternative strategies to recruit the host translational machinery (Dreher and Miller 2006). For example, animal picornaviruses contain only a poly(A) tail; however, RNA structures in their 5′ UTRs, called internal ribosome entry sites (IRESes), recruit ribosomes directly to the start codon, thereby obviating the need for a 5′-cap (Belsham 2009). Plant potyviruses also lack a 5′-cap but contain a 5′ covalently linked protein (termed Vpg) and an adjacent IRES, both of which bind to eIFs and facilitate translation (Gallie 2001; Khan et al. 2008). Conversely, animal flaviviruses have only a 5′-cap; however, PABP interacts with an internal sequence in their 3′ UTRs and this enhances translation (Polacek et al. 2009). Plant ilarviruses also do not contain poly(A) tails; however, their capsid protein binds to both a 3′-proximal RNA element and eIFs associated with their 5′-cap, thereby mimicking the translation-enhancing functions of PABP (Neeleman et al. 2001; Krab et al. 2005). Tobamo- and tymoviruses contain 5′-caps but possess tRNA-like structures for 3′ termini (Dreher 2009). Both viruses contain 3′ translational enhancers (TEs) that correspond, respectively, to an internal RNA element in the 3′ UTR (Leathers et al. 1993) and a terminal tRNA-like structure (Matsuda and Dreher 2004). These examples illustrate some of the diverse strategies used when one of the two conventional terminal mRNA structures is absent in a viral RNA genome.
Members of the large plus-strand RNA plant virus family Tombusviridae, as well as luteoviruses and umbraviruses, possess RNA genomes that are neither 5′-capped nor 3′-polyadenylated (Kneller et al. 2006). Instead, these viruses contain RNA elements in or near the 3′ UTRs of their genomes that facilitate the translation of viral proteins. These RNA structures have been termed 3′-cap-independent translational enhancers (3′CITEs) (Miller et al. 2007). Functional 3′CITEs have been reported for the aureusvirus Cucumber leafspot virus (CLSV) (Xu and White 2009); the carmoviruses Turnip crinkle virus (TCV) (Qu and Morris 2000; Stupina et al. 2008), Hibiscus chlorotic ringspot virus (HCRSV) (Koh et al. 2002), and Melon necrotic spot virus (MNSV) (Truniger et al. 2008); the dianthovirus Red clover necrotic mosaic virus (RCNMV) (Mizumoto et al. 2003); the necroviruses Tobacco necrosis virus (TNV) (Shen and Miller 2004; Meulewaeter et al. 2004) and Satellite tobacco necrosis virus (STNV) (Danthinne et al. 1993; Timmer et al. 1993; Meulewaeter et al. 1998); the panicovirus Panicum mosaic virus (PMV) (Batten et al. 2006); the tombusviruses Tomato bushy stunt virus (TBSV) (Wu and White 1999), Carnation Italian ringspot virus (CIRV) (Nicholson and White 2008), and Maize necrotic streak virus (MNeSV) (Scheets and Redinbaugh 2006); the luteovirus Barley yellow dwarf virus (BYDV) (Wang and Miller 1995; Wang et al. 1997); and the umbravirus Pea enation mosaic virus (PEMV) (Wang et al. 2009). Of these, detailed RNA structure/function analyses of 3′CITEs in BYDV (Guo et al. 2000), TBSV (Fabian and White 2004, 2006), TCV (Stupina et al. 2008; Zuo et al. 2010), and PEMV (Wang et al. 2009) have led to sound RNA secondary structure models for these elements. Based on these models, and the predicted RNA structures for the others, seven classes of 3′CITEs have been proposed (Miller et al. 2007). An additional class has been reported for the nepovirus Blackcurrant reversion virus (BRV); however, unlike those mentioned above, this uncapped virus contains a 3′-poly(A) tail (Karetnikov et al. 2006; Karetnikov and Lehto 2008). 3′CITEs are also present and active in viral subgenomic (sg) mRNAs (Miller et al. 2007), which are smaller 3′-coterminal viral messages transcribed during infections that template the translation of certain viral proteins (Miller and Koev 2000). However, some 3′CITE-containing viral sg RNAs are not translated and instead act as regulatory competitive inhibitors of translation for specific viral messages, as shown for BYDV (Shen et al. 2006) and RCNMV (Iwakawa et al. 2008). Consequently, 3′CITEs can mediate both positive and negative effects on viral translation.
The identification of protein factors that interact directly with 3′CITEs has been reported in a limited number of cases: STNV (Gazo et al. 2004), BYDV (Treder et al. 2008), PEMV (Wang et al. 2009), and TCV (Stupina et al. 2008). The 3′CITE of TCV binds directly to 60S and 80S ribosomal subunits and this binding correlates with translational activity in vivo (Stupina et al. 2008) and inhibition of RNA synthesis in vitro (Yuan et al. 2009). Alternatively, the 3′CITEs of STNV (Gazo et al. 2004), BYDV (Treder et al. 2008), and PEMV (Wang et al. 2009) interact directly with eIF4F. The 3′CITE-bound protein components are proposed to facilitate translation initiation at the 5′-end of the viral messages (Miller et al. 2007). However, for this to occur, some form of communication between the 5′ UTRs and the 3′CITEs would be required. This interaction could be mediated by a protein factor(s), as suggested for STNV (Gazo et al. 2004), or, alternatively, by an RNA–RNA base-pairing interaction between the 3′CITE and the 5′ UTR, as supported experimentally for BYDV (Guo et al. 2001; Rakotondrafara et al. 2006), TBSV (Fabian and White 2004, 2006), CIRV (Nicholson and White 2008), CLSV (Xu and White 2009), and BRV (Karetnikov et al. 2006; Karetnikov and Lehto 2008). For BYDV, a model incorporating the 3′CITE binding to both eIF4Fs and the 5′ UTR (i.e., a 5′ UTR/3′CITE/eIF interaction) has been proposed to explain how 3′CITE-bound eIFs could access the 5′-end of a viral message, where they likely mediate recruitment of the 43S ribosomal subunit (Guo et al. 2001; Rakotondrafara et al. 2006; Treder et al. 2008). This type of RNA/RNA/protein (RRP) model for 3′CITE function has also been suggested for several other virus genera (Gazo et al. 2004; Meulewaeter et al. 2004; Shen and Miller 2004; Karetnikov et al. 2006; Karetnikov and Lehto 2008; Truniger et al. 2008; Xu and White 2009), including tombusviruses (Fabian and White 2006). However, the factor(s) that presumably binds to tombusvirus 3′CITEs has yet to be identified. Moreover, the RRP model for 3′CITE function has not been validated in any virus, since critical components of it, namely, formation of the key tripartite RRP complex, eIF4F functioning from a distance via an RNA–RNA interaction, and the 3′CITE facilitating the initiation step of translation, have not been demonstrated experimentally.
Tombusviruses are among the best characterized viruses with respect to virus–host interactions (Nagy and Pogany 2006; Nagy 2008). CIRV and MNeSV are tombusviruses that encode five proteins in their comparably organized plus-strand RNA genomes (Fig. 1A, only the CIRV genome is shown; White and Nagy 2004). In its 3′ UTR, CIRV contains a branched, or Y-shaped, 3′CITE (Nicholson and White 2008), which is the class of 3′CITE present in most tombusviruses (Fig. 1A; Fabian and White 2004). In contrast, two tombusviruses, Cucumber Bulgarian virus (CBV) and MNeSV, possess a different class of 3′CITE (Fabian and White 2004; Scheets and Redinbaugh 2006; Miller et al. 2007), which we herein term an I-shaped 3′CITE, since its predicted structure is an extended RNA stem–loop (Scheets and Redinbaugh 2006). This I-shaped class of 3′CITE is also present in the carmovirus MNSV (Truniger et al. 2008), where it determines host range (Díaz et al. 2004) and is implicated as the viral determinant of nsv-mediated resistance in melon plants (Nieto et al. 2006; Truniger et al. 2008).
FIGURE 1.
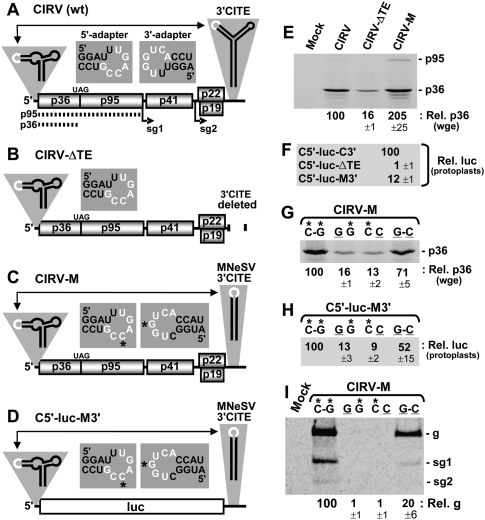
In vitro and in vivo analysis of Tombusvirus 3′CITE activity. (A–D) Schematic depictions of the RNA genomes of (A) CIRV, (B) CIRV-ΔTE, (C) CIRV-M, and (D) reporter mRNA C5′-luc-M3′. Encoded proteins are shown as boxes and noncoding regions are represented by the thick horizontal line. In CIRV and its derivatives, p36 and its read-through product p95 are translated directly from the viral genome as shown by the dotted lines in A. Sg1 and sg2 correspond to the initiation sites for sg mRNAs that are transcribed during infections. Relevant RNA structures in the 5′ and 3′ UTRs are depicted schematically above the message termini with the adapter segments denoted in white. Double-headed arrows represent 5′-3′ RNA–RNA interactions involving base-pairing between the adapter sequences in the terminal structures. The actual sequences of the 5′ and 3′ adapters in these terminal structures are shown in the center just above the messages. The nucleotides in white represent complementary residues, and those with asterisks in C and D correspond to the residues targeted for compensatory mutational analysis in G−I. The luc ORF corresponds to firefly luciferase and the message is not to scale. (E) SDS-PAGE analysis of proteins generated from 0.5 pmol of in vitro-generated transcripts of viral genome during a 1-h incubation at 25°C in wge in the presence of 35S-Met. The identities of the viral genomes are indicated above each lane (“mock” indicates that no RNA was added to the reaction). Relative p36 levels, quantified by radioanalytical scanning of the gel and normalized to WT CIRV (set to 100), are indicated below with standard errors of the mean (SEM) from three experiments. (F) Relative luciferase activity measured following a 5-h incubation of protoplasts transfected with the indicated viral mRNAs. Values (±SEM) are expressed as a percentage relative to that for C5′-luc-C3′ (set to 100). (G) SDS-PAGE analysis of p36 generated from CIRV-M and its compensatory mutants in wge to assess the importance of complementarity between the 5′ and 3′ adapters. Above each lane, the left nucleotide of the pair is in the 5′ adapter, while the right nucleotide is in the 3′ adapter. The asterisks indicate WT nucleotides and correspond to those shown in the adapter sequences in C. (H) Relative luciferase activity in protoplasts from C5′-luc-M3′ and its compensatory mutants with substitutions in the 5′ and 3′ adapters at the positions indicated by asterisks in (D). (I) Northern blot analysis detecting viral RNAs isolated 22 h posttransfection from protoplasts inoculated with CIRV-M and its compensatory mutants described in G. The positions of the viral genomic (g) and subgenomic (sg1 and sg2) RNAs are indicated to the right, while relative viral genome levels, calculated as a percentage (±SEM) of WT CIRV-M accumulation (set to 100), are shown below the blot.
Here we have investigated the structure and function of the I-shaped class of 3′CITE present in the tombusvirus MNeSV. Our results show that the 3′CITEMNeSV forms a complex RNA structure that binds to both the eIF4E and eIF4G subunits of eIF4F, with eIF4E contributing specificity to the interaction. The eIF4F-bound 3′CITEMNeSV also simultaneously interacts with the 5′ UTR via RNA–RNA base-pairing, and this tripartite complex mediates the initiation step of translation by a mechanism that shares some features with cap-dependent translation. The relevance of these findings is discussed in relation to the unique structure/function properties of this 3′CITE, what is known about other 3′CITEs, the proposed role for the I-shaped class 3′CITEs in host resistance, and an RRP model for 3′CITE function.
RESULTS
In vitro and in vivo activities of Tombusvirus 3′CITEs
The Y-shaped class 3′CITE in the CIRV RNA genome (Fig. 1A) was previously shown to mediate efficient in vitro production of p36 in a wheat germ extract (wge) translation system by a process requiring complementarity between the adapter sequences in its 3′CITE and 5′ UTR (Fig. 1A; Nicholson and White 2008). Indeed, when a portion of the 3′ UTR containing the 3′CITE was deleted in uncapped in vitro-generated transcripts of CIRV-ΔTE (Fig. 1B) the level of p36 accumulation, assessed by 35S-Met labeling and electrophoretic analysis, dropped significantly (approximately sixfold) compared with that for uncapped transcripts of the full-length CIRV genome (Fig. 1E). To determine if the I-shaped class 3′CITE from the related tombusvirus MNeSV could functionally replace the Y-shaped 3′CITE, the latter was substituted with the former in the CIRV genome, creating CIRV-M (Fig. 1C). Importantly, the wild-type (WT) 3′CITEMNeSV used for the exchange contained a 3′-adapter sequence that was complementary to the 5′ adapter in the CIRV 5′ UTR and, thus, could potentially mediate 5′ UTR/3′CITEMNeSV communication (Fig. 1C, insets). CIRV-M mediated efficient p36 accumulation in wge (Fig. 1E), and a luciferase reporter mRNA containing the 5′ and 3′ UTRs of CIRV-M, C5′-luc-M3′ (Fig. 1D), showed 12-fold enhanced activity in cucumber protoplasts when compared with C5′-luc-ΔTE lacking a 3′CITE (Fig. 1F). Since MNeSV naturally infects monocotyledonous plants (i.e., grasses), its 3′CITE is likely better adapted to the translational machinery of wheat, thereby accounting for its more robust activity in wge (Fig. 1E). Conversely, CIRV naturally infects dicotyledonous plants (i.e., broadleaves), and its 3′CITE was more active than the 3′CITEMNeSV in cucumber protoplasts (Fig. 1F). For both CIRV-M and C5′-luc-M3′, augmented protein production was dependent upon complementarity between the adapter loops in their 5′ UTRs and 3′CITEs, as shown through compensatory mutational analysis with mutants in which base-pairing potential was reduced (mutants GG, CC) and then restored (GC) (Fig. 1G,H). Additionally, CIRV-M was able to replicate in transfected protoplasts and exhibited a similar requirement for complementarity between its 5′- and 3′-adapter loops for viral genome accumulation (Fig. 1I). Collectively, these results demonstrate that the 3′CITECIRV can be functionally replaced by the 3′CITEMNeSV both in vitro and in vivo, thereby providing an appropriate viral context to study the 3′CITEMNeSV. This surrogate system was necessary because we were not able to obtain the full-length clone of MNeSV. The fact that this class of 3′CITE is found in three different genera of virus supports the concept that its activity is largely context independent.
Delineating a minimal 3′CITEMNeSV that functions in trans
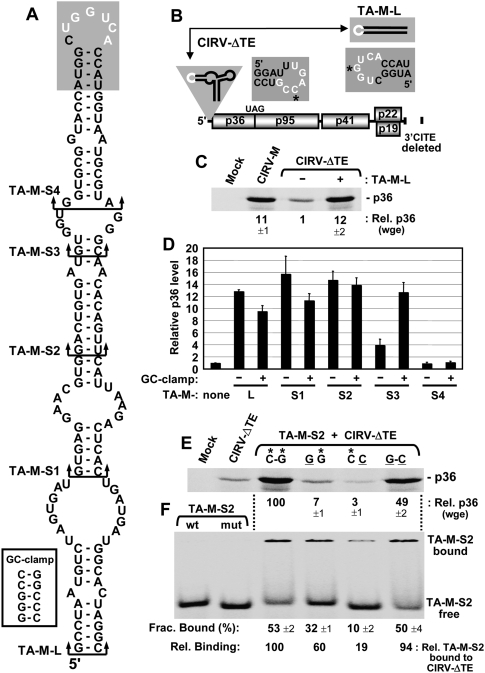
The high level of activity observed for the 3′CITEMNeSV in the CIRV genome prompted us to investigate whether this 3′CITE could function in trans (Fig. 2B). To address this, the MNeSV-derived RNA segment shown to function as a 3′CITE in cis in CIRV-M was used. Mfold analysis (Mathews et al. 1999; Zuker 2003) of this viral RNA, termed TA-M-L, predicted that it forms an extended stem–loop RNA structure (i.e., I-shaped), with its complementary adapter sequence located in its terminal loop (Fig. 2A). When CIRV-ΔTE was tested with a 10-fold molar excess of TA-M-L in wge, there was an ∼12-fold increase in the p36 level when compared with CIRV-ΔTE alone (Fig. 2C). A minimal functional trans-acting 3′CITE was then defined by removing increasingly larger segments from the bottom of the predicted TA-M-L structure, thereby generating TA-M-S1 through TA-M-S4 (Fig. 2A). Additionally, for each deletion mutant, a corresponding version was made that contained an added 5-base-pair-long GC-clamp at the base of the stem (Fig. 2A, inset). The rationale for this modification was that the clamp would aid in identifying functional regions near the bottom of the stem that required adjacent sequence-independent base-pairing. This strategy revealed that the reduced activity observed for TA-M-S3 could be fully restored by addition of the clamp (Fig. 2D). In contrast, no rescue was observed for TA-M-S4 when the clamp was appended to it (Fig. 2D). These results indicate that the lower activity for TA-M-S3 was related to a general requirement for a stable adjacent terminal stem and not due to the absence of specific sequence elements, such as the AA mismatch (Fig. 2A).
FIGURE 2.
Defining a minimal trans-acting I-shaped 3′CITE. (A) Mfold-predicted RNA secondary structure for the 3′CITE TA-M-L. Arrowed brackets indicate the bottom of truncated forms of TA-M-L, designated TA-M-S1 through -S4. The GC-clamp added to the bottom of the 3′CITEs is shown in the white box, while the 3′ adapter at the top is shaded with the complementary sequence depicted in white. (B) Schematic representation of the CIRV-ΔTE genome and TA-M-L. The residues in the 5′ and 3′ adapters targeted for compensatory mutational analysis in E and F are indicated by asterisks. (C) SDS-PAGE analysis of p36 levels from CIRV-M or CIRV-ΔTE in the absence (−) or presence (+) of a 10-fold molar excess of TA-M-L in wge. Values (±SEM) for p36 levels were normalized to that from CIRV-ΔTE alone (set to 1). (D) Relative p36 levels in wge from CIRV-ΔTE in the presence of TA-M-L or its truncated mutants depicted in (A) and specified below the graph, with (+) or without (−) the GC-clamp. Error bars indicate SEM. (E) SDS-PAGE analysis of p36 levels in wge reactions containing CIRV-ΔTE and TA-M-S2 harboring compensatory mutations as described in Fig. 1G. (F) RNA–RNA EMSA by nondenaturing PAGE assessing CIRV-ΔTE binding to TA-M-S2 using the WT and compensatory mutants analyzed in E. The left-most lanes show the positions of free WT (containing *G) and mutant (containing C) TA-M-S2 used in the EMSAs in the lanes to the right. Five nanomolars of 32P-labeled WT or mutant TA-M-S2 was incubated with 500 nM of WT or mutant CIRV-ΔTE for 30 min at 25°C. Complexes were then separated in a nondenaturing acrylamide gel, and the fraction of bound TA-M-S2 was quantified (±SEM) by radioanalytical scanning of the gel. Relative binding values were normalized to that for WT (C–G) binding (set to 100).
Additional studies were performed using TA-M-S2, which was the smallest element that contained only virus-derived RNA sequence and retained high levels of trans activity (Fig. 2A,D). TA-M-S2 enhancement of p36 production was shown by compensatory mutational analysis to be dependent on complementarity between its adapter sequence and that in the 5′ UTR of CIRV-ΔTE (Fig. 2E). Correspondingly, RNA–RNA EMSAs revealed lower levels of relative binding between TA-M-S2 and CIRV-ΔTE when the adapter sequences had reduced base-pairing potential (Fig. 2F). These latter findings indicate that optimal trans-mediated production of p36 requires an efficient RNA-based bimolecular interaction between the 3′CITEMNeSV and the mRNA template.
Solution structure analysis of the 3′CITE
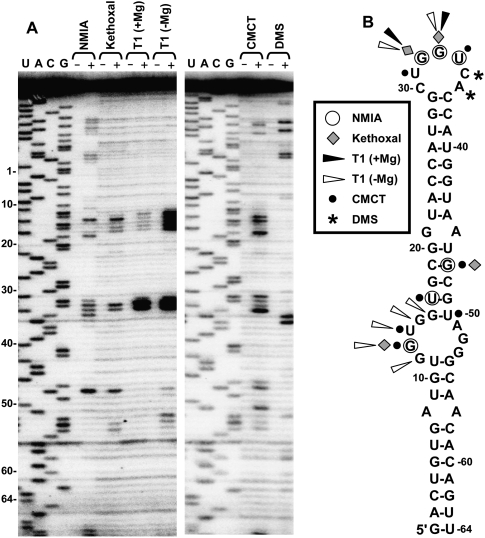
Having localized the functional region of the 3′CITEMNeSV, we next sought to gain an understanding of its structure by probing it with single-strand–specific modifying agents in solution (Fig. 3). Modified residues were mapped by primer extension (Fig. 3A), and their positions were plotted onto an mfold-predicted structure for TA-M-S2 (Fig. 3B). Initially, the 3′CITE was analyzed by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (Merino et al. 2005) using the chemical N-methylisatoic anhydride (NMIA), which modifies flexible and likely single-stranded nucleotides in a sequence-independent manner. As anticipated, some of the residues in the terminal loop were efficiently acylated (32GGU34); however, additional reactive sites mapped both within (G13) and above (U17, G47) the predicted internal loop (Fig. 3B). G13 and G47 were also preferentially modified by kethoxal and 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT), and U17 by CMCT (Fig. 3). In addition, CMCT reacted well with U14 in the predicted internal loop and with U50 located adjacent to this loop (Fig. 3). In the absence of Mg++, RNase T1 efficiently cleaved residues in the 5′ half of the predicted internal loop, but these residues were largely nonreactive in the presence of Mg++, which can facilitate the formation of higher-order RNA interactions (Fig. 3).
FIGURE 3.
Solution structure probing of the 3′CITE. (A) Denaturing acrylamide gel showing products of primer extension of TA-M-L after treatment with the enzyme RNase T1 (T1) plus or minus Mg++ or the chemicals N-methylisatoic anhydride (NMIA), kethoxal, 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfate (CMCT), or dimethyl sulfate (DMS). The (+) and (−) indicate treated and untreated RNA, respectively. Sequencing ladders generated with the same primer were run in the gels and relative coordinates for TA-M-S2 are indicted at the left. (B) Solution structure probing results were mapped onto the mfold-predicted secondary structure for TA-M-S2. Nucleotides in the structure that were reactive with the enzyme or chemicals are denoted by symbols that are defined in the box.
The absence of significant reactivity with all modifying agents in the majority of the RNA sequence is in general agreement with the extensively base-paired structure predicted by mfold. As well, efficient modification of most residues in the terminal loop and certain residues in the internal loop is also consistent with the predicted structure. However, the notable signals obtained for U17, G47, and U50 located in a predicted stem region above the internal loop and the general lack of reactivity of some of the residues within the internal loop bring into question the secondary structure predicted by mfold in this region. Based on the probing results it seems probable that these sequences in the central section engage in noncanonical interactions that may be Mg++ dependent.
Mutational analysis of the 3′CITE
We next utilized site-directed mutagenesis to delineate the functional elements in the 3′CITEMNeSV. These studies were performed using the trans system (i.e., TA-M-S2 and CIRV-ΔTE), since it offered several advantages. First, TA-M-S2 lacks additional flanking sequences that could differentially influence the structure and function of its mutant forms. Second, it is well known that RNA elements in the 3′ UTRs of mRNAs can influence message stability (Grzybowska et al. 2001); however, in the trans system, the 3′CITE is not part of the CIRV-ΔTE message, thus this concern is alleviated. Third, since identical CIRV-ΔTE mRNA is used in each assay, message stability and other mRNA-related effects are standardized. Fourth, the same functional or nonfunctional trans-acting 3′CITEs identified in the translation assay can also be used for binding studies to identify protein partners that correlate with function.
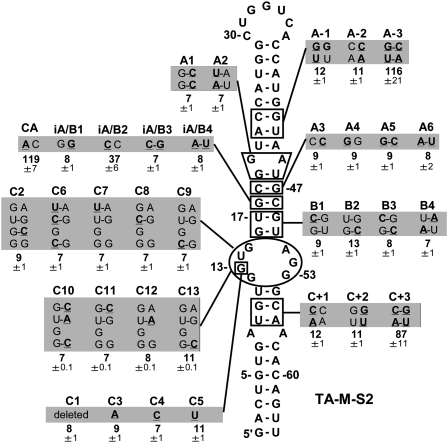
WT and mutant forms of TA-M-S2 were assessed for translational activity by incubating them with CIRV-ΔTE in wge and monitoring p36 production. Mutational analysis of TA-M-S2 revealed that base-pairing in the upper (mutants A−1 to A−3) and lower (C+1 to C+3) portion of the predicted structure was important for activity (Fig. 4). In contrast, no functional compensatory mutants were identified in four different sets of mutants (B1−B4, iA/B1−iA/B4, A3−A6, and A1−A2) that targeted the predicted stem region above the internal loop (Fig. 4). Similarly, all mutants were inactive when individual (C7−C9, C11−C13) or sets of residues (C6, C10) in the predicted internal loop were substituted with nucleotides that could form Watson–Crick base pairs with corresponding residues in the opposing strand (Fig. 4). Additionally, deletion (C1), substitution (C3−C5), or potential pairing (C2) of G13 (which was consistently reactive in probing studies; Fig. 3) resulted in a nonfunctional 3′CITE. Thus, other than iA/B2 (a CC mismatch) that showed ∼40% activity and CA (an AC mismatch) exhibiting slightly higher than WT activity, all modifications in the central region of the structure led to inactive 3′CITEs. The extremely strict requirement for particular residues in this central section indicates a complex functional structure that is highly sensitive to perturbation.
FIGURE 4.
Mutational analysis of the 3′CITE. TA-M-S2 mutants are presented with names at the top, and nucleotide modifications are underlined and in bold in the shaded boxes. The relative p36 levels (±SEM) in wge from coincubations of CIRV-ΔTE and a 10-fold molar excess of the different TA-M-S2 mutants are shown below (normalized to a value of 100 for WT TA-M-S2).
eIF4F facilitates 3′CITE activity
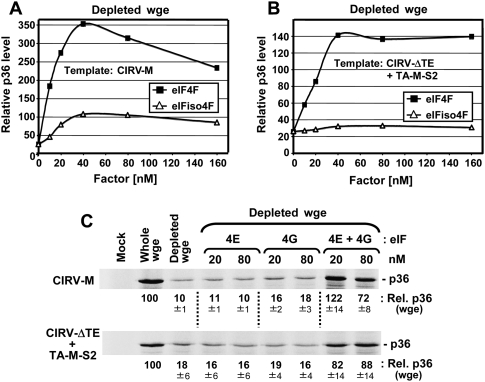
To investigate the possible involvement of eIFs in 3′CITEMNeSV function, we employed wge depleted of cap-associated eIFs (e.g., eIF4F and eIFiso4F), which was prepared using cap structure-conjugated sepharose resin to sequester the factors (Gallie 2007). Depleted wge was then supplemented with either eIF4F or eIFiso4F to determine the effect on p36 accumulation levels in translation assays. For CIRV-M, the reduced level of p36 in depleted wge was more efficiently restored by the addition of purified recombinant eIF4F versus eIFiso4F (Fig. 5A), and similar recovery profiles were observed in the trans system with CIRV-ΔTE and TA-M-S2 (Fig. 5B). The more efficient restoration of 3′CITE activity by eIF4F prompted us to examine the role of its individual subunits in the process. In depleted wge, the addition of either purified recombinant eIF4E or eIF4G did not notably affect the level of p36 production in either the cis or trans system (Fig. 5C). On the contrary, when both subunits were added together there was substantial recovery of p36 levels (Fig. 5C). This latter finding suggests that the individual subunits used in these assays were also biologically active and indicates that 3′CITE activity likely requires both subunits of eIF4F.
FIGURE 5.
3′CITE activity in eIF-depleted wheat germ extract supplemented with eIFs. (A) and (B) Representative data set from in vitro translation assays using eIF-depleted wge. The depleted extract was supplemented with the indicated amount of purified recombinant eIF4F or eIFiso4F before translation assays were performed. Relative p36 levels for (A) CIRV-M or (B) CIRV-ΔTE plus a 10-fold molar excess of TA-M-S2 are plotted and are relative to those for whole (i.e., nondepleted) wge (set at 100). (C) SDS-PAGE analysis of relative p36 levels (±SEM) from CIRV-M (upper panel) or CIRV-ΔTE plus TA-M-S2 (lower panel) in eIF-depleted wge supplemented with the indicated amounts of eIF4E, eIF4G, or both.
eIFs bind to the 3′CITE
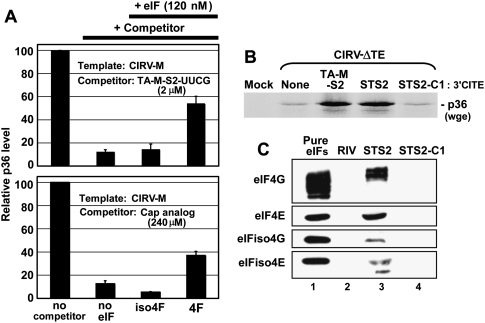
If the 3′CITEMNeSV interacts with eIFs, then it should be possible to inhibit, via sequestration of the eIFs, the cis function of a 3′CITE within an mRNA by adding free 3′CITE that is not able to interact with the mRNA. To test this idea, a TA-M-S2-based 3′CITE, TA-M-S2-UUCG, containing a modified noncomplementary adapter sequence (i.e., its terminal loop sequence was changed to UUCG) was used as a competitor against CIRV-M in a wge assay (Fig. 6A, upper graph). The addition of 2 μM of TA-M-S2-UUCG (a 400-fold molar excess) caused an ∼10-fold decrease in p36 production, suggesting that the free 3′CITE is able to sequester a factor(s) important for activity. Supplementing the extract with eIFiso4F did not notably change p36 accumulation levels, whereas the addition of eIF4F led to an approximately fourfold recovery (Fig. 6A, upper graph). When a cap analog was used as a competitor, an ∼120-fold greater concentration was required (i.e., 240 μM) to attain a similar level of inhibition (Fig. 6A, lower graph). An approximately threefold recovery of the cap-inhibited p36 level was mediated by supplementation with eIF4F, but not with eIFiso4F (Fig. 6A, lower graph). These results are consistent with our earlier data (Fig. 5) indicating a more prominent role for eIF4F in 3′CITE-mediated activity and implicating this eIF as a 3′CITE-binding factor. If the latter notion is correct, binding of the 3′CITE and cap structure to eIF4F may be mutually exclusive, since cap analog inhibited 3′CITE activity.
FIGURE 6.
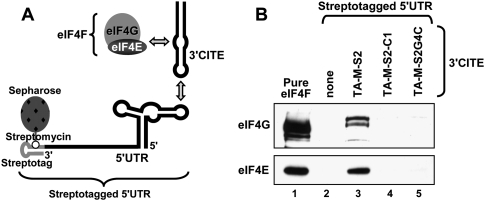
Interaction of the 3′CITE with eIFs. (A) Analysis of p36 levels in wge containing 5 nM CIRV-M and 2 μM TA-M-S2-UUCG (upper panel) or 5nM CIRV-M and 240 μM cap analog m7GTP (lower panel). The reactions containing the competitors were supplemented with 120 nM of the eIF indicated below the graphs. Relative p36 levels (±SEM) were normalized to that in the uninhibited reaction (set to 100). (B) SDS-PAGE analysis of p36 levels from CIRV-ΔTE in the presence of TA-M-S2, its streptotagged counterpart STS2 or streptotagged mutant STS2-C1. (C) Western blots showing the detection of eIFs (indicated on the left) in eluates from columns with different streptotagged RNAs (indicated above the blots) immobilized on streptomycin-conjugated sepharose and incubated with wge. The negative control, RIV, is a streptotagged version of the 3′-terminal 82 nt of TBSV. After immobilization of the tagged viral RNAs in the column, wge was added to the columns and incubated for 1 h at 4°C, followed by washing. Streptotagged RNAs and bound proteins were eluted with free streptomycin, separated by SDS-PAGE, and analyzed by Western blotting using antisera against individual eIF subunits. The eIF standards in lane 1 contained 1.8 pmol of each subunit.
To determine if the 3′CITE could associate with eIF4F in wge, TA-M-S2 was engineered with a StreptoTag RNA aptamer (Windbichler and Schroeder 2006) at its 3′ end and used for RNA-based affinity chromatography. Importantly, the tagged TA-M-S2, termed STS2, was still functional and able to trans-activate p36 production from CIRV-ΔTE in wge (Fig. 6B). In contrast, the tagged mutant TA-M-S2-C1 (with the critical G13 deleted), termed STS2-C1, was not able to enhance p36 accumulation (Fig. 6B). Streptotagged RNAs bind efficiently to streptomycin-conjugated sepharose (Windbichler and Schroeder 2006). Accordingly, STS2 was immobilized in a column containing this matrix, wge was then added, the column was subsequently washed, and STS2 and bound proteins were eluted with free streptomycin. Western blot analysis revealed that STS2 was able to form complexes that included the 4E and 4G subunits of both eIF4F and eIFiso4F (Fig. 6C, lane 3). Conversely, the tagged nonfunctional mutant STS2-C1 or a tagged 3′-terminal replication-related tombusvirus RNA element, termed RIV (White and Nagy 2004), was not able to form corresponding complexes (Fig. 6C, lanes 2,4). When compared with equimolar amounts of protein standards for the different eIF subunits, the relative levels of STS2 binding to eIF4F components were consistently greater than those of eIFiso4F (Fig. 6C). This is significant as there is at least five times more eIFiso4F in wge than eIF4F (Browning et al. 1990), which, in turn, suggests preferential association of the 3′CITE with eIF4F.
eIF4F and subunit interactions with the 3′CITE
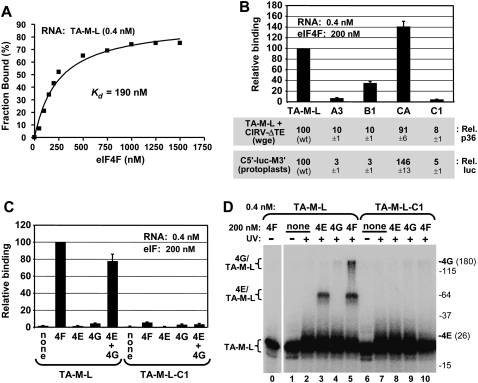
Having established that the 3′CITEMNeSV is able to form a complex with eIF4F in wge, we next wanted to determine if this interaction was direct. Preliminary studies (data not shown) revealed that binding of the 3′CITE to purified recombinant factors was most efficient with the longer TA-M-L (Fig. 2); thus this RNA and its modified versions were used in these analyses. In filter-binding assays, eIF4F bound to TA-M-L with an apparent Kd of ∼190 nM (Fig. 7A), and similar values were obtained using an EMSA-based assay (data not shown). The interaction was specific, as the TA-M-L-based mutants A3, B1, and C1 bound poorly (refer to Fig. 4 for modifications), while mutant TA-M-L-CA bound well (Fig. 7B, graph). Importantly, the relative eIF4F binding efficiencies of the TA-M-L-based mutants correlated well with their ability to enhance protein accumulation when tested in wge assays in trans with CIRV-ΔTE or in protoplast assays in cis with C5′-luc-M3′ and mutants containing the equivalent 3′CITE modifications (Fig. 7B, bottom panels). The binding activity of TA-M-L with the subunits of eIF4F was also tested. Individually, neither eIF4E nor eIF4G bound well to TA-M-L (Fig. 7C). In contrast, when both subunits were present, binding to TA-M-L, but not to mutant TA-M-L-C1, was efficient (Fig. 7C). Consequently, 3′CITE binding (Fig. 7C) and its ability to enhance protein accumulation (Fig. 5C) require both subunits of eIF4F.
FIGURE 7.
Binding of the 3′CITE to eIF4F and its subunits. (A) Representative binding curve of the TA-M-L/eIF4F complex determined by filter-binding analysis and used to determine the Kd. 32P-labeled TA-M-L (0.4 nM) was incubated with the indicated amounts of eIF4F for 60 min at 25°C, and the fraction of bound RNA was determined by quantifying the radioactivity of protein–RNA complexes retained on a nitrocellulose membrane. (B) eIF4F binding and translation activities of TA-M-L and mutants. Relative binding levels of TA-M-L and its mutants to eIF4F were determined by filter-binding assays at the indicated concentrations (upper panel). Relative binding values were normalized to binding of WT TA-M-L to eIF4F (set to 100). TA-M-L and its mutants were assayed for translation activity via p36 levels in wge in coincubations with CIRV-ΔTE (middle panel), and corresponding 3′CITE mutants in C5′-luc-M3′ were assayed for translational activity via relative luciferase levels in protoplasts (lower panel). (C) Binding of TA-M-L and mutant TA-M-L-C1 to purified recombinant eIF4E, eIF4G, or both, was assayed by filter-binding at the concentrations indicated. Relative binding values were normalized to binding of WT TA-M-L to eIF4F (set to 100). (D) UV cross-linking analysis of TA-M-L and TA-M-L-C1 interactions with eIF4F and its subunits. Radiolabeled RNA (0.4 nM) was incubated with 200 nM eIF4F or its individual subunits for 60 min at 25°C and then irradiated with 254 nm light for 15 min, followed by separation by 10% SDS-PAGE. The positions of molecular weight markers in the gel are indicated to the right (in kDa) along with the locations of free eIF4E (26 kDa) and eIF4G (180 kDa) in the gel. The positions of bands corresponding to free RNA (TA-M-L) or RNA cross-linked with eIF4G (4G/TA-M-L) or eIF4E (4E/TA-M-L) are indicated to the left of the gel.
Next, UV cross-linking studies were performed to identify the subunit(s) of eIF4F in close contact with the 3′CITE. In UV-treated mixtures containing radiolabeled TA-M-L and unlabeled eIF4G, a faint shifted eIF4G-dependent complex, termed 4G/TA-M-L, was observed, whereas comparable mixtures with TA-M-L and eIF4E generated a smaller but far more abundant complex, termed 4E/TA-M-L (Fig. 7D, lanes 3,4). Interestingly, equilibrium binding assays at these same concentrations did not indicate any notable association between eIF4E and TA-M-L (Fig. 7C), yet cross-linking revealed that eIF4E on its own does indeed interact with TA-M-L and that this contact is specific; since mutant TA-M-L-C1 did not generate a corresponding complex (Fig. 7D, lane 8). When TA-M-L and eIF4F incubations were analyzed, both 4E/TA-M-L and 4G/TA-M-L complexes were observed (Fig. 7D, lane 5), which were also specific (Fig. 7D, lane 10). Compared with results of the individual subunits, there was a significant increase in the 4G/TA-M-L complex and a modest increase in the 4E/TA-M-L complex (Fig. 7D, cf. lane 4 and lane 5, and lane 3 and lane 5, respectively). This suggests that when present in the eIF4F heterodimer, the subunits, particularly eIF4G, are able to more efficiently contact the 3′CITE. Taken together, these results indicate that 3′CITE interacts directly with both subunits in eIF4F.
Recapitulating 3′CITE activity with a cap structure
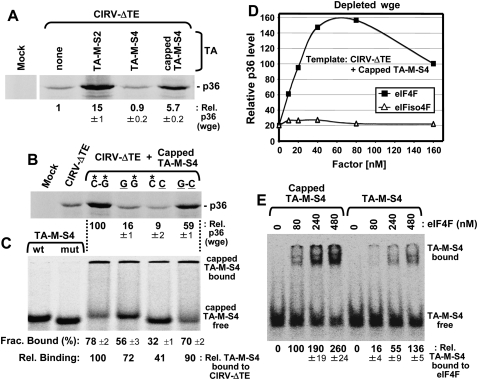
At a fundamental level, the binding of the 3′CITE to eIF4F is akin to the binding of the 5′-cap structure to eIF4F. Therefore, if the 3′CITE is in fact functioning to recruit eIF4F to the mRNA, it should be possible to reproduce its mode of action, including the mechanism of delivery via an RNA–RNA interaction, using a 5′-cap structure. To this end, we assessed whether a nonfunctional 3′CITE that retained its terminal loop adapter sequence, TA-M-S4 (Fig. 2A,D), could be converted to a functional trans-acting enhancer by adding a 5′-cap to it. Including a 5′-cap structure in TA-M-S4 resulted in an approximately sixfold increase in p36 levels from CIRV-ΔTE in wge (Fig. 8A). This enhanced activity was dependent on complementarity between adapter sequences in the capped TA-M-S4 and CIRV-ΔTE, as shown by the analysis of compensatory mutants in wge (Fig. 8B) and corresponding RNA–RNA EMSAs (Fig. 8C). Like for the functional TA-M-S2 3′CITE (Fig. 5B), capped TA-M-S4 activity was efficiently restored in depleted wge by the addition of eIF4F, but not eIFiso4F (Fig. 8D). Also, in RNA–protein EMSAs, capped TA-M-S4 bound to eIF4F up to approximately sixfold more efficiently (at 80 nM eIF4F) than its uncapped counterpart, TA-M-S4 (Fig. 8E). Collectively, these data validate the concept that eIF4F can be functionally delivered to an mRNA via an RNA–RNA interaction and establish that a 5′-capped RNA can stimulate translation in trans via base-pairing.
FIGURE 8.
Recapitulating 3′CITE activity with a cap structure. (A) SDS-PAGE analysis of relative p36 levels from wge reactions containing CIRV-ΔTE in the presence of a 10-fold molar excess of TA-M-S2, TA-M-S4, or capped TA-M-S4. (B) SDS-PAGE analysis of compensatory mutants for capped TA-M-S4 and CIRV-ΔTE in wge reactions analyzed by SDS-PAGE. The base pair between the 5′ adapter in CIRV-ΔTE and 3′ adapter in TA-M-S4 was disrupted (GG, CC) and then restored (GC) as described in Figure 2E. (C) RNA–RNA EMSA by nondenaturing PAGE was used to assess CIRV-ΔTE binding with radiolabeled TA-M-S4 using the WT and compensatory mutants analyzed in (B) and described in Figure 2F. (D) Assessment of p36 levels from coincubations of CIRV-ΔTE and capped TA-M-S4 in eIF-depleted wge supplemented with the indicated amounts of eIF4F or eIFiso4F. Values are relative to those for whole (i.e., nondepleted) wge (set at 100). (E) RNA–protein EMSA by nondenaturing 4% PAGE to determine relative binding levels of radiolabeled capped or uncapped TA-M-S4 (0.4 nM) to the indicated amounts of eIF4F.
Formation of a 5′ UTR/3′CITE/eIF4F complex in wge
If initiation of translation is mediated by the 3′CITE-bound eIF4F associating with the 5′ UTR, then it should be possible to identify a complex containing all of these components (Fig. 9A). To this end, we utilized streptotagged WT CIRV 5′ UTR that contained an additional 158 viral nucleotides downstream from the start codon (Fig. 9A). This extra viral segment was required to provide a downstream priming site for corresponding toe-printing analyses described in the next section. The tagged 5′ UTR segment that was immobilized on streptomycin-conjugated sepharose was not able to bind to eIF4F subunits when wge was applied to the column (Fig. 9B, lane 2). However, when the 3′CITE TA-M-S2 was applied to the 5′ UTR-immobilized column prior to the application of wge, both eIF4E and eIF4G were detected in eluates (Fig. 9B, lane 3). Conversely, when the identical protocol was performed with TA-M-S2-C1 (containing an inactive 3′CITE with G13 deleted) or TA-M-S2G4C (containing a mutated adapter sequence resulting in a CC mismatch between the adapters), no notable eIF4F subunit binding was detected (Fig. 9B, lanes 4,5). Since complex formation was dependent on the same 3′CITE properties already shown to be necessary for direct interactions between the bipartite subsets of the complex, i.e., 3′CITE/eIF4F (Figs. 6C, 7, mutant C1) and 5′ UTR/3′CITE (Fig. 2E,F, mutant CC), it follows that the formation of the tripartite 5′ UTR/3′CITE/eIF4F complex was also mediated by these same direct contacts.
FIGURE 9.
Formation of a 5′ UTR/3′ CITE/eIF4F complex in wge. (A) Cartoon depicting the formation of a 5′ UTR/3′CITE/eIF4F complex in a streptomycin-conjugated sepharose column. (B) Western blot analysis showing detection of eIF4F subunits in eluates from affinity columns. The 5′ UTR segment was immobilized in columns via its streptotag and TA-M-S2, its mutant TA-M-S2-C1 (with the critical G13 deleted), or mutant TA-M-S2G4C (containing the same 3′-adapter substitution as mutant CC in Fig. 2E,F) or no 3′CITE were subsequently applied. Following a 30-min incubation, the column was washed. Next, wge was added to the columns and allowed to incubate for 60 min, followed by washing. Eluates from the columns, released by adding streptomycin, were analyzed by Western blotting with antisera to eIF4G and eIF4E. The eIF standards in lane 1 contained 0.6 pmol of each subunit.
The 3′CITE mediates the initiation step of translation
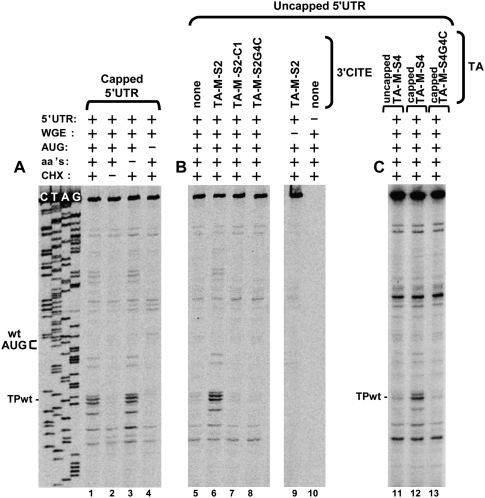
To investigate whether the 3′CITE facilitates the initiation step of translation, we carried out toe-printing analysis of translation reactions in wge. Using this approach, the position and relative abundance of ribosomes stalled at a start codon in an mRNA (due to treatment with cycloheximide) can be determined by primer extension (Kozak 1998). This assay utilized the same CIRV 5′ UTR segment used in the preceding section. This 5′-terminal viral RNA sequence contained the WT start codon (position 78 for the A in AUG) and 158 downstream nucleotides, which provided an appropriately located priming site for 32P-labeled oligonucleotide annealing and extension. Initially, we analyzed a 5′-capped version of this 5′ UTR segment to validate the utility of the system. When incubated in wge in the presence of cycloheximide (which stalls the 80S ribosomal initiation complexes at the start codon by inhibiting elongation) prominent primer extension products, i.e., toe-prints, were observed 16–18 nucleotides (nt) downstream from the WT start codon that correspond to the leading edges of stalled ribosomes (i.e., TPwt) (Fig. 10A, lane 1; Kozak 1998). The same toe-prints were also present in reactions when no exogenous amino acids were added; thus they likely do not represent stalled elongating ribosomes (Fig. 10A, lane 3). Moreover, in other controls, when either cycloheximide was excluded from the reaction or the start codon was mutated to AAG, no corresponding toe-prints were observed (Fig. 10A, lanes 2,4, respectively).
FIGURE 10.
Toe-printing analysis of 3′CITE-dependent ribosome loading on viral 5′ UTRs. Denaturing PAGE showing the products of primer extension generated by reverse transcription of the CIRV 5′ UTR segment. 5′ UTR segments were incubated in translationally competent wge containing cycloheximide (CHX) for 20 min at 25°C. A radiolabeled oligonucleotide, complementary to a downstream region in the 5′ UTR fragment (i.e., 84-nt 3′ to the WT start codon), was then added to the reaction and extended by reverse transcriptase. Primer extension products corresponding to toe-prints from stalled ribosomes were then assessed under different conditions, which are indicated at the top of the gels. Toe-print analysis of (A) the capped 5′ UTR segment, (B) the uncapped 5′ UTR segment in the presence of WT or mutant TA-M-S2, and (C) the uncapped 5′ UTR segment in the presence of capped and uncapped TA-M-S4. The positions of the WT start codon (WT AUG) and its corresponding toe-print (TPwt) are indicated.
Having confirmed the functionality of the system with a 5′-capped message, we next assessed an uncapped version of the 5′ UTR segment, which is the version present in tombusviruses. On its own, the uncapped 5′ UTR did not efficiently load ribosomes at the initiation codon (Fig. 10B, lane 5), consistent with its inability to interact with eIF4Fs (Fig. 9). However in the presence of TA-M-S2, ribosomes were loaded on the uncapped 5′ UTR as efficiently as they were loaded on the capped 5′ UTR (Fig. 10, cf. lanes 6 and 1), correlating with the ability of uncapped 5′ UTR to form a TA-M-S2–dependent complex that includes eIF4F (Fig. 9). Also, as observed for tripartite complex formation (Fig. 9), ribosome loading was dependent on the 3′CITE being able to bind to eIF4F (Fig. 10B, lane 7) and complementarity between the adapters in the 3′CITE and 5′ UTR (Fig. 10B, lane 8). In all cases, similar levels of the 5′ UTRs were observed (as monitored by the primer extension products at the top of the gel), indicating comparable message stabilities. The positive correlation between tripartite complex formation (Fig. 9) and ribosome loading (Fig. 10B) establishes an important role for the 3′CITE in facilitating the initiation step of translation via assembly of the 5′ UTR/3′CITE/eIF4F complex. The ability to promote initiation via this mechanism was further supported by the similar results observed when the cap structure was provided to the 5′ UTR segment in trans via capped TA-M-S4 (Fig. 10C, lanes 11,12). Here again, disruption of the RNA–RNA interaction by mutating the adapter in TA-M-S4 inhibited ribosome loading (Fig. 10C, lane 13).
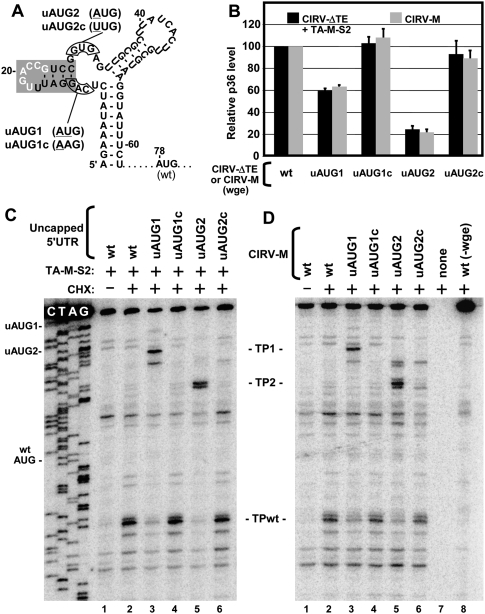
We also investigated whether the 3′CITE delivered ribosomes directly to the start codon (i.e., IRES-like) or to the 5′ end of the message from which scanning in a 3′ direction could occur (i.e., cap-dependent-like). To address this, we assessed start site selection in messages, CIRV-ΔTE and CIRV-M, that included an additional upstream start codon (uAUG), which was out of frame with the WT AUG at position 78 (Fig. 11A). The introduction of individual uAUGs at two different positions in CIRV-ΔTE, i.e., mutants CIRV-ΔTE-uAUG1 and -uAUG2 (Fig. 11A), resulted in decreased levels of production of p36 from the WT AUG in wge when TA-M-S2 was provided to these messages in trans (Fig. 11B, black bars). Conversely, modifications at the same upstream positions in control mutants CIRV-ΔTE-uAUG1c and -uAUG2c that did not introduce start codons (Fig. 11A) had little effect on p36 levels (Fig. 11B, black bars). In corresponding toe-printing assays with 5′ UTR templates that contained the same uAUG or control modifications, only the uAUG-containing mutants generated new toe-prints (i.e., TP1 and TP2), indicating ribosome recognition of uAUG1 and uAUG2 (Fig. 11C, lanes 3–6). Additionally, in both cases, there were concurrent reductions of ribosomes at the WT AUGs (i.e., TPwt) (Fig. 11C, lanes 3,5). Comparable results were also obtained when CIRV-M (containing a cis-acting 3′CITE), and its corresponding uAUG and control mutants were tested for p36 production in wge (Fig. 11B, gray bars) and analyzed by toe-printing (Fig. 11D). The efficient recognition of the uAUGs with consequential decreases in WT AUG recognition and protein production suggests that 3′CITEs, in cis or in trans, facilitate 43S ribosome subunit loading in wge at or near the 5′ end of the mRNA, followed by scanning in a net 5′-to-3′ direction.
FIGURE 11.
Translation and toe-print analyses of viral 5′ UTRs containing upstream AUGs. (A) RNA secondary structure of the 5′-terminal segment of the CIRV 5′ UTR showing the positions where upstream AUGs (in uAUG1 and uAUG2) or control mutations (in uAUG1c and uAUG2c) were introduced relative to the WT AUG. Mutated nucleotides are underlined and the shaded stem–loop corresponds to the 5′ adapter. (B) Translation assay monitoring p36 levels from WT, uAUG, and control mutants of CIRV-M (gray bars) or CIRV-ΔTE plus TA-M-S2 (black bars) in wge. (C) Toe-printing analysis of WT, uAUG, and control mutant 5′ UTR segments in the presence of TA-M-S2 in wge. (D) Toe-printing analysis of WT, uAUG, and control mutants of CIRV-M in wge. The toe-prints corresponding to uAUG1 and uAUG2 are labeled TP1 and TP2 and are located 17 and 18-20 nt, respectively, downstream from their cognate uAUGs.
DISCUSSION
A variety of plus-strand RNA viruses use 3′CITEs to mediate efficient production of their encoded viral proteins (Miller et al. 2007). Here we investigated one class of 3′CITE that is present in tombusviruses, aureusviruses, and carmoviruses and provide structural and functional data that illuminate how this type of RNA element mediates efficient protein translation.
Structural features of the 3′CITE
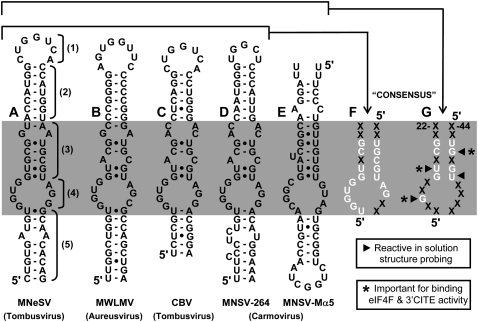
When compared with members of the other known classes of 3′CITE (Miller et al. 2007), the I-shaped 3′CITEMNeSV appears to be the smallest. The 64-nt-long TA-M-S2 mediated efficient translation in trans in wge, as did the smaller GC-clamped TA-M-S3, which is 56-nt long (Fig. 2). The latter length is about half the size of the 3′CITEs defined in wge for BYDV (∼109 nt) (Wang et al. 1997), PEMV (∼102 nt) (Wang et al. 2009), and STNV (∼93 nt) (Gazo et al. 2004). At the secondary structure level, the 3′CITEMNeSV is most similar to the 3′CITESTNV, which is also predicted to form an extended SL structure (Gazo et al. 2004; van Lipzig et al. 2002). However, other than this general likeness, there is no other obvious structural relationship. Similarly, the 3′CITEMNeSV does not share apparent structural features with those in TBSV, BYDV, PEMV, TCV, or BRV, all of which possess branched RNA structures (Miller et al. 2007). Instead, the 3′CITEMNeSV resembles other members of its I-shaped class, such as the 3′CITEs in the monocot-infecting aureusvirus Maize white line mosaic virus (MWLMV) (Carstens 2010) and also the dicot-infecting tombusvirus CBV (Scheets and Redinbaugh 2006) and carmovirus MNSV-264 (Díaz et al. 2004; Fig. 12A–D). Accordingly, it appears that the I-shaped class of 3′CITE is able to function in nature in both grasses and broadleaves and is used by viruses in at least three different genera of Tombusviridae.
FIGURE 12.
Sequence and structure comparisons of I-shaped class 3′CITEs. (A–E) Mfold-predicted RNA secondary structures of I-shaped class 3′CITEs from different viruses, as indicated below. The numbered brackets in (A) correspond to the different regions (region 1–5) assigned to the 3′CITEMNeSV and the shaded band (i.e., region 3 and 4) highlights the central core that is very sensitive to mutation and has components conserved in all members of this class of 3′CITE, as revealed by the consensus in (F) that is based on A−D and the more limited consensus in (G) that is based on A−E. “X” represents an undefined nucleotide and the arrowheads and asterisks are defined in the accompanying boxes.
Mutational analyses allowed us to define functional elements in the 3′CITEMNeSV (Figs. 2, 4) that can be divided into five regions (Fig. 12A, region 1–5). (1) A terminal loop that base-pairs with the complementary adapter in the 5′ UTR. (2) A sequence-independent upper stem that supports formation of the terminal loop and possibly structural elements below it. (3) An mfold-predicted stem between the upper stem and internal loop that contains sequence-specific determinants of function. (4) An mfold-predicted internal loop that also contains sequence-specific determinants. (5) A sequence-independent lower stem that presumably supports formation of the internal loop structure. The sequence-independent nature of the upper and lower helices, regions 2 and 5, respectively, is further supported by nucleotide mono- and covariations in the other 3′CITEs that maintain these stems (Fig. 12A−D), while sequence-specific regions 3 and 4 define a central core (Fig. 12, shaded region) in which many residues are conserved (Fig. 12A−D) and form a nucleotide consensus (Fig. 12F). Adding the 3′CITEMNSV-Mα5 to the comparison, which is orientated antiparallel to the others (Fig. 12E), defines a more limited consensus (Fig. 12G). Four of the residues in this latter consensus were highly reactive to chemical probing (Fig. 12G, arrowheads), and, of these, three mapped to the predicted stem in region 3. This, combined with the inability to generate functional compensatory mutants in region 3 and its tolerance of mismatches at one position (Fig. 4, mutants AC and CC; note, the AC mismatch is naturally present in MWLMV, CBV, and MNSV-264 in Fig. 12), suggests that this region forms a structure that is more complex than the “standard” helix predicted by mfold. This notion is further supported by the mismatches or the bulge in region 3 in other 3′CITEs (Fig. 12B–E). The structure and/or interactions formed by the predicted internal loop in region 4 are equally elusive, with the notable exception of the invariant and critical G13 (Fig. 12G). Despite such divergence, the central cores in CBV (a tombusvirus), MNSV-264 (a carmovirus) and Johnsongrass chlorotic stripe mosaic virus (JCSMV) (an aureusvirus) (Carstens 2010) are identical (Fig. 12C,D; data not shown for JCSMV), suggesting that this particular core in three different genera is compatible with both grass and broadleaf hosts. Deciphering the complex higher-order structure(s) of the central core will likely require methods that are able to resolve atomic arrangements.
Binding partners of the 3′CITE
The 3′CITEMNeSV has two different types of binding partners, RNA and protein. Our results indicate that direct physical contact between the 3′CITE and 5′ UTR, whether in cis or in trans, is required for efficient protein production. Similar 5′–3′ RNA–RNA interactions are also required in tombusviruses that contain the Y-shaped class 3′CITE (i.e., TBSV and CIRV) (Fabian and White 2004, 2006; Nicholson and White 2008), in BYDV containing the BTE class 3′CITE (Guo et al. 2001; Rakotondrafara et al. 2006), and those in BRV (Karetnikov et al. 2006; Karetnikov and Lehto 2008). For its protein partner(s), several lines of experimental evidence indicate that the 3′CITEMNeSV requires eIF4F for optimal activity and directly interacts with this protein complex (Figs. 5–7). The isoform of eIFiso4F also facilitated 3′CITE activity, but at a reduced level, and its subunits interacted less efficiently with the 3′CITE. Compared with eIFiso4F, eIF4F is known to mediate more efficient translation of mRNAs that are uncapped and have stable secondary structures in their 5′ UTRs (Gallie and Browning 2001). Both of these features are characteristic of the comparably structured 5′ UTRs in tombusvirus genomes (Ray et al. 2003); thus, it is possible that the stronger specific binding to eIF4F was preferentially selected for over time to best deal with these structural properties. Regardless, the 3′CITE-binding eIFs identified in this study are the first host factors shown to be required for tombusvirus translation.
Individually, neither eIF4E nor eIF4G was able to efficiently interact with the 3′CITEMNeSV (Fig. 7C). This is in contrast with the 3′CITEBYDV that binds well to eIF4G (Treder et al. 2008) and the 3′CITEPEMV that binds efficiently to eIF4E (Wang et al. 2009). The strict requirement for the eIF4F complex by the 3′CITEMNeSV for proficient binding (Kd ≈190 nM) is instead more akin to the 3′CITESTNV, which exhibits much higher affinity for eIF4F (Kd ≈17 − 30 nM) than for eIF4E (Kd ≈780 nM) (Gazo et al. 2004). For these 3′CITEs, the greater affinity for eIF4F is not unexpected, since this complex is presumably the major form of eIF4E and eIF4G in cells and, thus, would represent the prevalent structure to which the 3′CITEs would adapt. This concept also seems to apply to the 3′CITEBYDV, which binds with approximately fivefold greater affinity to eIF4F (Kd ≈37 nM) than to eIF4G (Kd ≈177 nM) (Treder et al. 2008). Nonetheless, there do appear to be exceptions to this trend, as the 3′CITEPEMV binds to eIF4E (Kd ≈58 nM) and eIF4F (Kd ≈48 nM) with essentially identical affinities (Wang et al. 2009).
The 3′CITEMNeSV is able to form a weak but specific interaction with eIF4E alone, which was only detectable by UV-induced covalent linkage of the transiently formed complex (Fig. 7D). Specific binding to eIF4E is also observed for the 3′CITEPEMV (Wang et al. 2009) and the 3′CITESTNV (Gazo et al. 2004). However, there are no obvious structural similarities between any of these 3′CITEs, and they bind with quite different affinities (3′CITEPEMV > 3′CITESTNV > 3′CITEMNeSV); thus, their specific interactions with eIF4E likely occur via distinct contacts. Formation of the eIF4F complex also facilitated an interaction between eIF4G and the 3′CITEMNeSV (Fig. 7D). This could have been mediated by allosteric effects within the eIF4F complex that caused structural rearrangements in eIF4G allowing for a more productive, and possibly specific, interaction with this 3′CITE.
Within the eIF4F complex, interactions with both eIF4E and eIF4G contribute to 3′CITEMNeSV binding. The 3′CITE interaction with eIF4G could be mediated by the latter's RNA-binding domain (Kim et al. 1999). As for eIF4E, it is possible that the 3′CITE interaction involves the cap-binding pocket, as cap analog did inhibit 3′CITE activity (Fig. 6A). Indeed, at least two essential and solvent accessible guanylates (G13 and G47) in the consensus sequence (Fig. 12G) could potentially act as cap mimics by interacting via stacking forces with the cap-binding pocket. A similar possibility has also been entertained for the binding of the 3′CITEPEMV to eIF4E (Wang et al. 2009). Conversely, the 3′CITEMNeSV inhibition observed in the presence of free cap analog could have been indirect and caused by allosteric effects in eIF4E (and/or eIF4G) induced by cap binding (Rutkowska-Wlodarczyk et al. 2008). For MNSV, amino acid 228 in melon eIF4E is a determinant of 3′CITEMNSV activity (Nieto et al. 2006; Truniger et al. 2008); therefore this residue is a potential candidate for direct, or allosterically mediated, binding to 3′CITEMNSV and potentially to 3′CITEMNeSV.
In terms of binding sites in the 3′CITEMNeSV, the region responsible for binding to eIF4F likely includes the essential central core that is also found in other 3′CITEs (Fig. 12). Indeed, three nucleotides in this region (G13, U17, and G47) are both important for eIF4F binding and critical for efficient 3′CITE activity in vitro and in vivo (Fig. 12G, asterisks). Additionally, all three are solvent accessible and present in the minimal consensus (Fig. 12G). Moreover, since the G13 is also essential for binding to either eIF4E alone or the eIF4F complex (Fig. 7D), this nucleotide represents a key determinant of eIF4F binding via specific contact with eIF4E. Thus, although 3′CITEMNeSV binds to both eIF4E and eIF4G in the eIF4F complex, eIF4E plays a key role in the specificity of this interaction.
The 3′CITE and nsv-mediated resistance
Translation initiation factors have been identified as key determinants of recessive resistance in important crop plants (Robaglia and Caranta 2006) and 3′CITEs have been linked to this phenomenon (Nieto et al. 2006). The 3′CITEMNeSV belongs to the same I-shaped class of 3′CITE as those in the carmoviruses MNSV-264 and MNSV-Mα5 (Fig. 12D,E, note that these two structures are oriented antiparallel to each other). Interestingly, MNSV-Mα5 is unable to infect melon cultivars exhibiting nsv-mediated resistance, which is conferred by a point mutation, H228L, in eIF4E (Nieto et al. 2006). Conversely, MNSV-264, which has a modified 3′CITE, is able to infect nsv-cultivars (Nieto et al. 2006). Based on these and other observations a model was proposed where binding compatibility/incompatibility between melon eIF4Es and MNSV 3′CITEs specifies efficient/impaired translation of viral proteins, which, in turn, confers plant susceptibility/resistance to infection (Truniger et al. 2008). However, a direct interaction between these host and viral components has not yet been demonstrated; thus this model remains to be validated. The findings of our study are consistent with the proposed nsv-based resistance model in that (1) eIF4E interacts specifically and directly with 3′CITEMNeSV, which is the same I-shaped class of 3′CITE as those in MNSV; (2) the 3′CITE–eIF4E interaction correlates positively with 3′CITE activity mediating efficient protein production in vitro and in vivo; and (3) previous sequence exchanges mapped the virulent and avirulent determinants to RNA segments (Truniger et al. 2008) that correspond to the critical and conserved central core delineated by our mutational and comparative sequence analyses (Fig. 12F,G).
Mechanism of 3′CITE-mediated translational enhancement
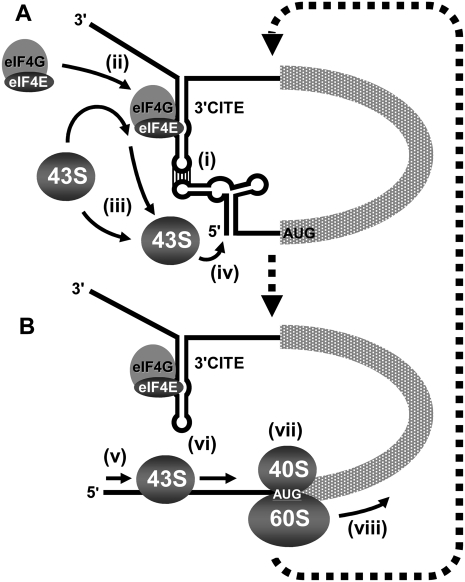
The prevailing model for 3′CITE function in STNV (Gazo et al. 2004), BYDV (Treder et al. 2008), MNSV (Truniger et al. 2008), and tombusviruses (Fabian and White 2006) is enhancement of translation via delivery of 3′CITE-bound eIFs to the 5′ UTR via a 5′-3′ RNA–RNA interaction (i.e., an RNA/RNA/protein [RRP] interaction). The 5′-localized eIF4F is envisioned to recruit the 43S ribosomal subunit and facilitate assembly of the 80S complex at the start codon (i.e., the initiation step of translation); however, direct evidence for this is lacking. Our data support key components of this RRP model including the 3′CITE interacting with the 5′ UTR (via complementary base-pairing) [Fig. 13A, (i)] and binding (mediated by its core region) to both eIF4E and eIF4G subunits of eIF4F (facilitated by specificity determinants in eIF4E) [Fig. 13A, (ii)]. Interestingly, the 3′CITE/5′ UTR interaction also functions in trans, thus base-pairing between the two adapter sequences appears sufficient to facilitate activity and there is no strict requirement for covalent tethering of the 3′CITE to the message that it acts on. The trans activity also suggests that ribosome recycling, from the 3′ to the 5′ end of the message, is not required for cap-independent translation stimulation by the 3′CITE.
FIGURE 13.
The RRP model for 3′CITE-mediated translational enhancement. Model depicting how the 3′CITEMNeSV is proposed to enhance viral translation. The viral genome is shown as a simplified linear mRNA that includes relevant higher order RNA structural elements in its 5′ and 3′ UTRs. (A) Initial steps in the proposed mechanism are shown, including (i) the 3′CITE interacting with the 5′ UTR via RNA–RNA base-pairing; (ii) 3′CITE simultaneously binding to eIF4F, thereby bringing it into proximity of the 5′ end of the genome; (iii) the 43S ribosomal subunit being recruited to the 5′ UTR via 3′CITE-bound eIF4F; and (iv) the 43S ribosome entering at or near the 5′ terminus. (B) Subsequent steps include (v) scanning of the 43S in a net 5′-3′ direction; (vi) concurrent disruption of the 3′CITE/5′ UTR interaction; (vii) recognition of the start codon and joining of the 60S subunit to form the 80S initiation complex; and (viii) the ribosome transitioning into the elongation stage of translation. (dotted arrows) Presumed cyclical nature of the process that would mediate repeated loading and initiation of ribosomes. See the text for additional details.
The modularity of the trans system allowed us to demonstrate that an RRP interaction corresponding to the 5′ UTR/3′CITE/eIF4F complex can form in a translationally relevant milieu [Fig. 13A, (i), (ii)]. This is a critical step in the RRP model as the 5′ UTR, on its own, cannot interact with eIF4F (Fig. 9); thus the tripartite complex is required to localize eIF4F to the 5′ end of the message. Conversely, it is clear that the 3′CITE can interact with eIF4F independent of the 5′ UTR and that the 5′ UTR/3′CITE interaction does not require eIF4F to occur. Accordingly, there are two possible pathways for assembly of the 5′ UTR/3′CITE/eIF4F complex. The 3′CITE is normally located in cis with respect to the message, and this connected arrangement would facilitate the RNA–RNA interaction forming efficiently after release of genomes from virions or viral replication complexes. Thus, based on constant proximity, it is possible that the intramolecular RNA–RNA interaction precedes eIF4F binding. The feasibility of this assembly pathway is supported by formation of the tripartite complex under conditions where the RNA–RNA interaction was allowed to occur before the RNA–protein interaction (Fig. 9). The presence of the 3′CITE within the viral message would also promote rapid reformation (after disruption) of the RNA–RNA constituents of the RRP complex. This is likely relevant as toe-printing analyses (Fig. 11) indicated that 3′CITE-mediated ribosome loading [Fig. 13A, (iii)] occurs near the very 5′ end of the 5′ UTR [Fig. 13A, (iv)] followed by scanning in a net 3′ direction [Fig. 13B, (v)], a process that would presumably disrupt the RNA–RNA interaction [Fig. 13B, (vi)], as also proposed for TBSV (Fabian and White 2006) and BYDV (Rakotondrafara et al. 2006). Also, since the 3′CITE is able to interact with eIF4F independent of its interaction with the 5′ UTR, it is possible that it maintains this association with eIF4F if displaced from the 5′ UTR by ribosomes [Fig. 13B, (vi)]. This retention of a subset of the RRP complex (i.e., 3′CITE/eIF4F) would presumably accelerate reformation of the tripartite complex and potentially allow bound eIF4F to service multiple rounds of initiation (Fig. 13, thick dashed arrows).
The RRP model for 3′CITE function centers on enhancement of the initiation step of translation via efficient ribosome loading, but no direct evidence for this has been reported (Miller et al. 2007). It is well established that eIF4F facilitates this step when recruited directly to the 5′ UTR by the 5′-cap structure (Dreher and Miller 2006). However, it was unclear whether eIF4F could do so if recruited indirectly to the 5′ UTR via an RNA–RNA interaction. The recapitulation of 3′CITE activity, using the cap structure as a ligand to recruit eIF4F to the 5′ UTR by the same indirect means as the 3′CITE (Fig. 8), validates the proposed RNA-based localization mechanism for eIF4F [Fig. 13A, (i)]. Additionally, the recognized role of the cap structure in mediating initiation of translation, combined with its ability to facilitate protein production in trans with the same preferential dependence on eIF4F as the 3′CITE, supports the 3′CITE operating by a functionally equivalent mechanism in which eIF4F in the viral RRP complex, via its eIF4G subunit, recruits the 43S subunit to the 5′ UTR [Fig. 13A, (iii), (iv)]. This 3′CITE-mediated enhancement of the initiation step of translation was substantiated by the results of toe-printing analyses of uncapped messages that were similar to those observed for the cap functioning in cis or in trans (Fig. 10). The recruited 43S subunits are envisioned to scan in a net 3′ direction [Fig. 13B, (v)] followed by assembly with 60S complexes into 80S initiation complexes at the first start codon that they encounter [Fig. 13B, (vii)]; events supported by the preferential assembly of ribosomes at more 5′-proximal AUGs (Fig. 11). These initiation complexes are translationally competent and subsequently enter the elongation step of translation [Fig. 13B, (viii)], as corroborated by the positive correlations observed between the efficiency of complex formation and corresponding level of protein production (Fig. 11). Thus, there is now compelling evidence for an RRP model in which the 3′CITEMNeSV mediates the initiation step of translation by a mechanism that shares several fundamental features with cap-dependent translation. Collectively, our results provide new insights into this distinctive viral mechanism of hijacking the host translational machinery, and we anticipate that aspects of the RRP model for the 3′CITEMNeSV will be relevant to other viruses that utilize this kind of RNA element.
MATERIALS AND METHODS
Plasmid construction
The full-length infectious cDNA clone of CIRV has been described previously (Burgyan et al. 1996). All mutants were constructed by standard recombinant DNA cloning techniques and PCR-based oligonucleotide-mediated mutagenesis (Sambrook et al. 1989). PCR-derived regions that were introduced into constructs were sequenced completely to ensure that only the desired modifications were present. For small modifications, the relevant changes in mutants are shown in the figures in this study. CIRV-M containing the MNeSV 3′CITE was made by substituting nucleotides 4410–4673 of the CIRV genome with nucleotides 3892–4003 from MNeSV (GenBank accession number NC_007729). C5′-luc-C3′ and C5′-luc-M3′ were generated by inserting the luciferase ORF from reporter plasmid pSP-luc+ (Promega) into pUC19-based constructs containing the 5′ UTR of CIRV and the 3′ UTR of CIRV or CIRV-M. TA-M-L and TA-M-S1, -S2, -S3, and -S4 were created by PCR amplification of the corresponding segments of the MNeSV 3′CITE present in CIRV-M and cloning into pUC19. Constructs containing a streptotag were generated by PCR amplification of the desired viral sequence with a reverse primer containing the streptomycin aptamer described previously (Dangerfield et al. 2006), which has the RNA sequence 5′-GGAUCGCAUUUGGACUUCUGCCCGCAAGGGCACCACGGUCGGAUCC-3′. The TA-M-S2 mutants analyzed in Figure 4 were transcribed directly from synthetic DNA oligonucleotides (Sigma) containing a T7 promoter.
Computer-aided analysis of RNA
The genome sequences of MNeSV (NC_007729), MWLMV (NC_009533), CBV (NC_004725), MNSV-264 (AY330700) and -Mα5 (AY122286), and JCSMV (NC_005287) were obtained from the NCBI GenBank database. RNA secondary structures were predicted at 37°C using Mfold version 3.2 (Mathews et al. 1999; Zuker 2003).
In vitro transcription
Uncapped transcripts were generated from linearized DNA plasmids or synthetic DNA oligonucleotides with a T7 promoter as described previously (White and Morris 1994) using AmpliScribe T7-Flash transcription kits (Epicenter Technologies). Capped transcripts were made using AmpliCap T7 High-Yield Message Maker kits (Epicenter Technologies). The capped and poly(A) tailed Renilla luciferase mRNA (used as a standard for transfection efficiency in luciferase translation assays in protoplasts) was transcribed by an mScript mRNA Production System kit (Epicenter Technologies) from the plasmid pRL-CMV (Promega) after linearization with XbaI.
Protein expression and purification
Non-affinity-tagged versions of eIF4F, eIFiso4F, eIF4G, and eIF4E were expressed and purified as described previously (Mayberry et al. 2007). The purity of the individual recombinant factors, eIF4E and eIF4G, was similar to the purity of the individual subunits of recombinant eIF4F prepared as described by Mayberry et al. 2007.
In vitro translation
Translation of in vitro transcripts in nuclease-treated wheat germ extract (wge; Promega) was carried out as described previously in the presence of 35S-Met (Nicholson and White 2008). Translation products were separated by SDS-PAGE in 12% polyacrylamide gels and quantified by radioanalytical scanning using a PharosFX Plus Molecular Imager (Bio-Rad) and QuantityOne Software (Bio-Rad). For reactions including free 3′CITE in trans, 5 pmol of the 3′CITE RNA was added to the reaction containing 0.5 pmol of cognate mRNA. These RNAs were not preannealed to each other prior to the assay, and a 10-fold excess of free 3′CITE over cognate mRNA was used, as this yielded similar accumulation levels as when the 3′CITE was present in cis. Cap-binding factor-depleted wge used for translation assays was generated by incubation with 7-methyl GTP-sepharose 4B (GE Healthcare) and supplemented with purified factors as described previously (Gallie 2007).
RNA and protein analysis in protoplasts
Cucumber cotyledon protoplasts were prepared and transfected using polyethylene glycol as described previously (White and Morris 1994). For Northern blot analysis, viral genomic RNAs (3 μg) were transfected into ∼3 × 105 protoplasts and incubated for 22 h at 22°C under constant light. Total nucleic acid was then extracted, separated in a nondenaturing 1% agarose gel, and transferred to membrane and hybridized with a 32P-ATP-labeled probe complementary to the 3′ end of the CIRV genome. Bands on membranes were quantified by radioanalytical scanning. For translation assays, 3 × 105 protoplasts were transfected with 2 pmol of the firefly luciferase RNAs (experimental message) and 0.2 pmol of the Renilla luciferase mRNA (control message). Following a 5-h incubation at 22°C under constant light luciferase assays were performed using a Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase levels were determined relative to those for the Renilla luciferase control mRNA, and Firefly/Renilla luciferase ratios were then expressed as percentages relative to the ratio for C5′-luc-C3′, which was set to 100.
RNA–RNA gel shift assay
RNA–RNA EMSAs were carried out essentially as described previously (Fabian and White 2006), with some modifications. Briefly, uncapped 3′CITE RNAs were 32P-end labeled using polynucleotide kinase (for TA-M-S2) or internally labeled by incorporation of α32P-UTP into the RNA during transcription (for capped TA-M-S4). Labeled 3′CITE (5 nM) was incubated for 30 min at 25°C with 500 nM CIRV-ΔTE in RNA binding buffer (5 mM HEPES at pH 7.8, 100 mM KCl, 6 mM MgCl2, 3.8% glycerol) in a total volume of 4 μL. Samples were then cooled on ice for 10 min, mixed with loading buffer (50% glycerol, 50% RNA binding buffer), and separated at 4°C in a nondenaturing 5% polyacrylamide gel containing 6 mM MgCl2 using 1 × TBE running buffer containing 6 mM MgCl2. After running, gels were dried and the bands were quantified by radioanalytical scanning.
Solution structure mapping
Prior to chemical or enzyme treatment, transcript of TA-M-L was folded by incubating for 3 min at 95°C, immediately followed by a 2-min incubation on ice and a 20-min incubation at 25°C in the following reaction-specific buffers: N-methylisatoic anhydride (NMIA) folding buffer (100 mM HEPES at pH 8.0, 5 mM MgCl2, 100 mM NaCl); RNase T1 folding buffer (10 mM Tris at pH 7.5, 100 mM NaCl, with or without 2.1 mM MgCl2); kethoxal folding buffer (50 mM sodium borate at pH 7.5, 2.1 mM MgCl2, 100 mM KCl); dimethyl sulfate (DMS) folding buffer (50 mM sodium cacodylate at pH 7.5, 2.1 mM MgCl2, 100 mM KCl); and 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfate (CMCT) folding buffer (50 mM sodium borate at pH 8.0, 2.1 mM MgCl2, 100 mM KCl). Chemical or enzyme treatments were then carried out on aliquots of RNA in buffer, under the following conditions: 24 mM NMIA for 45 min at 37°C; 10−4 U/μL RNase T1 for 20 min at 37°C; 14 mM kethoxal for 25 min at 37°C; 0.3% DMS for 10 min at 37°C; and 3.8 mg/mL CMCT for 15 min at 37°C. Treated RNAs were then ethanol precipitated and used as templates in primer extension reactions as described previously (Wilkinson et al. 2006). Products of primer extension were separated in 7% polyacrylamide-7 M urea sequencing gels and visualized by radioanalytical scanning of the dried gel.
StreptoTag affinity chromatography
Streptomycin-conjugated sepharose matrix was prepared as described previously (Windbichler and Schroeder 2006). Before conducting assays with wheat germ extract, it was confirmed that all streptotagged RNAs bound to the matrix with similar efficiencies by taking aliquots of RNA before loading and after elution from the column, and comparing their levels on ethidium bromide-stained agarose gels. Streptotag affinity chromatography was carried out as described (Windbichler and Schroeder 2006) with some modifications. Streptotagged RNA (1.5 nmol) was refolded in 1 mL of column buffer (50 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 250 mM NaCl) by incubating for 5 min at 80°C, 5 min at 50°C, and 10 min at 37°C, and then placing on ice. The streptotagged RNA was applied to a column containing 3 mL of column buffer-equilibrated streptomycin-conjugated sepharose matrix, and incubated at room temperature for 30 min, followed by washing three times with 3 mL column buffer. Four hundred microliters of wge (Promega) mixed with 1.2 mL column buffer was then applied to the column at 4°C in two batches of 800 μL, with a 30-min incubation at 4°C after application of each batch. For the samples in Figure 9 only, an additional step was added prior to addition of wge. Specifically, 7.5 nmol of 3′CITE RNA was added to the column in 1.5 mL RNA binding buffer, followed by washing with 3 mL of RNA binding buffer, and subsequently, wge was added and incubated in the column as described above. After incubation with wge, the column was washed five times at 4°C with 3 mL column buffer. Next, the RNA–protein complexes were eluted at room temperature with 1 mM streptomycin, and concentrated using a centrifugal filter (Amicon). Western blotting of the samples was performed using antisera to eIF4E, eIF4G, eIFiso4E, and eIFiso4G, and visualized using a goat anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase (Sigma) and ECL Plus Western blotting detection reagents (Amersham).
Filter-binding assays
A filter-binding assay was used, essentially as described previously (Treder et al. 2008). Briefly, 0.4 nM α-32P UTP internally labeled RNA was mixed with eIFs in 25 μL of 4F binding buffer (28 mM HEPES-KOH at pH 7.6, 57 mM KCl, 2.3 mM MgAc2, 0.114 mg/mL BSA, 85 μg/mL yeast RNA, 2.8% glycerol, 2.4 mM DTT) and incubated at 25°C for 1 h. Seven microliters of each sample was vacuum-filtered through nitrocellulose membrane and N+ nylon membrane (Amersham) on a microfiltration apparatus (Bio-Rad, Bio-Dot SF). Both membranes were dried and analyzed by radioanalytical scanning. The fraction of RNA bound to protein was determined by comparing the amount of RNA–protein complexes retained on the nitrocellulose membrane with the amount of free RNA retained on the underlying nylon membrane. Complete binding was not achievable, probably because a fraction of the RNA was inactive due to misfolding. The apparent Kd was thus defined as half the maximal binding observed. Data were fitted using GraphPad PRISM software and Kd values were calculated from the best-fit curves generated by nonlinear regression analysis.
RNA–protein EMSAs
RNA internally labeled with α-32P UTP was incubated with protein factors in 4F binding buffer as described for filter-binding assays (0.4 nM RNA; 80, 240, or 480 nM protein). Ten microliters of the binding mixture was mixed with 3 μL loading buffer (50% glycerol, 0.075% bromophenol blue) separated in a nondenaturing 4% polyacrylamide gel (80:1 acrylamide:bis) at 4°C, dried, and analyzed by radioanalytical scanning.
UV cross-linking assays
RNA internally labeled with α-32P UTP was incubated with protein factors in 4F binding buffer as described for filter-binding assays (0.4 nM RNA; 200 nM protein). Fifteen microliters of the binding mixture was transferred to a Petri dish on ice and exposed to 254 nm UV light at a distance of 8 cm for 15 min (Spectrolline UV Crosslinker: Select XLE-1000). Samples were then mixed with loading buffer (1×X concentration: 45 mM Tris-HCl at pH 6.8, 1% SDS, 10% glycerol, 147 mM β-mercaptoethanol, 0.05% bromophenol blue), and separated by 10% SDS-PAGE (29:1 acrylamide:bis), dried, and analyzed by radioanalytical scanning.
Toe-printing assays
Toe-printing assays were adapted from those described previously (Sachs et al. 2002; Matsuda and Dreher 2006). Wheat germ extract reactions were prepared as they were for use in in vitro translation assays, except that a complete amino acid mix was used and no 35S-Met was included. Additionally, reactions were supplemented with 2.8 mM cycloheximide. RNAs were incubated in the wge in a total volume of 10 μL for 20 min at 25°C. One microliter of the translation reaction was then diluted in 20 μL of primer extension buffer (1 × SSIII FS buffer [Invitrogen], 20 mM dithiothreitol, 5 mM MgCl2, 0.4 mM deoxynucleotide triphosphates, 1 U/μL RNaseOUT [Invitrogen], 2.8 mM cycloheximide) and incubated for 2 min at 55°C. One picomole of 32P-end-labeled primer (complementary to the positions 164–184 nt of the CIRV genome) was then added to the sample and allowed to anneal at 37°C for 2 min. SuperScript III reverse transcriptase (Invitrogen) (2.5 μL) was then added, and the reaction was incubated at 52°C for 10 min. The reactions were stopped by mixing with 35 μL acid stop solution (4:25 [v/v] mixture of 1 M Tris-HCl and stop dye [85% formamide, 0.5 × TBE, 50 mM EDTA at pH 8.0, bromophenol blue, xylene cyanol]) and then separated in a 6% polyacrylamide/7 M urea sequencing gel alongside a sequencing ladder generated using the same primer. The sequencing ladder and toe-prints were visualized by radioanalytical scanning of the dried gel.
ACKNOWLEDGMENTS
We thank Karen Browning for purified eIFs and corresponding antisera and for reviewing the manuscript. We also thank members of our laboratory for comments on the manuscript. This work was supported by NSERC, CFI, and a Canada Research Chair to K.A.W.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2135210.
REFERENCES
- Batten JS, Desvoyes B, Yamamura Y, Scholthof KB 2006. A translational enhancer element on the 3′-proximal end of the Panicum mosaic virus genome. FEBS Lett 580: 2591–2597 [DOI] [PubMed] [Google Scholar]
- Belsham GJ 2009. Divergent picornavirus IRES elements. Virus Res 139: 183–192 [DOI] [PubMed] [Google Scholar]
- Browning KS 1996. The plant translational apparatus. Plant Mol Biol 32: 107–144 [DOI] [PubMed] [Google Scholar]
- Browning KS, Humphreys J, Hobbs W, Smith GB, Ravel JM 1990. Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J Biol Chem 265: 17967–17973 [PubMed] [Google Scholar]
- Browning KS, Webster C, Roberts JK, Ravel JM 1992. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J Biol Chem 267: 10096–10100 [PubMed] [Google Scholar]
- Burgyan J, Rubino L, Russo M 1996. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J Gen Virol 77: 1967–1974 [DOI] [PubMed] [Google Scholar]
- Carstens EB 2010. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol 155: 133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangerfield JA, Windbichler N, Salmons B, Gunzberg WH, Schroder R 2006. Enhancement of the StreptoTag method for isolation of endogenously expressed proteins with complex RNA binding targets. Electrophoresis 27: 1874–1877 [DOI] [PubMed] [Google Scholar]
- Danthinne X, Seurinck J, Meulewaeter F, Van Montagu M, Cornelissen M 1993. The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol Cell Biol 13: 3340–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz JA, Nieto C, Moriones E, Truniger V, Aranda MA 2004. Molecular characterization of a Melon necrotic spot virus strain that overcomes the resistance in melon and nonhost plants. Mol Plant Microbe Interact 17: 668–675 [DOI] [PubMed] [Google Scholar]
- Dreher TW 2009. Role of tRNA-like structures in controlling plant virus replication. Virus Res 139: 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Miller WA 2006. Translational control in positive strand RNA plant viruses. Virology 344: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, White KA 2004. 5′-3′ RNA–RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: A potential common mechanism for tombusviridae. J Biol Chem 279: 28862–28872 [DOI] [PubMed] [Google Scholar]
- Fabian MR, White KA 2006. Analysis of a 3′-translation enhancer in a tombusvirus: A dynamic model for RNA-RNA interactions of mRNA termini. RNA 12: 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR 1998. A tale of two termini: A functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 216: 1–11 [DOI] [PubMed] [Google Scholar]
- Gallie DR 2001. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J Virol 75: 12141–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR 2007. Use of in vitro translation extract depleted in specific initiation factors for the investigation of translational regulation. Methods Enzymol 429: 35–51 [DOI] [PubMed] [Google Scholar]
- Gallie DR, Browning KS 2001. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem 276: 36951–36960 [DOI] [PubMed] [Google Scholar]
- Gazo BM, Murphy P, Gatchel JR, Browning KS 2004. A novel interaction of cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J Biol Chem 279: 13584–13592 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Grzybowska EA, Wilczynska A, Siedlecki JA 2001. Regulatory functions of 3′UTRs. Biochem Biophys Res Commun 288: 291–295 [DOI] [PubMed] [Google Scholar]
- Guo L, Allen E, Miller WA 2000. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA 6: 1808–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Allen EM, Miller WA 2001. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol Cell 7: 1103–1109 [DOI] [PubMed] [Google Scholar]
- Hernández G, Vazquez-Pianzola P 2005. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech Dev 122: 865–876 [DOI] [PubMed] [Google Scholar]
- Imataka H, Gradi A, Sonenberg N 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 17: 7480–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa HO, Mizumoto H, Nagano H, Imoto Y, Takigawa K, Sarawaneeyaruk S, Kaido M, Mise K, Okuno T 2008. A viral noncoding RNA generated by cis-element-mediated protection against 5′→3′ RNA decay represses both cap-independent and cap-dependent translation. J Virol 82: 10162–10174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karetnikov A, Lehto K 2008. Translation mechanisms involving long-distance base pairing interactions between the 5′ and 3′ nontranslated regions and internal ribosomal entry are conserved for both genomic RNAs of Blackcurrant reversion nepovirus. Virology 371: 292–308 [DOI] [PubMed] [Google Scholar]
- Karetnikov A, Keränen M, Lehto K 2006. Role of the RNA2 3′ nontranslated region of Blackcurrant reversion nepovirus in translational regulation. Virology 354: 178–191 [DOI] [PubMed] [Google Scholar]
- Khan MA, Miyoshi H, Gallie DR, Goss DJ 2008. Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: Interactions with translation initiation factors eIF4F and eIFiso4F. J Biol Chem 283: 1340–1349 [DOI] [PubMed] [Google Scholar]
- Kim CY, Takahashi K, Nguyen TB, Roberts JK, Webster C 1999. Identification of a nucleic acid binding domain in eukaryotic initiation factor eIFiso4G from wheat. J Biol Chem 274: 10603–10608 [DOI] [PubMed] [Google Scholar]
- Kneller EL, Rakotondrafara AM, Miller WA 2006. Cap-independent translation of plant viral RNAs. Virus Res 119: 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DC, Liu DX, Wong SM 2002. A six-nucleotide segment within the 3′ untranslated region of hibiscus chlorotic ringspot virus plays an essential role in translational enhancement. J Virol 76: 1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1998. Primer extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res 26: 4853–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krab IM, Caldwell C, Gallie DR, Bol JF 2005. Coat protein enhances translational efficiency of Alfalfa mosaic virus RNAs and interacts with the eIF4G component of initiation factor eIF4F. J Gen Virol 86: 1841–1849 [DOI] [PubMed] [Google Scholar]
- Leathers V, Tanguay R, Kobayashi M, Gallie DR 1993. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol Cell Biol 13: 5331–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW 2004. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology 321: 36–46 [DOI] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW 2006. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA 12: 1338–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberry LK, Dennis MD, Allen ML, Ruud Nitka K, Murphy PA, Campbell L, Browning KS 2007. Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F, and eIF5. Methods Enzymol 430: 397–408 [DOI] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231 [DOI] [PubMed] [Google Scholar]
- Meulewaeter F, Van Montagu M, Cornelissen M 1998. Features of the autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA 4: 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulewaeter F, van Lipzig R, Gultyaev AP, Pleij CW, Van Damme D, Cornelissen M, van Eldik G 2004. Conservation of RNA structures enables TNV and BYDV 5′ and 3′ elements to cooperate synergistically in cap-independent translation. Nucleic Acids Res 32: 1721–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Koev G 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273: 1–8 [DOI] [PubMed] [Google Scholar]
- Miller WA, Wang Z, Treder K 2007. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem Soc Trans 35: 1629–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto H, Tatsuta M, Kaido M, Mise K, Okuno T 2003. Cap-independent translational enhancement by the 3′ untranslated region of red clover necrotic mosaic virus RNA1. J Virol 77: 12113–12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PD 2008. Yeast as a model host to explore plant virus-host interactons. Annu Rev Phytopathol 46: 217–242 [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J 2006. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344: 211–220 [DOI] [PubMed] [Google Scholar]
- Neeleman L, Olsthoorn RC, Linthorst HJ, Bol JF 2001. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc Natl Acad Sci 98: 14286–14291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BL, White KA 2008. Context-influenced cap-independent translation of Tombusvirus mRNAs in vitro. Virology 380: 203–212 [DOI] [PubMed] [Google Scholar]
- Nieto C, Morales M, Orjeda G, Clepet C, Monfort A, Sturbois B, Puigdomènech P, Pitrat M, Caboche M, Dogimont C, et al. 2006. An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J 48: 452–462 [DOI] [PubMed] [Google Scholar]
- Polacek C, Friebe P, Harris E 2009. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J Gen Virol 90: 687–692 [DOI] [PubMed] [Google Scholar]
- Qu F, Morris TJ 2000. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J Virol 74: 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotondrafara AM, Polacek C, Harris E, Miller WA 2006. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA 12: 1893–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Wu B, White KA 2003. A second functional RNA domain in the 5′ UTR of the Tomato bushy stunt virus genome: Intra- and interdomain interactions mediate viral RNA replication. RNA 9: 1232–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Caranta C 2006. Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci 11: 40–45 [DOI] [PubMed] [Google Scholar]
- Rutkowska-Wlodarczyk I, Stepinski J, Dadlez M, Darzynkiewicz E, Stolarski R, Niedzwiecka A 2008. Structural changes of eIF4E upon binding to the mRNA 5′ monomethylguanosine and trimethylguanosine Cap. Biochemistry 47: 2710–2720 [DOI] [PubMed] [Google Scholar]
- Sachs MS, Wang Z, Gaba A, Fang P, Belk J, Ganesan R, Amrani N, Jacobson A 2002. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods 26: 105–114 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: A laboratory manual 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Scheets K, Redinbaugh MG 2006. Infectious cDNA transcripts of Maize necrotic streak virus: infectivity and translational characteristics. Virology 350: 171–183 [DOI] [PubMed] [Google Scholar]
- Shen R, Miller WA 2004. The 3′ untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J Virol 78: 4655–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Rakotondrafara AM, Miller WA 2006. Trans regulation of cap-independent translation by a viral subgenomic RNA. J Virol 80: 10045–10054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N 2008. eIF4E, the mRNA cap-binding protein: From basic discovery to translational research. Biochem Cell Biol 86: 178–183 [DOI] [PubMed] [Google Scholar]
- Stupina VA, Meskauskas A, McCormack JC, Yingling YG, Shapiro BA, Dinman JD, Simon AE 2008. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA 14: 2379–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer RT, Benkowski LA, Schodin D, Lax SR, Metz AM, Ravel JM, Browning KS 1993. The 5′ and 3′ untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5′ cap structure. J Biol Chem 268: 9504–9510 [PubMed] [Google Scholar]
- Treder K, Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA 2008. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 14: 134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger V, Nieto C, González-Ibeas D, Aranda M 2008. Mechanism of plant eIF4E-mediated resistance against a Carmovirus (Tombusviridae): Cap-independent translation of a viral RNA controlled in cis by an (a)virulence determinant. Plant J 56: 716–727 [DOI] [PubMed] [Google Scholar]
- van Lipzig R, Gultyaev AP, Pleij CW, van Montagu M, Cornelissen M, Meulewaeter F 2002. The 5′ and 3′ extremities of the satellite tobacco necrosis virus translational enhancer domain contribute differentially to stimulation of translation. RNA 8: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Miller WA 1995. A sequence located 4.5 to 5 kilobases from the 5′ end of the barley yellow dwarf virus (PAV) genome strongly stimulates translation of uncapped mRNA. J Biol Chem 270: 13446–13452 [DOI] [PubMed] [Google Scholar]
- Wang S, Browning KS, Miller WA 1997. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J 16: 4107–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Treder K, Miller WA 2009. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J Biol Chem 284: 14189–14202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2: 135–140 [DOI] [PubMed] [Google Scholar]
- White KA, Morris TJ 1994. Nonhomologous RNA recombination in tombusviruses: Generation and evolution of defective interfering RNAs by stepwise deletions. J Virol 68: 14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Nagy PD 2004. Advances in the molecular biology of tombusviruses: Gene expression, genome replication and recombination. Prog Nucleic Acid Res Mol Biol 78: 187–226 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616 [DOI] [PubMed] [Google Scholar]
- Windbichler N, Schroeder R 2006. Isolation of specific RNA-binding proteins using the streptomycin-binding RNA aptamer. Nat Protoc 1: 637–640 [DOI] [PubMed] [Google Scholar]
- Wu B, White KA 1999. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J Virol 73: 8982–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, White KA 2009. RNA-based regulation of transcription and translation of aureusvirus subgenomic mRNA1. J Virol 83: 10096–10105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Shi K, Meskauskas A, Simon AE 2009. The 3′ end of Turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA 15: 1849–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Wang J, Yu P, Eyler D, Xu H, Starich MR, Tiede DM, Simon AE, Kasprzak W, Schwieters CD, et al. 2010. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus. Proc Natl Acad Sci 107: 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]