Abstract
The analysis of protein–RNA association in vitro commonly involves radiolabeled in vitro transcribed RNAs. Nucleotides labeled with near-infrared (NIR) dyes provide promising alternatives for studying protein–RNA binding in vitro. However, it remained elusive whether random labeling of RNA probes by NIR dyes interferes with protein binding. Here, we demonstrate that infrared scanning allows the detection of randomly NIR-labeled RNA probes in the low femtomole range. The analyses of eight distinct protein–RNA complexes by electrophoretic mobility shift assay, filter binding, or UV crosslinking revealed that protein binding specificity remains unaffected by random NIR labeling. Accordingly, NIR probes allowed the rapid identification of the short noncoding Y3-RNA as a novel RNA target of ZBP1 (zipcode binding protein). Whereas binding of ZBP1 to the ACTB-zipcode and Y3 was exclusive, the protein formed a trimeric complex with the La protein and Y3. This was dissociated in the presence of Y5 RNA, resulting in the formation of ZBP1/Y3 and La/Y5 complexes. Hence, ZBP1 apparently resides in at least two distinct cellular RNPs: mRNA-containing mRNPs or Y3-containing yRNPs. In conclusion, our findings indicate that randomly labeled NIR probes provide a powerful tool for the rapid and sensitive analysis of protein–RNA binding in vitro. In contrast to radiolabeled RNAs, NIR probes remain stable for months, do not pose any safety considerations, and enable the significantly expedited analysis of experimental data due to fast read technologies available. The most prominent advantage of probes labeled by NIR dyes is the option to color-code distinct transcripts, allowing the unbiased identification of distinct protein–RNA complexes in one sample.
Keywords: La, NIR dye, Y-RNA, ZBP1, protein–RNA binding
INTRODUCTION
The analysis of protein–RNA interactions in vitro is traditionally conducted by using radiolabeled RNA probes that are applicable in a variety of assay systems. However, radiolabeled RNAs remain dissatisfactory due to: (1) their short half-life; (2) safety considerations; (3) the delayed analysis of experimental data due to time-consuming sample processing and prolonged exposure times; and (4) the fact that only one isotope labeling can be analyzed in most assays, thus limiting studies of complex protein–RNA mixtures. Therefore, we set out to validate randomly near infrared (NIR) dye-labeled RNAs (further referred to as NIR probes or NIR-RNAs) for studying protein–RNA association in vitro. NIR-labeled nucleotides with excellent fluorescent properties, in particular high signal-to-noise ratios and quantum yields, have become available recently (e.g., Jena Bioscience Inc.). Combined with infrared scanning using an Odyssey Scanner (LI-COR Inc.) allowing the rapid and sensitive analysis of gels, blots, tissue samples, and multiwell plate-based assays, NIR-labeled nucleotides were expected to yield RNA probes detectable at a high dynamic range and improved signal-to-noise ratio. However, it remained elusive if and how random labeling of probes with NIR dyes affects protein–RNA binding.
To evaluate if NIR probes are suitable for studying protein–RNA interactions in vitro, we initially analyzed binding of Atto680- or DY776-labeled MS2 to the MS2-binding protein fused to the maltose-binding protein (further referred to as MS2BP). The MS2BP/MS2 association was used since binding had been investigated extensively using conventional approaches and this protein–RNA complex is widely used for tethering analyses (Lim and Peabody 1994; Peabody and Lim 1996; Bertrand et al. 1998; Parrott et al. 2000; Zhou and Reed 2003; Keryer-Bibens et al. 2008). Moreover, the MS2BP/MS2 association requires formation of a stem–loop structure with at least one uridine in the loop that is essential for protein binding and two uridines involved in stem formation (Supplemental Fig. 1A; Lim and Peabody 1994; Parrott et al. 2000; Powell and Peabody 2001). Therefore, NIR-labeled UTP was used for in vitro transcription to validate how NIR dye modification affects both the structural properties and protein binding of NIR probes. By now, several mutations have been introduced in MS2BP that prevent phage assembly but retain RNA binding of MS2BP as a dimer with low nanomolar KD values (Peabody and Lim 1996; Peabody and Al-Bitar 2001; Lima et al. 2006). Thus, the MS2BP/MS2 association was also expected to allow analyzing if NIRdye labeling of probes affects RNA-binding-dependent protein oligomerization.
The suitability of NIR-RNAs for the identification and characterization of “biologically relevant” protein–RNA associations was evaluated by analyzing binding of ZBP1 (zipcode binding protein) to human Y-RNAs. ZBP1 (IGF2BP1 in human) associates with specific target mRNAs and directs their transport, stability, and/or translation (Leeds et al. 1997; Oleynikov and Singer 2003; Huttelmaier et al. 2005). Recent findings indicate that the protein facilitates these functions via cytoplasmic mRNPs by associating with several other RNA-binding proteins including NCL, TROVE2 (Ro60), and PTBs (Jonson et al. 2007; Weidensdorfer et al. 2009). Next to other RNA targets, these three ZBP1 ligands associate with a specific class of small noncoding RNAs (ncRNAs), termed Y-RNAs (Hendrick et al. 1981; Fabini et al. 2001; Fouraux et al. 2002). In humans, four different Y-RNAs have been identified so far: Y1, Y3, Y4, and Y5 (Hendrick et al. 1981). Short noncoding Y-RNAs (83–112 nucleotides [nt]) share a common stem–loop structure with a protruding single-stranded U-rich 3′ end. Although their function remains largely elusive, it has been postulated that Y-RNAs are involved in replication and 5S-rRNA quality control (Christov et al. 2006; Hogg and Collins 2007). Whether ZBP1 associates with small ncRNAs, in particular Y-RNAs, has not been investigated so far.
RESULTS
Analysis of randomly NIR dye-labeled RNAs by infrared scanning
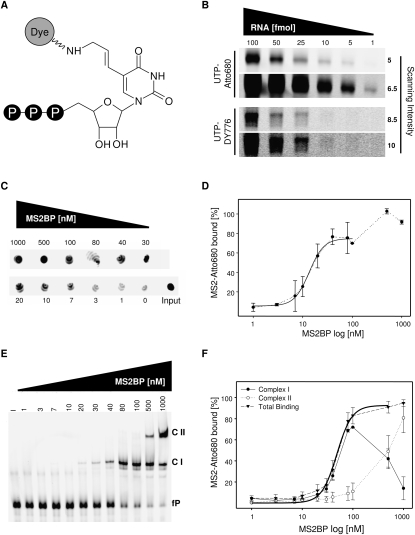
To analyze NIR probes by infrared scanning, MS2 RNA (further referred to as MS2) was randomly labeled by UTP-Atto680 or UTP-DY776 during in vitro transcription (Fig. 1A; Supplemental Fig. 1A). Labeled and unlabeled MS2 were analyzed by spectroscopic methods and native TBE-PAGE followed by ethidium bromide staining. This demonstrated that the efficiency of T7-driven transcription was decreased approximately fivefold in the presence of NIR-labeled UTPs (data not shown). The mobility of NIR-labeled RNA probes in native TBE-PAGE remained unaffected (Supplemental Fig. 1B). The interference of NIR dyes with UV absorption at 260 nm was insignificant upon efficient abscission of unincorporated nucleotides by phenol extraction. Therefore, the concentration of NIR probes was conveniently determined by UV absorption, as confirmed in comparison to unlabeled probes by ethidium bromide staining (Supplemental Fig. 1B).
FIGURE 1.
Analysis of MS2BP/MS2 binding by NIR-labeled RNA probes. (A) NIR-labeled UTPs (Jena Bioscience) were used for in vitro transcription. Dyes (Atto680 or DY776) were covalently bound to UTP (C-5 of pyrimidine ring) by a flexible aminoallyl linker. (B) Analysis of in vitro transcribed MS2 labeled with UTP-Atto680 (upper panel) or UTP-DY776 (lower panel). Indicated amounts of labeled RNA probes (fmol) were analyzed by infrared scanning of 4%–12% native TBE gels using a LI-COR Odyssey scanner. Used scanning intensities (maximum of 10) are indicated (right). (C) Filter binding analyses of the MS2BP/MS2-Atto680 association. RNA spotted on nitrocellulose filters (Input: 100%, 15 fmol) or immobilized on filters upon binding to indicated amounts of MS2BP was analyzed by infrared scanning. (D) The percentage of MS2-Atto680 RNA bound by indicated protein amounts was determined by input-normalized fluorescence intensities (C). The Hill equation was used for fitting (solid black line) of data up to 100 nM MS2BP and KD calculation. (Dotted line) Increased binding at protein concentrations >100 nM. (E) EMSA analysis of the MS2BP/MS2–Atto680 association. Complex formation of indicated amounts of MS2BP and MS2-Atto680 (15 fmol) was analyzed by EMSA in native 4%–12% TBE gels and infrared scanning. (fP) Free probe, (CI) Complex 1, (CII) Complex 2. (F) The percentage of MS2-Atto680 RNA bound by indicated MS2BP amounts was determined by input-normalized (I) fluorescence intensities (E) observed for CI (filled circle, solid line), CII (open circle, dotted line), or total binding (CI+CII: triangle, dashed line). The Hill equation was used for fitting of total binding (solid thick black line) using data up to 1 μM MS2BP. Error bars indicate SD from three independent analyses.
Detection limits for NIR probes were analyzed by native TBE-PAGE and infrared scanning using MS2 amounts of 1–100 fmol (Fig. 1B). While Atto680-labeled probes were detected at 1 fmol using moderate scanning intensities, the detection limit of DY776-labeled RNAs was determined at ∼25 fmol, applying the maximum scanning intensity (10) available. Hence, NIR probes provide an excellent signal-to-noise ratio with a detection limit beyond 1 fmol for Atto680-labeled RNAs.
Analysis of MS2BP/MS2 binding by NIR dye-labeled RNAs
The MS2BP/MS2 association was first analyzed by nitrocellulose filter binding assays using Atto680-labeled probes and standard binding protocols. MS2BP-associated probe was determined by infrared scanning of filters after extensive washing. Signal intensities for each filter in one scan were quantified by the Odyssey software (LI-COR). Samples without protein but with RNA served as background controls. RNA input levels (set to 100%) were determined using buffer-treated filters and RNA spotting without further washing. These analyses demonstrated that MS2BP bound Atto680-labeled MS2 within the low nanomolar range (Fig. 1C). Fitting of data by the Hill equation and assuming dimerization of MS2BP upon RNA binding determined a KD of ∼13.5 (±1.1) nM (Fig. 1D, solid line). This was in good agreement with previous analyses that proposed KD values of 0.4–3 nM using radiolabeled probes in filter binding studies as well as a KD value of ∼40 nM determined by tryptophan quenching analyses using unlabeled RNAs (Lim and Peabody 1994; Peabody and Lim 1996; Parrott et al. 2000). Protein concentrations above ∼100 nM apparently led to the formation of larger complexes indicated by increased RNA binding, as previously proposed (Fig. 1D, dotted line; Lim and Peabody 1994).
To get a more detailed view of complex formation, binding of MS2BP to Atto680-labeled MS2 was analyzed by electrophoretic mobility shift assay (EMSA). Complex formation was monitored by infrared scanning of 4%–12% native TBE gels (Fig. 1E). In agreement with the filter binding studies presented here as well as previous analyses using radiolabeled probes, significant amounts of a second protein–RNA complex were observed at MS2BP concentrations >100 nM (Lim and Peabody 1994). Binding affinity for both complexes (total binding) was determined by Hill fitting revealing a KD of ∼48.3 ± 3.4 nM (Fig. 1F). Hence, the affinity of MS2BP to MS2 in EMSA analyses was decreased three- to fourfold compared with filter binding studies, but was in good agreement with previously reported tryptophan quenching studies (Parrott et al. 2000). Discrepancies in KD values determined by EMSA versus filter binding or tryptophan quenching are frequently observed. These presumably reflect distinct properties of each assay, for instance the determination of distinct protein–RNA complexes in EMSA in contrast to filter binding. We assume that the first complex formed upon RNA binding is the MS2BP–Dimer/RNA complex (Complex I [CI]). At higher protein concentrations a second complex (Complex II [CII]) with reduced mobility was observed, indicating the association of additional protein or RNA molecules to CI that accordingly decreased with increasing amounts of CII (Fig. 1E,F). For cooperative binding of a protein dimer to RNA, a Hill coefficient of approximately two has to be assumed. Therefore, it appears likely that more than two MS2BP molecules can bind one RNA in the same complex, since the determined Hill coefficient was n = 2.9 ± 0.5 in both the filter binding and EMSA analyses.
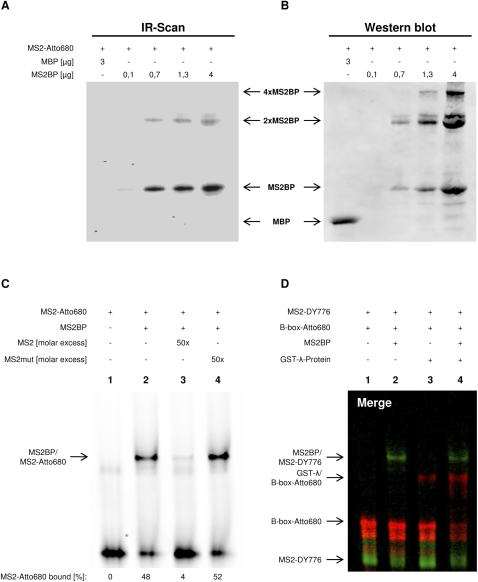
Another widely used method to determine the binding of proteins to RNAs is based on covalent coupling via UV light (UV crosslinking). Since various fluorophores are susceptible to UV light exposure, we analyzed if UV crosslinking can be used to study the association of MS2BP with Atto680-labeled MS2. Increasing amounts of MS2BP were incubated with MS2 followed by UV irradiation, RNAse treatment, and SDS-PAGE (Fig. 2A). Scanning of gels demonstrated that Atto680-labeled MS2 crosslinked with MS2BP in a concentration-dependent manner, whereas no association was observed with MBP, indicating specificity. Hence, UV irradiation did not severely affect the RNA-coupled dye or protein-binding properties of the NIR-labeled RNA. Notably, crosslinking of MS2BP in the presence or absence of MS2 resulted in the formation of large complexes with an apparent molecular mass corresponding to MS2BP dimers or tetramers, respectively (Fig. 2B; Supplemental Fig. 2A). This oligomerization was independent of MBP that remained monomeric even after exposure to UV light. In agreement with EMSA studies, more than one protein–RNA complex was observed. However, the size and ratio of protein complexes cannot be compared adequately between both assays due to RNAse treatment and the denaturing conditions used in UV crosslinking studies.
FIGURE 2.
Analysis of distinct protein–RNA complexes in one sample by NIR probes. (A) UV crosslinking of MS2-Atto680 (200 fmol) to MBP (3 μg) or indicated amounts of MS2BP after RNase A treatment and SDS-PAGE. Complex formation was analyzed by infrared scanning of the SDS-gel (A) followed by Western blotting with anti-MBP (B). (4×MS2BP) Tetramer, (2×MS2BP) dimer, (MS2BP) monomer, (MBP) maltose binding protein. (C) Competition of the MS2BP/MS2 association analyzed by EMSA. Binding of MS2-Atto680 (15 fmol) to MS2BP (80 nM) was competed by a 50-fold molar excess of unlabeled MS2 (lane 3) or MS2mut (lane 4). The percentage of bound probe (bottom panel) was determined as described in Figure 1. Sequence and stem–loop structure of used probes are indicated in Supplemental Figure 1A. (D) EMSA analysis of the MS2BP/MS2 and GST-λ/B-box association in one sample. Complex formation of MS2BP and/or GST-λ with MS2-DY776 (green, 25 fmol) and/or B-box-Atto680 (red, 15 fmol) was analyzed by EMSA. Note that both NIR RNAs were present in all samples. (Lane 1) Buffer control, (lane 2) MS2BP (80 nM), (lane 3) GST-λ (500 nM), (lane 4) MS2BP (80 nM) and GST-λ (500 nM). Free NIR probes and protein–RNA complexes are indicated. For color-separated scans, see Supplemental Figure 2C.
To test if binding of Atto680-labeled RNAs is specific and thus can be selectively competed, complex formation of MS2BP with Atto680-MS2 was challenged by unlabeled MS2 in EMSA studies (Fig. 2C). At a 50-fold molar excess of unlabeled MS2, binding of the Atto680-labeled probe was significantly reduced. In contrast, binding remained essentially unaffected by the addition of a binding-deficient MS2 mutant (MS2mut) in which UC was replaced by AA in the protein binding loop (Supplemental Fig. 1A; Lim and Peabody 1994).
One major advantage of the NIR dye technology is that at least two different dyes can be analyzed in the same sample by infrared scanning. Hence, we expected that two distinct protein–RNA interactions can be monitored in the same sample by color-coding the RNA probes with Atto680 or DY776. Therefore, a second protein–RNA interaction was analyzed: the association of GST-tagged Lambda peptide (GST-λ) with the B-box RNA. First, the affinity of GST-λ for Atto680-labeled B-box RNA was determined by EMSA (Supplemental Fig. 2B). Compared with the MS2BP/MS2 association (KD ∼50 nM in EMSA), at least a 10-fold reduced affinity was observed for the GST-λ/B-box association (KD >500 nM). This was surprising, since previous analyses using nontagged synthetic λ-peptide indicated association to the B-box RNA with KD values in the low nanomolar range (Keryer-Bibens et al. 2008). To analyze if the specific formation of distinct MS2BP/MS2 and GST-λ/B-box complexes can be monitored in one sample despite the observed differences in the determined KD values, binding of both proteins to Atto680-labeled B-box or DY776-labeled MS2 was analyzed by EMSA (Fig. 2D). MS2- (Fig. 2D, green) and B-box-specific (Fig. 2D, red) complexes were observed in samples comprising both proteins and labeled RNAs. In agreement with the lower molecular weight and presumably distinct charge of the GST-λ (∼30 kDa) compared with the MBP-fused MS2BP protein (∼53 kDa), the GST-λ/B-box complex showed a faster migration in EMSA. Specific complex formation was observed by: (1) the lack of shifted B-box RNA in the absence of GST-λ (Fig. 2D, lane 2); (2) the lack of shifted MS2 in samples not containing MS2BP (Fig. 2D, lane 3); (3) reduced levels of free probes in the presence of binding proteins; and (4) the lack of complexes associated with both RNAs that should result in yellow fluorescence (Supplemental Fig. 2C, color-separated scans). Surprisingly, binding of GST-λ to the B-box was promoted in the presence of MS2BP/MS2-DY776 (Fig. 2D, cf. lanes 1–3 and lane 4). However, no supershifted complex or MS2/B-Box binding was observed. This suggests that the GST-λ/B-box association was promoted at elevated total protein and/or RNA concentration. Despite the unexpected promotion of GST-λ/B-box binding in the presence of MS2BP/MS2-DY776, we conclude that the random labeling of RNAs by NIR dyes allows the analysis of at least two distinct protein–RNA complexes in one sample.
NIR probes identify Y1 and Y3 as novel target RNAs of ZBP1
After validating the suitability of randomly labeled NIR probes for studying the MS2BP/MS interactions, we intended to apply the method to a “biologically relevant” association. Therefore, binding of ZBP1 to the coding region instability determinant (CRD) of MYC was analyzed (Supplemental Fig. 3A). In contrast to previous studies suggesting a KD of ∼500 nM for the ZBP1/CRD association, filter binding revealed KD values of 116.5 ± 13.4 nM for NIR-labeled and 147.1 ± 10.5 nM for radiolabeled CRD (Sparanese and Lee 2007). Hence, as observed for the MS2BP/MS2 association, RNA binding of ZBP1 remained unaffected by NIR labeling.
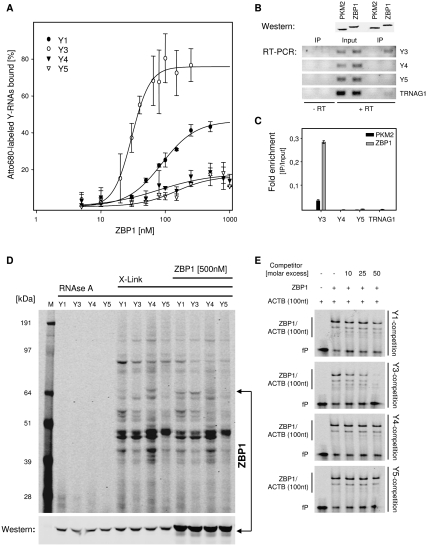
To investigate if NIR labeling allows the rapid identification of novel target RNAs of ZBP1, we analyzed binding of the protein to Y-RNAs. Previous studies revealed that at least three proteins found to associate with ZBP1 also bind Y-RNAs: NCL, TROVE2, and PTBs (Hendrick et al. 1981; Fabini et al. 2001; Fouraux et al. 2002; Jonson et al. 2007; Weidensdorfer et al. 2009). Direct association of ZBP1 with Y-RNAs was first probed in vitro by filter binding (Fig. 3A). As observed for its protein ligands NCL and PTB, ZBP1 selectively bound Y3 (KD = 30.2 ± 1.5 nM) and Y1 (KD = 88.1 ± 7.2 nM). No significant association was observed with Y4 and Y5. Binding of ZBP1 to endogenous Y-RNAs was analyzed in 293 cells transfected with Flag-tagged ZBP1 or PKM2 (pyruvate kinase muscle 2), serving as a negative control (Fig. 3B). The analyses of RNAs coimmunopurified with Flag-tagged proteins by RT-PCR demonstrated a selective coenrichment of Y3 with ZBP1 compared with input levels. In contrast, no significant copurification with ZBP1 was observed for Y4, Y5, or TRNAG1 (encoding a tRNA for glycine). Y1 expression in 293 cells was beyond detection limits (data not shown). Specific binding of ZBP1 to Y3 in 293 cells was further validated via quantitative RT-PCR by comparing RNA levels between the input and immunopurified samples (Fig. 3C). Confirming the semiquantitative analyses, selective enrichment upon ZBP1 immunopurification was observed only for Y3.
FIGURE 3.
ZBP1 associates with Y1 and Y3 RNAs. (A) Filter binding analysis of ZBP1 and Y-RNAs. Filter binding was performed as described in Figure 1, C and D. For fitting, the Hill equation was used, assuming RNA-dependent dimerization of ZBP1 (Nielsen et al. 2004; Chao et al. 2010). (B) Association of Flag-ZBP1 with Y3 in 293 cells. Flag-tagged ZBP1 or PKM2 were immunopurified from transiently transfected 293 cells as confirmed by Western blotting with anti Flag-M2 (upper panel, Western). Copurification of Y-RNAs or TRNAG1 was analyzed by semiquantitative RT-PCR using 26 amplification cycles for Y4, Y5, and TRNAG1 or 30 amplification cycles for Y3 (lower panels, RT-PCR). PCR analyses of IP samples without preceding reverse transcription (−RT) served as controls. (C) Enrichment of indicated RNAs upon ZBP1 or PKM2 immunopurification was determined by quantitative RT-PCR using the ΔCt-method and normalization to input samples. (D) Binding of ZBP1 to exogenous Y-RNAs in 293 cell lysates. Lysates were supplemented with Atto680-labeled Y-RNAs (500 fmol) and recombinant ZBP1 (500 nM) before UV crosslinking as indicated (X-Link). Complex formation was analyzed by infrared scanning of SDS gels and Western blotting with anti-ZBP1. (E) Y3 competes for binding of ZBP1 to the ACTB-3′UTR in EMSA studies. Binding of ZBP1 (120 nM) to the ACTB-3′UTR(1–100 nt) (50 fmol) was competed by indicated molar excess of unlabeled Y-RNAs. Free probe (fP) and the ZBP1/ACTB(100 nt) complex are indicated. Error bars indicate SD from three independent experiments.
To determine if ZBP1 associates with NIR-labeled Y3 in cell extracts, Atto680-labeled Y-RNAs were UV-crosslinked in 293 cell lysates (Fig. 3D). Surprisingly, a ZBP1-specific crosslink to Y1 and Y3 was only observed by supplementing extracts with recombinant ZBP1. This suggested that the addition of 200 fmol of Atto680-labeled Y-RNAs was insufficient to compete for the association of endogenous ZBP1 with mRNAs and/or endogenous Y3.
Whether the association of ZBP1 with Y-RNAs and other target transcripts is exclusive was analyzed by competition of the ZBP1/ACTB-3′UTR association using EMSA (Fig. 3E). Binding of ZBP1 to Atto680-ACTB-3′UTR(1–100 nt) comprising the zipcode, previously identified as a ZBP1-specific binding motif, was first analyzed in the absence of Y-RNAs (Supplemental Fig. 3B; Ross et al. 1997). In the presence of Y-RNAs, a significant competition was observed only by Y3. Although Y1 was found to associate with ZBP1 in vitro and in cell extracts, competition of the ZBP1/ACTB-3′UTR(1–100 nt) association remained modest even at a 200-fold molar excess (data not shown). Therefore, we further analyzed competition of Y-RNAs for the binding of ZBP1 to the chicken zipcode comprising the first 54 nt of the ACTB-3′UTR (Supplemental Fig. 3C). As observed for the association with ACTB-3′UTR(1–100 nt), significant competition of the ZBP1–zipcode interaction was only observed for Y3. These findings suggested that either binding of ZBP1 to Y1 is unspecific or it remains unaffected by association of the protein with other target RNAs. In contrast, the presented findings indicate that binding of ZBP1 to Y3 versus other RNA targets is exclusive, as demonstrated here for the ZBP1/ACTB association.
Analysis of Y3- versus Y5-associated protein complexes
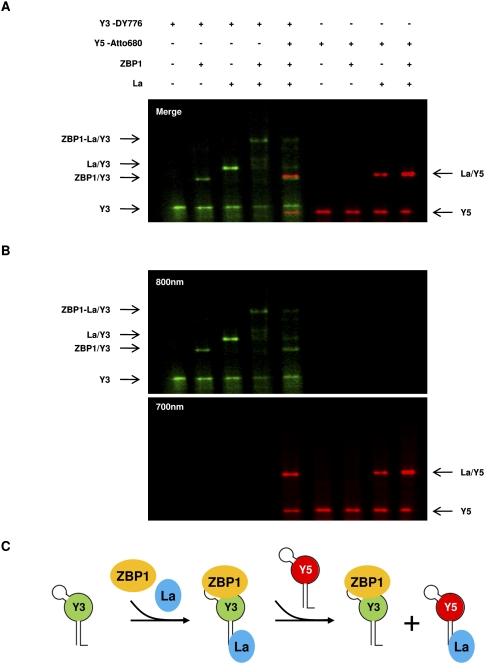
Previous studies revealed that the La protein associates with all Y-RNAs via their extending poly-U stretch at the 3′ end (Stefano 1984). In contrast, PTB and nucleolin associate with the Y3 RNA via the variable loop region (Fabini et al. 2001; Fouraux et al. 2002). This raised the question if ZBP1 and La can associate via Y3.
As demonstrated here, NIR labeling of RNAs provides a convenient approach to identify distinct protein–RNA complexes in one sample by the color-coding of RNA probes. Therefore, binding of ZBP1 and La to DY776-labeled Y3 and/or Atto680-labeled Y5-RNA was analyzed by EMSA (Fig. 4A,B). In agreement with previous findings, Y3 was bound by both proteins, resulting in the formation of distinct complexes. Indicating the formation of a trimeric complex consisting of ZBP1, La, and Y3, a supershifted complex was observed upon the incubation of both proteins with the DY776-labeled Y-RNA. In agreement with filter binding (Fig. 3A), no association of ZBP1 was observed with Atto680-labeled Y5. In contrast, Y5 associated with La resulting in the formation of a complex migrating at the almost same position as the ZBP1/Y3 complex in EMSA. This suggested that an excess of Atto680-labeled Y5 should compete for the formation of a trimeric ZBP1-La/Y3 complex. As expected, the trimeric complex was dissociated by a 25-fold molar excess of Atto680-Y5.
FIGURE 4.
Formation of a trimeric ZBP1-La-Y3 complex. (A,B) Binding of ZBP1 and/or La protein to DY776-labeled Y3 (25 fmol) and/or Atto680-labeled Y5 (625 fmol) was analyzed by EMSA as indicated. Specific protein–RNA complexes are indicated. (B) Color-separated scans for DY776 (800 nm) or Atto680 (700 nm). Note that the supershifted complex comprising ZBP1, La, and Y3 is dissociated in the presence of Y5. (C) Schematic of complexes observed in A. ZBP1 and La form a trimeric complex with Y3. La is dissociated in the presence of excess Y5. This results in the formation of two complexes of similar migration in EMSA: ZBP1/Y3 and La/Y5.
In conclusion, these findings reveal that ZBP1 can form a trimeric complex with Y3 and La. Formation of this complex is specific for Y3, since it is dissociated by Y5, resulting in the formation of a ZBP1/Y3 and a La/Y5 complex with almost identical migration in EMSA (Fig. 4C).
DISCUSSION
The presented analyses indicate that RNAs randomly labeled by NIR dyes can be detected in the low femtomole range. NIR probes are suitable for analyzing protein–RNA binding in vitro, since the secondary RNA structure as well as protein contact formation remain essentially unaffected by the random insertion of NIR dye modifications, as demonstrated here for eight distinct protein–RNA complexes (Supplemental Table 1). Notably, the binding specificity as well as affinity of NIR probes remained unbiased even when NIR-modified bases were presumably involved in protein contact formation, as demonstrated for the MS2BP/MS2, La/Y3, and La/Y5 complexes.
In contrast to radiolabeled probes, random NIR labeling provides transcripts that have an almost unlimited shelf-life when stored properly, allows working without any safety restrictions, and enables the significantly accelerated analyses of experiments. The latter is due to the fact that the scanning of samples is completed within minutes and does not require long exposure times or the drying of gels. However, the most prominent advantage of NIR probes is that distinct protein–RNA complexes can be resolved by color-coding. Due to the excellent signal-to-noise properties of the dyes used, NIR probes even allowed the discrimination of complexes migrating at similar or identical velocity in EMSA. This significantly amplifies options for analyzing protein–RNA complexes that cannot be accomplished by using radiolabeled probes.
Applied to “biologically relevant” protein–RNA associations, NIR-labeled probes allowed the validation of two known RNA targets of ZBP1, MYC-CRD and ACTB-zipcode. Moreover, the applied use of NIR probes identified the short noncoding Y3-RNA as a novel target RNA of ZBP1. The latter selectively associates with Y3 and at lower affinity with Y1 in vitro. While binding of ZBP1 to Y3 versus mRNAs appears to be exclusive, this could not be demonstrated conclusively for Y1. Interestingly, ZBP1 forms a trimeric complex with Y3 and La that is dissociated in the presence of Y5, resulting in the formation of ZBP1/Y3 and La/Y5 complexes (Fig. 4C). These findings suggest that ZBP1 associates with at least two distinct RNP-like complexes in cells: (1) mRNPs—comprising ZBP1 and mRNAs; and (2) yRNPs —comprising ZBP1 and Y3. Future analyses have to reveal the composition and role of ZBP1-containing yRNPs and how these direct the regulatory function of ZBP1 in modulating the transport, stability, or translation of its target mRNAs.
MATERIALS AND METHODS
Plasmids
To generate PCR templates for in vitro transcription, DNA templates of the MS2, MS2mut, ACTB-Zipcode (chicken), or a double B-box (referred to as “B-box” in the text) aptamer were cloned with NheI/HindIII (MS2, MS2mut, and Zipcode) and XhoI/XbaI (2xB-box) in pcDNA 3.1(+) (Invitrogen) using the indicated DNA oligonucleotides (Supplemental Table 2). The expression plasmid for MBP-MS2BP was provided by addgene (plasmid 11246 [Zhou and Reed 2003]). Human Y-RNAs were amplified on randomly primed cDNA (293T) with PCR Mix (Promega) using the indicated oligonucleotides and cloned into the pGEM-T vector (Promega). GST-ZBP1 and Flag-ZBP1 were purified as described (Farina et al. 2003; Huttelmaier et al. 2005). The cDNA encoding human La protein was kindly provided by Ger J.M. Pruijn (Nijmegen Center for Molecular Life Sciences). This cDNA served as a template for PCR-based cloning of La into the pGEX6p1 (GE Healthcare) vector via EcoRI/XhoI using the indicated primers. The cDNA for human PKM2 was obtained from Open Biosystems (Clone ID 3859987) and served as the template for PCR-based cloning into pcDNA3.1-Flag via BamHI/EcoRI. PCR templates for MYC-CRD and human ACTB-3′UTR were as previously described (Huttelmaier et al. 2005; Weidensdorfer et al. 2009).
In vitro transcription
DNA templates for in vitro transcription of MS2, chicken zipcode, and B-box RNAs were generated by PCR (2×Taq PCR master mix, Promega) using T7 and BGH reverse primers. Y-RNA templates were generated by digesting pGEM-T vectors with NcoI/SalI followed by Taq-PCR using T7 and the indicated antisense primers. After purification of PCR products (Wizard SV Kit, Promega) RNAs were in vitro transcribed by T7 RNA polymerase according to the manufacturer's protocols (0.5 mM rNTP end concentration; Promega). For NIR dye labeling, in vitro transcription reactions (0.5 mM rATP, rCTP, and rGTP) were supplemented with 12.5 μM rUTP and 20 μM Atto680- or DY776-UTP (final concentration; Jena Bioscience). Synthesized RNAs were purified using phenol extraction. Labeling efficiencies were estimated to be ∼14%–20%. This was calculated according to the ARES Kit manual (Invitrogen) assuming CF260 = 0.3 and εdye = 125.000 cm−1 M−1 for Atto680. A correction factor for DY776 is not available yet. C-myc CRD-RNA and ACTB-3′UTR were transcribed as previously described using the indicated primer (Huttelmaier et al. 2005; Weidensdorfer et al. 2009) in the presence of UTP-Atto680 (see above) or CTP-[α-32P] (Perkin Elmer). RNA concentrations of labeled and unlabeled RNAs were determined by OD260 absorption. Where indicated, unlabeled RNAs served for calibration in ethidium bromide-stained TBE gels to validate the concentration of NIR dye-labeled probes. NIR dye-labeled RNAs can be stored at −20°C with no loss of activity for up to at least 18 mo.
Protein purification
MBP-MS2BP was essentially purified as previously described (Jurica et al. 2002). The fusion protein was affinity purified using amylose resin (NEB) in 20 mM Tris at pH 7.2, 50 mM NaCl; 1 mM EDTA (MBP-buffer) and eluted by 10 mM maltose. The protein was further purified in MBP buffer by linear NaCl elution from a heparin column using an Äkta purifier (GE Healthcare). Eluted fractions were combined and dialyzed in RNA binding buffer (TMK buffer, see below) for further analyses.
GST-λ-His was affinity purified using GSH resin (GE Healthcare) in GST buffer: 50 mM Tris at pH 8.0, 500 mM NaCl, 5% glycerol, 0.1% Triton, 1 mM DTT. Protein eluted by 20 mM glutathione was dialyzed to GST buffer and further purified by a second affinity purification using Ni-NTA (elution with 250 mM imidazole) and dialyzed in TMK buffer. ZBP1 was purified as described previously (Farina et al. 2003). Essentially the same procedure was used for the purification of La.
Nitrocellulose filter binding assay
For complex formation, different amounts of protein, pre-diluted NIR-labeled RNAs (if not stated otherwise: 15 fmol for Atto680- or 25 fmol for DY776-RNAs) and pre-diluted yeast t-RNA (final concentration: 50 μg/mL; Sigma) were mixed in TMK buffer for MS2BP/GST-λ-proteins (25 mM Tris/HCl at pH 7.4, 100 mM KCl, 5 mM MgCl2) or RBB for ZBP1/La-proteins (50 mM Tris-acetate at pH 7.6, 200 mM KOAc, 2 mM MgOAc) to a final volume of 15 μL. Samples were incubated for 30 min on ice before spotting on pre-washed (5 mL of TMK/RBB buffer) nitrocellulose filters (Millipore). Filters were washed twice with 5 mL of TMK/RBB buffer and analyzed by infrared scanning using an Odyssey scanner (LI-COR). RNA inputs were determined by spotting pre-diluted NIR-RNA on a prewashed filter and scanning without any further washing.
Electrophoretic mobility shift assay (EMSA)
Samples were essentially assembled as for filter binding assays and 10× loading buffer (250 mM Tris at pH 7.4, 0.2% Bromphenol blue, 40% sucrose) was added just before TBE-PAGE (15 mA for ∼90 min) performed at 4°C in TBE buffer. TBE gels were analyzed by infrared scanning as described above. Note that the scanning of TBE “mini-gels” is completed within 4 min. For competition studies binding samples were pre-incubated for 15 min on ice before the addition of competitor RNAs. After an additional incubation for 15 min on ice, samples were analyzed by TBE-PAGE.
UV crosslinking
Samples containing pre-diluted Atto680-RNA (200 fmol), yeast t-RNA (final concentration 25 μg/mL; Sigma), indicated amounts of MS2BP or MBP in TMK buffer (final volume 30 μL) were incubated for 30 min on ice. 293T lysates (2 mg/mL protein) for Y-RNA crosslinks were prepared in TEN100 (20 mM Tris at pH 7.4, 0.1 mM EDTA, 100 mM NaCl, 0.1% NP-40, 2 mM MgCl2) and assembled as above. Pre-incubated samples were irradiated in a Stratalinker 2400 (Stratagene) for 20 min in an ice bath before RNAse A treatment and analyses by SDS-PAGE. For Western blotting transfer to nitrocellulose membranes was performed using NUPAGE standard protocols (Invitrogen). Western blots were analyzed by infrared scanning (Odyssey scanner, LI-COR) using an IRDye 800CW-linked secondary antibody.
Cell culture and transfection
HEK-293T cells were cultured in DMEM containing 10% fetal bovine serum. Flag-ZBP1 and Flag-PKM2 were transfected with Lipofectamine 2000 (Invitrogen), as previously described (Huttelmaier et al. 2005).
Immunoprecipitation and Q-RT-PCR
Immunopurification was essentially performed as described previously using Flag-beads (Sigma) (Huttelmaier et al. 2005). Cells were lysed in BB buffer (20 mM Tris at pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 8.7% glycerol, 0.05% NP-40). Bound proteins and RNAs were eluted by heating beads to 65°C in BB containing 1% (w/v) SDS. RNAs were extracted by phenol and precipitated with isopropanol. RT-PCR was performed using indicated primers (Supplemental Table 2), as previously described (Stohr et al. 2006).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
The expression plasmid encoding GST-λ-His was kindly provided by Kent Duncan (The European Molecular Biology Laboratory [EMBL]). The expression plasmid for MBP-MS2 was provided by Addgene. The cDNA clone encoding human La was kindly provided by Ger J.M. Pruijn (Nijmegen Center for Molecular Life Sciences). This work was supported by Deutsche Forschungsgemeinschaft (DFG) funding to S.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2152710.
REFERENCES
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM 1998. Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH 2010. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev 24: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christov CP, Gardiner TJ, Szuts D, Krude T 2006. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol 26: 6993–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabini G, Raijmakers R, Hayer S, Fouraux MA, Pruijn GJ, Steiner G 2001. The heterogeneous nuclear ribonucleoproteins I and K interact with a subset of the ro ribonucleoprotein-associated Y RNAs in vitro and in vivo. J Biol Chem 276: 20711–20718 [DOI] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouraux MA, Bouvet P, Verkaart S, van Venrooij WJ, Pruijn GJ 2002. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J Mol Biol 320: 475–488 [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA 1981. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: Further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol 1: 1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Collins K 2007. Human Y5 RNA specializes a Ro ribonucleoprotein for 5S ribosomal RNA quality control. Genes Dev 21: 3067–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH 2005. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515 [DOI] [PubMed] [Google Scholar]
- Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC 2007. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics 6: 798–811 [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer-Bibens C, Barreau C, Osborne HB 2008. Tethering of proteins to RNAs by bacteriophage proteins. Biol Cell 100: 125–138 [DOI] [PubMed] [Google Scholar]
- Leeds P, Kren BT, Boylan JM, Betz NA, Steer CJ, Gruppuso PA, Ross J 1997. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene 14: 1279–1286 [DOI] [PubMed] [Google Scholar]
- Lim F, Peabody DS 1994. Mutations that increase the affinity of a translational repressor for RNA. Nucleic Acids Res 22: 3748–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SM, Vaz AC, Souza TL, Peabody DS, Silva JL, Oliveira AC 2006. Dissecting the role of protein–protein and protein–nucleic acid interactions in MS2 bacteriophage stability. FEBS J 273: 1463–1475 [DOI] [PubMed] [Google Scholar]
- Nielsen J, Kristensen MA, Willemoes M, Nielsen FC, Christiansen J 2004. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: A cooperative mechanism providing RNP stability. Nucleic Acids Res 32: 4368–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH 2003. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol 13: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott AM, Lago H, Adams CJ, Ashcroft AE, Stonehouse NJ, Stockley PG 2000. RNA aptamers for the MS2 bacteriophage coat protein and the wild-type RNA operator have similar solution behavior. Nucleic Acids Res 28: 489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS, Al-Bitar L 2001. Isolation of viral coat protein mutants with altered assembly and aggregation properties. Nucleic Acids Res 29: e113 doi: 10.1093/nar/29.22.e113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS, Lim F 1996. Complementation of RNA binding site mutations in MS2 coat protein heterodimers. Nucleic Acids Res 24: 2352–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AJ, Peabody DS 2001. Asymmetric interactions in the adenosine-binding pockets of the MS2 coat protein dimer. BMC Mol Biol 2: 6 doi: 10.1186/1471-2199-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparanese D, Lee CH 2007. CRD-BP shields c-myc and MDR-1 RNA from endonucleolytic attack by a mammalian endoribonuclease. Nucleic Acids Res 35: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano JE 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145–154 [DOI] [PubMed] [Google Scholar]
- Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S 2006. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol 175: 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidensdorfer D, Stohr N, Baude A, Lederer M, Kohn M, Schierhorn A, Buchmeier S, Wahle E, Huttelmaier S 2009. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15: 104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Reed R 2003. Purification of functional RNA-protein complexes using MS2-MBP. Curr Protoc Mol Biol 27: 27.3 doi: 10.1002/0471142727.mb2703s63 [DOI] [PubMed] [Google Scholar]