Abstract
MicroRNAs are small noncoding RNAs that serve as important regulators of eukaryotic gene expression and are emerging as novel diagnostic and therapeutic targets for human diseases. Robust and reliable detection of miRNAs is an essential step for understanding the functional significance of these small RNAs in both physiological and pathological processes. Existing methods for miRNA quantification rely on fluorescent probes for optimal specificity. In this study, we developed a high-performance real-time reverse transcriptase-polymerase chain reaction (RT-PCR) assay that allows specific and rapid detection of mature miRNAs using a fast thermocycling profile (10 sec per cycle). This assay exhibited a wide dynamic range (>7 logs) and was capable of detecting miRNAs from as little as 1 pg of the total RNA or as few as 10 cells. The use of modified reverse-transcription oligonucleotides with a secondary structure and hemi-nested reverse PCR primers allowed excellent discrimination of mature miRNAs from their precursors and highly homologous family members using SYBR Green I. Using a novel approach involving uracil-DNA glycosylase treatment, we showed that carryover of the reverse transcription oligonucleotide to the PCR can be successfully eliminated and discrimination between miRNA homologs could be further enhanced. These assays were further extended for multiplexed detection of miRNAs directly from cell lysates without laborious total RNA isolation. With the robust performance of these assays, we identified several miRNAs that were regulated by glial cell-line-derived neurotrophic factor in human glioblastoma cells. In summary, this method could provide a useful tool for rapid, robust, and cost-effective quantification of existing and novel miRNAs.
Keywords: microRNA, quantification, real-time RT-PCR, hemi-nested primer, deoxyuridine, SYBR Green I
INTRODUCTION
MicroRNAs (miRNAs) are small noncoding RNAs (∼22 nucleotides [nt]) that were discovered as important post-transcriptional regulators of gene expression in metazoans (Bartel 2004). While expression of miRNAs is critical in various physiological processes (Harfe 2005; Miska 2005; Carthew 2006; Lindsay 2008), dysregulation of miRNAs is implicated in pathologies of many human diseases, such as cancer (Visone and Croce 2009), muscle disorders (Chen et al. 2009a), and neurodegeneration (Hebert and De Strooper 2009). Recently, mature miRNAs were found to be remarkably stable in blood, and thus hold great promise as potential noninvasive biomarkers of human diseases (Chen et al. 2008; Mitchell et al. 2008). To date, hundreds of unique miRNAs have been identified in many species and each of these is predicted to regulate diverse target genes (Bartel 2009). With the continual discovery of more miRNAs by both in silico prediction and in vivo validation (Mendes et al. 2009), profiling of miRNA expression remains an essential tool not only for assessment of the distribution and regulation of miRNAs, but also for identification of novel biomarkers and potential therapeutic targets.
Currently, mature miRNAs can be detected by either direct or indirect methods. Although direct detection methods (e.g., fluorescent, colorimetric, and electrical-based methods) can minimize variations introduced during sample measurements, these methods are limited by low assay sensitivity and poor discrimination among the miRNA homologs (for review, see Hunt et al. 2009). Indirect detection methods primarily include Northern blotting, microarray, and reverse transcription-polymerase chain reaction (RT-PCR). Although widely used, both the Northern blotting and microarray methods are semiquantitative and suffer from poor sensitivity and they require large amounts of the starting RNA. Although microarrays offer high-throughput detection of miRNAs and the potential capability of absolute quantification (Bissels et al. 2009), a recent study showed that in comparison real-time PCR remains superior in sensitivity and specificity (Chen et al. 2009b). Recent attempts to measure miRNAs with isothermal methods have met with some success, but are labor intensive (Cheng et al. 2009; Yao et al. 2009). To date, real-time RT-PCR remains the most sensitive and efficient method for quantification of RNA species. The TaqMan probe-based real-time RT-PCR has been reported and widely used for efficient and specific detection of miRNAs. However, due to an additional probe hydrolysis step, TaqMan assays were not compatible with fast thermocycling protocols for rapid detection of miRNAs. Furthermore, with the escalating identification of hundreds of candidate miRNAs by deep sequencing (Bar et al. 2008; Goff et al. 2009), the design of the TaqMan probe for each of the novel miRNAs is not only cost prohibitive, but also technically challenging, and it faces practical difficulties (Varkonyi-Gasic et al. 2007). Attempts have been made to improve miRNA detection without reliance on fluorescent probes (Raymond et al. 2005; Shi and Chiang 2005; Sharbati-Tehrani et al. 2008); however, these assays usually involved multiple sample processing steps (Shi and Chiang 2005) and suffered from a limited dynamic range of detection (Raymond et al. 2005) and/or poor specificity against homologous miRNAs (Raymond et al. 2005; Shi and Chiang 2005; Sharbati-Tehrani et al. 2008). It has been noticed that a consensus strategy of these assays and some other TaqMan assays (Varkonyi-Gasic et al. 2007; Yang et al. 2009) is to use a universal or common reverse PCR primer and/or a fluorescent probe for amplification and detection of multiple miRNAs. In these assays, specificity of real-time PCR is only achieved by the forward PCR primer, which is not sufficient for discrimination of many homologous miRNAs. Furthermore, it is yet to be determined whether these assays are capable of rapid, multiplexed, and direct detection of miRNAs without RNA isolation.
In this study, we present a novel approach by coupling a deoxyuridine-incorporated RT oligonucleotide with a secondary structure and a hemi-nested real-time PCR for the rapid and robust quantification of mature miRNAs directly from cultured cells. This miRNA assay showed a high dynamic range, efficiency, and sensitivity of detection and excellent discrimination of the target mature miRNA against its precursor form and homologous family members. Instead of using a fluorescent probe, we found that a hemi-nested reverse PCR primer could drastically enhance the assay specificity and allow quantification of subzeptomole amounts of miRNA using SYBR Green I under rapid thermocycling conditions. Using multiplexed miRNAs assays, we identified six miRNAs up-regulated by GDNF through MEK-ERK1/2 pathway in human glioblastoma cells. Furthermore, we showed that miRNAs can be efficiently quantified directly from ten to 1000 cells using this multiplexed assay without laborious RNA isolation.
RESULTS
Overview of a hemi-nested real-time RT-PCR assay
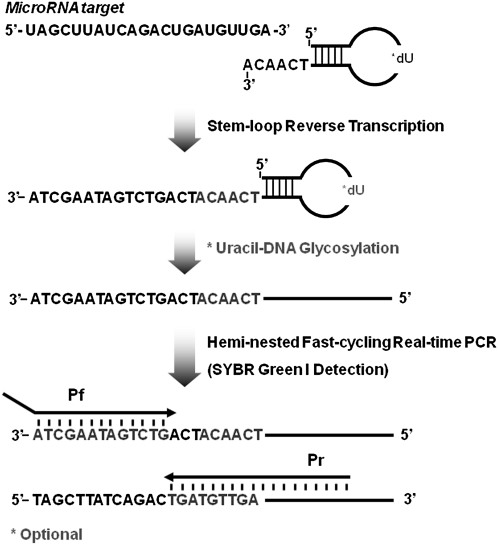
In the present study, we have designed a facile method for specific detection of mature miRNAs by hemi-nested real-time RT-PCR (Fig. 1). Initially, we noticed that in a number of previous reports (Chen et al. 2005; Raymond et al. 2005; Shi and Chiang 2005; Duncan et al. 2006; Varkonyi-Gasic et al. 2007; Sharbati-Tehrani et al. 2008; Yang et al. 2009), the reverse PCR primers were designed to anneal directly to the sequences in the RT oligonucleotide. To achieve specificity, a unique miRNA-specific fluorescent probe is required to discriminate the targets from the nonspecific amplicons (Chen et al. 2005). In the case of some TaqMan-based assays, this specificity was compromised for the convenience of throughput by using one common probe for detection. We hypothesized that increased specificity can be achieved by designing a hemi-nested reverse PCR primer instead of using a common or universal reverse PCR primer. To test this hypothesis, mature miRNA was first reverse transcribed at 42°C by the RT oligonucleotide, which adopted a stable stem–loop secondary structure during RT. The cDNA sample was then amplified using a tagged forward primer (Pf) and a hemi-nested reverse primer (Pr), where 3–5 nt extend beyond the RT oligonucleotide. Amplification of the cDNA sample was monitored in real-time PCR using SYBR Green I. Optionally, the RT oligonucleotide can be incorporated with deoxyuridine (dU) residues and the cDNA generated was subsequently treated with uracil-DNA glycosylase (UDG) at 37°C before amplification. To assist in the user-defined design of any real-time RT-PCR assay, a flowchart was presented using hsa-miR-21 (hereafter miR-21) as an example (Supplemental Fig. 1).
FIGURE 1.
Schematics for the design of a hemi-nested real-time RT-PCR assay. Pf, forward primer; Pr, reverse primer; dU, deoxyuridine.
Performance of the hemi-nested real-time RT-PCR assay
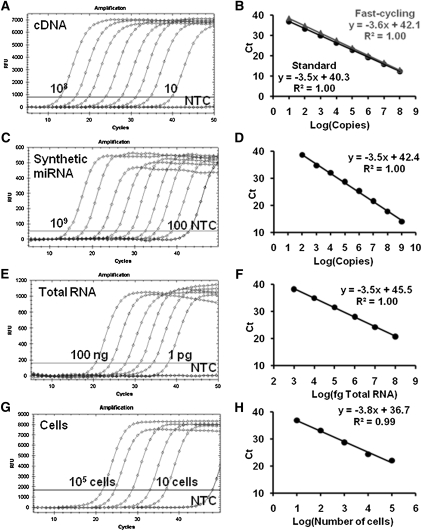
The performance of the hemi-nested real-time PCR assay was first evaluated using miR-21 cDNA dilutions under both fast (10 sec per cycle) and standard (75 sec per cycle with a 15-sec denaturation and 60-sec annealing/extension) thermocycling profiles. The assay exhibited excellent dynamic range and linearity under both cycling profiles (Fig. 2A,B), demonstrating the robustness of this assay for rapid detection of miRNAs. To evaluate the performance of both RT and PCR, synthetic miR-21 was diluted over seven orders of magnitude (from 109 to 100 copies per RT) and quantified by hemi-nested real-time RT-PCR. The excellent linearity of the standard curve suggested that the miR-21 assay had a wide dynamic range of at least 7 logs and was able to detect as few as 100 copies (subzeptomoles) per RT reaction (Fig. 2C,D).
FIGURE 2.
Performance of the miR-21 hemi-nested real-time RT-PCR assay. (A,B) miR-21 cDNA (108–10 copies) were amplified by real-time PCR assay under fast-cycling conditions compared with standard cycling protocol. (C,D) Dilutions of the synthetic miR-21 miRNA (109–102 copies), (E,F) dilutions of total RNA from A172 cells (from 100 ng to 1 pg), and (G,H) total RNA isolated from dilutions of U251 cells (100,000–10 cells) were reverse transcribed with the miR-21 RT oligonucleotide. The cDNA samples (10% v/v) were amplified by real-time PCR along with the nontemplate control (NTC). Amplification plots (A,C,E,G) and standard curves (B,D,F,H) of the assay are shown. Standard curves are plotted as Ct versus Log (starting material per RT).
We next examined the performance of this assay in detecting miRNA from total RNA samples. Total RNA dilutions from 100 ng to 1 pg (Fig. 2E,F) and the total RNA isolated from 100,000 to 10 cells (Fig. 2G,H) were quantified using a miR-21 hemi-nested real-time RT-PCR assay. The standard curves again showed excellent linearity (Fig. 2F,H), suggesting that this assay was capable of detecting miRNAs with high reliability from a minute amount (1 pg) of total RNA or from as few as 10 cells. The capability of this assay in quantifying mature miRNA from total RNA without RNA fractionation was further supported by total RNA spike-in experiments. Here, 100 ng of total RNA from U251 cells, which did not contain let-7d and let-7e (data not shown), was spiked with varying amounts of synthetic let-7d or let-7e miRNAs. The presence of total RNA did not affect the quantification of mature let-7d/let-7e miRNAs (Supplemental Fig. 2).
Hemi-nested real-time RT-PCR assays for three other miRNAs (miR-24, miR-92, and miR-218) were also evaluated (Supplemental Fig. 3). All three assays showed excellent dynamic range (at least 7 logs), sensitivity (103–104 copies per RT), and RT-PCR efficiency (88%–100%). Furthermore, these assays showed specific amplifications of target miRNAs from both synthetic standards and total RNA samples. The sizes of the amplicons were verified by gel electrophoresis (Supplemental Fig. 4).
Specific detection of mature against precursor miRNAs
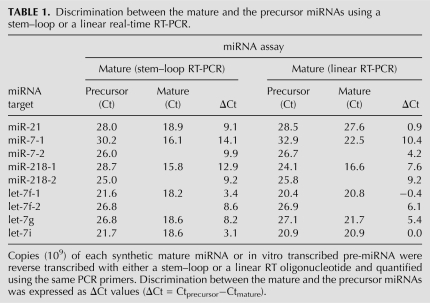
The capability of the hemi-nested real-time RT-PCR assay in discriminating mature against precursor miRNAs was investigated using nine mature miRNAs and their corresponding precursors (miR-21, miR-7-1, miR-7-2, miR-218-1, miR-218-2, let-7f-1, let-7f-2, let-7g, and let-7i). For comparison, we also evaluated linear RT oligonucleotides that do not form favorable secondary stem–loop structures during RT (ΔG ≥ −0.5 kcal/mol). For better comparison, these stem–loop RT oligonucleotides and their corresponding linear RT oligonucleotides were designed to (1) differ by only four to five bases at 5′; (2) have identical GC content and similar Tm (<1°C difference); and (3) have an identical 3′ sequence so that the same primers can be used in PCR.
The same amount (109 copies per RT) of mature miRNAs and pre-miRNAs was individually quantified using either stem–loop or linear RT and hemi-nested real-time PCR. Depending on the nine pre-miRNAs examined, mature miRNAs were detected three to 14 cycles (average ΔCt of 8.7 cycles) earlier than pre-miRNAs using stem–loop RT oligonucleotides. In contrast, the assays using linear RT oligonucleotides were less discriminative (average ΔCt of 4.8 cycles) and failed in discriminating some of the mature miRNAs from their precursors, such as pre-miR-21, pre-let-7f-1, and pre-let-7i (Table 1).
TABLE 1.
Discrimination between the mature and the precursor miRNAs using a stem–loop or a linear real-time RT-PCR.
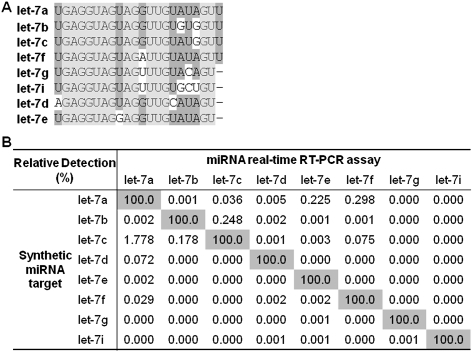
Discrimination of human let-7 miRNA homologs
Several miRNA families (e.g., hsa-let-7 and hsa-miR-30) consist of highly homologous miRNAs that differ by only a single or a few nucleotides. The eight let-7 family miRNAs share up to 63.6% overall sequence identity, among which let-7a and let-7c, let-7a and let-7f, and let-7b and let-7f differ only by a single nucleotide (Fig. 3A). In this study, hemi-nested real-time RT-PCR assays for each let-7 miRNA were designed. Each assay was used to amplify all eight synthetic let-7 miRNAs and the relative detection was compared. Briefly, the specific reverse transcription of each let-7 miRNA (109 copies per RT) was first achieved using the stem–loop RT oligonucleotide targeting the less homologous 3′ region. Discrimination of let-7 miRNAs was further improved by a miRNA-specific forward PCR primer and a hemi-nested reverse primer, preferably with a miRNA-specific 3′-terminal sequence. All eight of the let-7 assays showed excellent discrimination against the homologous miRNAs with less than 1% nonspecific detection, except for let-7a assay with 1.778% relative detection against let-7c (Fig. 3B). For let-7 miRNAs that differ by 2 nt or more, these assays were able to specifically detect the target miRNA with less than 0.3% cross-target amplification. This result suggests that the hemi-nested real-time RT-PCR assay was capable of discriminating the highly homologous miRNAs at levels comparable to fluorescent probe-based real-time RT-PCR assays (Chen et al. 2005), with inexpensive SYBR Green I detection chemistry.
FIGURE 3.
Discrimination of human let-7 homologs. (A) Sequence alignment of the eight let-7 family miRNAs. (B) Relative detection (%) of each let-7 miRNA by specific hemi-nested real-time RT-PCR assays.
Improved assay performance by the dU-incorporated RT oligonucleotide and the UDG treatment
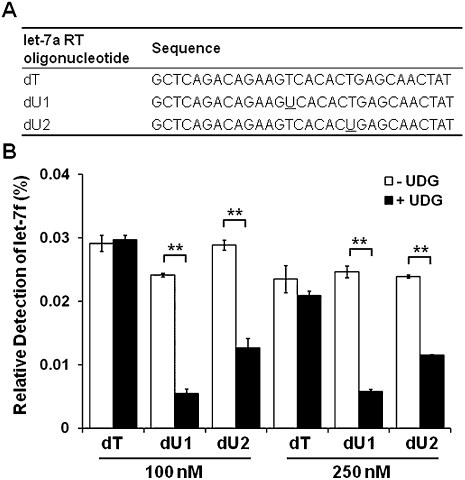
Discrimination between certain highly homologous miRNAs can be further improved using a novel strategy involving a UDG treatment. In this study, we observed that the amount of RT oligonucleotides carried over was able to serve as amplification primers during PCR, although with poorer efficiency (Supplemental Fig. 5). Interestingly, after the treatment with UDG, the dU-incorporated RT (dU-RT) oligonucleotide, but not the standard RT (dT-RT) oligonucleotide, was not able to serve as a reverse PCR primer (Supplemental Fig. 5B,C). We then hypothesized that discrimination between let-7 miRNA homologs may be further improved with a similar strategy. Indeed, while the standard (dT) RT oligonucleotide for let-7a showed a relative detection of 0.03% against let-7f miRNA; let-7a RT oligonucleotides with dU incorporation at either the loop (dU1) or the stem (dU2) region were significantly less capable of cross-amplifying let-7f after treatment with UDG (Fig. 4). Both the let-7a dT and dU RT oligonucleotides were able to prime reverse transcription of let-7a equally well (data not shown).
FIGURE 4.
Specificity of the hemi-nested real-time RT-PCR assay using dU-incorporated RT oligonucleotides. (A) Sequence comparison of dT and dU RT oligonucleotides. The dU residues are underlined. (B) Relative detection of the let-7f miRNA by the let-7a assay. Copies (109) of let-7a or let-7f were reverse transcribed using 100 or 250 nM of the dT, dU1, or dU2 RT oligonucleotide. The cDNA samples (10% v/v) were treated with or without UDG and amplified using let-7a-specific PCR primers. Nonspecific amplification of the let-7f miRNA was calculated and expressed as a percentage of relative detection. Error bars indicate standard deviations of quadruplicate measurements. Significant differences in relative detection between the UDG treated and nontreated samples were calculated by Student's t-test; **P < 0.001.
Application of multiplexed assays to identify GDNF-induced miRNA expressions in U251 cells
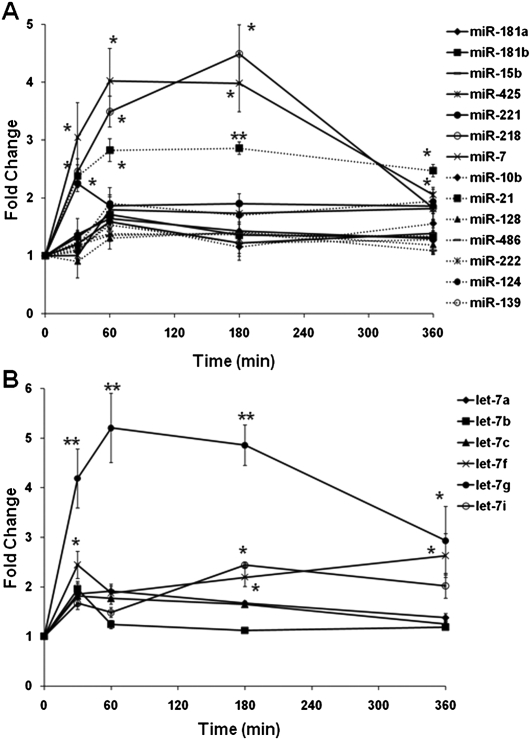
To evaluate the robustness of this miRNA detection method, hemi-nested real-time RT-PCR assays were designed and applied to identify GDNF-regulated miRNAs in U251 cells. A total of 26 miRNAs were examined including 18 miRNAs (miR-7, -10b, -15b, -21, -124, -128, -137, -139, -146b, -181a, -181b, -181c, -218, -221, -222, -425, -451, and -486) reported to be dysregulated in the human glioblastoma (Pang et al. 2009) and eight let-7 family miRNAs. Twenty miRNAs (except for miR-137, -146b, -181c, -451, let-7d, and let-7e) were found to be expressed in U251 cells (data not shown). Stimulation of the cells with GDNF induced time-dependent activation of ERK1/2 MAPK, which was inhibited by the pretreatment of MEK inhibitor U0126 (Supplemental Fig. 6). We then quantified the temporal regulation of expressions of these 20 miRNAs in the U251 cells by GDNF. Interestingly, GDNF stimulation induced time-dependent up-regulation of miR-7, -21, -218 (Fig. 5A), and let-7f, -7g, -7i (Fig. 5B). No significant regulation of the other miRNAs was observed.
FIGURE 5.
GDNF regulated miRNA expressions in U251 cells. Regulation of selected mature miRNAs (A,B) were quantified by hemi-nested real-time RT-PCR and expressed as fold changes to nonstimulated control samples. Error bars indicate standard deviations of triplicate measurements. Significant differences in gene expression between treated and control samples were calculated by Student's t-test; *P < 0.01; **P < 0.001.
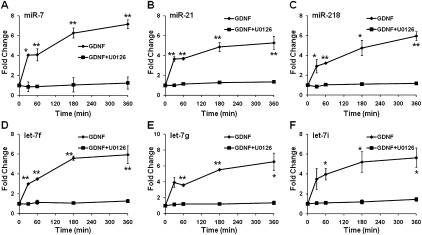
We then developed a multiplexed assay for simultaneous reverse transcription of the 26 miRNAs (corresponding to 24 RT oligonucleotides) and subsequent detection of individual miRNAs by hemi-nested real-time PCR. Using this 24-plex assay, we found that up-regulation of the six miRNAs by GDNF was effectively inhibited by the pretreatment of the cells with U0126, suggesting that the MEK-ERK1/2 signaling pathway was required for GDNF-induced miRNA expressions (Fig. 6). A similar result was obtained by a standard single-plex assay using miR-218 and let-7i as examples (Supplemental Fig. 7).
FIGURE 6.
Multiplexed quantification of the six GDNF regulated miRNAs. Expressions of miR-7 (A), miR-21 (B), miR-218 (C), let-7f (D), let-7g (E), and let-7i (F) were quantified by multiplexed real-time RT-PCR assays of 24 miRNAs. Regulation of these miRNAs was expressed as fold changes to nonstimulated control samples. Error bars indicate standard deviations of triplicate measurements. Significant differences in gene expression between treated and control samples were calculated by Student's t-test; *P < 0.01; **P < 0.001.
Direct and multiplex detection of miRNAs in cell lysates
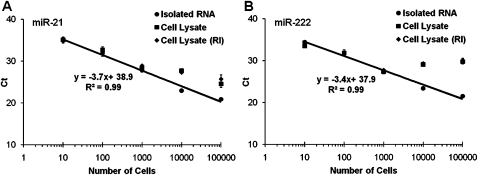
Direct detection of miRNAs from cell lysates can avoid the time-consuming multistep RNA isolation process and dramatically increase the throughput of the assay. We then further investigated the capability of the hemi-nested real-time RT-PCR assay in the direct and multiplex quantification of miRNAs from 10 to 100,000 cultured U251 cells in 96 wells. The isolated total RNAs from these cells were used as controls for quantification. Similar to Figure 2H, the excellent linearity of the miR-21 and miR-222 standard curves of the isolated RNA controls suggested that miRNAs can be reliably quantified from the total RNA extracted from as few as 10 cells (Fig. 7). Importantly, both miR-21 and miR-222 can be detected equally well from the lysates of 10–1000 cells using this multiplex assay, as compared with the isolated RNA controls. However, it is worthy to note that the direct quantification of miRNAs was compromised with the lysates from near-confluent (104 cells per well) or over-confluent (105 cells per well) cell densities. Interestingly, no significant difference was observed in the detection of miRNAs from the cell lysates with or without the RNase inhibitors, suggestive of the remarkable stability of mature miRNAs.
FIGURE 7.
Direct and multiplexed detection of miRNAs from cell lysates. U251 cells cultured in 96-wells at various densities (10, 100, 1000, 10,000 and 100,000 cells per well) were directly lysed and reverse transcribed in a reaction mixture containing 24 miRNA RT oligonucleotides. The cDNA samples (2.5% v/v) generated from isolated RNA or cell lysates were amplified by (A) miR-21 and (B) miR-222 real-time PCR assays. Standard curves for isolated RNA were plotted as Ct versus Log (cells per RT); RI, RNase inhibitor.
DISCUSSION
It is now known that miRNAs play important roles in modulating gene expression and it has been suggested that up to 60% of human genes are targeted by miRNAs (Friedman et al. 2009). With the increased interest in the expression profiles of miRNAs, rapid, robust, and cost-effective methods for the detection of mature miRNAs are highly desirable. The hemi-nested real-time RT-PCR assays described herein were simple to design, showed excellent performance, and provided the flexibility for the design of any miRNAs that may be identified in the future.
With synthetic miRNA targets, the hemi-nested real-time RT-PCR assay showed a wide dynamic range of at least 7 logs, high sensitivity of as few as 100 molecules per RT (subzeptomoles), and high RT-PCR efficiency of >90%. Besides the relative quantification of miRNA expression, the synthetic miRNA standards could also allow the absolute quantification of miRNA expression from total RNA samples. In contrast to the previously reported miRNA real-time PCR assays that were performed under standard thermocycling profiles of 45–75 sec per cycle (Chen et al. 2005; Raymond et al. 2005; Shi and Chiang 2005; Sharbati-Tehrani et al. 2008; Yang et al. 2009), our assay was capable of fast thermocycling (10 sec per cycle) without any modification of the reaction mixtures. The fast-cycling capability of this assay may, in part, be attributed to the short amplicon (<50 base pairs) generated by hemi-nested primers and the rapid fluorescence acquisition of SYBR Green I without the necessity of probe hydrolysis.
Specific and sensitive quantification of mature miRNA from total RNA samples usually requires size fractionation and preamplification, respectively. It has been reported that besides additional sample handling steps, size-fractionation can result in a consistent loss of miRNAs (Wang et al. 2007) and preamplification efficiency is significantly affected by the number of A bases in miRNAs (Mestdagh et al. 2008). With our miRNA quantitative assay, small amounts of the total RNA (from 1 pg to 100 ng) and the total RNA isolated from as few as 10 cells can be efficiently detected without the need for fractionation or preamplification. Previously, Megaplex miRNA assays for simultaneous reverse transcription of 220–450 miRNAs have been applied to quantify miRNA expression from a minute amount of the total RNA or a single cell (Tang et al. 2006; Mestdagh et al. 2008). Because a much lower concentration of the RT oligonucleotide was used for each miRNA, preamplification was required prior to real-time PCR, especially when the starting total RNA was less than 350 ng. In this study, we have shown that a multiplex of 24 miRNAs resulted in conclusions that were comparable to the single-plexed assays without reduction of RT oligonucleotide concentration or the necessity of preamplification. Therefore, we suggest that a multiplex of a relatively small number of miRNAs, with this method, could reduce the sample and reagent requirements and yet allow reliable quantification of miRNAs. These multiplexed assays can further be used for the direct quantification of miRNAs from cultured cells in 96 wells. Therefore, the method reported herein is capable of reliable, rapid, and high-throughput quantitative profiling of miRNA expressions, not only from minute amounts of isolated RNAs, but also directly from lysed cells without the need for laborious, time-consuming methods of RNA isolation.
In this study, specific detection of mature miRNAs from precursors is achieved by RT oligonucleotides with a stem–loop secondary structure. Although the expression levels of precursor miRNAs often correlate with mature miRNAs (Calin et al. 2004; Schmittgen et al. 2004), it is not uncommon that the maturation of miRNA can be regulated, whereby precursor miRNAs were expressed, but their mature forms were undetectable (Ambros et al. 2003; Michael et al. 2003; Wulczyn et al. 2007; Schmittgen et al. 2008). In order to discriminate between the regulation of miRNA processing and maturation, specific assays for both mature and precursor miRNAs are highly desirable. We found that the stem–loop, but not the linear RT oligonucleotides preferentially reverse transcribed the mature but not the precursor miRNAs. This strategy has been previously applied successfully for the specific reverse transcription of the sense strand of the replicating retrovirus (Anwar et al. 2006). It has been suggested that linear RT oligonucleotides should contain at least 7 nt of miRNA-specific sequence for efficient reverse transcription of mature miRNAs (Raymond et al. 2005). The poor discrimination of mature against pre-miRNAs by the linear RT oligonucleotides (with six miRNA-specific nucleotides) used in this study is likely due to the inefficient reverse transcription of mature miRNAs, consistent with the previous findings (Chen et al. 2005). These results suggested that the stem–loop secondary structure may stabilize the short base-pairing between the RT oligonucleotide and the mature miRNA during reverse transcription.
miRNAs are grouped into families (e.g., let-7 family) based on an identical seed sequence that spans 2–7 nt at 5′ of miRNAs, which is a critical determinant of target recognition (Lewis et al. 2005). Although miRNAs of the same family are likely to share some common targets and functions, specific members of the family may be involved in certain physiological processes and disease states. For instance, let-7b was found to be specifically up-regulated with age and responsible for the declined stem cell self-renewal (Nishino et al. 2008). Low expression of let-7d was suggested as a prognostic marker for head and neck squamous cell carcinoma (Childs et al. 2009) and let-7i was identified as a novel biomarker for human epithelial ovarian cancer (Yang et al. 2008). As such, specific and quantitative detection of these miRNAs is required for the development of biomarkers and for studies on the biogenesis of miRNAs. Previously, discrimination of let-7 homologs was achieved with TaqMan real-time RT-PCR assays, whereby after a miRNA-specific RT, a common reverse primer was used for PCR and a miRNA-specific TaqMan probe was required for discrimination (Chen et al. 2005). Assays with a similar strategy, but without the use of fluorescent probes, suffered from a significantly poorer discrimination of the let-7 homologs (Raymond et al. 2005; Shi and Chiang 2005; Sharbati-Tehrani et al. 2008). We hypothesized and showed that the specificity of the assays can be dramatically enhanced by a hemi-nested miRNA-specific reverse PCR primer and do not require the use of a fluorescent probe. Using this method, all the let-7 miRNAs can be specifically detected using the inexpensive SYBR Green I, with excellent discrimination, comparable, and in some cases, superior to the TaqMan real-time RT-PCR (Chen et al. 2005).
The presence of excess RT oligonucleotides can serve as reverse primers during PCR, which will contribute to nonspecific amplification. This is problematic, especially when the forward primer in PCR can also hybridize to shared sequences (e.g., cDNA from homologous miRNAs). A common practice to mitigate this problem is to reduce the carry-over of the RT oligonucleotides by diluting the cDNA samples after RT. However, this approach will invariably reduce the assay sensitivity, especially with low abundance target miRNAs. For low abundance miRNAs, the assay is further improved by modifying the RT oligonucleotides with deoxyuridine residues and treating the cDNA with UDG prior to real-time PCR. UDG was first purified from Escherichia coli and found to be able to cleave uracil from uracil-containing DNA (Lindahl et al. 1977). Release of uracil residues results in apyrimidinic sites on the DNA, which can block the replication by the DNA polymerase during PCR (Longo et al. 1990). It is likely that the UDG treatment disabled the unused dU RT oligonucleotides from serving as PCR primers. This is a simple and attractive approach in situations where a maximum specificity is desired or discrimination is difficult to achieve during RT (e.g., let-7a and let-7f share identical 3′ sequences for RT priming).
It is known that miRNAs are differentially expressed in glioblastoma as compared with normal brain and many of these dysregulated miRNAs are involved in glioblastoma growth, invasion, and chemoresistance (Lawler and Chiocca 2009; Li et al. 2009). Using the hemi-nested real-time RT-PCR assays for selected miRNAs, GDNF was found to regulate the expressions of miR-7, miR-21, miR-218, let-7f, let-7g, and let-7i through the MEK-ERK1/2 MAPK signaling pathway. The functional significance of these GDNF-induced miRNAs remains to be investigated. Using the rapid and high-throughput method developed herein, it is now feasible for drug screening by profiling changes in miRNA expression.
The number of miRNAs in the public domain is currently more than 10,000 (Griffiths-Jones et al. 2008) and expected to increase with both in silico prediction and in vivo validation, such as deep sequencing. With the continuous demand for miRNA expression analysis, the hemi-nested real-time RT-PCR assay presented in this study provides a high-performance method for the rapid and reliable detection of functional mature miRNAs.
MATERIALS AND METHODS
Mature and precursor miRNA templates and sequence analysis
Mature miRNAs were synthesized by Proligo (Sigma) or Integrated DNA Technologies (Coralville). T7 promoter-sequenced tagged PCR primers were used to clone the precursor miRNAs (Supplemental Table 1). The purified PCR products were validated by sequencing and subjected to the MEGAscript T7 Transcription Kit (Ambion) for in vitro transcription of the precursor miRNAs according to the manufacturer's instructions. Both mature and precursor miRNAs were quantified by spectrophotometry and diluted to the desired concentration to serve as the standard. The sequence alignment and phylogenetic analysis of the miRNA homologs were performed using Vector NTI software version 8.0 (Invitrogen).
Cell culture and total RNA isolation
Human glioblastoma cell lines A172 and U251 were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Sigma) and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin) in a 5% CO2 humidified incubator at 37°C. The U251 cells were seeded at 200,000 cells per well in a six-well plate for 24 h and incubated with a FBS-free DMEM medium for 12–16 h. The cells were then stimulated with 100 ng/mL of GDNF for defined periods of time in DMEM. For the MEK inhibitor studies, the cells were preincubated with 5 μM U0126 (Promega) for 45 min in DMEM prior to the GDNF stimulations. The total RNA from the untreated or the GDNF-treated glioblastoma cells was isolated using TRIzol reagent (Invitrogen) in the presence of 20 μg/mL linear acrylamide (Ambion), according to the manufacturer's instructions. The extracted RNA samples were then quantified using spectrophotometry and the integrity was examined by denaturing RNA gel electrophoresis.
Reverse transcription (RT)
Total RNA samples were treated with RNase-free DNase I (Promega) at 37°C for 30 min. The DNase was inactivated at 80°C for 5 min. For the RT of the primary and precursor miRNAs, 100 ng of DNase I-treated total RNA or dilution of the in vitro transcribed precursor miRNA standards was initially heated at 80°C in the presence of 150 nM of gene-specific reverse primers for 5 min, snapped chilled on ice, and reverse transcribed (1X buffer, dNTPs [10 mM], dithiothreitol [DTT], RNase inhibitor, Thermoscript [15 U]), as specified by the manufacturer (Invitrogen). The reverse transcription was carried out at 60°C for 45 min and terminated by a further incubation at 85°C for 5 min. For detection of the precursors, both RT and real-time PCR were carried out with prevalidated gene-specific primers as reported previously (Jiang et al. 2005).
For RT of mature miRNAs, 100 ng of DNase I-treated total RNA or dilutions of synthetic human mature miRNA standards was reverse transcribed using 100 U of Improm II (Promega) and 100 nM of either the stem–loop or the linear RT oligonucleotide in a total volume of 10 μL for 30 min at 42°C. The reaction was terminated by heating at 70°C for 5 min. For multiplex RT of 24 mature miRNAs, 100 ng of DNase I-treated total RNA samples was reverse transcribed using 400 U of Improm II and 100 nM of each RT oligonucleotide (2.4 μM of total) in a total volume of 40 μL for 30 min at 42°C. The cDNA samples were then used for real-time PCR using miRNA-specific primers. Primers for both RT and real-time PCR are listed in Supplemental Table 2. The linear RT oligonucleotides are listed in Supplemental Table 3.
Uracil-DNA Glycosylase (UDG) treatment
The RT oligonucleotides with deoxyuridine (dU) incorporation were synthesized by Integrated DNA Technologies. Reverse transcription was performed as described above. The cDNA samples were then treated with 5 U UDG (New England Biolabs) at 37°C for 10 min. The reaction was inactivated at 95°C for 10 min and subjected to real-time PCR.
Detection of miRNA without RNA isolation
U251 cells in a 96-well plate at various cell densities (10–105 cells per well) were washed once with PBS, lysed, and reverse transcribed directly in the wells with 40 μL of the RT mixture containing 0.5% Triton X-100 (Thermo Fisher Scientific), 400 U Improm II (Promega), and 100 nM of each of the RT oligonucleotides (2.4 μM of total) in the presence or absence of RNase inhibitor SUPERase-In (1 U/μL, Ambion) for 30 min at 42°C. For comparison, the total RNA from the identical densities of cells was isolated and reverse transcribed in the same RT mixture. The cDNA samples were then used for real-time PCR.
Real-time PCR
Real-time PCR was performed on the CFX96 system (Bio-Rad) using SYBR Green I. Real-time PCR for the precursor miRNA and the U6 snRNA was performed according to previous reports (Schmittgen et al. 2004; Jiang et al. 2005). Fast thermocycling of miRNA cDNAs was performed after 10 min of initial denaturation at 95°C followed by 50 cycles of 5-sec denaturation at 95°C and 5-sec annealing/extension at 60°C. One microliter of each cDNA sample was subjected to real-time PCR in a total volume of 25 μL in 1X XtensaMix-SG (BioWORKS), containing 2.5 mM MgCl2, 100 nM of each primer, and 1.25 U KlearTaq DNA polymerase (KBiosciences). The threshold cycles (Ct) were calculated automatically using the CFX manager software (Bio-Rad). Where applicable, all miRNA expression levels were normalized to the U6 snRNA expression.
Calculation of real-time RT-PCR assay efficiency and specificity
The efficiency and specificity of real-time RT-PCR were determined as previously described (Too 2003). Briefly, 10-fold dilutions of miRNA were subjected to real-time RT-PCR. The standard curve of the miRNA was obtained by plotting Ct versus Log (copies) of the synthetic miRNA dilutions. The assay efficiency was calculated by (101/S – 1) × 100%, where S is the slope of the standard curve. For the calculation of specificity, identical amounts (109 copies per RT) of perfectly matched miRNAs and mismatched miRNAs were quantified by real-time RT-PCR. The change in the threshold cycles, ΔCt, was then calculated as Ctperfectly matched miRNA – Ctmismatched miRNA. The relative detection of the mismatched miRNAs was then calculated by 10ΔCt/S × 100%.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENT
We thank the Department of Biochemistry (National University of Singapore) for providing the necessary facilities to carry out this study.
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2001610.
REFERENCES
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13: 807–818 [DOI] [PubMed] [Google Scholar]
- Anwar A, August JT, Too HP 2006. A stem–loop-mediated reverse transcription real-time PCR for the selective detection and quantification of the replicative strand of an RNA virus. Anal Biochem 352: 120–128 [DOI] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. 2008. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26: 2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, Bosio A 2009. Absolute quantification of microRNAs by using a universal reference. RNA 15: 2375–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. 2004. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci 101: 11755–11760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW 2006. Gene regulation by microRNAs. Curr Opin Genet Dev 16: 203–208 [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res 33: e179 doi: 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. 2008. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997–1006 [DOI] [PubMed] [Google Scholar]
- Chen JF, Callis TE, Wang DZ 2009a. microRNAs and muscle disorders. J Cell Sci 122: 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Gelfond JA, McManus LM, Shireman PK 2009b. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics 10: 407 doi: 10.1186/1471-2164-10-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhang X, Li Z, Jiao X, Wang Y, Zhang Y 2009. Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification. Angew Chem Int Ed Engl 48: 3268–3272 [DOI] [PubMed] [Google Scholar]
- Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB, et al. 2009. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol 174: 736–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan DD, Eshoo M, Esau C, Freier SM, Lollo BA 2006. Absolute quantitation of microRNAs with a PCR-based assay. Anal Biochem 359: 268–270 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Davila J, Swerdel MR, Moore JC, Cohen RI, Wu H, Sun YE, Hart RP 2009. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS One 4: e7192 doi: 10.1371/journal.pone.0007192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD 2005. MicroRNAs in vertebrate development. Curr Opin Genet Dev 15: 410–415 [DOI] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B 2009. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci 32: 199–206 [DOI] [PubMed] [Google Scholar]
- Hunt EA, Goulding AM, Deo SK 2009. Direct detection and quantification of microRNAs. Anal Biochem 387: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Lee EJ, Gusev Y, Schmittgen TD 2005. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res 33: 5394–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler S, Chiocca EA 2009. Emerging functions of microRNAs in glioblastoma. J Neurooncol 92: 297–306 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu H, Chen J, He J, Yu H 2009. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res 1286: 13–18 [DOI] [PubMed] [Google Scholar]
- Lindahl T, Ljungquist S, Siegert W, Nyberg B, Sperens B 1977. DNA N-glycosidases: Properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem 252: 3286–3294 [PubMed] [Google Scholar]
- Lindsay MA 2008. MicroRNAs and the immune response. Trends Immunol 29: 343–351 [DOI] [PubMed] [Google Scholar]
- Longo MC, Berninger MS, Hartley JL 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93: 125–128 [DOI] [PubMed] [Google Scholar]
- Mendes ND, Freitas AT, Sagot MF 2009. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res 37: 2419–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J 2008. High-throughput stem–loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res 36: e143 doi: 10.1093/nar/gkn725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael MZ, van Holst SMOC, Pellekaan NG, Young GP, James RJ 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 1: 882–891 [PubMed] [Google Scholar]
- Miska EA 2005. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 15: 563–568 [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci 105: 10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ 2008. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 135: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JC, Kwok WK, Chen Z, Ng HK 2009. Oncogenic role of microRNAs in brain tumors. Acta Neuropathol 117: 599–611 [DOI] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM 2005. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11: 1737–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Jiang J, Liu Q, Yang L 2004. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32: e43 doi: 10.1093/nar/gnh040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C 2008. Real-time PCR quantification of precursor and mature microRNA. Methods 44: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbati-Tehrani S, Kutz-Lohroff B, Bergbauer R, Scholven J, Einspanier R 2008. miR-Q: A novel quantitative RT-PCR approach for the expression profiling of small RNA molecules such as miRNAs in a complex sample. BMC Mol Biol 9: 34 doi: 10.1186/1471-2199-9-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Chiang VL 2005. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39: 519–525 [DOI] [PubMed] [Google Scholar]
- Tang F, Hajkova P, Barton SC, Lao K, Surani MA 2006. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res 34: e9 doi: 10.1093/nar/gnj009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Too HP 2003. Real time PCR quantification of GFRα-2 alternatively spliced isoforms in murine brain and peripheral tissues. Brain Res Mol Brain Res 114: 146–153 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP 2007. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12 doi: 10.1186/1746-4811-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Croce CM 2009. MiRNAs and cancer. Am J Pathol 174: 1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ach RA, Curry B 2007. Direct and sensitive miRNA profiling from low-input total RNA. RNA 13: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R 2007. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 21: 415–426 [DOI] [PubMed] [Google Scholar]
- Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L, et al. 2008. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res 68: 10307–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Schmuke JJ, Flagg LM, Roberts JK, Allen EM, Ivashuta S, Gilbertson LA, Armstrong TA, Christian AT 2009. A novel real-time polymerase chain reaction method for high throughput quantification of small regulatory RNAs. Plant Biotechnol J 7: 621–630 [DOI] [PubMed] [Google Scholar]
- Yao B, Li J, Huang H, Sun C, Wang Z, Fan Y, Chang Q, Li S, Xi J 2009. Quantitative analysis of zeptomole microRNAs based on isothermal ramification amplification. RNA 15: 1787–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]