Abstract
Porous polyoxometalate nanocapsules of Keplerate type are known to exhibit the functionality of biological ion channels, however, their use as artificial ion channel is tempered by the high negative charge of the capsules, which renders their spontaneous incorporation into a lipid bilayer membrane unlikely. In this letter we report coarse-grained molecular dynamics simulations that demonstrate a route for embedding negatively charged nanocapsules into lipid bilayer membranes via self-assembly. A homogeneous mixture of water, cationic detergent, and phospholipid was observed to spontaneously self-assemble around the nanocapsule into a layered, liposome-like structure, where the nanocapsule was enveloped by a layer of cationic detergent followed by a layer of phospholipid. Fusion of such a layered liposome with a lipid bilayer membrane was observed to embed the nanocapsule into the lipid bilayer. The resulting assembly was found to remain stable even after the surface of the capsule was exposed to electrolyte. In the latter conformation, water was observed to flow into and out of the capsule as Na+ cations entered, suggesting that a polyoxometalate nanocapsule can form a functional synthetic ion channel in a lipid bilayer membrane.
Transport of ions across the cell's boundaries is a fundamental process underlying many cellular functions, from the secretion of hormones to the beating of a heart. A lipid bilayer membrane encapsulating a biological cell is virtually impervious to ions. Therefore, a special class of proteins residing in the cell's membranes is required to direct traffic of ions into and out of the cell and cellular organelles.1,2 The structures of some ion channel proteins are known in atomic detail.3–7 However, to fully understand the physico-chemical processes underlying the function of these remarkable systems, their synthetic analogs with functionality embedded by design have to be synthetized and characterized.8–11 In this manuscript we investigate the feasibility of assembling a biological lipid membrane with a particular class of compounds which exhibit the functionality of ion channels. Our molecular dynamics simulations suggest a route for transforming these systems into hybrid bio-inorganic assemblies which can model ion channels in biological settings.
A route to creating novel, synthetic models of ion channels may lie in the chemistry of polyoxomolybdates (POMs). Here we consider a porous POM nanocapsule:12–15
| (1) |
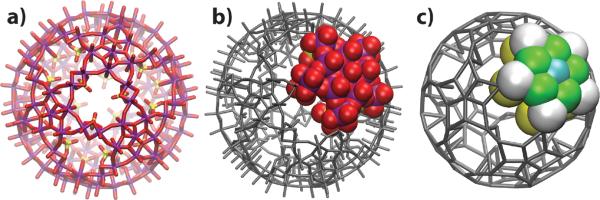
made of twelve pentagonal molybdenum oxide regions, each linked to five other regions by binuclear molybdenum oxide-ligand type groups. The connection of the 12 pentagonal units forms 20 circular pores on the surface of the capsule. The structure of the capsule is shown in Fig. 1 a; the total charge of the capsule is −72e, where e is the charge of a proton. The atomic coordinates of the capsule were taken from an earlier publication.16
Figure 1.
Mo132 type Keplerate capsule. a) All-atom structure of the capsule, oriented with a pore in the center. Three sulfate groups line each pore, forming a triangle with sides of 5.6 Å.17 Molybdenum atoms are shown in purple, oxygen in red and sulphur in yellow. b–c) All-atom (b) and CG (c) models of the porous nanocapsule. The capsule considered here is made of 12 repeats of a pentagonal unit. Thus, specifying the mapping of one all-atom pentagonal unit and its linker units to the CG representation is sufficient to describe the mapping of the entire capsule. The colors in the CG representation highlight the four different types of beads used to model the capsule. The detailed description of the CG model is provided in Supporting Information.
The high charge of the porous nanocapsules makes them soluble in water and attractive to cations. Studies have shown that water and small inorganic cations can enter the capsule cavity through the pores.12,17,18 Moreover, some organic cations, such as guanidinium, can reversibly block the passage of smaller ions through the pores, mimicking ligand-induced gating of biological ion channels.17,19 The structure of the groups lining the pore and hence the coordination of the passing ions can be adjusted, which provides a route for customizing the selectivity of the ion transport.12,17 Thus, it appears that the porous nanocapsules can serve as models of biological ion channels, whose structure can be engineered at the atomic level. To fully reproduce typical physiological conditions, a porous nanocapsule would, ideally, be placed in a lipid bilayer membrane. An underlying difficulty is, however, that the nonpolar, hydrophobic interior of the latter is not compatible with the high charge of the capsule.
In this manuscript, we detail molecular dynamics (MD) simulations demonstrating a route for embedding POM porous nanocapsules in lipid bilayer membranes via self-assembly. It was previously shown that such porous nanocapsules can be encapsulated with dimethyldioctadecylammonium (DODA) surfactants, amphiphilic cations consisting of a cationic ammonium group attached to two long nonpolar tails.14,20 Here, we show that the DODA–capsule interaction can be exploited to create a liposome-like, water soluble structure that can fuse with a lipid bilayer membrane and thereby embed a POM porous nanocapsule in the bilayer.
State of the art, all-atom MD simulations are limited to the timescale of hundreds of nanoseconds, which is considerably less than the timescales of liposome formation or fusion with a lipid bilayer membrane. Recently, a reliable coarse-grained (CG) model of lipid bilayers became available, which greatly extended the timescale and the scope of the MD method.21–26 In a CG model, groups of atoms are replaced by representative `beads' that interact much like atoms in the corresponding all-atom model. Due to the simplification of the potential energy function (the force field), CGMD simulations can employ larger timesteps (20 fs), which, in addition to the reduction of the number of particles, dramatically increases the span of an MD simulation, up to 1,000 times.22
To model interactions of our porous nanocapsule with a mixture of detergent, lipid and water we extended the Marrink CG model of phospholipids.21 DODA was coarse-grained using existing bead types available to describe lipids, with the addition of two angle parameters taken from similar lipid configurations. For the CG model of the porous nanocapsule, we created new bead types using parameters derived from the capsule's shape. Strong bond and angle forces were introduced to keep the capsule rigid; the equilibrium values of the bonds and angles were derived from the all-atom structure. Nonbonded forces between the capsule and other beads were taken from the Marrink model. Thus, the beads comprising our porous nanocapsule were either charged, polar or apolar.21 Figures 1 b and c illustrate the all-atom and CG models of the porous nanocapsule (1), respectively. When six copies of the porous nanocapsule were placed in a 120 Å ×120 Å ×120 Åbox filled a 0.25 M solution of NaCl and simulated for 0.4 μs, no aggregation of the capsules was observed. Complete details of our CG model can be found in Supporting Information.
To test our parameters for the porous nanocapsule and DODA detergent, we simulated self-assembly of DODA with the nanocapsule in octane, a simple nonpolar solvent. Our simulations confirmed that DODA molecules, starting from a random distribution, would aggregate to form a steady-state hydrophobic spheroid. In accord to previous modeling work20 on a porous CH3COO-linker nanocapsule (the total charge of which is −42e), the concentration of DODA was observed to decrease with the radial distance from the capsule's surface. In our case, the total charge of the capsule–DODA spheroid varied from −2 to −10e in the steady state. These simulations are discussion in more detail in Supporting Information.
Once we were satisfied with our description of the system, we investigated the feasibility of obtaining a water-soluble, liposome-like structure encapsulating our porous nanocapsule by spontaneous self-assembly. Several systems were prepared, each containing a porous nanocapsule, DODA detergent, palmitoyloleoylphosphatidylcholine (POPC) lipid, water and enough ions to neutralize the system. The DODA to POPC ratio was held constant at 1:2.2; several concentrations of DODA/POPC were tested. In the starting conformation, DODA, POPC, water and ions were randomly distributed around the capsule, as would result from sonication in experiment. A summary of all runs performed is given in Table S4 in Supporting Information.
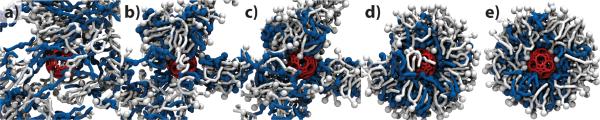
Our simulations revealed that a DODA/POPC mixture can spontaneously form a liposome-like structure around the porous nanocapsule. Fig. 2 illustrates a typical self-assembly trajectory. In the capsule-liposome assembly, DODA forms a shell surrounding the capsule with its ammonium groups facing the capsule's surface. The charge of this inner layer is about 68±3e. DODA's hydrophobic tails intermix with the hydrophobic tails of POPC, while the polar headgroups of POPC face the water, forming the outer layer of the liposomal structure. The presence of DODA molecules in the outer shell of the liposome renders the overall charge of the capsule liposome at about +50e. The structural details, as well as the kinetics of self-assembly at different conditions, are provided in Fig. 3.
Figure 2.
Self-assembly of a capsule liposome. The snapshots illustrate the molecular configuration obtained after 0 (a) 3.36 (b) 7.36 (c) 41.68 (d) 100.24 (e) ns of CGMD simulation. The porous nanocapsule is shown in red, DODA in blue, POPC in white, water and ions are not shown. An animation illustrating this simulation is available in Supporting Information.
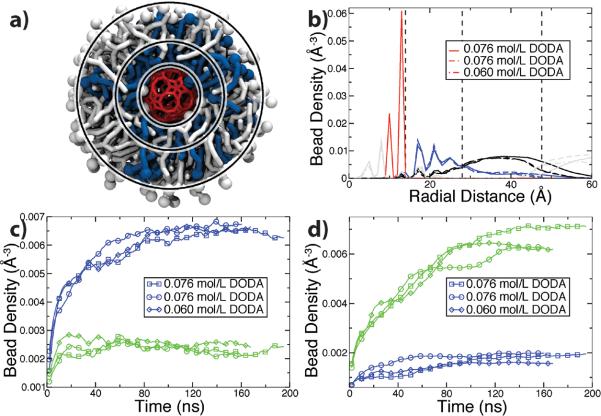
Figure 3.
Structure and kinetics of capsule-liposome assembly. (a) Structure of the capsule liposome. The porous nanocapsule (red) is surrounded by a shell of DODA (blue) and a secondary shell of POPC (white). The three rings separate the capsule from DODA (inner), DODA from POPC (middle) and POPC from water (outer) according to the density plots, panel b. (b) Density of the porous nanocapsule (red), DODA (blue), POPC (black) and water (grey) versus radial distance from the center of the capsule. Dashed lines correspond to the location of the rings in panel a. Results of three independent runs are shown. (c,d) Density of DODA (blue) and POPC (green) beads versus time in three simulations. The densities were computed by averaging over the volume between the inner and middle (c) and middle and outer (d) rings.
As detailed in Supporting Information, some simulations did not result in a liposome-like assembly. Successful assembly was observed for the range of DODA concentration between 0.060 to 0.076 M. For lower DODA concentrations, there was not enough DODA/POPC molecules to completely cover the porous nanocapsule. For higher DODA concentrations, an interconnected network of POPC and DODA was formed instead of discrete liposomes. Furthermore, increasing ion concentrations to 0.25 M was enough to arrest the growth of the liposomal aggregate, because of the competition between the ammonium groups of DODA and the Na+ ions for binding the porous nanocapsule. Figure S4 in Supporting Information shows an example of such an outcome, where Na+ ions surround the capsule, preventing DODA from approaching the capsule's surface.
Next, we tested whether a capsule liposome could deliver our porous nanocapsule into a lipid bilayer membrane. Fusion of two liposomes was previously studied using all-atom27 and coarse-grained systems.28–30 Note that even using a CG model, spontaneous fusion is too slow of a process to be observed in an MD simulation starting from a random conformation.30 To make the observation of liposome fusion possible within the time scale of an MD simulation, several methods have been used, for example, restraining the two liposomes in close proximity to one another through either external force28 or by introducing a linker molecules,30 applying external forces to move the liposomes into contact29 or manually removing water between adjacent liposome headgroups.27
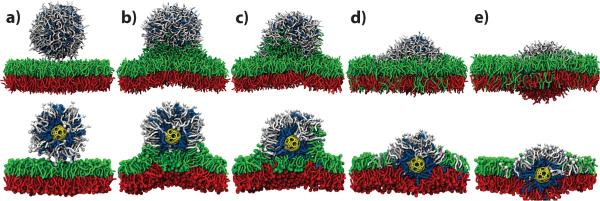
We performed CGMD simulations starting from a model of a fully formed capsule liposome and a patch of pre-equilibrated POPC bilayer membrane solvated in 0.25 M aqueous solution of NaCl. Fig. 4a shows one initial configuration. To induce liposome fusion, we tried four different methods. In our first method, we accelerated the approach of the liposome to the lipid membrane by pulling the center-of-mass of the capsule with a constant velocity towards the membrane using the standard protocol of Steered Molecular Dynamics (SMD).31 When the distance between the center of the capsule and the center of the membrane approached 60 Å (Fig. 4b), the SMD force was turned off. Alternatively, we placed the liposome a certain distance away from the surface of the membrane and let the system evolve freely. This method was successful for a closest-approach distance of 7 Å between the phosphate groups of the liposome and the bilayer. Both SMD and free equilibration methods led to essentially the same conformation (Fig. 4b), as a fusion stalk quickly formed, connecting the liposome to the bilayer. At distances greater than 12 Å, the liposome did not fuse with a lipid bilayer within the time scale of our simulation, even when a half of the lipid headgroups were replaced with negatively charged moieties or when a voltage bias was imposed across the system.
Figure 4.
Fusion of a liposome with a lipid bilayer. Snapshots illustrate the state of the CGMD simulation after 0 (a), 125.2 (b), 431.6 (c), 607.36 (d) and 940.4 (e) ns. The porous nanocapsule is shown in yellow, DODA in blue, POPC in white, the top and bottom leaflets of the POPC bilayer in green and red, respectively. For each panel, the image in the bottom row is a cut-away view of the same configuration as shown in the top row. An animation illustrating this simulation is available in Supporting Information.
Once fusion was initiated, our systems were allowed to evolve under constant pressure. During this period of equilibration, lipid and detergent from the capsule liposome mixed with the lipids from the upper leaflet of the membrane. This process slowed the fusion and the bilayer became bowed because of the imbalance of the number of lipids in the leaflets of the bilayer. This effect is an artifact of the periodic boundary conditions used in our simulations, which do not permit the lipid bilayer to form a curved membrane with the radius of curvature greater than half the system size. A larger patch of a lipid bilayer is expected to flatten out, distributing the lipids from the liposome over a large area and thereby effectively equalizing the density of lipids in both leaflets.
To eliminate the effect of the periodic boundary conditions in our simulations of the liposome-bilayer fusion, we used the following method to equalize the number of lipids in the leaflets. The coordinates of randomly selected lipids from the top leaflet at the periphery of the system were reflected with respect to the plane passing through center of the bilayer, which effectively inserted them into the bottom leaflet. Following a short period of relaxation where all beads of the capsule, DODA and POPC, except for those moved to the bottom leaflet, were harmonically constrained, the system was equilibrated until the area of the bilayer became steady. This procedure was then repeated until the number of lipids in the leaflets became equal. This allowed the membrane to flatten out and expand, leaving the capsule in the middle of the bilayer, surrounded by DODA and POPC, Fig. 4e. An animation illustrating the fusion process is available in Supporting Information.
At the end of the fusion simulations, the capsule remained trapped inside the bilayer, not directly connected to the solvent. To test whether the capsule would be stable in a configuration open to water on either side of the membrane, we created a model of such a system by removing POPC and DODA above and below the capsule and filled the empty space with water and ions. Following a brief period where the capsule, DODA and POPC were constrained while the water and ions were allowed to assume a new equilibrium conformation, the system was simulated free of restraints under constant pressure conditions. Fig. 5 illustrates one system obtained at the end of a 351-ns simulation. In the equilibrium configuration, water was observed to flow into and out of the capsule as Na+ cations entered, showing that the capsule can be both stable in this conformation and transport ions across the membrane, even though the capsule's charge is reduced by the presence of the cationic DODA. An animation illustrating this simulation is available in Supporting Information. Inside the capsule, water beads were seen to form highly ordered configurations. From time to time, they were observed to rearrange themselves in quick cascades resulting in a net flow of water. From these simulations, we estimated the osmotic permeability to water of the porous nanocapsule at 3.5×10−15 cm3/s, which is roughly three orders of magnitude less than that of the α-hemolysin channel.32 This estimate should be taken with caution, as it is not known how accurate the CGMD method is for simulating ion and water conductance through narrow pores. Furthermore, based on the present isolated capsule (1) it has been proven that an equilibrium between Li+ cation uptake and release exists, and that encapsulated Li+ cations can be exchanged by Na+.17,18,33–36 This means there is the option to study counterion transport systematically.17 Furthermore, temperature dependent 7Li NMR studies provided a rough estimate of the rate of Li+ ion exchange between the inside and outside of the capsule: ~3×104 per minute at 343K.37
Figure 5.
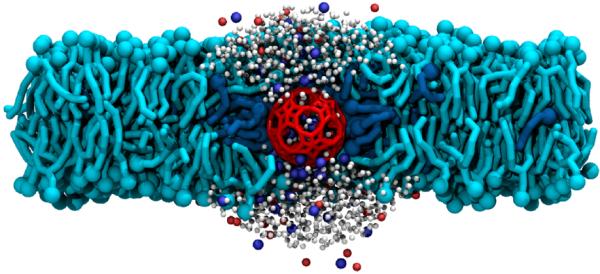
Porous nanocapsule as an ion channel. The snapshot illustrates the outcome of a CGMD simulation of a porous nanocapsule (red) held in a POPC bilayer membrane (cyan) by DODA (blue). The top of the capsule is open to water (white) and ions (blue Na+, red Cl−). Note the ions inside and around the capsule.
Here we have detailed and simulated a method for embedding porous POM nanocapsules in lipid bilayer membranes. By combining the highly charged porous nanocapsule (1) with a cationic amphiphilic surfactant (DODA) and a model phospholipid (POPC) we demonstrated that liposome-like, water soluble structures form spontaneously around the capsule. These liposome-like structures were shown to insert the porous nanocapsule into lipid bilayer membranes. This process can be further optimized by using specific lipid types that facilitate membrane fusion and by covalently linking DODA-like moieties to the porous nanocapsule to prevent competition between DODA and smaller cations which can destabilize the capsule/bilayer assembly. Future studies are required to investigate the long-time stability of the capsule/membrane system and develop a method for exposing the surface of the capsule to the electrolyte via self-assembly. The latter will require employing lipids of specific spontaneous curvature, charge and hydrophobic tail length. The successful inclusion of porous nanocapsules in lipid bilayer membranes is the first step to creating a synthetic model of ion channels that would allow studies of ionic transport in greater detail than it is currently possible. In this context it is important that the capsule functionalities can be tuned by changing the internal ligands like sulphates (see formula above) and that the nanocapsules are stable in aqueous solutions [23].
Supplementary Material
Acknowledgement
This work was supported by the grant from the National Institutes of Health (PHS 5 P41-RR05969). Authors gladly acknowledge supercomputer time provided by the National Center for Supercomputing Applications (NCSA) via Large Resources Allocation Committee grant MCA05S028.
References
- 1.Hille B. Ionic channels of excitable membranes. 3rd ed. Sinauer Associates; Sunderland, MA: 2001. [Google Scholar]
- 2.Gouaux E, MacKinnon R. Nature. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- 3.Doyle DA, ao Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 4.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 5.Hilf R, Dutzler R. Nature. 2008;452:375. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 6.Long SB, Campbell EB, MacKinnon R. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 7.Jasti J, Furukawa H, Gonzales E, Gouaux E. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 8.Fyles T. Chem. Soc. Rev. 2007;36:335–347. doi: 10.1039/b603256g. [DOI] [PubMed] [Google Scholar]
- 9.Koert U, Al-Momani L, Pfeifer JR. Synthesis. 2004;2004:1129–1146. [Google Scholar]
- 10.Matile S, Som A, Sordé N. Tetrahedron. 2004;60:6405–6435. [Google Scholar]
- 11.Gokel G, Mukhopadhyay A. Chem. Soc. Rev. 2001;30:274–286. [Google Scholar]
- 12.Mueller A, Das SK, Talismanov S, Roy S, Beckmann E, Boegge H, Schmidtmann M, Merca A, Berkle A, Allouche L, Zhou Y, Zhang L. Angew. Chem. Int. Ed. 2003;42:5039–5044. doi: 10.1002/anie.200352358. [DOI] [PubMed] [Google Scholar]
- 13.Müller A, Roy S. J. Mater. Chem. 2005;15:4673–4677. [Google Scholar]
- 14.Müller A, Roy S. Eur. J. Inorg. Chem. 2005:3561–3570. [Google Scholar]
- 15.Müller A, Roy S. The Chemistry of Nanomaterials: Synthesis, Properties and Applications. Wiley-VCH; 2004. [Google Scholar]
- 16.Müller A, Krickemeyer E, Bögge H, Schmidtmann M, Botar B, Talismanova MO. Angew. Chem. Int. Ed. 2003;42:2085–2090. doi: 10.1002/anie.200351126. [DOI] [PubMed] [Google Scholar]
- 17.Merca A, Haupt ETK, Mitra T, Bögge H, Rehder D, Müller A. Chem. Eur. J. 2007;13:7650–7658. doi: 10.1002/chem.200700294. [DOI] [PubMed] [Google Scholar]
- 18.Rehder D, Haupt ETK, Bögge H, Müller A. Chem. Asian J. 2006;1–2:76–81. doi: 10.1002/asia.200600035. [DOI] [PubMed] [Google Scholar]
- 19.Müller A, Zhou Y, Bögge H, Schmidtmann M, Mitra T, Haupt E, Berkle A. Angew. Chem. Int. Ed. Engl. 2006;45:460. doi: 10.1002/anie.200501994. [DOI] [PubMed] [Google Scholar]
- 20.Volkmer D, Du Chesne A, Kurth D, Schnablegger H, Lehmann P, Koop M, Muller A. J. Am. Chem. Soc. 2000;122:1995–1998. [Google Scholar]
- 21.Marrink SJ, de Vries AH, Mark AE. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
- 22.Shih AY, Arkhipov A, Freddolino PL, Schulten K. J. Phys. Chem. B. 2006;110:3674–3684. doi: 10.1021/jp0550816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 24.Lopez CF, Nielsen SO, Moore PB, Klein ML. Proc. Natl. Acad. Sci. USA. 2004;101:4431–4434. doi: 10.1073/pnas.0400352101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace J, Sansom M. Chem. Phys. of Lipids. 2007;149:89–89. [Google Scholar]
- 26.Wong-Ekkabut J, Baoukina S, Triampo W, Tang I, Tieleman D, Monticelli L. Nature Nanotech. 2008;3:363. doi: 10.1038/nnano.2008.130. [DOI] [PubMed] [Google Scholar]
- 27.Knecht V, Marrink S. Biophys. J. 2007;92:4254. doi: 10.1529/biophysj.106.103572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marrink SJ, Mark AE. J. Am. Chem. Soc. 2003;125:15233–42. doi: 10.1021/ja0352092. [DOI] [PubMed] [Google Scholar]
- 29.Stevens MJ, Hoh JH, Woolf TB. Phys. Rev. Lett. 2003;91:188102. doi: 10.1103/PhysRevLett.91.188102. [DOI] [PubMed] [Google Scholar]
- 30.Kasson PM, Kelley NW, Singha N, Vrljic M, Brunger AT, Pande VS. Proc. Natl. Acad. Sci. USA. 2006;103:11916–11921. doi: 10.1073/pnas.0601597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isralewitz B, Izrailev S, Schulten K. Biophys. J. 1997;73:2972–2979. doi: 10.1016/S0006-3495(97)78326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aksimentiev A, Schulten K. Biophys. J. 2005;88:3745–3761. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller A, Rehder D, Haupt E, Merca A, Bogge H, Schmidtmann M, Heinze-Bruckner G. Angew. Chem. Int. Ed. 2004;43:4466–4470. doi: 10.1002/anie.200453762. [DOI] [PubMed] [Google Scholar]
- 34.Haupt E, Wontorra C, Rehder D, Müller A. Chemical Communications. 2005;2005:3912–3914. doi: 10.1039/b506087g. [DOI] [PubMed] [Google Scholar]
- 35.Haupt ETK, Wontarra C, Rehder D, Merca A, Müller A. Chem. Eur. J. 2008 doi: 10.1002/chem.200801122. In Press. [DOI] [PubMed] [Google Scholar]
- 36.Rehder D, Haupt ETK, Müller A. Magn. Res. Chem. 2008 In Press. [Google Scholar]
- 37.Haupt ETK, Müller A. To be published.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.