Abstract
TolC is a multifunctional outer-membrane protein (OMP) of Escherichia coli that folds into a unique α/β-barrel structure. Previous studies have shown that unlike the biogenesis of β-barrel OMPs, such as porins, TolC assembles independently from known periplasmic folding factors. Yet, the assembly of TolC, like that of β-barrel OMPs, is dependent on BamA and BamD, two essential components of the β-barrel OMP assembly machinery. We have investigated the folding properties and cellular trafficking of a TolC derivative that lacks the entire signal sequence (TolCΔ2–22). A significant amount of TolCΔ2–22 was found to be soluble in the cytoplasm, and a fraction of it folded and trimerized into a conformation similar to that of the normal outer membrane-localized TolC protein. Some TolCΔ2–22 was found to associate with membranes, but failed to assume a wild-type-like folded conformation. The null phenotype of TolCΔ2–22 was exploited to isolate suppressor mutations, the majority of which mapped in secY. In the secY suppressor background, TolCΔ2–22 resumed normal function and folded like wild-type TolC. Proper membrane insertion could not be achieved upon in vitro incubation of cytoplasmically folded TolCΔ2–22 with purified outer membrane vesicles, showing that even though TolC is intrinsically capable of folding and trimerization, for successful integration into the outer membrane these events need to be tightly coupled to the insertion process, which is mediated by the Bam machinery. Genetic and biochemical data attribute the unique folding and assembly pathways of TolC to its large soluble α-helical domain.
INTRODUCTION
Escherichia coli has four distinct compartments – the cytoplasm, the inner membrane, the periplasm, and the outer membrane – each with a unique protein composition. Although all proteins are synthesized in the cytoplasm, they follow different trafficking pathways to reach their ultimate destination where they can function. Periplasmic and outer-membrane proteins (OMPs) are synthesized as preproteins with a cleavable N-terminal signal sequence. Preproteins are generally targeted to the inner membrane SecYEG translocon by cytoplasmic chaperones, such as SecB and the molecular ATPase motor SecA (Danese & Silhavy, 1998). Whereas the mechanism by which proteins are targeted to and through the inner membrane by the Sec machinery is relatively well understood, it is still unclear how proteins are subsequently assembled and inserted into the outer membrane. Studies involving the multiprotein Bam (β-barrel assembly machinery) complex indicate a general pathway by which β-barrel OMPs are targeted and inserted into the outer membrane (Voulhoux et al., 2003; Werner & Misra, 2005; Wu et al., 2005; Charlson et al., 2006; Malinverni et al., 2006; Jain & Goldberg, 2007). Following inner membrane translocation and cleavage of the signal sequence, unfolded mature β-barrel OMPs are released into the periplasm. There, a number of periplasmic folding factors act to maintain OMPs in a conformation competent for assembly and final outer membrane insertion and to minimize misfolded species (Danese & Silhavy, 1998; Mogensen & Otzen, 2005; Sklar et al., 2007a). Such factors include the general chaperone Skp (Missiakas et al., 1996; Schafer et al., 1999; Bulieris et al., 2003), the major peptidyl prolyl cis–trans isomerase SurA (Missiakas et al., 1996; Rouvière & Gross, 1996), the periplasmic protease/chaperone DegP (Strauch et al., 1989; Krojer et al., 2008), and lipopolysaccharide (de Cock & Tommassen, 1996; Kloser et al., 1996; Bulieris et al., 2003). Final assembly/insertion sites are represented by the hetero-oligomeric outer membrane complex BamABCDE (formerly YaeT-YfgL-NlpB-YfiO-SmpA) (Wu et al., 2005; Sklar et al., 2007b; Misra, 2007).
TolC is a minor but functionally important OMP of E. coli. It is the outer membrane component of various type I secretion systems and multidrug efflux pumps (Andersen, 2003), and is exploited by colicin E1 (Nagel de Zwaig & Luria, 1967; Masi et al., 2007), colicin 10 (Pilsl & Braun, 1995) and phage TLS (German & Misra, 2001) for import. The crystal structure of TolC from E. coli shows a distinctive and previously unknown fold (Koronakis et al., 2000). Three TolC protomers assemble to form a single conduit called a ‘channel-tunnel’ that extends across the outer membrane and into the periplasm. In contrast to porins, which are exclusively transmembrane β-barrel proteins, ∼70 % of the structure of TolC is made of water-soluble α-helices, resulting in an α/β-barrel. Little is known about the assembly pathway of TolC. For example, the hallmark of outer membrane porins is the presence of an aromatic amino acid at their C-terminal end, the removal of which has a dramatic impact on the folding of the porin monomers and their assembly into trimers (Struyve et al., 1991). TolC and channel-tunnel homologues lack such a conserved C-terminal residue. The essential proteins BamA and BamD are required for the final assembly and outer membrane insertion of the majority of β-barrel proteins, including TolC (Werner & Misra, 2005; Malinverni et al., 2006). A search for additional factors for TolC assembly in our laboratory has been unsuccessful (Werner et al., 2003; J. Werner & R. Misra, unpublished results). These observations raise the possibility that TolC could fold and trimerize spontaneously with the help of its large soluble α-helical domain. Here, we report studies of the folding, trimerization and membrane insertion of a signal sequence-less TolC (henceforth referred to as TolCΔ2–22). We found that a population of TolCΔ2–22 can fold and trimerize in the cytoplasm of E. coli into a conformation similar to that of the outer membrane-localized wild-type TolC. However, the TolCΔ2–22 trimers formed in the cytoplasm were not competent for insertion into purified outer membrane vesicles. These results are consistent with the view that in vivo folding/trimerization and subsequent membrane insertion are tightly coupled processes, with the latter being catalysed by the Bam machinery.
METHODS
Chemicals, bacterial strains and media.
For analytical ultracentrifugation, ultrapure sucrose and Histodenz were obtained from Boehringer Mannheim and Sigma, respectively. Detergents used were β-octyl-d-glucopyranoside (Glycon) and Triton X-100 (Calbiochem). All strains used in this study were derivatives of E. coli MC4100 (Casadaban, 1976). Strain RAM1129 (MC4100 ΔtolC : : Kmr) (Augustus et al., 2004) carrying plasmids pTrc99A-TolCS350F or pTrc99A-TolCP246R, S350C was used for PCRs. Strain RAM1330 (MC4100 Δara ΔtolC : : Kmr) was used for plasmid expression. Cultures were grown in Luria broth (LB) at 37 °C. When necessary, media were supplemented with ampicillin (100 μg ml−1). Arabinose (0.2 %) was used to induce the expression of the tolC genes under PBAD control.
Plasmid construction.
tolC DNA was amplified by PCR from a single colony of the appropriate strain, with primers 5′-GAGCCAGGTCATGA(BspHI)ACCTGATGC-3′ and 5′-GCTCTAGAAGCTTA(HindIII)GTGATGGTGATGGTGATGGTTACGGAAAGGGTTATGACC-3′, and cloned in-frame with the arabinose-inducible PBAD promoter in pBAD24 (Guzman et al., 1995). All plasmid-borne TolC proteins contained a His6 tag (indicated by italic type) at the C terminus to allow protein purification by immobilized-nickel affinity chromatography.
Cell fractionation.
Cultures of E. coli were grown to OD600 ∼0.2 and expression of plasmid-borne TolC proteins was induced with 0.2 % arabinose for 2 h at 37 °C. Routinely, cell pellets from 100 ml cultures were washed once and resuspended in 3 ml lysis buffer containing 10 mM Tris/HCl, pH 7.5, 1 mM MgCl2, 100 μg DNase I ml−1. Bacterial cells were broken by one passage through a French pressure cell at 750 p.s.i. (5167.5 kPa). Unlysed cells were removed by low-speed centrifugation (8000 g for 20 min) and supernatant containing the whole-cell extract was centrifuged at 105 000 g for 1 h. The resulting supernatant contained soluble cytoplasmic and periplasmic proteins. Final pellets, corresponding to the whole-cell envelopes, were resuspended in 200 μl 10 mM Tris/HCl, pH 7.5. Preparation of spheroplasts was carried out as described elsewhere (Betton & Hofnung, 1996).
Outer and inner membranes were separated by ultracentrifugation of whole-cell envelopes on a two-step sucrose gradient. The sucrose gradient was prepared by placing 3.8 ml 53 % (w/v) sucrose on top of 1 ml 70 % (w/v) sucrose. A 200 μl membrane sample was layered on top and gradients were centrifuged for 6 h at 120 000 g using an SW55 rotor (Beckman Coulter). Fractions (200 μl) were collected from top to bottom. Flotation gradient experiments using Histodenz, a non-toxic substitute for metrizamide, were conducted essentially as described previously (Misra et al., 1991).
Conformational assays.
Proteinase K treatment of whole cells in vivo has been described previously (Werner et al., 2003). The presence of high-molecular-mass TolC oligomers was tested in protein samples mixed with 2 % SDS loading buffer without boiling.
Purification of TolC and TolCΔ2–22.
Cultures of E. coli RAM1330 containing pBAD24-TolC and pBAD24-TolCΔ2–22 were grown as described above. Cells were resuspended in 20 mM sodium phosphate buffer (pH 7.5) and lysed by passage through a French pressure cell, and membranes were separated from soluble proteins by high-speed centrifugation as described above. The soluble fraction from RAM1330 (pBAD24-TolCΔ2–22) was adjusted to 150 mM NaCl and 20 mM imidazole. Membranes from RAM1330 (pBAD24-TolC) were homogenized in buffer A (20 mM sodium phosphate, pH 7.5, 150 mM NaCl, 5 % Triton X-100, 20 mM imidazole) and stirred for 1 h at room temperature. Insoluble material was then removed by centrifugation at 100 000 g for 1 h. Protein samples were applied onto a 5 ml HiTrap chelating column (Amersham) charged with Ni2+ and equilibrated with buffer B (20 mM sodium phosphate, pH 7.5, 250 mM NaCl) containing 1 % Triton X-100 and 20 mM imidazole. The column was washed with buffer B containing 1 % Triton X-100 and 100 mM imidazole, and then with buffer B containing 1 % β-octyl-d-glucopyranoside and 100 mM imidazole. TolC and TolCΔ2–22 were eluted with buffer B containing 1 % β-octyl-d-glucopyranoside and 300 mM imidazole. Fractions containing protein peaks from the affinity column were analysed by SDS-PAGE.
Electrophysiological analysis in planar lipid bilayers.
In a 0.5 % solution of 1,2-diphytanoyl-sn-glycero-3-phosphocholine in pentane, virtually solvent-free planar lipid bilayers were formed by the apposition of two monolayers on a 100 μm diameter hole from a thin Teflon film (10 μm) sandwiched between two Teflon half-cells. Voltage was applied through an Ag/AgCl electrode in the cis side with the trans side grounded. The electrolyte solution was 5 mM HEPES, pH 7.2, 1 M KCl. TolC and TolCΔ2–22 were added to the cis compartment at a final concentration of about 5 ng ml−1. All experiments were performed at room temperature.
SDS-PAGE and Western blot analyses.
Unless otherwise noted, all samples were boiled for 5 min in 2 % SDS loading buffer before proteins were separated on 11 % SDS-polyacrylamide mini gels. After electrophoresis, proteins were either stained with Coomassie blue or silver nitrate (SilverQuest, Invitrogen) or electrotransferred onto PVDF membranes (Immobilon-P, Millipore). Polyclonal rabbit antisera against TolC-MalE (1 : 5000), LamB (1 : 5000) and MalE (1 : 10 000) were from our laboratory stock. Polyclonal rabbit antibodies against GroEL were obtained from Sigma. Anti-rabbit alkaline horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence Western blotting reagents (Bio-Rad) were used for protein detection. For detection of DnaK, membranes were probed with a primary monoclonal mouse antibody against DnaK (Stressgen) and anti-mouse alkaline phosphatase-coupled secondary antibodies (Sigma) and developed by enhanced chemiluminescence (Amersham).
RESULTS
A large fraction of TolCΔ2–22 is soluble in the cytoplasm
In an effort to better understand the unusual assembly pathway of TolC, we constructed a plasmid from which TolC is expressed without its signal sequence (TolCΔ2–22). Additionally, we introduced the substitutions S350F and P246R–S350C, which had been previously shown to cause severe TolC assembly defects (Werner et al., 2003; Gerken & Misra, 2004), to see whether their presence also influences the folding pathway of TolCΔ2–22.
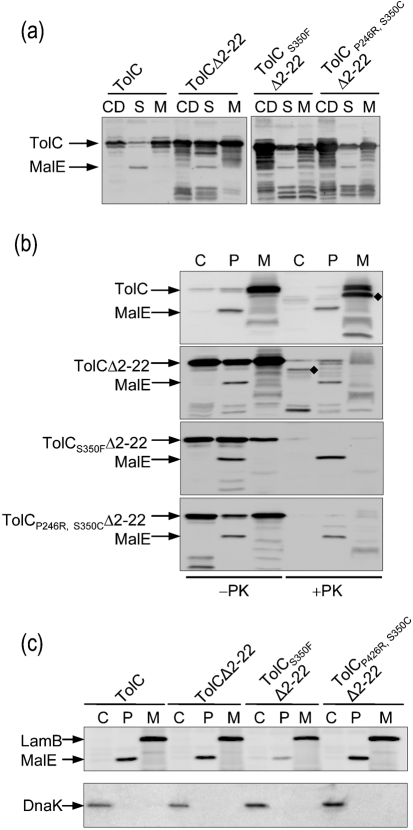
Fates of various TolC derivatives were examined from cells that had been fractionated into soluble (cytoplasm plus periplasm) and insoluble (membrane) fractions. These fractions, including the cell debris or protein aggregates obtained after low-speed centrifugation, were analysed by SDS-PAGE and TolC was detected after immunoblotting. As expected, wild-type TolC was present primarily in the membrane fraction and was barely detectable in the soluble whole-cell extract fraction obtained after high-speed centrifugation (105 000 g for 1 h; Fig. 1a). The detection of wild-type TolC in the low-speed pellet reflected the presence of either unlysed cells or membrane fragments; however, absence of the periplasmic maltose binding protein (MalE) from pellets obtained after low- and high-speed centrifugation showed that they were largely devoid of unbroken cells and soluble proteins. In contrast to wild-type TolC, a significant amount of TolCΔ2–22 was present in the soluble whole-cell extract fraction (Fig. 1a). Almost equal amounts of TolCΔ2–22 were also detected in pellets obtained after low- and high-speed centrifugation; again, absence of MalE from these pellets showed a lack of cross-contamination from soluble proteins and unbroken cells. The majority of TolCΔ2–22S350F and TolCΔ2–22P246R, S350C was recovered with cell debris obtained after low-speed centrifugation, reflecting elevated aggregation of these mutant proteins (Fig. 1a). Like wild-type TolC, all mutant TolCs were also readily detected from the high-speed membrane pellet fraction (Fig. 1a). The folding status of these populations and those present in the soluble fraction was investigated below.
Fig. 1.
Cellular fate of TolCΔ2–22 and influence of assembly-defective mutations. (a) Cell lysates were fractionated into cell debris (CD) after low-speed centrifugation, and soluble (S) and membrane (M) fractions after high-speed centrifugation. Protein samples were analysed by SDS-PAGE and Western blotting using antibodies raised against TolC fused to maltose binding protein (MalE). (b) Subcellular fractionation analysis of TolC from spheroplasts. Cytoplasmic (C), periplasmic (P) and membrane (M) fractions were obtained from cultures induced for 2 h and prepared via spheroplasts as described in Methods. The cell fractions were either untreated (−) or treated (+) with proteinase K (10 μg ml−1) at 30 °C for 10 min. TolC was detected by Western blotting. The diamonds (⧫) mark the location of the 46 kDa protease-resistant TolC fragment. (c) The presence of the cytoplasmic marker DnaK, the periplasmic marker MalE and the membrane marker LamB in each fraction isolated in (b) was analysed by immunoblotting with specific antibodies.
Fractionation and folding analyses of TolCΔ2–22 from spheroplasts
The detection of a significant amount of TolCΔ2–22 from soluble whole-cell extracts prompted us to carry out detailed cellular localization studies and assess the folding status of the protein. TolC, TolCΔ2–22 and TolCΔ2–22-derivatives were examined after fractionating spheroplasts into cytoplasm, periplasm, and membranes (Fig. 1b). As expected, wild-type TolC fractionated almost exclusively with the membranes (Fig. 1b). However, TolCΔ2–22 and its two mutant derivatives partitioned almost evenly to the cytoplasm, the periplasm and the membranes (Fig. 1b). In all of our fractionation experiments, the cytoplasmic (DnaK), periplasmic (MalE) and membrane (LamB) protein markers were properly localized to their respective cellular compartments, showing no cross-contamination and thus no fractionation artefacts (Fig. 1c). Moreover, the absence of DnaK in the spheroplast supernatants showed that these fractions were pure periplasmic fractions and that the cytoplasmic content had not been released into the periplasm (Fig. 1c). It is conceivable that some membrane-associated TolCΔ2–22 molecules are spontaneously flipped and released into the periplasm, just as has been found for the signal sequence-less PhoA and MalE variants (Derman et al., 1993).
The folding status of TolC molecules can be assessed by examining their sensitivity towards proteinase K (Werner et al., 2003). The C-terminal end of the 471-residue mature TolC is accessible from the periplasm, and treatment of membranes or permeabilized cells with proteinase K generates a stable membrane-bound N-terminal fragment (residues 1–451 of the mature TolC protein) of 46 kDa (Koronakis et al., 1997; Werner et al., 2003). Additionally, it has been shown that only fully assembled, membrane-inserted TolC shows this characteristic proteinase K digestion pattern, while periplasmic TolC assembly intermediates are completely degraded (Werner et al., 2003). Thus, the appearance of the 46 kDa TolC fragment signals that unfolded TolC monomers have trimerized and inserted into the outer membrane. Treatment of various cell fractions with proteinase K showed that membrane-localized wild-type TolC produced a 46 kDa fragment (Fig. 1b). In contrast, TolCΔ2–22 and its derivatives present in the membrane fractions were completely degraded by proteinase K, showing that they did not attain the correct folding status (Fig. 1b). The same was true for ‘periplasmically localized’ TolCΔ2–22 and its derivatives (Fig. 1b). These data showed that despite the fact that TolCΔ2–22 and mutant derivatives somehow associate with the membrane and find their way to the periplasm, these anomalously localized species are largely unfolded or incorrectly folded. Interestingly, however, while the two mutant derivatives of TolCΔ2–22 in the cytosol were also completely degraded by proteinase K, a significant population of cytosolic TolCΔ2–22 produced the 46 kDa fragment, showing that this population had achieved a folding status similar to that of fully assembled TolC in the outer membrane (Fig. 1b, +PK lanes).
Spontaneous tendency of TolCΔ2–22 to associate with the membranes
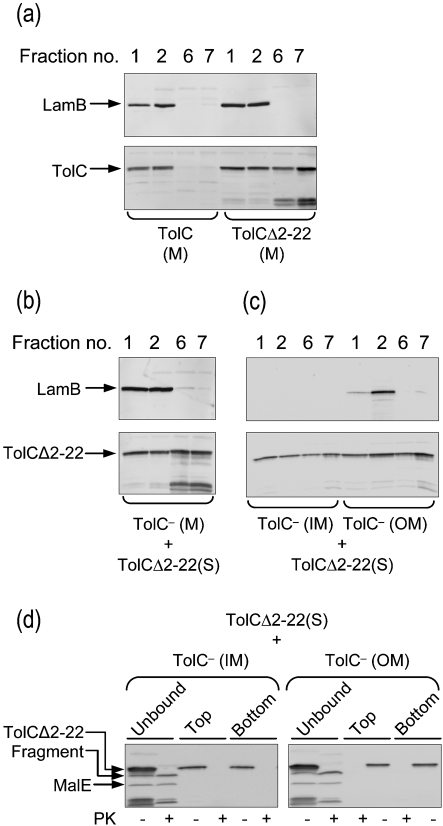
Fractionation of whole cells (Fig. 1a) and spheroplasts (Fig. 1b) consistently showed partitioning of a significant amount of TolCΔ2–22 with the membranes. To investigate in detail the nature of this membrane association and the folding status of the membrane-bound TolCΔ2–22, isolated membranes were floated in Histodenz gradients as described previously by Misra et al. (1991). In this procedure, membranes are placed at the bottom of the gradient solution, and after centrifugation, contaminant soluble proteins, protein aggregates and proteins that are loosely adhering to the membranes tend to remain at the bottom of the gradient, while integral membrane proteins or proteins that are strongly associating with the membranes tend to float to the top in the lower-density region of the gradient. Immunodetection of LamB served as a control for an integral membrane protein. As expected, LamB floated with the membranes in the top two fractions and was absent from the dense bottom fractions 6 and 7 (Fig. 2a). Wild-type TolC was also present only in the top fractions [Fig. 2a, TolC (M); fractions 1 and 2] and not in the bottom fractions [Fig. 2a, TolC (M); fractions 6 and 7]. In contrast, TolCΔ2–22 fractionated in roughly equal proportions in the top and bottom fractions of the gradient [Fig. 2a, TolCΔ2–22 (M)], showing the existence of both weakly and strongly membrane-associated populations. To test whether TolCΔ2–22 would spontaneously stick to membranes, soluble TolCΔ2–22 was incubated with purified membranes obtained from RAM1330 (ΔtolC). After 1 h incubation at room temperature, the mixture was subjected to flotation gradient analysis [Fig. 2b; TolC− (M) and TolCΔ2–22 (S)]. The fractionation profile was similar to that observed with TolCΔ2–22 (M) (Fig. 2a). The flotation of soluble TolCΔ2–22 with the TolC− membranes showed the tendency of TolCΔ2–22 to strongly and spontaneously associate with the membranes.
Fig. 2.
In vitro membrane association of TolCΔ2–22. (a) The membranes (M) from cells producing TolC or TolCΔ2–22 were separated from soluble proteins (S) and analysed by Histodenz density flotation gradients. Fractions were collected from top (1 and 2) to bottom (6 and 7) of the gradient. Note the presence of unknown TolC degradation products associated with the production of TolCΔ2–22. (b) Soluble TolCΔ2–22 was incubated with membranes obtained from RAM1330 (ΔtolC) and incubated for 1 h, after which the mixture was analysed by flotation gradient centrifugation. (c) Membranes of RAM1330 (ΔtolC) were separated into inner and outer membranes by sucrose density gradients. Fractions of equal volume of inner and outer membranes were mixed with the soluble fraction from cells producing TolCΔ2–22. After 1 h incubation at room temperature, the membranes were harvested by ultracentrifugation. Membrane-unbound proteins were withdrawn and membrane-bound proteins were subjected to flotation gradient centrifugation. (d) Proteinase K treatment of the membrane-unbound proteins and those in the top and bottom fractions from (c). LamB, TolCΔ2–22 and MalE were detected from various fractions by immunoblotting.
The tight partitioning of some TolCΔ2–22 with the membrane fraction raised the question of whether trimers of TolCΔ2–22 formed in the cytoplasm were competent for membrane insertion. If so, it was expected that trimeric TolCΔ2–22 could only insert into the outer membrane, and not the inner membrane, because the latter lacks the essential Bam complex. To test this, envelopes from RAM1330 (ΔtolC) were separated into inner and outer membranes by sucrose density gradients and fractions corresponding to the inner and outer membranes were then incubated with a soluble fraction containing TolCΔ2–22. After incubation for 1 h at room temperature, membranes were reisolated by ultracentrifugation and subjected to flotation gradient centrifugation, as described above (Fig. 2c). Immunodetection of cytoplasmic TolCΔ2–22 revealed it to be bound to both the inner and outer membrane (Fig. 2c, bottom panel), showing a tendency of soluble TolCΔ2–22 to ‘stick’ indiscriminately to the membranes. Immunoblot analysis of LamB from membrane fractions showed the presence of LamB exclusively in the outer membrane fractions, thus demonstrating that there was no cross-contamination of the outer membrane fractions with the inner membrane fractions (Fig. 2c, top panel). Next, proteinase K sensitivity assays were conducted on soluble TolCΔ2–22 (membrane-unbound) and the top and bottom flotation gradient fractions (Fig. 2d). Only the soluble species of TolCΔ2–22 that had been harvested from the supernatant after incubation with membranes (i.e. the membrane-unbound TolCΔ2–22 species) showed a proteinase K-resistant pattern, while the membrane-associated species, regardless of whether they floated upward or remained at the bottom of the gradient, were completely degraded (Fig. 2d).
Taken together, these results showed that a fraction of cytosolic TolCΔ2–22 folded in a proteinase K-resistant form identical to that of the wild-type TolC trimers isolated from the outer membrane. However, the membrane-associated TolCΔ2–22 could neither insert properly nor assume a folding status like that of the soluble TolCΔ2–22 species. Thus, trimers of TolCΔ2–22 once formed in the cytoplasm are not competent for subsequent insertion into the outer membrane.
In vivo folding of TolCΔ2–22
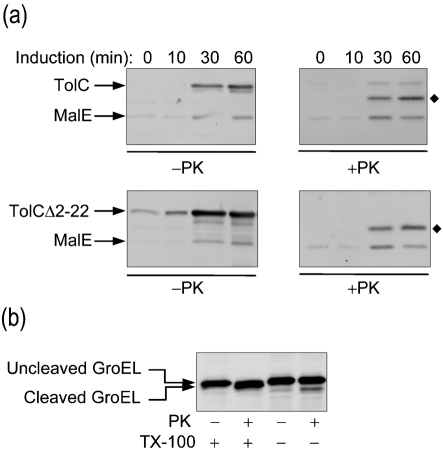
We wanted to follow up on our striking finding above, that a fraction of cytosolic TolCΔ2–22 can assume the folded conformation of normal TolC. We conducted a time-course experiment to monitor folding of TolCΔ2–22 from unfractionated cells. The expression of TolC or TolCΔ2–22 in RAM1330 (ΔtolC) was induced by the addition of 0.2 % arabinose. Cells were withdrawn at various time points, pelleted, permeabilized in a buffer containing EDTA and Triton X-100, and half of each sample was treated with proteinase K. The proteinase K-resistant TolC fragments were visualized by SDS-PAGE and immunoblotting (Fig. 3a). In the untreated samples, TolCΔ2–22 consistently appeared sooner, after the addition of the inducer, and at a higher level at the end of the 1 h induction than did wild-type TolC (Fig. 3a, −PK). The reason for a higher total TolCΔ2–22 level compared with wild-type TolC is unknown, but may involve uncoupling of a potential feedback regulatory loop. This may result from interactions of cytoplasmic factors with the signal sequence or with the nascent precursor chain, slowing down the biogenesis of precursor proteins in the cytoplasm. The absence of the signal sequence in TolCΔ2–22 would allow it to bypass the proof-reading steps of the normal protein translocation pathway. The proteinase K-treated samples showed that both TolC and TolCΔ2–22 were converted to a 46 kDa fragment, reflecting that just like wild-type TolC, TolCΔ2–22 is capable of adopting a folded conformation (Fig. 3a, +PK). Proteinase K-mediated cleavage of the cytoplasmic chaperone GroEL (Weissman et al., 1995) indicated that the externally added protease reached the cytoplasm during the cell permeabilization step of the experimental procedures (Fig. 3b). Therefore, the proteinase K-cleaved 46 kDa fragment of TolCΔ2–22 is likely derived from the soluble cytosolic molecules, as shown by the cell fractionation experiment above (Fig. 1b).
Fig. 3.
Examination of TolC folding from unfractionated cells. (a) Strains expressing TolC and TolCΔ2–22 were induced with arabinose. Cells were collected at t=0, 10, 30 and 60 min, permeabilized and treated with (+) or without (−) proteinase K (PK) as described in Methods. Loaded samples were normalized to a cell density of OD600 0.5. The diamonds (⧫) indicate the 46 kDa proteinase K-resistant fragment of TolC. (b) Proteinase K-mediated cleavage of cytoplasmic GroEL in whole cells resuspended in buffer supplemented with (+) or without (−) 0.5 % Triton X-100 prior to the addition of proteinase K.
Mutations in secY suppress non-functional TolCΔ2–22
Cells producing TolCΔ2–22 display a tolC-null phenotype (Table 1), showing that the population of TolCΔ2–22 that either attains folding status resembling that of wild-type (Figs 1 and 3) or anomalously localizes with the membranes (Fig. 1) is non-functional. TolC must form trimers in the outer membrane to expel antimicrobials, including bile salt deoxycholate (DOC), and to serve as a unique receptor for phage TLS and ColE1. Consequently, cells lacking TolC or expressing non-functional TolC, such as TolCΔ2–22, are hypersensitive to DOC and resistant to TLS and ColE1. We sought suppressors of the export-defective TolCΔ2–22 in an attempt to uncover a mechanism by which the signal sequence-deficient TolC protein can escape the cytoplasm and assemble properly in the outer membrane. Suppressors were isolated by selecting for growth in the presence of DOC. DOCr revertants appeared with an average frequency of 2×10−7. Eight DOCr colonies were analysed further.
Table 1.
Phenotypes of strains expressing TolCΔ2–22
| Strain | Plasmid | Suppressor mutation* (alteration) | Zone of inhibition† | Sensitivity‡ | |||
|---|---|---|---|---|---|---|---|
| SDS | DOC | NB | TLS | ColE1 | |||
| RAM1330 | pBAD24 | wt | 26 | 16 | 16 | R | R |
| RAM1330 | pBAD24-TolC | wt | 6 | 6 | 10 | S | S |
| RAM1330 | pBAD24-TolCΔ2–22 | wt | 25 | 14 | 16 | R | R |
| S1 | pBAD24-TolCΔ2–22 | secY/prlA9 (D69G) | 19 | 8 | 13 | S | S |
| S7 | pBAD24-TolCΔ2–22 | secY (R74C) | 21 | 13 | 16 | S | S |
| S14 | pBAD24-TolCΔ2–22 | Unknown | 21 | 14 | 17 | S | S |
| S15 | pBAD24-TolCΔ2–22 | Unknown | 22 | 14 | 17 | S | S |
| S24 | pBAD24-TolCΔ2–22 | secY/prlA666 (S67F) | 23 | 11 | 14 | S | S |
| S27 | pBAD24-TolCΔ2–22-Bla42-TolCΔ2–22 | wt | 6 | 6 | 10 | S | S |
| S29 | pBAD24-TolCΔ2–22 | Unknown | 26 | 14 | 17 | S | S |
| S31 | pBAD24-TolCΔ2–22-Bla42-TolCΔ2–22 | wt | 6 | 6 | 10 | S | S |
*All strains are derived from RAM1330 (MC4100 Δara ΔtolC : : Kmr); wt, wild-type allele; prlA, suppressor mutations in secY.
†Growth inhibition diameters are in mm. The diameter of the disks was 6 mm. SDS (10 μl of a 10 % solution); DOC, sodium deoxycholate (10 μl of a 10 % solution); NB, novobiocin (30 μg).
‡Sensitivities to phage TLS and ColE1 were determined by spotting dilutions at 10−4.
All eight DOCr colonies became sensitive to TLS and ColE1 (Table 1), indicating that suppression involved correct localization and folding of TolCΔ2–22 instead of establishing a TolC-independent pathway. In two revertants, the suppressor mutation moved with the plasmid that expressed the mutant TolC protein. DNA sequence analysis showed no changes within the tolC gene; instead, in both cases we discovered an unusual DNA rearrangement resulting in the expression of a bla42-tolCΔ2–22 chimera. The 42 N-terminal amino acids of the plasmid-encoded β-lactamase served as the signal sequence for TolCΔ2–22, allowing it to be exported normally.
In the remaining six revertants, suppressor mutations did not move with the plasmid, thus indicating their chromosomal location. Efficient export of signal sequence-less PhoA and MalE has been shown to occur in prlA (secY) mutants (Derman et al., 1993). Accordingly, we proceeded to map suppressor mutations in the six revertants by P1 transductions using a chloramphenicol resistance marker (acrE : : Cmr) that is 60 % linked to wild-type secY (prlA+). Based on TLS sensitivity of Cmr transductants, suppressor mutations in three revertants were most likely present in or near secY. DNA sequence analysis of secY from these three revertants confirmed the presence of suppressor mutations in this gene: in two revertants, S1 and S24, the mutation was identical to the previously characterized prlA9 (Flower et al., 1994; Osborne & Silhavy, 1993) and prlA666 (Puziss et al., 1992) alleles of secY. prlA9 changes D69 to G and prlA666 changes S67 to F. secY from S24 bears a secondary mutation that converts L267 to M. Evidence presented in earlier studies showed that the S67F is responsible for suppression in this mutant (Puziss et al., 1992; Osborne & Silhavy, 1993). The prlA allele in the third revertant, S7, bears a novel mutation that results in an R74 to C substitution. Suppression of TolCΔ2–22 was also tested using signal sequence suppressor alleles of secY identified elsewhere: prlA4 (Emr et al., 1981), prlA208 (Osborne & Silhavy, 1993) and prlA726 (Flower et al., 1994). On the basis of phenotypic assays, these three characterized prlA alleles were able to suppress TolCΔ2–22 (data not shown).
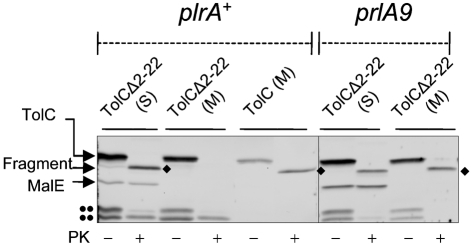
The folding status of TolCΔ2–22 in prlA suppressor backgrounds was examined by a proteinase K sensitivity assay. In prlA+ cells, the membrane-associated TolCΔ2–22 species was highly sensitive to proteolysis (Fig. 4). In contrast, in a strain expressing prlA9, which was the most effective suppressor of TolCΔ2–22, a significant amount of membrane-bound TolCΔ2–22 was converted to the characteristic 46 kDa proteinase K-resistant fragment (Fig. 4). Thus, consistent with the phenotypic data, this assay indicates that the suppressor mutation allows TolCΔ2–22 to be exported and to assemble properly in the outer membrane. In the remaining three DOCr revertants, the suppressor mutations did not map in secY. By P1 transductional crosses, we concluded that they are not prlG (secE), prlH (secG) or prlD (secA) alleles. The locations of these suppressor mutations are presently unknown.
Fig. 4.
Suppression of TolCΔ2–22 by prlA9. The folding status of TolC and TolCΔ2–22 was examined in wild-type (prlA+) and S1 (prlA9) backgrounds. Membranes (M) were separated from the soluble lysates (S), as described in Fig. 1. These fractions were treated with (+) or without (–) proteinase K (PK). TolC was detected by SDS-PAGE and immunoblotting. The diamonds (⧫) indicate the 46 kDa proteinase K-resistant fragment of TolC. The double dots (••) indicate unknown TolC degradation products. Note that only the membrane fraction of cells expressing wild-type TolC was used.
Purification and in vitro channel activity of TolCΔ2–22
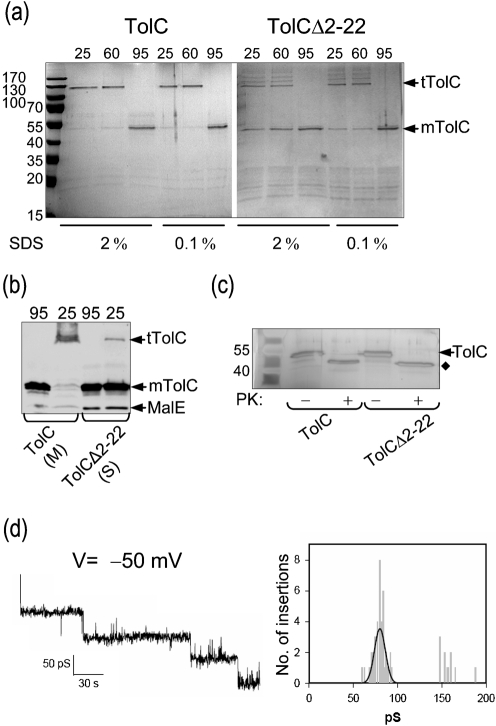
Preliminary characterization of purified TolCΔ2–22 provided evidence that it can fold into native trimers. We conducted electrophysiological analysis to see whether purified TolCΔ2–22 can produce channel activity like that of wild-type TolC. His-tagged TolC and TolCΔ2–22 were expressed in fresh cultures after arabinose induction. Wild-type His-tagged TolC was extracted from the membranes using 5 % Triton X-100. This extract and the detergent-free soluble fraction obtained from cells expressing TolCΔ2–22 were passed through nickel affinity columns in the presence of β-octyl-d-glucopyranoside. Purified TolC migrated on SDS-PAGE as a trimer unless boiled in the presence of SDS (Fig. 5a). In contrast, TolCΔ2–22 did not migrate exclusively as a trimer even when the loading buffer contained only 0.1 % SDS and the samples were unheated (Fig. 5a). We noted that purified TolCΔ2–22 displayed more resistance toward heat and SDS than unpurified soluble TolCΔ2–22 (compare protein samples in Figs 5a, b). It is conceivable that the presence of detergent at a concentration above the critical micellar concentration during affinity purification could have induced further folding and trimerization of TolCΔ2–22. Treatment of purified TolCΔ2–22 with proteinase K also yielded a stable 46 kDa fragment, similar to the fragment generated by wild-type TolC (Fig. 5c).
Fig. 5.
Purification of TolC and TolCΔ2–22 and electrophysiological analysis. (a) TolC was solubilized from membranes with 5 % Triton X-100. TolCΔ2–22 was recovered from the soluble protein fraction without any detergent. His-tagged TolC and TolCΔ2–22 were purified by immobilized-nickel affinity chromatography. A normalized sample from each elution peak was mixed with SDS loading buffer containing either 0.1 or 2 % SDS and heated at the indicated temperature prior to electrophoresis. Proteins were visualized by Coomassie blue staining; mTolC, monomers of TolC; tTolC, trimers of TolC. The migration of molecular mass standards is shown to the left. (b) Membrane (M) and soluble (S) fractions from the experiment described in Fig. 1(a) were mixed with loading buffer containing 2 % SDS and were unheated (25 °C) or heated (95 °C) for 10 min prior to electrophoresis. (c) The peak elution fractions were either untreated (−) or treated (+) with proteinase K. Proteins were separated by SDS-PAGE and visualized by silver staining. (d) Channel-forming activity of purified TolCΔ2–22. Shown are sequential insertions and an associated amplitude histogram of purified TolCΔ2–22 in planar lipid bilayers at –50 mV in 1 M KCl.
To determine whether the TolCΔ2–22 trimers are structurally and functionally similar to those of wild-type TolC, single channel activity of purified TolCΔ2–22 was examined after reconstitution in black lipid bilayers. The resultant pores were stable and showed properties similar to those of wild-type TolC. Data in Fig. 5(d) show a representative current recording at an applied transmembrane potential of –50 mV in 1 M KCl. From the associated current amplitude histogram of the insertion events, a mean conductance value of 80.23±0.69 pS was measured. Under the same conditions, the single channel conductance of wild-type TolC was 76±7.0 pS (data not shown). These conductance values are in the published range (Andersen et al., 2002). These striking in vitro results, together with the in vivo data, show that TolC can adopt a native trimeric conformation in the cytoplasm of E. coli.
DISCUSSION
Studies involving the localization and folding of an export-deficient TolC protein that lacks its entire N-terminal signal sequence (TolCΔ2–22) provided evidence that despite being retained in the cytoplasm, it could fold and attain a conformation resembling that of the outer membrane-localized wild-type TolC. Moreover, the oligomeric states of purified TolCΔ2–22 and wild-type TolC were alike and both could efficiently form functional channels in lipid bilayers. Since the single channel conductances of TolCΔ2–22 and wild-type TolC were very similar, it is reasonable to assume that TolCΔ2–22 had achieved the correct folding and quaternary structure. Most of the soluble cytoplasmic TolCΔ2–22, however, failed to fold correctly. Therefore, either TolC requires an unidentified folding factor found only in the periplasm or its final folding is tightly coupled to proper insertion into the outer membrane, which the cytoplasmic TolCΔ2–22 cannot achieve.
A population of TolCΔ2–22 that fractionates with the membranes or associates with purified inner membrane or outer membrane vesicles after in vitro incubation fails to achieve proper folding. We think that the absence of the Bam complex, which mediates the final stages of folding and membrane insertion of β-barrel OMPs and TolC (Werner & Misra, 2005; Wu et al., 2005), precludes cytoplasmic TolCΔ2–22 from inserting into the inner membrane. Instead, most of the TolCΔ2–22 misfolds and/or associates spuriously with the membrane and become insoluble. Based on pulse–chase experiments, it has been proposed that the assembly of TolC occurs in three steps: (i) folding of the nascent monomers, (ii) trimerization and (iii) membrane insertion (Werner et al., 2003). Detection of monomers but not trimers in the periplasm indicates that either membrane insertion instantly follows trimerization or that trimerization occurs in the outer membrane. The fact that trimerization of TolCΔ2–22 can occur in the cytoplasm shows that the Bam complex or a periplasmic factor is not obligatory for TolC trimerization. Interestingly, suppressor mutations in secY allowed a significant amount of TolCΔ2–22 to escape the cytoplasm and assemble correctly in the outer membrane, an observation similar to that reported for signal sequence-less PhoA and MalE (Derman et al., 1993).
The vast majority of E. coli OMPs require periplasmic folding factors for their proper assembly (de Cock et al., 1996; Missiakas et al., 1996; Rouvière & Gross, 1996; Schafer et al., 1999; Bulieris et al., 2003; Krojer et al., 2008). As a result, if OMPs are retained in the cytoplasm, they are unable to fold into their native conformation and then aggregate. These OMPs can be extracted from aggregated cytoplasmic inclusion bodies with denaturing agents, and then refolded in vitro in the presence of detergents (de Cock et al., 1996; Eisele & Rosenbusch, 1990). The refolding efficiency is generally enhanced by the addition of chaperones and LPS (Bulieris et al., 2003). TolC differs from all other model E. coli OMPs such as porins in that its biogenesis occurs in the absence of known periplasmic assembly factors (Werner et al., 2003). This independence from periplasmic factors could be a reason why TolC can adopt its ultimate native and active conformation outside of its physiological environment while other integral E. coli OMPs cannot.
We think that the presence of the large soluble α-helical domain renders TolC assembly independent of known periplasmic folding factors that are critical for the folding of other OMPs comprised exclusively of β-barrel domains, such as OmpF and LamB (Cowan et al., 1992). Consistent with this view, we find that substitutions at mature TolC residue S350 in the α-helical domain that interfere with the trimerization and membrane insertion steps of normal TolC (Gerken & Misra, 2004; Werner et al., 2003) also block the folding of TolCΔ2–22 in the cytoplasm. Thus, proper interactions between the α-helices of adjacent monomers are required for the assembly of TolCΔ2–22 in the cytoplasm, just as they are required for the assembly of wild-type TolC in the outer membrane. Based on these observations it is tempting to speculate that folding of the α-helical domain in the periplasm or in the cytoplasm, in the case of TolCΔ2–22, triggers folding of the rest of the molecule. However, since the final assembly of the β-barrel domain likely depends on the Bam complex, the α-helical domain-mediated folding is not sufficient to achieve full folding and proper membrane insertion. Recently, a study on a TolC homologue, TdeA, from Actinobacillus actinomycetemcomitans showed that cytosolically expressed recombinant TdeA containing just the periplasmic α-helical domain can fold and trimerize efficiently (Kim et al., 2008). This further underscores our assertion that the soluble α-helical domain of TolC is capable of triggering trimerization, but that proper membrane insertion would require interaction of the β-barrel domain of TolC with the Bam complex.
Recent studies on PulD, a secretin from Klebsiella oxytoca, have also revealed the ability of a signal sequence-less variant of PulD to multimerize like the normal PulD (Guilvout et al., 2008). However, unlike TolCΔ2–22, the signal sequence-less PulD confers acute cell toxicity, presumably due to an attempt by this protein to insert into and disrupt the inner membrane from the cytoplasmic side. Thus, whereas both PulD and TolC share the common property of being able to oligomerize outside their physiological environment, they differ in their mechanism of membrane insertion by being either independent (PulD; Collin et al., 2007) or dependent (TolC; Werner & Misra, 2005) on BamA. Based on the cellular requirements of integral OMPs for their folding and insertion, there appear to be at least three distinct pathways of OMP assembly. In one pathway, OMPs, such as OmpF and LamB, rely heavily on both periplasmic and outer membrane factors for their folding and membrane insertion. In the second pathway, exemplified by TolC, and perhaps other OMPs with large periplasmic domains, including BamA itself, OMPs display independence from periplasmic folding factors but rely on the outer membrane machinery for their insertion. The last pathway appears to be followed by secretins, such as PulD, which are largely independent of both periplasmic and outer membrane factors for their folding and insertion. Instead, their correct membrane targeting seems to require a cognate pilotin protein (Guilvout et al., 2006). It is quite conceivable that different assembly pathways are chosen based on the intrinsic folding properties of the various OMPs, leading to distinct folded structures that either are composed exclusively of membrane-embedded β-barrels or in addition contain large amounts of soluble (periplasmic) α-helical domains.
Acknowledgments
We are indebted to Phu Vuong and Drew Bennion for their contribution in cloning experiments and for critical reading of the manuscript. This work was supported by grant R01 GM48167 from the National Institutes of Health.
Abbreviations
Bam, β-barrel assembly machinery
DOC, deoxycholate
OMP, outer-membrane protein
References
- Andersen, C. (2003). Channel-tunnels: outer membrane components of type I secretion systems and multidrug efflux pumps of Gram-negative bacteria. Rev Physiol Biochem Pharmacol 147, 122–165. [DOI] [PubMed] [Google Scholar]
- Andersen, C., Hughes, C. & Koronakis, V. (2002). Electrophysiological behavior of the TolC channel-tunnel in planar lipid bilayers. J Membr Biol 185, 83–92. [DOI] [PubMed] [Google Scholar]
- Augustus, A. M., Celaya, T., Husain, F., Humbard, M. & Misra, R. (2004). Antibiotic-sensitive TolC mutants and their suppressors. J Bacteriol 186, 1851–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betton, J. M. & Hofnung, M. (1996). Folding of a mutant maltose-binding protein of Escherichia coli which forms inclusion bodies. J Biol Chem 271, 8046–8052. [DOI] [PubMed] [Google Scholar]
- Bulieris, P. V., Behrens, S., Holst, O. & Kleinschmidt, J. H. (2003). Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem 278, 9092–9099. [DOI] [PubMed] [Google Scholar]
- Casadaban, M. J. (1976). Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol 104, 541–555. [DOI] [PubMed] [Google Scholar]
- Charlson, E. S., Werner, J. N. & Misra, R. (2006). Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol 188, 7186–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin, S., Guilvout, I., Chami, M. & Pugsley, A. P. (2007). YaeT-independent multimerization and outer membrane association of secretin PulD. Mol Microbiol 64, 1350–1357. [DOI] [PubMed] [Google Scholar]
- Cowan, S. W., Schirmer, T., Rummel, G., Steiert, M., Ghosh, R., Pauptit, R. A., Jansonius, J. N. & Rosenbusch, J. P. (1992). Crystal structures explain functional properties of two E. coli porins. Nature 358, 727–733. [DOI] [PubMed] [Google Scholar]
- Danese, P. N. & Silhavy, T. J. (1998). Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet 32, 59–94. [DOI] [PubMed] [Google Scholar]
- de Cock, H. & Tommassen, J. (1996). Lipopolysaccharides and divalent cations are involved in the formation of an assembly-competent intermediate of outer-membrane protein PhoE of E. coli. EMBO J 15, 5567–5573. [PMC free article] [PubMed] [Google Scholar]
- de Cock, H., van Blokland, S. & Tommassen, J. (1996). In vitro insertion and assembly of outer membrane protein PhoE of Escherichia coli K-12 into the outer membrane: role of Triton X-100. J Biol Chem 271, 12885–12890. [DOI] [PubMed] [Google Scholar]
- Derman, A. I., Puziss, J. W., Bassford, P. J. & Beckwith, J. (1993). A signal sequence is not required for protein in prlA mutants of Escherichia coli. EMBO J 12, 879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele, J. L. & Rosenbusch, J. P. (1990). In vitro folding and oligomerization of a membrane protein: transition of bacterial porin from random coil to native conformation. J Biol Chem 265, 10217–10220. [PubMed] [Google Scholar]
- Emr, S. D., Hanley-Way, S. & Silhavy, T. J. (1981). Suppressor mutations that restore export of a protein with a defective signal sequence. Cell 23, 79–88. [DOI] [PubMed] [Google Scholar]
- Flower, A. M., Doebele, R. C. & Silhavy, T. J. (1994). PrlA and PrlG suppressors reduce the requirement for signal sequence recognition. J Bacteriol 176, 5607–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken, H. & Misra, R. (2004). Genetic evidence for functional interactions between TolC and AcrA proteins of a major antibiotic efflux pump of Escherichia coli. Mol Microbiol 54, 620–631. [DOI] [PubMed] [Google Scholar]
- German, G. J. & Misra, R. (2001). The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. J Mol Biol 308, 579–585. [DOI] [PubMed] [Google Scholar]
- Guilvout, I., Chami, M., Engel, A., Pugsley, A. P. & Bayan, N. (2006). Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J 25, 5241–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilvout, I., Chami, M., Berrier, C., Ghazi, A., Engel, A., Pugsley, A. P. & Bayan, N. (2008). In vitro multimerization and membrane insertion of bacterial outer membrane secretin PulD. J Mol Biol 382, 13–23. [DOI] [PubMed] [Google Scholar]
- Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, S. & Goldberg, M. B. (2007). Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J Bacteriol 189, 5393–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Yum, S., Jo, W. S., Lee, B. L., Jeong, M. H. & Ha, N. C. (2008). Expression and biochemical characterization of the periplasmic domain of bacterial outer membrane porin TdeA. J Microbiol Biotechnol 18, 845–851. [PubMed] [Google Scholar]
- Kloser, A. W., Laird, M. W. & Misra, R. (1996). asmB, a suppressor locus for assembly-defective OmpF mutants of Escherichia coli, is allelic to envA (lpxC). J Bacteriol 178, 5138–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis, V., Li, J., Koronakis, E. & Stauffer, K. (1997). Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol Microbiol 23, 617–626. [DOI] [PubMed] [Google Scholar]
- Koronakis, V., Sharff, A., Koronakis, E., Luisi, B. & Hughes, C. (2000). Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919. [DOI] [PubMed] [Google Scholar]
- Krojer, T., Sawa, J., Schäfer, E., Saibil, H. R., Ehrmann, M. & Clausen, T. (2008). Structural basis for the regulated protease and chaperone function of DegP. Nature 453, 885–890. [DOI] [PubMed] [Google Scholar]
- Malinverni, J. C., Werner, J., Kim, S., Sklar, J. G., Kahne, D., Misra, R. & Silhavy, T. J. (2006). YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61, 151–164. [DOI] [PubMed] [Google Scholar]
- Masi, M., Vuong, P., Humbard, M., Malone, K. & Misra, R. (2007). Initial steps of Colicin E1 import across the outer membrane of Escherichia coli. J Bacteriol 189, 2667–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, R. (2007). First glimpse of the crystal structure of the YaeT's POTRA domains. ACS Chem Biol 2, 649–651. [DOI] [PubMed] [Google Scholar]
- Misra, R., Peterson, A., Ferenci, T. & Silhavy, T. J. (1991). A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J Biol Chem 266, 13592–13597. [PubMed] [Google Scholar]
- Missiakas, D., Betton, J. M. & Raina, S. (1996). New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol 21, 871–884. [DOI] [PubMed] [Google Scholar]
- Mogensen, J. E. & Otzen, D. E. (2005). Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol 57, 326–346. [DOI] [PubMed] [Google Scholar]
- Nagel de Zwaig, R. & Luria, S. E. (1967). Genetics and physiology of colicin-tolerant mutants of Escherichia coli. J Bacteriol 94, 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, R. S. & Silhavy, T. J. (1993). PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J 12, 3391–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsl, H. & Braun, V. (1995). Novel colicin 10: assignment of four domains to TonB- and TolC-dependent uptake via the Tsx receptor and to pore formation. Mol Microbiol 16, 57–67. [DOI] [PubMed] [Google Scholar]
- Puziss, J. W., Strobel, S. M. & Bassford, P. J., Jr (1992). Export of maltose-binding protein species with altered charge distribution surrounding the signal peptide hydrophobic core in Escherichia coli cells harboring prl suppressor mutations. J Bacteriol 174, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière, P. E. & Gross, C. A. (1996). SurA, a periplasmic protein with peptidyl–prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev 10, 3170–3182. [DOI] [PubMed] [Google Scholar]
- Schafer, U., Beck, K. & Muller, M. (1999). Skp, a molecular chaperone of Gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem 274, 24567–24574. [DOI] [PubMed] [Google Scholar]
- Sklar, J. G., Wu, T., Kahne, D. & Silhavy, T. J. (2007a). Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21, 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar, J. G., Wu, T., Gronenberg, L. S., Malinverni, J. C., Kahne, D. & Silhavy, T. J. (2007b). Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 104, 6400–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch, K. L., Johnson, K. & Beckwith, J. (1989). Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol 171, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struyve, M., Moons, M. & Tommassen, J. (1991). Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol 218, 141–148. [DOI] [PubMed] [Google Scholar]
- Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M. & Tommassen, J. (2003). Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265. [DOI] [PubMed] [Google Scholar]
- Weissman, J. S., Hohl, C. M., Kovalenko, O., Kashi, Y., Chen, S., Braig, K., Saibil, H. R., Fenton, W. A. & Horwich, A. L. (1995). Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell 83, 577–587. [DOI] [PubMed] [Google Scholar]
- Werner, J. & Misra, R. (2005). YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol 57, 1450–1459. [DOI] [PubMed] [Google Scholar]
- Werner, J., Augustus, A. M. & Misra, R. (2003). Assembly of TolC, a structurally unique and multifunctional outer membrane protein of Escherichia coli K-12. J Bacteriol 185, 6540–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T., Malinverni, J., Ruiz, N., Kim, S., Silhavy, T. J. & Kahne, D. (2005). Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245. [DOI] [PubMed] [Google Scholar]