Abstract
The humoral response to hepatitis C virus (HCV) may contribute to controlling infection. We previously isolated human monoclonal antibodies to conformational epitopes on the HCV E2 glycoprotein. Here, we report on their ability to inhibit infection by retroviral pseudoparticles incorporating a panel of full-length E1E2 clones representing the full spectrum of genotypes 1–6. We identified one antibody, CBH-5, that was capable of neutralizing every genotype tested. It also potently inhibited chimeric cell culture-infectious HCV, which had genotype 2b envelope proteins in a genotype 2a (JFH-1) background. Analysis using a panel of alanine-substitution mutants of HCV E2 revealed that the epitope of CBH-5 includes amino acid residues that are required for binding of E2 to CD81, a cellular receptor essential for virus entry. This suggests that CBH-5 inhibits HCV infection by competing directly with CD81 for a binding site on E2.
Hepatitis C virus (HCV) is a small, enveloped, positive-strand RNA virus that infects an estimated 170 million individuals worldwide (Anonymous, 1999). The virus evades the immune system so successfully that most of those who contract it remain chronically infected and may eventually develop serious liver disease. HCV disables the innate antiviral response by blocking pathways of interferon induction and signalling (Gale & Foy, 2005; Meylan & Tschopp, 2006) and escapes the adaptive immune response largely by means of its genetic variability (Bowen & Walker, 2005; Brown et al., 2005; Farci et al., 2000; Shimizu et al., 1994; Thimme et al., 2006). There are six distinct genotypes of HCV, each divided into subtypes (Simmonds et al., 2005), and within a single individual the virus exists as a constantly evolving quasispecies (Bukh et al., 1995).
An effective T-cell response is essential for controlling HCV infection (Bowen & Walker, 2005; Shoukry et al., 2004), but neutralizing antibodies also play an important role. It was observed long ago that immunoglobulin prophylaxis protected against HCV infection (Conrad & Lemon, 1987; Knodell et al., 1976) and it has recently been shown that this protective effect correlates with the presence of antibodies that can neutralize retroviral pseudoparticles bearing HCV envelope glycoproteins (HCVpp) (Yu et al., 2004). Studies of acutely infected cohorts show that rapid induction of neutralizing antibodies may afford protection (Lavillette et al., 2005a; Pestka et al., 2007). Moreover, antibodies present during the acute phase of infection can neutralize an infectious dose of HCV administered to chimpanzees (Farci et al., 1994).
The envelope glycoproteins E1 and E2 are the natural targets for neutralizing antibodies (Bartosch et al., 2003a; Rosa et al., 1996). E2 binding to CD81 on the cell surface is necessary, but not sufficient, for infection (Cocquerel et al., 2006; Kapadia et al., 2007; Koutsoudakis et al., 2007). The first hypervariable region (HVR-1) within the amino-terminal part of E2 is involved in virus binding and entry (Bartosch et al., 2003c) and is immunodominant. Antibodies to HVR-1 neutralize infection, but they are typically isolate-specific, with little or no recognition of other isolates or genotypes. In contrast, conformational epitopes on E2 outside HVR-1 are less prone to variation and are able to elicit more broadly neutralizing antibodies (Habersetzer et al., 1998; Hadlock et al., 2000; Johansson et al., 2007; Perotti et al., 2008; Schofield et al., 2005).
We previously reported the generation of a panel of IgG1 human monoclonal antibodies (HmAbs) from the peripheral B cells of an individual with asymptomatic HCV genotype 1b infection (Hadlock et al., 2000). They recognize conformational epitopes on E2 of more than one genotype, and have been mapped to distinct domains, designated A, B and C (Keck et al., 2004). The HmAbs that recognize domains B and C block E2 binding to CD81 and neutralize genotype 1a HCVpp and genotype 2a JFH-1 cell culture-infectious HCV virions (HCVcc), whereas HmAbs to domain A do not block CD81 binding and are non-neutralizing (Hadlock et al., 2000; Keck et al., 2004, 2005, 2007; Op De Beeck et al., 2004).
Recent success in growing the genotype 2a JFH-1 isolate of HCV in cultured cells to produce infectious virions (Lindenbach et al., 2005; Wakita et al., 2005; Zhong et al., 2005) has yet to be replicated for all of the other genotypes. However, various aspects of HCV binding and entry, including antibody neutralization, can be studied by using HCVpp (Bartosch et al., 2003b; Hsu et al., 2003). We have assembled a panel of functional E1E2 glycoprotein sequences representing all major genotypes (Lavillette et al., 2005b; Owsianka et al., 2005; Tarr et al., 2006), allowing us to extend the characterization of our previously described HmAbs.
HCVpp displaying E1E2 of genotypes 1–6 and carrying a luciferase reporter gene were generated in HEK293T cells and purified as described previously (Bartosch et al., 2003b; Owsianka et al., 2005). Aliquots of the HCVpp were mixed with 100 μg ml−1 of each HmAb, incubated for 1 h at 37 °C and used to infect Huh-7 cells (Nakabayashi et al., 1982). Luciferase activity in the infected cells was measured after 4 days.
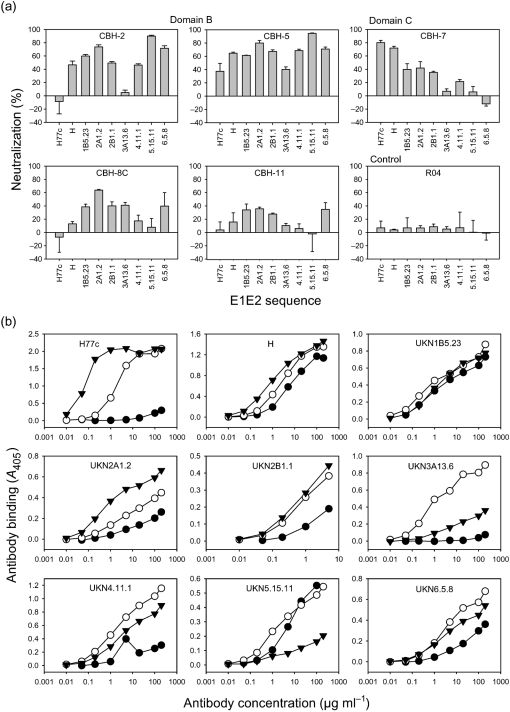
Domain B (CBH-2, -5, -8C and -11) and domain C (CBH-7) HmAbs each had a distinct spectrum of neutralizing activity against the genotype panel (Fig. 1a), whereas domain A HmAbs did not reduce the infectivity of any genotype by more than 25 % (not shown). An isotype-matched control HmAb against HCMV p64 (R04) did not reduce the infectivity of any genotype by more than 10 % (Fig. 1a).
Fig. 1.
(a) Neutralization by HmAbs of HCVpp pseudotyped with E1E2 sequences of genotypes 1–6. This dataset is representative of results obtained in at least three independent experiments. (b) Reactivity of HmAbs CBH-2 (•), CBH-5 (○) and CBH-7 (▾) with purified HCVpp in GNA ELISA. The following E1E2 sequences were used: genotype 1a, H77c (GenBank accession no. AF011751) and H (Bartosch et al., 2003b; Helle et al., 2007); genotype 1b, UKN1B5.23 (AY734976); genotype 2a, UKN2A1.2 (AY734977); genotype 2b, UKN 2B1.1 (AY734982); genotype 3a, UKN3A13.6 (AY894683); genotype 4, UKN4.11.1 (AY734986); genotype 5, UKN5.15.11 (AY894682); genotype 6, UKN6.5.8 (EF427671).
CBH-5 reduced infectivity of HCVpp representing all genotypes by at least 40 %, whilst the other domain B HmAbs each inhibited a more restricted range of genotypes. CBH-2 neutralized HCVpp of genotypes 2a, 5 and 6 by at least 70 %, and genotypes 2b and 4 by just under 50 %. It did not neutralize genotype 3a or genotype 1a clone H77c, although it neutralized the closely related clone H by almost 50 % (Fig. 1). These two clones differ at five amino acid positions within E2, which may account for the difference in CBH-2 neutralization. CBH-8C and CBH-11 did not neutralize either of the 1a isolates, consistent with previous findings that these HmAbs do not recognize E2 of genotype 1a strain H (Keck et al., 2004). CBH-8C neutralized genotypes 1b, 2a, 2b, 3a and 6, whilst CBH-11 showed modest neutralization of genotypes 1b, 2a, 2b and 6. The domain C HmAb CBH-7 neutralized both the 1a strain H clones by more than 70 %, genotypes 1b, 2a and 2b by about 40 % and genotype 4 by 20 %, but had no effect on genotypes 3a, 5 or 6.
To examine the correlation between binding and neutralization, we tested the binding of antibodies CBH-2, -5 and -7 to E2 displayed on HCVpp. After sucrose-density equilibrium centrifugation in 20–60 % sucrose at 270 000 g for 18 h at 4 °C, the infectious gradient fractions were pooled, captured onto GNA (Galanthus nivalis agglutinin)-coated plates and probed with each HmAb over a range of concentrations. Bound antibodies were detected with alkaline phosphatase-conjugated goat anti-human IgG followed by p-nitrophenylphosphate disodium hexahydrate, and A405 was measured.
CBH-5 was the most broadly reactive HmAb, giving a robust, concentration-dependent signal with each genotype (Fig. 1b). This broad reactivity clearly underpins its ability to neutralize HCVpp across the spectrum of genotypes. CBH-2 gave a concentration-dependent signal with E2 of all genotypes except 1a H77c and 3a (Fig. 1b), which agrees with its lack of ability to neutralize HCVpp bearing these two sequences. Binding to genotype 4 was detectable only at high CBH-2 concentrations. CBH-7 gave a strong concentration-dependent signal with genotypes 1 and 2, and a weaker one with genotypes 3–6, in agreement with its neutralization profile. However, the relationship between binding and neutralization was more complex for CBH-7, as it recognized genotype 6 E2, but did not neutralize genotype 6 HCVpp. It may be that greater saturation with CBH-7 is required for neutralization or that there is a difference in the mechanism of neutralization between CBH-7 and the domain B antibodies.
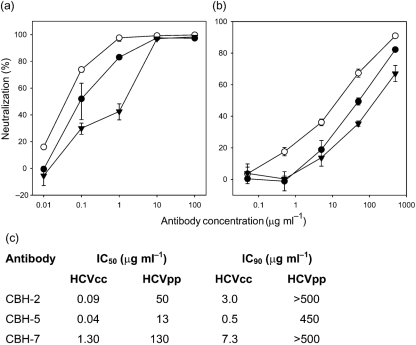
We showed previously that CBH-5 and the other domain B HmAbs potently neutralize genotype 2a (JFH-1) HCVcc, whilst CBH-7 has modest HCVcc-neutralizing activity (Keck et al., 2007). The half-maximal inhibitory concentration (IC50) of these HmAbs is considerably lower for HCVcc than for HCVpp (Keck et al., 2007). To see whether this holds true for other isolates, we generated an intragenotypic JFH-1 chimera carrying genotype 2b (UKN2B1.1; Owsianka et al., 2005) E1E2 glycoproteins. The 2b HCVcc were pre-incubated with HmAbs CBH-2, -5 and -7 over a range of concentrations before infecting Huh-7 cells. After 4 days, the levels of viral RNA in these cells were determined by real-time PCR (qRT-PCR) using relative quantification, where each sample was normalized to an endogenous control gene (glyceraldehyde-3-phosphate dehydrogenase).
All three HmAbs were very effective at reducing infectivity (Fig. 2a), and the order of neutralization potency of these antibodies for the 2b chimeric virus was the same as for the genotype 2a virus: CBH-5>CBH-2>CBH-7. A similar titration carried out with HCVpp displaying UKN2B1.1 E1E2 glycoproteins showed that all three HmAbs inhibited infection at concentrations approximately two orders of magnitude higher than those required for HCVcc neutralization (Fig. 2b, c).
Fig. 2.
Neutralization by HmAbs CBH-2 (•), CBH-5 (○) and CBH-7 (▾) of (a) genotype 2b HCVcc and (b) genotype 2b HCVpp. (c) IC50 and IC90 of each HmAb for genotype 2b HCVcc and HCVpp.
The difference between the IC50 (and IC90) values of all three antibodies for HCVcc and HCVpp is striking. This could be related to altered exposure or accessibility of antibody epitopes, possibly resulting from differences in glycosylation of E2 in the two systems, the lack of NS2 in HCVpp or the size difference between HCVpp and authentic HCV virions (Sandrin et al., 2005). However, the simplest explanation is that there is more non-infectious E2 relative to infectious particles in a preparation of HCVpp than of HCVcc, and so more antibody is required to neutralize HCVpp. Ideally, one would compare the neutralization of equal numbers of particles or of equal amounts of E2 presented on the two types of particles, but it would be very hard to achieve this with any degree of accuracy. The HCVpp system has been used in several studies to determine virus neutralization by patient sera (Farci et al., 1994; Lavillette et al., 2005a; Pestka et al., 2007; Yu et al., 2004). Our results indicate that modest inhibition of HCVpp may translate into more substantial neutralization of infectious virus.
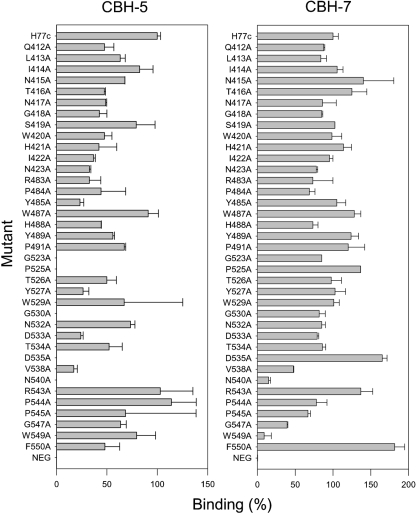
Domain B and C HmAbs all have neutralization-of-binding (NOB) activity (Hadlock et al., 2000), so it is likely that their epitopes overlap with regions of E2 involved in CD81 binding. Antibody blocking experiments have implicated several regions of E2 in CD81 binding (Clayton et al., 2002; Flint et al., 1999; Owsianka et al., 2001). We identified conserved amino acid residues within these regions and, using the 1a H77c sequence as a wild type, made a panel of mutants in which each conserved residue in turn was replaced by alanine. Some additional mutants, available from other studies, were also included. The mutations did not affect the overall conformation of the E2 protein or its ability to form non-covalent heterodimers with E1 (Owsianka et al., 2006). The mutant proteins were expressed transiently in HEK 293T cells; equivalent amounts of monomeric E2 protein were captured on GNA plates and probed with antibodies CBH-5 and CBH-7 at 2 μg ml−1. CBH-5 binding was abrogated completely by mutations G523A, P525A, G530A, D535A and N540A, indicating that these amino acid residues may be involved in its recognition of E2 (Fig. 3). Residues G523, P525, G530 and D535 are completely conserved in functional sequences across all genotypes, whereas N540 is not (Owsianka et al., 2006). G530 and D535 are essential for CD81 interaction with E2, and G523 is also likely to be involved (Owsianka et al., 2006). The residue corresponding to D535 in the JFH-1 virus is essential for infectivity, as a point mutation at this position renders HCVcc non-infectious (J. Witteveldt, unpublished results). These data indicate that the epitope of CBH-5 includes four highly conserved amino acid residues, three of which are involved in CD81 binding, suggesting that CBH-5 exerts its potent neutralization of HCV infectivity by competing directly with CD81 for binding to E2. This is likely to account for its broad spectrum of activity, as any changes in the CBH-5 epitope would be likely to affect CD81 binding and virus entry.
Fig. 3.
Alanine-replacement mutagenesis. Mutant proteins are annotated according to the amino acid in the H77c sequence, the amino acid position relative to the start of the H77 polyprotein chain and the substitution introduced. Binding of antibody to each mutant protein is expressed as a percentage of binding to wild-type H77c protein.
N540 is important for recognition by both antibodies because it is a glycosylation site, mutation of which probably induces a local conformational change in E2, which does not impair E2 folding and function severely, but reduces the affinity of various neutralizing antibodies including CBH-5 and CBH-7 (Goffard et al., 2005; Helle et al., 2007). The only other mutation that reduced the binding of CBH-7 substantially was W549A (Fig. 3). W549 is a highly conserved residue that is essential for HCVpp infectivity, but not for CD81 binding (Owsianka et al., 2006), which suggests that CBH-7 may act by steric hindrance rather than by direct competition with CD81. Our panel of E2 proteins did not include mutants in all regions of E2 that have been implicated in CD81 binding; other studies have demonstrated the involvement of residues 436–443 (Drummer et al., 2006) and 613–618 (Roccasecca et al., 2003). Further work is in progress to define the epitopes of the domain B and C HmAbs more completely, and how these relate to regions involved in CD81 binding. It was not possible to use the same panel of mutant E2 proteins to map the epitopes of CBH-2, -8C and -11, as these bind poorly to the wild-type H77c sequence.
In this study of five conformation-sensitive anti-E2 HmAbs, tested for their ability to neutralize HCVpp pseudotyped with glycoproteins of HCV genotypes 1–6, only one antibody, CBH-5, was found to be active against every E2 sequence. This highlights the need to use E1E2 from the full range of genotypes to identify truly broadly neutralizing antibodies. Cross-reactive HmAbs specific for conformational epitopes on E2 have been isolated and characterized in several laboratories (Allander et al., 2000; Bugli et al., 2001; Eren et al., 2006; Perotti et al., 2008; Schofield et al., 2005), but such broad reactivity across all genotypes has rarely been found (Johansson et al., 2007).
Broadly neutralizing antibodies are potentially of direct clinical relevance. They could be used prophylactically to reduce the risk of HCV infection after needlestick or other accidental exposure. In the liver-transplant setting, it would be very desirable to reduce the incidence of graft reinfection by passive immunotherapy with antibodies such as CBH-5. The characteristics of CBH-5 suggest that there is at least one highly conserved neutralizing epitope on the E2 glycoprotein to which the human immune system is capable of mounting a response. This is encouraging for future vaccine development, although focusing the immune response on this epitope will be challenging, given its conformational nature. Greater knowledge of the structure of the HCV glycoproteins, coupled with detailed mapping of the epitopes of other neutralizing antibodies, should bring us closer to this goal.
Acknowledgments
We thank Duncan McGeoch for critically reading the manuscript, T. Wakita for the JFH-1 clone and F.-L. Cosset for the MLV pseudoparticle system. This work was supported by the Medical Research Council (UK), European Union FP5 and FP6 contracts QLK2-CT-2001-01120 and MRTN-CT-2006-035599, the University of Nottingham Biomedical Research Committee and by National Institutes of Health grants HL079381 and AI47355 to S. K. H. F.
References
- Allander, T., Drakenberg, K., Beyene, A., Rosa, D., Abrignani, S., Houghton, M., Widell, A., Grillner, L. & Persson, M. A. (2000). Recombinant human monoclonal antibodies against different conformational epitopes of the E2 envelope glycoprotein of hepatitis C virus that inhibit its interaction with CD81. J Gen Virol 81, 2451–2459. [DOI] [PubMed] [Google Scholar]
- Anonymous (1999). Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J Viral Hepat 6, 35–47. [PubMed] [Google Scholar]
- Bartosch, B., Bukh, J., Meunier, J. C., Granier, C., Engle, R. E., Blackwelder, W. C., Emerson, S. U., Cosset, F. L. & Purcell, R. H. (2003a). In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci U S A 100, 14199–14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch, B., Dubuisson, J. & Cosset, F. L. (2003b). Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med 197, 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch, B., Vitelli, A., Granier, C., Goujon, C., Dubuisson, J., Pascale, S., Scarselli, E., Cortese, R., Nicosia, A. & Cosset, F. L. (2003c). Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-BI scavenger receptor. J Biol Chem 278, 41624–41630. [DOI] [PubMed] [Google Scholar]
- Bowen, D. G. & Walker, C. M. (2005). Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436, 946–952. [DOI] [PubMed] [Google Scholar]
- Brown, R. J., Juttla, V. S., Tarr, A. W., Finnis, R., Irving, W. L., Hemsley, S., Flower, D. R., Borrow, P. & Ball, J. K. (2005). Evolutionary dynamics of hepatitis C virus envelope genes during chronic infection. J Gen Virol 86, 1931–1942. [DOI] [PubMed] [Google Scholar]
- Bugli, F., Mancini, N., Kang, C. Y., Di Campli, C., Grieco, A., Manzin, A., Gabrielli, A., Gasbarrini, A., Fadda, G. & other authors (2001). Mapping B-cell epitopes of hepatitis C virus E2 glycoprotein using human monoclonal antibodies from phage display libraries. J Virol 75, 9986–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh, J., Miller, R. H. & Purcell, R. H. (1995). Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis 15, 41–63. [DOI] [PubMed] [Google Scholar]
- Clayton, R. F., Owsianka, A., Aitken, J., Graham, S., Bhella, D. & Patel, A. H. (2002). Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J Virol 76, 7672–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerel, L., Voisset, C. & Dubuisson, J. (2006). Hepatitis C virus entry: potential receptors and their biological functions. J Gen Virol 87, 1075–1084. [DOI] [PubMed] [Google Scholar]
- Conrad, M. E. & Lemon, S. M. (1987). Prevention of endemic icteric viral hepatitis by administration of immune serum gamma globulin. J Infect Dis 156, 56–63. [DOI] [PubMed] [Google Scholar]
- Drummer, H. E., Boo, I., Maerz, A. L. & Poumbourios, P. (2006). A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J Virol 80, 7844–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren, R., Landstein, D., Terkieltaub, D., Nussbaum, O., Zauberman, A., Ben-Porath, J., Gopher, J., Buchnick, R., Kovjazin, R. & other authors (2006). Preclinical evaluation of two neutralizing human monoclonal antibodies against hepatitis C virus (HCV): a potential treatment to prevent HCV reinfection in liver transplant patients. J Virol 80, 2654–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farci, P., Alter, H. J., Wong, D. C., Miller, R. H., Govindarajan, S., Engle, R., Shapiro, M. & Purcell, R. H. (1994). Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A 91, 7792–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farci, P., Shimoda, A., Coiana, A., Diaz, G., Peddis, G., Melpolder, J. C., Strazzera, A., Chien, D. Y., Munoz, S. J. & other authors (2000). The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288, 339–344. [DOI] [PubMed] [Google Scholar]
- Flint, M., Maidens, C., Loomis-Price, L. D., Shotton, C., Dubuisson, J., Monk, P., Higginbottom, A., Levy, S. & McKeating, J. A. (1999). Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol 73, 6235–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M., Jr & Foy, E. M. (2005). Evasion of intracellular host defence by hepatitis C virus. Nature 436, 939–945. [DOI] [PubMed] [Google Scholar]
- Goffard, A., Callens, N., Bartosch, B., Wychowski, C., Cosset, F. L., Montpellier, C. & Dubuisson, J. (2005). Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol 79, 8400–8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habersetzer, F., Fournillier, A., Dubuisson, J., Rosa, D., Abrignani, S., Wychowski, C., Nakano, I., Trepo, C., Desgranges, C. & Inchauspe, G. (1998). Characterization of human monoclonal antibodies specific to the hepatitis C virus glycoprotein E2 with in vitro binding neutralization properties. Virology 249, 32–41. [DOI] [PubMed] [Google Scholar]
- Hadlock, K. G., Lanford, R. E., Perkins, S., Rowe, J., Yang, Q., Levy, S., Pileri, P., Abrignani, S. & Foung, S. K. (2000). Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol 74, 10407–10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle, F., Goffard, A., Morel, V., Duverlie, G., McKeating, J., Keck, Z. Y., Foung, S., Penin, F., Dubuisson, J. & Voisset, C. (2007). The neutralizing activity of anti-HCV antibodies is modulated by specific glycans on the E2 envelope protein. J Virol 81, 8101–8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, M., Zhang, J., Flint, M., Logvinoff, C., Cheng-Mayer, C., Rice, C. M. & McKeating, J. A. (2003). Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci U S A 100, 7271–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, D. X., Voisset, C., Tarr, A. W., Aung, M., Ball, J. K., Dubuisson, J. & Persson, M. A. (2007). Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc Natl Acad Sci U S A 104, 16269–16274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia, S. B., Barth, H., Baumert, T., McKeating, J. A. & Chisari, F. V. (2007). Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J Virol 81, 374–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck, Z. Y., Op De Beeck, A., Hadlock, K. G., Xia, J., Li, T. K., Dubuisson, J. & Foung, S. K. (2004). Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J Virol 78, 9224–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck, Z. Y., Li, T. K., Xia, J., Bartosch, B., Cosset, F. L., Dubuisson, J. & Foung, S. K. (2005). Analysis of a highly flexible conformational immunogenic domain A in hepatitis C virus E2. J Virol 79, 13199–13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck, Z. Y., Xia, J., Cai, Z., Li, T. K., Owsianka, A. M., Patel, A. H., Luo, G. & Foung, S. K. (2007). Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. J Virol 81, 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodell, R. G., Conrad, M. E., Ginsberg, A. L. & Bell, C. J. (1976). Efficacy of prophylactic gamma-globulin in preventing non-A, non-B post-transfusion hepatitis. Lancet 1, 557–561. [DOI] [PubMed] [Google Scholar]
- Koutsoudakis, G., Herrmann, E., Kallis, S., Bartenschlager, R. & Pietschmann, T. (2007). The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol 81, 588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavillette, D., Morice, Y., Germanidis, G., Donot, P., Soulier, A., Pagkalos, E., Sakellariou, G., Intrator, L., Bartosch, B. & other authors (2005a). Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol 79, 6023–6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavillette, D., Tarr, A. W., Voisset, C., Donot, P., Bartosch, B., Bain, C., Patel, A. H., Dubuisson, J., Ball, J. K. & Cosset, F. L. (2005b). Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41, 265–274. [DOI] [PubMed] [Google Scholar]
- Lindenbach, B. D., Evans, M. J., Syder, A. J., Wolk, B., Tellinghuisen, T. L., Liu, C. C., Maruyama, T., Hynes, R. O., Burton, D. R. & other authors (2005). Complete replication of hepatitis C virus in cell culture. Science 309, 623–626. [DOI] [PubMed] [Google Scholar]
- Meylan, E. & Tschopp, J. (2006). Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol Cell 22, 561–569. [DOI] [PubMed] [Google Scholar]
- Nakabayashi, H., Taketa, K., Miyano, K., Yamane, T. & Sato, J. (1982). Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res 42, 3858–3863. [PubMed] [Google Scholar]
- Op De Beeck, A., Voisset, C., Bartosch, B., Ciczora, Y., Cocquerel, L., Keck, Z., Foung, S., Cosset, F. L. & Dubuisson, J. (2004). Characterization of functional hepatitis C virus envelope glycoproteins. J Virol 78, 2994–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsianka, A., Clayton, R. F., Loomis-Price, L. D., McKeating, J. A. & Patel, A. H. (2001). Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J Gen Virol 82, 1877–1883. [DOI] [PubMed] [Google Scholar]
- Owsianka, A., Tarr, A. W., Juttla, V. S., Lavillette, D., Bartosch, B., Cosset, F. L., Ball, J. K. & Patel, A. H. (2005). Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol 79, 11095–11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsianka, A. M., Timms, J. M., Tarr, A. W., Brown, R. J., Hickling, T. P., Szwejk, A., Bienkowska-Szewczyk, K., Thomson, B. J., Patel, A. H. & Ball, J. K. (2006). Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol 80, 8695–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotti, M., Mancini, N., Diotti, R. A., Tarr, A. W., Ball, J. K., Owsianka, A., Adair, R., Patel, A. H., Clementi, M. & Burioni, R. (2008). Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the HCV\E2 protein. J Virol in press [DOI] [PMC free article] [PubMed]
- Pestka, J. M., Zeisel, M. B., Blaser, E., Schurmann, P., Bartosch, B., Cosset, F. L., Patel, A. H., Meisel, H., Baumert, J. & other authors (2007). Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A 104, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccasecca, R., Ansuini, H., Vitelli, A., Meola, A., Scarselli, E., Acali, S., Pezzanera, M., Ercole, B. B., McKeating, J. & other authors (2003). Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J Virol 77, 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa, D., Campagnoli, S., Moretto, C., Guenzi, E., Cousens, L., Chin, M., Dong, C., Weiner, A. J., Lau, J. Y. & other authors (1996). A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci U S A 93, 1759–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrin, V., Boulanger, P., Penin, F., Granier, C., Cosset, F. L. & Bartosch, B. (2005). Assembly of functional hepatitis C virus glycoproteins on infectious pseudoparticles occurs intracellularly and requires concomitant incorporation of E1 and E2 glycoproteins. J Gen Virol 86, 3189–3199. [DOI] [PubMed] [Google Scholar]
- Schofield, D. J., Bartosch, B., Shimizu, Y. K., Allander, T., Alter, H. J., Emerson, S. U., Cosset, F. L. & Purcell, R. H. (2005). Human monoclonal antibodies that react with the E2 glycoprotein of hepatitis C virus and possess neutralizing activity. Hepatology 42, 1055–1062. [DOI] [PubMed] [Google Scholar]
- Shimizu, Y. K., Hijikata, M., Iwamoto, A., Alter, H. J., Purcell, R. H. & Yoshikura, H. (1994). Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol 68, 1494–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoukry, N. H., Cawthon, A. G. & Walker, C. M. (2004). Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol 58, 391–424. [DOI] [PubMed] [Google Scholar]
- Simmonds, P., Bukh, J., Combet, C., Deleage, G., Enomoto, N., Feinstone, S., Halfon, P., Inchauspe, G., Kuiken, C. & other authors (2005). Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42, 962–973. [DOI] [PubMed] [Google Scholar]
- Tarr, A. W., Owsianka, A. M., Timms, J. M., McClure, C. P., Brown, R. J., Hickling, T. P., Pietschmann, T., Bartenschlager, R., Patel, A. H. & Ball, J. K. (2006). Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43, 592–601. [DOI] [PubMed] [Google Scholar]
- Thimme, R., Lohmann, V. & Weber, F. (2006). A target on the move: innate and adaptive immune escape strategies of hepatitis C virus. Antiviral Res 69, 129–141. [DOI] [PubMed] [Google Scholar]
- Wakita, T., Pietschmann, T., Kato, T., Date, T., Miyamoto, M., Zhao, Z., Murthy, K., Habermann, A., Krausslich, H. G. & other authors (2005). Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11, 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M. Y., Bartosch, B., Zhang, P., Guo, Z. P., Renzi, P. M., Shen, L. M., Granier, C., Feinstone, S. M., Cosset, F. L. & Purcell, R. H. (2004). Neutralizing antibodies to hepatitis C virus (HCV) in immune globulins derived from anti-HCV-positive plasma. Proc Natl Acad Sci U S A 101, 7705–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, J., Gastaminza, P., Cheng, G., Kapadia, S., Kato, T., Burton, D. R., Wieland, S. F., Uprichard, S. L., Wakita, T. & Chisari, F. V. (2005). Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102, 9294–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]