Abstract
The genomes of commonly used variants of human cytomegalovirus (HCMV) strains Towne and AD169 each contain a substantial mutation in which a region (UL/b′) at the right end of the long unique region has been replaced by an inverted duplication of a region from the left end of the genome. Using high-throughput technology, we have sequenced HCMV strain Towne (ATCC VR-977) and confirmed the presence of two variants, one exhibiting the replacement in UL/b′ and the other intact in this region. Both variants are mutated in genes RL13, UL1, UL40, UL130, US1 and US9. We have also sequenced a novel AD169 variant (varUC) that is intact in UL/b′ except for a small deletion that affects genes UL144, UL142, UL141 and UL140. Like other AD169 variants, varUC is mutated in genes RL5A, RL13, UL36 and UL131A. A subpopulation of varUC contains an additional deletion affecting genes IRS1, US1 and US2.
Human cytomegalovirus (HCMV; species Human herpesvirus 5) was first isolated over 50 years ago (Craig et al., 1957; Rowe et al., 1956; Smith, 1956). The most widely used laboratory strains are Towne (Plotkin et al., 1975) and AD169 (Rowe et al., 1956). Both have been distributed widely and developed as vaccine candidates, and over the years their detailed histories have become obscure. Moreover, the fact that the biological properties of these strains are not conserved between stocks (Brown et al., 1995) demonstrates the existence of genetic variants, and this may affect the interpretation of experimental studies. This paper contributes to the characterization of variants present in the ATCC VR-977 stock of Towne and of commonly used variants of AD169 in comparison with a novel, genetically more intact variant.
As shown in Fig. 1, the commonly used variants of Towne and AD169 arose from wild-type HCMV genomes (236 kb) via replacement of a region (UL/b′) at the right end of UL (Cha et al., 1996; Davison et al., 2003b; Dolan et al., 2004; Dunn et al., 2003; Murphy et al., 2003; Prichard et al., 2001). These large-scale lesions were accompanied by additional mutations. AD169 has frameshifts in genes RL5A, RL13 and UL131A (Akter et al., 2003; Davison et al., 2003a, b; Yu et al., 2002) and a substitution in gene UL36 that inactivates the encoded inhibitor of apoptosis (Skaletskaya et al., 2001). Towne has frameshifts in genes RL13 and UL130 (Dolan et al., 2004).
Fig. 1.
Schema for the derivation of the major lesion in the genome of commonly used AD169 variants from a wild-type virus genome; lengths are shown to scale. In the wild-type genome, the long and short unique sequences (UL and US; shown as thinner structures) are flanked by inverted repeats (ba/b′a′ and ca/c′a′; shown as thicker structures). As indicated by the dashed lines, the AD169 genome was generated by replacing a region at the right end of UL (UL/b′; shaded grey) by an inverted duplication of a region from the left end of the genome (arrow). This resulted in UL becoming shorter by 15 kb (19 genes) and b/b′ becoming longer by 10 kb (six genes and part of another). A similar phenomen occurred in the derivation of the Towne varS genome, with UL becoming shorter by 13 kb (15 genes) and b/b′ becoming longer by 11 kb (seven genes and part of another).
The Towne genome sequence has been determined (Dunn et al., 2003; Murphy et al., 2003) from a bacterial artificial chromosome (BAC) constructed from plaque-purified ATCC VR-977 (Marchini et al., 2001). However, ATCC VR-977 is known to contain a mixture of two variants (Hahn et al., 2003). One (Towne varRIT3 or Towneshort; here called varS) is represented in the BAC and, as described above, lacks UL/b′. The other (Townelong; here called varL) is intact in UL/b′, and this region has been sequenced (Dolan et al., 2004).
We determined the sequence of ATCC VR-977 by aligning data obtained from an Illumina Genome Analyzer (http://www.illumina.com) against a constructed reference sequence. To obtain the reference, the more reliable of the two varS BAC sequences (AC146851; Murphy et al., 2003) was reorganized into a genome-equivalent arrangement after identifying the termini and removing insertions in gene UL32 and the origin of DNA replication. The varS reference was generated from this sequence by inserting the region containing genes IRS1–US12 from strain Merlin (AY446894; Dolan et al., 2004) in place of the plasmid vector, which had replaced these genes during generation of the BAC. The varL reference was then constructed by inserting the UL/b′ sequence (GenBank accession no. AY446869; Dolan et al., 2004) into the varS reference in place of the duplication that had originally replaced UL/b′.
The Illumina data were derived from whole cell DNA extracted from human fetal fibroblasts infected with ATCC VR-977, and assembled and viewed using Maq and Maqview (Li et al., 2008; http://maq.sourceforge.net). A total of 47.1 % of the 5 079 235 sequences (50 nt each) aligned with the derived varL consensus and the coverage was 516 reads per nt. Test assemblies using appropriate references confirmed that ATCC VR-977 contains both varL and varS, and showed that the whole population has frameshifts in genes RL13, UL1, UL130, US1 and US9. In addition, a 346 bp deletion in gene UL40 was present in almost the whole population, though detection of very sparse data from the deleted region indicated that a small proportion (much less than 1 %) of genomes might be intact. This deletion implies that varL and varS do not encode the gene UL40 signal peptide sequence, which contributes to natural killer (NK) cell evasion by upregulating human lymphocyte antigen-E (Tomasec et al., 2000). Eighteen clear single nucleotide polymorphisms (SNPs) were identified, but these could not be assigned to particular variants. In addition to the presence of UL/b′, and excluding the duplicate copies of the inverted repeats, ATCC VR-977 differed from the varS BAC at 40 loci consisting of 34 substitutions and six insertions. By referring to sequences available for other HCMV strains, it was possible to infer which sequence was mutated at 25 loci. The BAC was assessed as being mutated at 23 and ATCC VR-977 at one, with the remaining substitution corresponding to an SNP.
In contrast with ATCC VR-977, in which varL has retained UL/b′, all commonly used AD169 stocks lack this region. Therefore, it was of interest that one of us possessed an AD169 stock (varUC) that reputedly contained UL/b′ sequences. N. Lurain had received varUC at the University of Chicago from K. Thompson, who in turn had obtained it from M. Beem in 1981 at the same institution. No documentation was available on its history, but it was thought to have undergone at least 50 passages since its acquisition. Initial studies (N. Lurain, unpublished data) had demonstrated that varUC plaques were similar to those of strain Toledo (Kemble et al., 1996), which contains UL/b′ (Cha et al., 1996), appearing as clusters of refractile, rounded cells, rather than the well-separated, elongated cells characteristic of commonly used AD169 variants. Also, sequencing had revealed the presence of UL/b′ genes in varUC, specifically UL146 and a region containing the 3′ end of UL144 and the 5′ end of UL140 with a 3.2 kb deletion encompassing the intervening genes UL142 and UL141. Moreover, genotyping data from microarray experiments had indicated the presence of all UL/b′ genes except UL142 and UL141 (J. García-Ramírez, D. Foster, L. Buehler, N. Lurain & P. Ghazal, unpublished data). These findings implied that varUC is either an AD169 variant that has retained most of UL/b′ or another strain entirely.
In order to distinguish between these possibilities, several genes that are mutated in commonly used AD169 variants or that vary greatly between HCMV strains were amplified by PCR from DNA extracted from a stock of cell-released varUC and sequenced; UL/b′ was also sequenced in its entirety. These data were compared with the published genome sequences of two AD169 variants. One of these was varUK, for which the sequence (X17403) was derived by Chee et al. (1990) and updated (BK000394) by the correction of errors and the insertion of a 929 bp region that is absent from certain stocks (Dargan et al., 1997; Mocarski et al., 1997). The other was varATCC, for which the sequence (AC146999) was derived by Murphy et al. (2003) from a BAC generated from plaque-purified ATCC VR-538 (Yu et al., 2002). In the 16976 bp of the 28780 bp determined for varUC that were comparable with varUK and varATCC, all three genomes were closely similar. The presence of UL/b′ in varUC, except for the previously characterized 3.2 kb deletion, was confirmed. This deletion is predicted to result in lack of expression of the UL144, UL142 and UL141 proteins, and expression of the UL140 protein with the C-terminal eight residues replaced by 71 residues specified by a reading frame that overlaps UL144.
The partial information obtained was consistent with varUC being an AD169 variant, and formed the basis for deriving the complete genome sequence from Illumina data derived from DNA extracted from pelleted cell-free virions. A reference was constructed from the varUK sequence, utilizing the partial varUC data to amend differences, and inserting UL/b′ in place of the duplication that had originally replaced UL/b′. A total of 92.4 % of 6 264 332 sequences (50 nt each) aligned with the derived varUC consensus, and the coverage was 1267 reads per nt. Test assemblies and PCR experiments demonstrated the absence of the UL/b′ deleted form characteristic of varUK and varATCC and the 929 bp deleted form present in some varUK stocks. The 3.2 kb deletion in UL/b′ was confirmed as being a feature of the entire population, and a 3.7 kb deletion in c′/US, which affects genes IRS1, US1 and US2, was detected in the majority of the population. Test assemblies also showed that the entire varUC genome population contained the mutations in RL5A, RL13, UL36 and UL131A present in varUK and varATCC. Four clear SNPs were identified.
Given the apparent existence of a vast number of differentiable HCMV strains (e.g. Bradley et al., 2008; Rasmussen et al., 2003), the high degree of sequence similarity between the three variants and the sharing of several mutations in common, confirmed that varUC is an AD169 variant. Differences due to insertions included the presence of UL/b′ in varUC, length variations in the tandem repeat in a/a′ and heterogeneity in several non-coding polynucleotide tracts. Substitutions were identified at 54 loci (Table 1), with over half (29) in bac/b′a ′c′ and the adjacent sequence at the left end of UL, and 32 in protein-coding regions (five synonymous and 27 non-synonymous). By referring to sequences available for other HCMV strains, it was possible to infer which sequence was mutated at 42 loci. A total of 36 mutations were specific to a single variant: nine to varUC and 27 to varATCC. Each of the remaining six mutations was present in pairs of variants: one in varUC/varUK, two in varATCC/varUC and three in varATCC/varUK.
Table 1.
Nucleotide substitutions in AD169 variants
nd, Substitutions where the mutant and non-mutant residues could not be distinguished; cr, synonymous substitutions or substitutions not located in protein-coding regions; sub, non-coding and synonymous substitutions.
| Nucleotide* | Mutated variant | Region affected | Effect† |
|---|---|---|---|
| 971 | varUC | cr | sub |
| 1059 | varATCC | RL1 | L→S |
| 1080 | varATCC | RL1 | T→R |
| 1095 | varATCC | RL1 | Q→R |
| 1166 | varATCC | RL1 | C→S |
| 1270 | nd | RL1 | Q•H |
| 1403 | varATCC | RL1 | C→R |
| 1672 | varATCC | RL1 | I→M |
| 1749 | varATCC | RL1 | S→ς‡ |
| 1925 | varATCC | RL1 | W→R |
| 2337 | varATCC | cr | sub |
| 2781 | varATCC | cr | sub |
| 2898 | varATCC | cr | sub |
| 3337 | varATCC | cr | sub |
| 5080 | varUC | RL5A§ | sub |
| 8318 | varATCC/varUK | RL10 | N→D |
| 27980 | varATCC | UL23 | sub |
| 29374 | varATCC | UL24 | F→L |
| 42128 | varATCC | UL32 | E→G |
| 56856 | varUC | UL44 | A→S |
| 98223 | varATCC | cr | sub |
| 103266 | varUC | UL70 | H→Y |
| 108681 | varUC | UL74A | sub |
| 114378 | varATCC | UL78 | S→F |
| 118556 | varUC | UL82 | D→A |
| 118730 | varATCC/varUK | UL82 | R→P |
| 134412 | varATCC/varUK | UL89 | A→S |
| 136946 | varATCC | UL93 | A→D |
| 145916 | varATCC | UL99 | F→S |
| 155061 | varATCC | UL105 | sub |
| 160678 | varATCC | UL111A | K→R |
| 175085 | varUC | cr | sub |
| 191003 | nd | b′ | sub |
| 191021 | nd | b′ | sub |
| 191043 | nd | b′ | sub |
| 191071 | nd | b′ | sub |
| 191113 | nd | b′ | sub |
| 191146 | nd | b′ | sub |
| 191277 | nd | b′ | sub |
| 191334 | nd | b′ | sub |
| 191336 | nd | b′ | sub |
| 191588 | nd | a′ | sub |
| 191635 | nd | a′ | sub |
| 191743 | varATCC | a′ | sub |
| 192163 | varATCC | c′ | sub |
| 192244 | varUC/varUK | c′ (TRS1/IRS1) | sub |
| 192325 | varATCC/varUC | c′ (TRS1/IRS1) | sub |
| 192434 | varATCC | c′ (TRS1/IRS1) | S→G |
| 192723 | varATCC | c′ (TRS1/IRS1) | L→P |
| 192770 | varUC | c′ (TRS1/IRS1) | V→L |
| 192983 | varUC | c′ (TRS1/IRS1) | T→P |
| 197200 | varATCC | cr | sub |
| 215658 | varATCC/varUC | US23 | C→Y |
| 224225 | varATCC | US29US30 | L→P |
*Location in the varUC genome sequence (FJ527563; 231781 bp). This excludes the duplicate copies of the inverted repeats (i.e. ab and ca) and 12 substitutions (nt 222147–223855) in a region of the varATCC sequence that was replaced during construction of the BAC. This replacement appears to have originated from a strain other than AD169 (probably Toledo).
†For non-synonymous substitutions, an arrow indicates the change from the non-mutant to the mutant amino acid residue. Where it was not possible to distinguish the non-mutant and mutant residues, the alternatives are separated by a filled circle.
‡Termination codon indicated by ς.
§Frameshifted in all variants and therefore considered non-coding.
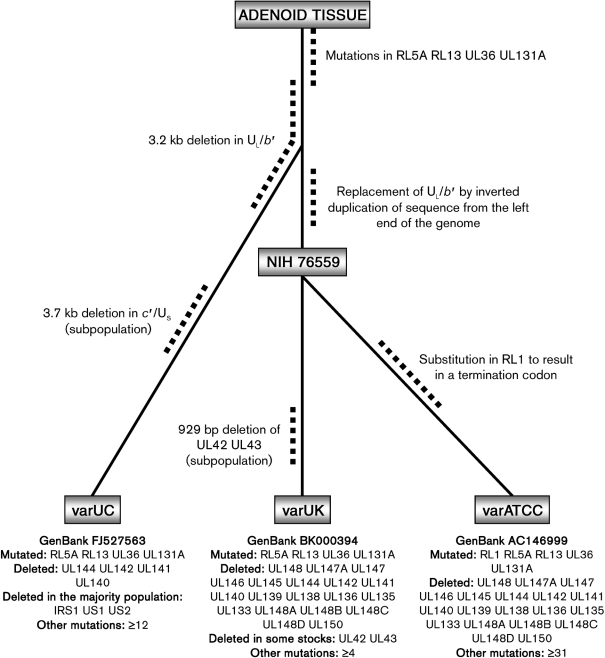
Given the lack of historical information, it is not possible to reconstruct fully the lineages that led to the three AD169 variants. AD169 was isolated at the National Institutes of Health (Bethesda, MD, USA) from the adenoids of a 7-year-old girl and passaged 14 times in human fibroblast cells to yield a stock named NIH 76559 (Rowe et al., 1956). The lineage that led to varUK was initiated by researchers at St George's Hospital Medical School (London, UK), who obtained NIH 76559 in 1960 and passaged it 40 times in human fibroblast cells. The resulting virus was used to make batches of a potential vaccine by 16–24 additional passages (Elek & Stern, 1974). The varUK sequence was determined from a set of plasmid clones generated from a plaque-purified derivative of one of these passages (Oram et al., 1982). The route by which varATCC was derived from NIH 76559 is less clear, but it seems likely that it originated from an exclusively US lineage, since a US researcher, W. A. Chappell, deposited the stock with the ATCC. The ATCC has declined to reveal to us when this occurred, but has distributed AD169 at least since 1973 (e.g. Smith & de Harven, 1973), and now markets it as ATCC VR-538. The relatively large number of mutations specific to varATCC is consistent with the impressive numbers of passages to which AD169 was subjected in early years in some US laboratories (e.g. 232 passages by Vonka & Benyesh-Melnick, 1966). If varATCC indeed originated from a purely US source, a schema of the type illustrated in Fig. 2 may be proposed. However, resolution of the details is confounded by the potential persistence of mutations (including those that may have arisen in the ancestor of any two or all three variants) to different extents in subsequent lineages, and to the fact that the varUK and varATCC sequences originated from molecular clones made from plaque-purified viruses and therefore do not necessarily represent whole populations.
Fig. 2.
Schema for the derivation of AD169 variants from the original clinical material, based on the assumption that varATCC was generated from a US source and not from importation of varUK. Mutations in RL5A, RL13, UL36 and UL131A (and perhaps other, undetected mutations resulting in amino acid substitutions or affecting non-coding sequences) arose during the 14 passages that led to NIH 76559 and were inherited by all three lineages. Later on during the 14 passages, the deletion of UL/b′ occurred, and this was inherited by varUK and varATCC from NIH 76559. The 3.2 kb deletion in UL/b′ that characterizes varUC occurred either at an intermediate stage as a precursor of the larger deletion or in the separated lineage. Genes that are detectably disabled, and other mutations whose effects are unknown (Table 1), are listed below each variant.
Like many other passaged HCMV strains, the Towne and AD169 variants are mutated in gene RL13 and one of the three genes in the UL128 locus (UL128, UL130 and UL131A) (Akter et al., 2003; Dolan et al., 2004; Hahn et al., 2004). This implies strong selection during passage in human fibroblasts against the encoded functions, which are involved in cell tropism (reviewed by Sinzger et al., 2008a). Towne varS and the AD169 variants are also mutated in UL/b′, as is strain TB40/E, which is frameshifted in UL141, with a derivative additionally lacking UL145 and UL144 (Dolan et al., 2004; Sinzger et al., 2008b; Tomasec et al., 2005). The patterns of mutation suggest that more than one gene in this region (UL145, UL144, UL142, UL141 or UL140) may be selected against, though not as strongly as RL13 and the UL128 locus. It is not immediately apparent why expression of these genes might be deleterious. The proteins encoded by UL142 and UL141 are involved in evasion of NK cell function, the former by downregulating MICA, which is a ligand for the activating receptor NKG2D (Chalupny et al., 2006; Wills et al., 2005), and the latter by sequestering CD155 (PVR), which is a ligand for the activating receptors CD226 (DNAM-1) and CD96 (TACTILE) (Tomasec et al., 2005). The UL144 protein activates NF-κB in a TRAF6-dependent manner, causing upregulation of the chemokine CCL22 (MDC) (Poole et al., 2008), and also inhibits T cell proliferation by binding CD272 (BTLA) (Cheung et al., 2005).
We have contributed towards the characterization of variants of HCMV strains Towne and AD169, so that biological data may be assessed with greater rigour. The sequence of Towne ATCC VR-977 confirmed the presence of two major variants (varL and varS) and extended knowledge of their shared mutations. A novel AD169 variant (varUC) was shown to be genetically more intact than varUK and varATCC and may be a new tool in the hands of HCMV researchers.
Acknowledgments
We are grateful to Duncan McGeoch for commenting on the manuscript. This work was supported by the Medical Research Council and the Wellcome Trust.
Footnotes
References
- Akter, P., Cunningham, C., McSharry, B. P., Dolan, A., Addison, C., Dargan, D. J., Hassan-Walker, A. F., Emery, V. C., Griffiths, P. D. & other authors (2003). Two novel spliced genes in human cytomegalovirus. J Gen Virol 84, 1117–1122. [DOI] [PubMed] [Google Scholar]
- Bradley, A. J., Kovács, I. J., Gatherer, D., Dargan, D. J., Alkharsah, K. R., Chan, P. K. S., Carman, W. F., Dedicoat, M., Emery, V. C. & other authors (2008). Genotypic analysis of two hypervariable human cytomegalovirus genes. J Med Virol 80, 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. M., Kaneshima, H. & Mocarski, E. S. (1995). Dramatic interstrain differences in the replication of human cytomegalovirus in SCID-hu mice. J Infect Dis 171, 1599–1603. [DOI] [PubMed] [Google Scholar]
- Cha, T.-A., Tom, E., Kemble, G. W., Duke, G. M., Mocarski, E. S. & Spaete, R. R. (1996). Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol 70, 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupny, N. J., Rein-Weston, A., Dosch, S. & Cosman, D. (2006). Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun 346, 175–181. [DOI] [PubMed] [Google Scholar]
- Chee, M. S., Bankier, A. T., Beck, S., Bohni, R., Brown, C. M., Cerny, R., Horsnell, T., Hutchison, C. A., III, Kouzarides, T. & other authors (1990). Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol 154, 125–169. [DOI] [PubMed] [Google Scholar]
- Cheung, T. C., Humphreys, I. R., Potter, K. G., Norris, P. S., Shumway, H. M., Tran, B. R., Patterson, G., Rochelle, J.-J., Yoon, M. & other authors (2005). Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc Natl Acad Sci U S A 102, 13218–13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, J. M., Macauley, J. C., Weller, T. H. & Wirth, P. (1957). Isolation of intranuclear inclusion producing agents from infants with illnesses resembling cytomegalic inclusion disease. Proc Soc Exp Biol Med 94, 4–12. [DOI] [PubMed] [Google Scholar]
- Dargan, D. J., Jamieson, F. E., MacLean, J., Dolan, A., Addison, C. & McGeoch, D. J. (1997). The published DNA sequence of human cytomegalovirus strain AD169 lacks 929 base pairs affecting genes UL42 and UL43. J Virol 71, 9833–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison, A. J., Akter, P., Cunningham, C., Dolan, A., Addison, C., Dargan, D. J., Hassan-Walker, A. F., Emery, V. C., Griffiths, P. D. & Wilkinson, G. W. G. (2003a). Homology between the human cytomegalovirus RL11 gene family and human adenovirus E3 genes. J Gen Virol 84, 657–663. [DOI] [PubMed] [Google Scholar]
- Davison, A. J., Dolan, A., Akter, P., Addison, C., Dargan, D. J., Alcendor, D. J., McGeoch, D. J. & Hayward, G. S. (2003b). The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol 84, 17–28. [DOI] [PubMed] [Google Scholar]
- Dolan, A., Cunningham, C., Hector, R. D., Hassan-Walker, A. F., Lee, L., Addison, C., Dargan, D. J., McGeoch, D. J., Gatherer, D. & other authors (2004). Genetic content of wild-type human cytomegalovirus. J Gen Virol 85, 1301–1312. [DOI] [PubMed] [Google Scholar]
- Dunn, W., Chou, C., Li, H., Hai, R., Patterson, D., Stolc, V., Zhu, H. & Liu, F. (2003). Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100, 14223–14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elek, S. D. & Stern, H. (1974). Development of a vaccine against mental retardation caused by cytomegalovirus infection in utero. Lancet 1, 1–5. [DOI] [PubMed] [Google Scholar]
- Hahn, G., Rose, D., Wagner, M., Rhiel, S. & McVoy, M. A. (2003). Cloning of the genomes of human cytomegalovirus strains Toledo, TownevarRIT3, and Townelong as BACs and site-directed mutagenesis using a PCR-based technique. Virology 307, 164–177. [DOI] [PubMed] [Google Scholar]
- Hahn, G., Revello, M. G., Patrone, M., Percivalle, E., Campanini, G., Sarasini, A., Wagner, M., Gallina, A., Milanesi, G. & other authors (2004). Human cytomegalovirus UL131–128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 78, 10023–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble, G., Duke, G., Winter, R. & Spaete, R. (1996). Defined large-scale alterations of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol 70, 2044–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Ruan, J. & Durbin, R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18, 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini, A., Liu, H. & Zhu, H. (2001). Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol 75, 1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski, E. S., Prichard, M. N., Tan, C. S. & Brown, J. M. (1997). Reassessing the organization of the UL42–UL43 region of the human cytomegalovirus strain AD169 genome. Virology 239, 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, E., Yu, D., Grimwood, J., Schmutz, J., Dickson, M., Jarvis, M. A., Hahn, G., Nelson, J. A., Myers, R. M. & Shenk, T. E. (2003). Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A 100, 14976–14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram, J. D., Downing, R. G., Akrigg, A., Dollery, A. A., Duggleby, C. J., Wilkinson, G. W. G. & Greenaway, P. J. (1982). Use of recombinant plasmids to investigate the structure of the human cytomegalovirus genome. J Gen Virol 59, 111–129. [DOI] [PubMed] [Google Scholar]
- Plotkin, S. A., Furukawa, T., Zygraich, N. & Huygelen, C. (1975). Candidate cytomegalovirus strain for human vaccination. Infect Immun 12, 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, E., Atkins, E., Nakayama, T., Yoshie, O., Groves, I., Alcami, A. & Sinclair, J. (2008). NF-κB-mediated activation of the chemokine CCL22 by the product of the human cytomegalovirus gene UL144 escapes regulation by viral IE86. J Virol 82, 4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard, M. N., Penfold, M. E. T., Duke, G. M., Spaete, R. R. & Kemble, G. W. (2001). A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev Med Virol 11, 191–200. [DOI] [PubMed] [Google Scholar]
- Rasmussen, L., Geissler, A. & Winters, M. (2003). Inter- and intragenic variations complicate the molecular epidemiology of human cytomegalovirus. J Infect Dis 187, 809–819. [DOI] [PubMed] [Google Scholar]
- Rowe, W. P., Hartley, J. W., Waterman, S., Turner, H. C. & Huebner, R. J. (1956). Cytopathogenic agent resembling human salivary gland virus recovered from tissue cultures of human adenoids. Proc Soc Exp Biol Med 92, 418–424. [PubMed] [Google Scholar]
- Sinzger, C., Digel, M. & Jahn, G. (2008a). Cytomegalovirus cell tropism. Curr Top Microbiol Immunol 325, 63–83. [DOI] [PubMed] [Google Scholar]
- Sinzger, C., Hahn, G., Digel, M., Katona, R., Sampaio, K. L., Messerle, M., Hengel, H., Koszinowski, U., Brune, W. & Adler, B. (2008b). Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89, 359–368. [DOI] [PubMed] [Google Scholar]
- Skaletskaya, A., Bartle, L. M., Chittenden, T., McCormick, A. L., Mocarski, E. S. & Goldmacher, V. S. (2001). A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc Natl Acad Sci U S A 98, 7829–7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. G. (1956). Propagation in tissue cultures of a cytopathogenic virus from human salivary gland virus (SGV) disease. Proc Soc Exp Biol Med 92, 424–430. [DOI] [PubMed] [Google Scholar]
- Smith, J. D. & de Harven, E. (1973). Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. I. Sequence of viral replication. J Virol 12, 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasec, P., Braud, V. M., Rickards, C., Powell, M. B., McSharry, B. P., Gadola, S., Cerundolo, V., Borysiewicz, L. K., McMichael, A. J. & Wilkinson, G. W. G. (2000). Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031–1033. [DOI] [PubMed] [Google Scholar]
- Tomasec, P., Wang, E. C., Davison, A. J., Vojtesek, B., Armstrong, M., Griffin, C., McSharry, B. P., Morris, R. J., Llewellyn-Lacey, S. & other authors (2005). Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol 6, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonka, V. & Benyesh-Melnick, M. (1966). Interactions of human cytomegalovirus with human fibroblasts. J Bacteriol 91, 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills, M. R., Ashiru, O., Reeves, M. B., Okecha, G., Trowsdale, J., Tomasec, P., Wilkinson, G. W. G., Sinclair, J. & Sissons, J. G. P. (2005). Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J Immunol 175, 7457–7465. [DOI] [PubMed] [Google Scholar]
- Yu, D., Smith, G. A., Enquist, L. W. & Shenk, T. (2002). Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol 76, 2316–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]