Abstract
Protein dynamics is intimately related to biological function. Core dynamics is usually studied with 2H spin relaxation of the 13CDH2 group, analyzed traditionally with the model-free (MF) approach. We showed recently that MF is oversimplified in several respects. This includes the assumption that the local motion of the dynamic probe and the global motion of the protein are decoupled, the local geometry is simple, and the local ordering has axial symmetry. Because of these simplifications MF has yielded a puzzling picture where the methyl rotation axis is moving rapidly with amplitudes ranging from nearly complete disorder to nearly complete order in tightly packed protein cores. Our conclusions emerged from applying to methyl dynamics in proteins the slowly relaxing local structure (SRLS) approach of Polimeno and Freed (J. Phys. Chem. 1995, 99, 1099), which can be considered the generalization of MF, with all the simplifications mentioned above removed. The SRLS picture derived here for the B1 immunoglobulin binding domain of peptostreptococcal protein L, studied over the temperature range of 15 − 45 °C, is fundamentally different from the MF picture. Thus, methyl dynamics is characterized structurally by rhombic local potentials with varying symmetries, and dynamically by tenfold slower rates of local motion. On average potential rhombicity decreases, mode-coupling increases and the rate of local motion increases with increasing temperature. The average activation energy for local motion is 2.0 ± 0.2 kcal/mol. Mode-coupling affects the analysis even at 15 oC. The accuracy of the results is improved by including in the experimental data set relaxation rates associated with rank 2 coherences.
Keywords: slowly relaxing local structure, methyl dynamics in proteins, NMR spin relaxation in proteins
I. Introduction
NMR spin relaxation is a powerful method for studying protein dynamics.1-9 The traditional probe for investigating backbone motion is the 15N-1H bond and the common probe for studying side chain motion is the uniformly 13C-labeled and fractionally deuterated methyl group, 13CH2D.5,6,10-12 In this study we focus on the latter. Methyl dynamics in proteins is analyzed typically with the model-free (MF) approach,13-15 that assumes that the global and local motions of the probe are decoupled due to the former being much slower than the latter. This is an approximation, and so are the high symmetries assigned implicitly to the diffusion, ordering and magnetic tensors involved, and the coincidence of their frames, which simplifies the local geometry. By virtue of these simplifications an analytical formula is obtained for the measurable spectral density,13 specific values of which enter the expressions for the experimental relaxation rates. The original MF spectral density13 is determined by an effective correlation time for local motion, τe, a squared generalized order parameter, S2, representing the spatial restrictions at the site of the motion of the probe, and the global motion correlation time, τm. The latter is usually determined independently.
For methyl dynamics MF considers two local motions including rotation about the C-CH3 axis and fluctuations of the C-CH3 axis.10 Moreover, the methyl rotation axis C-CH3 (to be denoted Mz, with M representing the local ordering/local diffusion frame) is tilted at βMQ = 110.5° from the magnetic quadrupolar frame, Q, which lies along the C-D bond (110.5o is the tetrahedral angle taking rCH = rCD = 1.115 Å)12. Yet, as pointed out above, the original MF formula,13 typically used in methyl dynamics analyses, features only one mode of local motion and has no provision for a “diffusion tilt”. These features entail further approximations (see below).
We have shown recently that the MF approach is oversimplified.16-23 This has been accomplished by applying to NMR spin relaxation in proteins16 the Slowly Relaxing Local Structure (SRLS) approach of Freed and co-workers.24-26 SRLS can be considered the generalization of MF, yielding the latter in asymptotic limits.16,20,21,24 Unlike MF the SRLS model takes into account rigorously the dynamical coupling between the global motion of the protein and the local motion of the dynamic probe, brought into effect by a rhombic coupling potential. It features explicitly local motional modes parallel and perpendicular to the methyl rotation axis.16-23,25,26 and accounts rigorously for the tilt between a rhombic local ordering frame, M, and the magnetic frame, Q. The Euler angles ΩC'M, which relate the M frame to a local director frame, C’ (e.g., the equilibrium C-CH3 orientation), are governed by the coupling/ordering potential. “Dynamical coupling” means that through the time dependence of ΩC'M the locally reorienting dynamic probe follows the slower motion of the protein.
The MF simplifications have far-fetched implications. For a M frame tilted relative to the Q frame the spectral density, JQQ(ω) (QQ denotes quadrupolar auto-correlated relaxation), comprises three generic spectral density functions, jK(ω), with K = 0, 1, 2 (refs. 16, 25, 26), by analogy with J(ω) for axial global diffusion of a rigid protein comprising three Lorentzian functions with K = 0, 1, 2. Yet, the MF spectral density consists of a single function which represents the K = 0 contribution. Various parameterizations of its form have been attempted to overcome the flaw of omission of the K = 1 and K = 2 contributions. If the local ordering frame, M, is rhombic, as we found it to be,23 cross-term functions jKK’(ω) will also enter the expression for JQQ(ω). This renders the parameterized MF spectral density to be very different from the actual spectral density. However, in many cases the experimental data can be reproduced by force-fitting, with the statistical criteria fulfilled, but the best-fit parameters (S2 and τe) highly inaccurate.23
The form of JQQ(ω) is parameterized in MF as follows. To accommodate two local motional modes S2 is factored into the product [P2(cos 110.5°)]2 × Saxis2 = 0.1 × Saxis2 . The term [P2(cos 110.5°)]2 = 0.1 represents the squared order parameter for methyl rotation about C−CH3, and Saxis2 the axial squared order parameter for motion of the C−CH3 axis.10,27a As outlined below in detail, factoring S2 as shown, with the meaning of the constituents as indicated, is only valid when τe is in the extreme motional narrowing limit.23,27b,c Yet, in practice finite values of τe are required in MF analyses to fit the experimental data.
Practical implications of the MF simplifications have been investigated recently23 using the B1 immunoglobulin binding domain of peptostreptococcal protein L (to be called below “protein L”)12 and ubiquitin28 as test cases. The respective data were subjected to SRLS analysis23 and the emerging dynamic pictures were compared with the corresponding previously obtained MF pictures.12,28
We found that rhombic local potential/local ordering is required to analyze methyl dynamics consistently and insightfully.23 MF analyses yield unduly large distributions in the value of Saxis2 ranging from nearly complete disorder (Saxis2 ~ 0.1) to nearly complete order (S axis2 ~ 1), often exhibiting three distinct maxima.6,28-31 The (pervasive) low Saxis2 values imply large-amplitude excursions of the C-CH3 axis in tightly packed protein cores.6 Interpretation in terms of limited excursions using the 1D and 3D Gaussian Axial Fluctuations (GAF) models32-34 is incompatible with axial symmetry around C−CH3, inherent in the definition of Saxis2 (ref. 27a). Contrary to the problematic MF picture, SRLS interprets the variations in the experimental data as variations in the symmetry, and to some extent the magnitude, of the local ordering potential (or local ordering tensor).23 The three categories of Saxis2 values correspond to different forms (symmetries) of the rhombic local potential.23 This is physically tenable, provides new and interesting site-specific structural information, and agrees with NMR J-coupling and reduced dipolar coupling,35,36 molecular-dynamics37,38 and molecular mechanics39 studies. All of these investigations have shown that local structural asymmetry prevails at methyl sites in proteins, contrary to the axial Saxis2 based MF picture.
The present paper is an extension of our previous study23 which was based on 2H T1 and T2 data acquired for protein L12 and ubiquitin28 at ambient temperature and magnetic fields of 11.7 and 14.1 T. Kay and co-workers developed pulse sequences for measuring relaxation rates associated with double-quantum, two-spin-order and antiphase rank 2 coherences,11 in addition to 2H T1 and T2.10 For protein L the Kay group acquired all five 2H relaxation rates at 5, 15, 25, 35 and 45 °C at a magnetic field of 11.7 T. At 25 (5) °C additional data were acquired at magnetic fields of 9.4, 14.1 and 18.8 (14.1) T. This is among the most extensive and robust data sets of autocorrelated 2H relaxation rates currently available. In the present study we used these data, kindly provided by Prof. L. E. Kay, to explore temperature, magnetic field and rank 2 coherence dependence, and treat several important aspects of methyl dynamics. The issue of relatively large uncertainties in the best-fit parameters, implied by the relatively narrow portion of JQQ(ω) sampled by the experimental data, is addressed. Note that unlike the case of proton-bound heteronuclei, where 1H contributes high-frequency J(ω) values through the NOE, for 2H relaxation only the ω = 0, ωD and 2ωD values, with ωD denoting the 2H resonance frequency, are sampled at any given magnetic field. In this context the benefit of using up to four-field data sets, including rank 2 coherences, is explored herein.
We find that the protein L methyl sites exhibit rhombic potentials of different forms in the SRLS scenario instead of amplitudes of C-CH3 motion of different extents in the MF scenario. The local motional modes are tenfold slower in the SRLS scenario. Mode-coupling is important even at 15 °C. On average potential rhombicity decreases, mode-coupling increases and the rate of local motion increases with increasing temperature. The average activation energy for local motion is 2.0 ± 0.2 kcal/mol. The accuracy of the results is improved by including in the experimental data set relaxation rates associated with rank 2 coherences.
The Theoretical Background appears in section II. The various topics mentioned above are treated under Results and Discussion in section III. Our conclusions appear in section IV.
II. Theoretical Background
The Theoretical Background relevant for this paper appears in ref. 23. For convenience a brief summary is presented below.
1. The Model-free (MF) approach
The original MF spectral density, J(ω), based on τe << τm (i.e., an effective local motion, τe, much faster than the global motion, τm), is given by:13
| (1) |
where 1/τe’ = 1/τm + 1/τe ~ 1/τe.
This equation has been adapted to methyl dynamics where two restricted local motions around and of the methyl averaging axis are considered10,27a by setting S2 equal to [P2(cos 110.5°)]2× Saxis2 = 0.1×Saxis2. The term 0.1 represents the squared order parameter associated with the motion around the C-CH3 axis, and Saxis2 the axial squared order parameter associated with motion of the C-CH3 axis. The effective correlation time for local motion, τe, has been associated with both local motional modes. This yields the spectral density:10,27a
| (2) |
2. The Slowly Relaxing Local Structure (SRLS) model
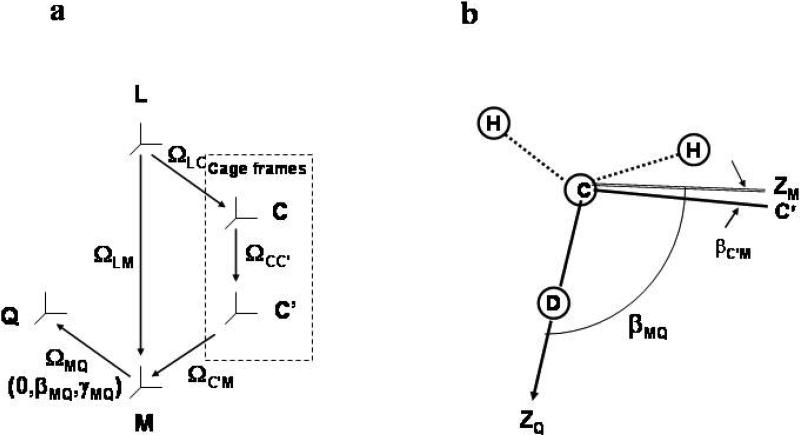
The fundamentals of the stochastic coupled rotator slowly relaxing local structure (SRLS) theory24,25 as applied to biomolecular dynamics26 have been developed recently for NMR spin relaxation in proteins.16-23 Two rotators, representing the global motion of the protein, RC, and the local motion of the probe (C-D bond in this case), RL, are treated. The motions of the protein and the probe are coupled by a local potential, U(ΩC'M), where C’ denotes the local director fixed in the protein, and M the local ordering/local diffusion frame fixed in the probe. The Euler angles ΩC'M are modulated by the local motion whereas the Euler angles ΩLC', with L denoting the fixed laboratory frame, are modulated by the overall tumbling of the protein.21 If the protein is considered axially symmetric then a global diffusion frame C tilted relative to the C’ frame will be defined. The site-specific angles, βCC’, are fixed in the protein. The various frames entering the SRLS/model and the magnetic quadrupoalr frame are shown in Figure 1.
Figure 1.
(a) Various reference frames which define the SRLS model: L – laboratory frame, C – global diffusion frame associated with protein shape, C’ – local director frame fixed in the protein, M – local ordering/local diffusion frame fixed in the C-D bond, Q – quadrupolar tensor frame along the C-D bond. (b) Application to methyl dynamics. The simple case of motion about the rotation axis of the methyl group is illustrated. The equilibrium orientation of the CH2D−C bond (Cα−Cβ for alanine, Cβ−Cγ1 and Cβ−Cγ2 for valine, etc.) is taken as the local director, C’. The local ordering frame, M, is assumed in this illustration for simplicity to be axially symmetric. ZM orients preferentially parallel to C’. It also represents the methyl rotation axis, and it is tilted relative to ZQ, i.e., the C−D bond, at βMQ = 110.5° (when rCH = rCD is set equal to 1.115Å − ref. 23). The angle βC'M is the stochastic angle between the instantaneous orientation of the M frame, ZM, and its equilibrium orientation, C’.
Formally the diffusion equation for the coupled system is given by:
| (3) |
where X is a set of coordinates completely describing the system.
| (4) |
where Ĵ(ΩC'M) and Ĵ(ΩLC’) are the angular momentum operators for the probe and the protein, respectively.
The Boltzmann distribution Peq = exp [−u (ΩC'M)]/ 〈exp [−u(ΩC'M)]〉 is defined with respect to the (scaled) probe-cage interaction potential given by:
| (5) |
This represents the expansion in the full basis set of Wigner rotation matrix elements, DLKM(ΩC'M), with only lowest order, i.e. L = 2, terms being preserved.21,23,25,26 The coefficient (given in units of kBT) is a measure of the orientational ordering of the C-D bond with respect to the local director, C’, whereas measures the asymmetry of the ordering around the director. Time correlation functions are calculated as:
| (6) |
Their Fourier-Laplace transforms yield the spectral densities jJMKK’(ω).
In the case of zero potential, , the solution of the diffusion operator associated to the time evolution operator features three distinct eigenvalues for the probe motion:
| (7) |
where RL∥ = 1/(6τ∥) and RL⊥ = 1/(6τ⊥) = 1/(6τ0). Only the diagonal terms, jK(ω) (the functions jKK’ denote the real part of j2M,KK’ – see ref. 25), are non-zero and they can be calculated analytically as Lorentzian spectral densities, each defined by width 1/τK . When the ordering potential is axially symmetric, , again only diagonal terms persist, but they are given by infinite sums of Lorentzian spectral densities which are defined in terms of eigenvalues 1/τi of the diffusion operator, and weighing factors cK,i, such that:
| (8) |
The eigevalues 1/τi represent modes of motion of the system, in accordance with the parameter range considered. Note that although in principle the number of terms in eq 8 is infinite in practice a finite number of terms is sufficient for numerical convergence of the solution.
Finally, when the local ordering potential is rhombic, , both diagonal jK(ω) and non-diagonal jKK'(ω) terms are different from zero and need to be evaluated explicitly according to expressions analogous to eq 8.
The spectral densities jKK’(ω) are defined in the M frame. If the M frame and the magnetic frame are tilted a Wigner rotation will be required to obtain the measurable auto-correlated spectral density, JQQ(ω), from the jK(ω) and jKK’(ω) spectral densities.40 Due to the additional symmetry jM,K,K′ = jM,−K,−K′ only the diagonal terms, jK(ω), with K = 0, 1, 2 and the non-diagonal terms, jKK’(ω), with KK’ = (−2,2), (−1,1), (−1,2), (0,1), (0,2), (1,2), need to be considered.
For an axial magnetic frame, Q, one has the explicit expression:
| (9) |
with only the diagonal terms, jK(ω), with K = 0, 1, 2, and the non-diagonal terms, jKK’(ω), with KK’ = (0, 2) , (−1,1) and, (−2, 2) contributing.
A convenient measure of the orientational ordering of the C−D bond is provided by the order parameters, and , which are related to the orienting potential (eq 5), hence and , via the ensemble averages:
| (10) |
One may convert to Cartesian ordering tensor components according to , , . Note that Sxx + Syy + Szz = 0.
For 2H relaxation the measurable quantities are JQQ(0), JQQ(ωD) and JQQ(2ωD). Together with the squared magnetic quadrupole interaction they determine the experimentally measured relaxation rates according to standard expressions for NMR spin relaxation.41,42
In the present study we allowed for at most four fitting parameters including the potential coefficients and , RC defined in units of RL (hence representing the time scale separation between the global and local motions) and the “diffusion tilt” βMQ. We used RC = 1/6τm with τm as determined in refs. 12 and 43 based on 15N T1/T2 ratios.44 When βMQ is set equal to zero then the SRLS spectral density is formally analogous with the original MF spectral density (eq 1).
The functions jK(ω) (eq 8) and jKK’(ω) (equations analogous to eq 8) are calculated during data fitting on the fly. In the methyl dynamics application the local potential (the equivalent of S2 in MF) is low, with and on the order of 1 − 2 (in units of kBT). The time scale separation, RC, is also not too large. The computational effort was found to be very reasonable in this case. Thus, it took about 40 minutes on a 3.2 GHz Pentium 4 processor to fit the relaxation data of a given methyl group of protein L. Our SRLS-based fitting program is similar to the MF fitting programs. The only extra requirement on the part of the user is to determine a truncation parameter, which determines the size of the matrix representation required for convergence of the solution (given by eq 8 or similar equations). Several trial and error calculations carried out for typical parameter sets suffice. Our current software is available upon request. The “Theoretical background” sections of this paper and of references 21 and 23 comprise the information required for ab initio programming.
3. MF as SRLS asymptote
Equation 1, from which eq 2 has been derived, represents the SRLS solution in the Born-Oppenheimer (BO) limit where τm >> τe.45,46a Equation 1 was obtained in early work45 as a perturbational expansion of SRLS in the limit of τm >> τe for axial local ordering, isotropic global (RC) and local (RL) diffusion, and collinear magnetic and ordering tensors.45 In this limit S2 represents (eqs 5 and 10 with ) and τe is given by τ0 of eq 7. When the coupling potential is very high then the phenomenon called renormalization46b,c becomes important. The renormalized correlation time, τren, is given approximately by where represents the potential coefficient.46c We found that in this limit τe agrees with τren and S2 agrees with (references 20 and 21). Outside of the BO limit eq 1 is not valid for diffusive motion (or wobblein-a-cone, if the latter is associated with a cosine squared potential), and will consequently lead to force-fitting.
As already noted, eq 2 features two dynamic modes associated with the axial order parameters, [P2(cos βMQ)]2 = [P2(cos 110.5°)]2 = 0.1 and Saxis2, for motion around and of the CCH3 axis, respectively, and a common correlation time, τe. A major inconsistency in eq 2 is having the local ordering/local diffusion axis, M, tilted at 110.5° from the magnetic axis, Q, but ignoring the K = 1 and K = 2 contributions (eq 8). Independent of the model assumed (e.g., see reference 27d where the Woessner model is being developed), these terms enter the calculation of the exact JQQ(ω) function as 3 sin2 (110.5°) cos2 (110.5°) j1(ω) = 0.323 j1(ω), and 0.75 sin4 (110.5°) j2(ω) = 0.577 j2(ω). Yet, JQQ(ω) of eq 2 comprises only the K = 0 term. This is only valid in the extreme motional narrowing limit23 where RL⊥ = RC and RL∥ → ∞ (eq 7, ref. 26; see also eq 31 of ref. 27b and pertinent discussion). The condition that RL∥ → ∞ (equivalent to τe → 0 in MF) renders the functions j1(ω) and j2(ω) so small that the K = 1 and K = 2 terms can be ignored. When τe → 0 the second term of eq 2 can be ignored to obtain JQQ(ω) = (1.5 cos2 110.5° – 0.5)2 j0(ω) = 0.1 j0(ω), where j0(ω) = Saxis2 τm (1 + ω2 τm2). It can be seen that in this limit the effect of the local motional modes consists of reducing the quadrupole interaction (featured by the relaxation rate expressions) consecutively by 0.1 and Saxis2.
In practice combined 2H T1 and T2 auto-correlated relaxation rates cannot be fit from a statistical point of view with τe set equal to zero in eq 2 because the extreme motional narrowing limit has not been attained. Technically the data can often be fit with τe ≠ 0. However, in this case the quantities S axis2 and τe have only vague physical meaning and, as shown below, they provide a distorted picture of the actual situation if taken seriously.
III. Results and discussion
In principle one should first consider axial local potentials in the SRLS fitting process. We showed previously23 that this leads to a physically problematic picture and implies inconsistencies between 2H auto-correlation in 13CDH2 (ref. 12) and HC-HH cross-correlation in 13CH3 (ref. 43). The problems mentioned have been resolved by allowing for rhombic potentials.23 Therefore in this study we allow for rhombic potentials from the start.
The form of JQQ(ω) for methyl dynamics
The SRLS model yields the generic spectral densities, jKK’(ω). The measurable spectral density, JQQ(ω), is given by linear combinations of the relevant jKK’(ω) functions. The symmetry of the local ordering (M frame) determines which KK’ quantum numbers are non-zero, and, together with the other physical parameters, the form of the jKK’(ω) functions. The orientation of the M frame with respect to the magnetic quadrupolar frame determines the coefficients in the linear combination yielding JQQ(ω) (eq 9). For a rhombic M frame and βMQ = 110.5° JQQ(ω) features large contributions from j00(ω), j22(ω) and j20(ω), and smaller contributions from j11(ω), j2-2(ω) and j1-1(ω) (eq 9). JQQ(0), JQQ(ωD) and JQQ(2ωD) enter the expressions for the 2H spin relaxation rates.
The appropriate representation of methyl dynamics by JQQ(ω) makes possible the determination of the physical parameters (in general, RL, RC, βMQ, and ) despite the fact that relatively few values of JQQ(ω) are available at any given magnetic field. As pointed out above, unlike heteronuclear 15N-1H and 13C-1H spin relaxation 2H spin relaxation does not feature high-frequency values of J(ω), with ω on the order of ωH. Choy and Kay47 have shown that even with twenty synthetic data points (2H T1, T2 and three relaxation rates associated with rank 2 coherences generated at 9.4, 11.7, 14.1 and 18.8 T) the results of fitting these data with a model-free spectral density were unacceptable. Only further parameterization of this function yielded statistically acceptable results by force fitting.
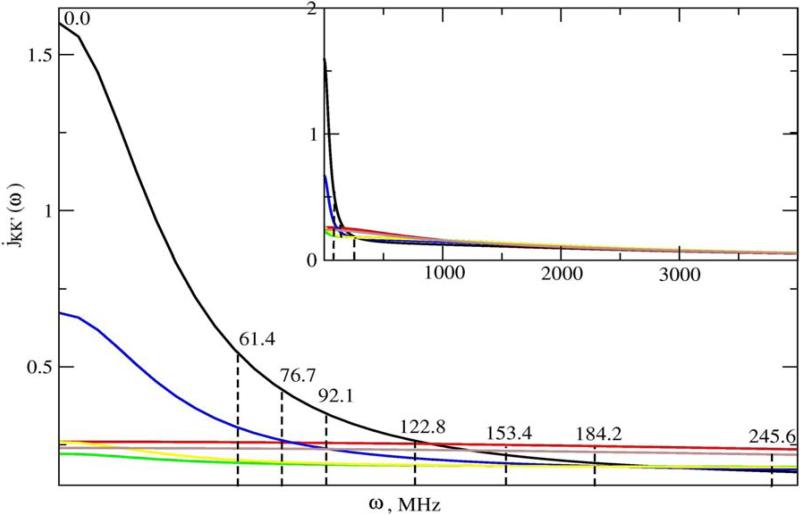
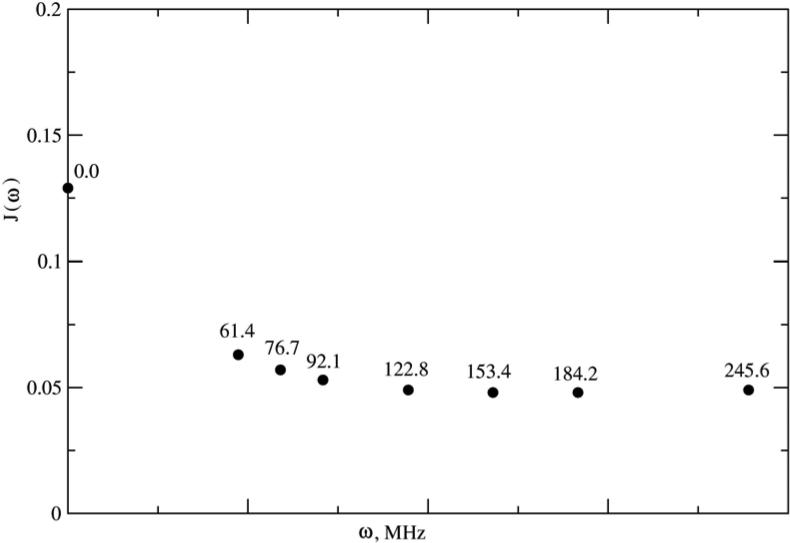
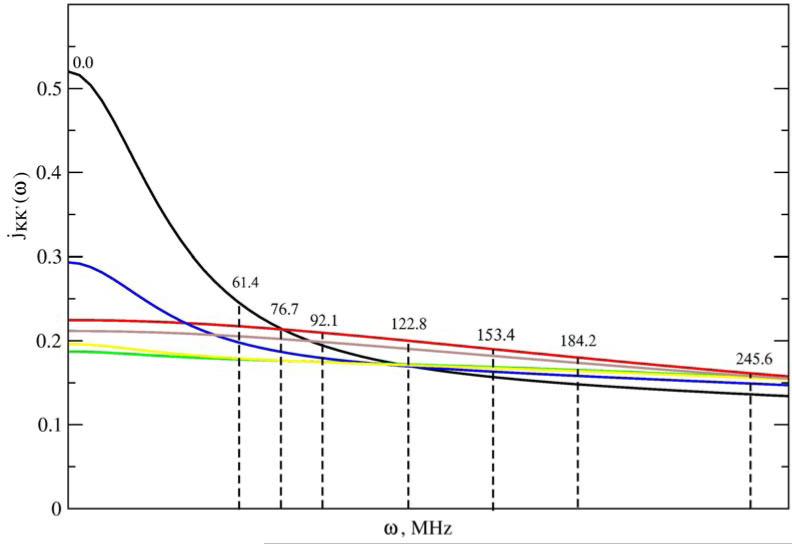
We illustrate below typical SRLS spectral densities used in methyl dynamics analysis (Figures 2–4). Figure 2 shows the jKK’(ω) functions calculated using a typical parameter set (obtained by analyzing the data acquired for methyl T23 of protein L at magnetic fields of 9.4, 11.7, 14.1 and 18.8 T, 25 °C) featuring , and RC = 0.017. Note that RC is given in units of RL. Hence 0.017 represents the ratio between the global and local motional rates. Since the global motional rate is known independently, the parameter RC actually determines the local motional rate. The inset shows a compressed ω-range extending from 0 to 4000 MHz. Clearly the portion of the jKK’(ω) functions sampled consist of a restricted region outside of which these functions are not defined experimentally. Note that the magnetic field range scanned is almost as large as feasible with currently available technology. Figure 3 shows the jKK’(ω) functions calculated for , and RC = 0.05, and Figure 4 shows the jKK’(ω) functions of Figure 3 assembled into JQQ(ω) for βMQ = 69.5° (the complement of 110.5°; the time correlation functions (eq 6) are the same for βMQ and (180° − βMQ)). While SRLS fitting needs to account for the various jKK’(ω) functions (e.g., Figs. 3 and 4) and their coefficients, model-free only needs to reproduce specific values of the JQQ(ω) function (e.g., Fig. 4) because the MF spectral density is a parameterized version of the actual spectral density. We found that SRLS and MF spectral densities, which are formally equivalent, yield best-fit parameters with significantly smaller uncertainties in the SRLS scenario.
Figure 2.
jKK’(ω) functions with KK’ = (0,0) – black, (1,1) – red, (2,2) – green, (2,0) – blue, (2,−2) – yellow, and (1,−1) – brown calculated with eq 8 and an analogous equation appropriate for cross-terms, using , and RC = 0.017, which are the best-fit SRLS parameters obtained for methyl T23 at 25°C. The inset shows a compressed ω-range extending from 0 to 4000 MHz. jKK’(ω) is given in units of 1/RL and ω is given in units of RL.
Figure 4.
JQQ(ω) function assembled from the jKK’(ω) functions shown in Figure 3, using βMQ = 69.5°. J(ω) is given in units of 1/RL and ω is given in units of RL.
Figure 3.
jKK’(ω) functions with KK’ = (0,0) – black, (1,1) – red, (2,2) – green, (2,0) – blue, (2,−2) – yellow, and (1,−1) – brown calculated with eq 8 and an analogous equation appropriate for cross-terms, using , and RC = 0.05. jKK’(ω) is given in units of 1/RL and ω is given in units of RL.
Fitting strategy and error estimation
Exhaustive grid searches are impractical with SRLS. To ascertain that the global minimum of the Least Squares Sum (LSS) “target” function has been reached in a given fitting process we tested various strategies. It was found effective to carry out a coarse grid search, where , , RC and βMQ were allowed to vary, with the starting value of βMQ in the vicinity of the tetrahedral angle, followed by a finer grid search. This strategy was superior to one where βMQ was fixed at the tetrahedral angle value. In general the minimization converged for numerical reasons to (180° – βMQ) (which, as indicated above, yields the same time correlation functions as βMQ). To enable meaningful comparison of potential coefficients among methyl groups we fixed βMQ at 69.5° in the final calculation of each methyl group. An alternative (more tedious) strategy, which yielded very similar but not identical results, consisted of allowing βMQ to vary freely and accepting only those sets of temperature-dependent fits where βMQ was within half a degree of 69.5°. Clearly it is necessary to devise an effective automated fitting protocol based on both statistical and physical criteria. This effort is underway.
Both procedures outlined above comprise error estimation capabilities, which can be used in different ways. The Monte Carlo-based error estimation methods used in MF-based fitting,48 which would involve hundreds of calculations of JQQ(ω), are not practical with SRLS. In reference 12 a strategy where part of the experimental data was eliminated systematically was used to evaluate uncertainties. Ultimately we used a combination of the various methods mentioned to estimate the errors in the best-fit parameters, found to be typically on the order of 10%.
Importance of the rank 2 coherences
Including the rank 2 coherences into the experimental data set increases the accuracy of the results, obviously with a higher but still acceptable reduced χ2 value. This is illustrated in Table 1, using for simplicity axial potentials. It can be seen that practically the same results are obtained independent of the starting values with 16 data points, 8 of which are relaxation rates associated with the rank 2 coherences (rows 1−6). Using 8 data points comprising only the 2H T and T relaxation rates yields values lower by 4.8% (rows 7−12). The discrepancies are parameter-range dependent (not shown).
Table 1.
Best-fit parameters, listed under "output", obtained with combined fitting of 16 relaxation rates (8 relaxation rates including 2H T1 and T2 acquired at 9.4, 11.7, 14.1 and 18.8 T T, and 6 relaxation rates associated with the three rank 2 coherences acquired at 11.7 and 14.1 T) measured at 25 °C for methyl A50, using the input parameters shown under "input" (rows 1–6). Same as above, using only 2H T1 and T2 data (rows 7–12). The τm value used was 4.05 ns.12 χ2red values are also shown. The local motional rate is given by RC × τm.
| Input | Output | ||||
|---|---|---|---|---|---|
| RC | RC | χ 2red | |||
| 1 | 0.5 | 0.0035 | 0.88 | 0.0038 | 7 |
| 2 | 0.89 | 0.0035 | 0.88 | 0.0037 | 7 |
| 3 | 1.2 | 0.0035 | 0.88 | 0.0038 | 7 |
| 4 | 0.5 | 0.008 | 0.88 | 0.0038 | 7 |
| 5 | 0.89 | 0.008 | 0.89 | 0.0037 | 7 |
| 6 | 1.2 | 0.008 | 0.88 | 0.0038 | 7 |
| 7 | 0.5 | 0.0035 | 0.84 | 0.0038 | 0.1 |
| 8 | 0.89 | 0.0035 | 0.84 | 0.0039 | 0.2 |
| 9 | 1.2 | 0.0035 | 0.84 | 0.0038 | 0.1 |
| 10 | 0.5 | 0.008 | 0.85 | 0.0038 | 0.1 |
| 11 | 0.89 | 0.008 | 0.85 | 0.0038 | 0.1 |
| 12 | 1.2 | 0.008 | 0.85 | 0.0038 | 0.1 |
A difference of 4.8% in Saxis2 implies differences in the potential coefficient, , exceeding 20%, due to the shape of the squared order parameter versus function for high values. This has been discussed in detail in ref. 21. Note that Table 1 features an illustrative example. In general the differences between corresponding best-fit parameters determined with rank 2 coherences included or excluded might be larger.
Qualitative MF-based information
We checked whether adequate qualitative information could be obtained with MF. The parameter used in MF to estimate the strength of the local spatial restrictions is Saxis2. The SRLS parameter, which serves the same purpose, is the potential coefficient, . Table 2 shows groups of methyl moieties with very similar Saxis2 values and the corresponding best-fit SRLS parameters. Ten data points (2H T1, T2 and the three relaxation rates associated with the rank 2 coherences acquired at 11.7 and 14.1 T) measured at 25 °C have been used for the SRLS calculations. The MF data shown in Table 2 were taken from ref. 12. We also show derived from S2 = 0.1×Saxis2 using the axial versions of eqs 5 and 10. The penultimate and ultimate columns on the right show and R(τ) = τ0(SRLS)/τe(MF), respectively. It can be seen that these ratios are larger than unity and in many cases vary considerably within a given group of similar Saxis2 values, indicating that the variations in Saxis2 (MF) and are likely to differ qualitatively.
Table 2.
Combined fitting of 10 relaxation rates (2H T1, T2 and the three relaxation rates associated with the rank 2 coherences) acquired at 11.7 and 14.1 T, 25 °C for the depicted methyl groups. The data under "MF" were taken from ref. 12. The penultimate and ultimate columns on the right show and R(τ) = τ0(SRLS)/τe(MF), respectively. The residues marked in bold face letters required an extended MF formula for data analysis.12
| Methyl | MF | SRLS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Saxis2 | τe, ps | RC | τ0, ps | RC | R(τ) | |||||
| V2γ1 | 0.73 | 1.22 | 54 | 0.013 | 1.77 | -0.82 | 101 | 0.025 | 1.5 | 1.9 |
| T37 | 0.74 | 1.23 | 50 | 0.012 | 1.89 | -0.92 | 97 | 0.024 | 1.5 | 1.9 |
| T55 | 0.98 | 1.41 | 51 | 0.012 | 1.83 | -0.95 | 113 | 0.028 | 1.3 | 2.2 |
| T17 | 0.97 | 1.42 | 45 | 0.011 | 1.56 | -0.82 | 117 | 0.029 | ||
| I9δ | 0.38 | 0.89 | 24 | 0.006 | 1.57 | -0.50 | 28 | 0.007 | 1.8 | 1.2 |
| L8δ1 | 0.30 | 0.79 | 35 | 0.009 | -0.29 | -0.50 | 53 | 0.013 | ||
| L8δ2 | 0.30 | 0.79 | 41 | 0.010 | -0.35 | -0.50 | 57 | 0.014 | ||
| T15 | 0.57 | 1.09 | 69 | 0.017 | 1.79 | -0.95 | 105 | 0.026 | 1.6 | 1.5 |
| L38δ1 | 0.56 | 1.08 | 34 | 0.008 | 1.68 | -0.74 | 61 | 0.015 | 1.6 | 1.8 |
| V47γ1 | 0.57 | 1.09 | 55 | 0.014 | 1.51 | -0.58 | 93 | 0.023 | 1.4 | 1.7 |
| I58δ | 0.58 | 1.10 | 17 | 0.004 | 1.86 | -0.56 | 32 | 0.008 | 1.7 | 1.9 |
| V49γ2 | 0.62 | 1.13 | 40 | 0.010 | 1.68 | -0.82 | 61 | 0.015 | 1.5 | 1.5 |
| L56δ1 | 0.61 | 1.12 | 70 | 0.017 | 1.89 | -1.09 | 117 | 0.029 | 1.7 | 1.7 |
| L56δ2 | 0.61 | 1.12 | 38 | 0.009 | 1.60 | -0.68 | 65 | 0.016 | 1.4 | 1.7 |
| A61 | 0.60 | 1.11 | 46 | 0.011 | 1.68 | -0.73 | 77 | 0.019 | 1.5 | 1.7 |
| T3 | 0.88 | 1.33 | 39 | 0.010 | 1.78 | -0.89 | 85 | 0.021 | 1.3 | 2.2 |
| T28 | 0.88 | 1.33 | 41 | 0.010 | 1.82 | -1.02 | 85 | 0.021 | 1.4 | 2.1 |
| I4γ | 0.87 | 1.32 | 24 | 0.006 | 1.84 | -0.78 | 73 | 0.018 | 1.4 | 3.1 |
| A33/A11 | 0.89 | 1.34 | 37 | 0.009 | 1.87 | -0.91 | 101 | 0.025 | 1.4 | 2.7 |
| A11/A33 | 0.82 | 1.29 | 49 | 0.012 | 1.98 | -1.02 | 134 | 0.033 | 1.5 | 2.7 |
| A18 | 0.81 | 1.28 | 57 | 0.014 | 1.90 | -1.00 | 109 | 0.027 | 1.5 | 1.9 |
| I58γ | 0.82 | 1.29 | 27 | 0.007 | 1.79 | -0.96 | 53 | 0.013 | 1.4 | 2.0 |
| T46 | 0.69 | 1.20 | 63 | 0.016 | 1.89 | -1.00 | 105 | 0.026 | 1.6 | 1.7 |
| V49γ1 | 0.68 | 1.18 | 34 | 0.008 | 1.72 | -0.68 | 69 | 0.017 | 1.5 | 2.0 |
| T23 | 0.84 | 1.31 | 39 | 0.010 | 1.84 | -0.94 | 81 | 0.020 | 1.4 | 2.1 |
| A50 | 0.84 | 1.31 | 24 | 0.006 | 1.81 | -0.93 | 53 | 0.013 | 1.4 | 2.2 |
| A31 | 0.83 | 1.30 | 77 | 0.019 | 1.94 | -1.15 | 134 | 0.033 | 1.5 | 1.7 |
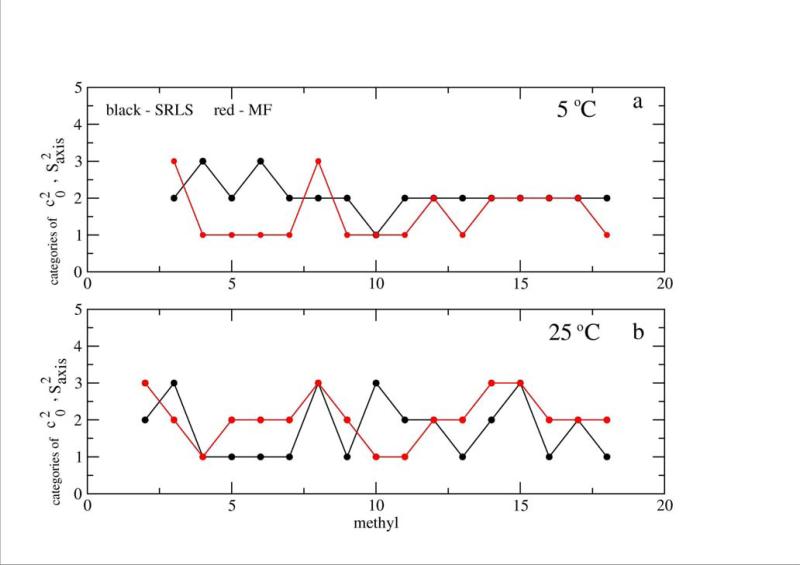
Profiles of (based on the best-fit parameters obtained with SRLS at 5 and 25 °C for the methyl groups of Tables 3 and 5) and the corresponding Saxis2 (MF) values (taken from ref. 12) are shown in Figure 5. For clarity the methyl groups have been classified as follows. SRLS categories 1, 2 and 3 correspond to , and , (, , and ) at 5 (25) °C. MF categories 1, 2 and 3 correspond to Saxis2 > 0.85, 0.6 ≤ Saxis2 ≤ 0.85 and Saxis2 < 0.6 (Saxis2 > 0.89, 0.6 ≤ Saxis2 ≤ 0.89 and Saxis2 < 0.6) at 5 (25) ° C. Clearly the Saxis2 profiles differ significantly, often exhibiting opposite trends at the same temperature, and different temperature dependences.
Table 3.
Best-fit parameters obtained with combined fitting of 5 relaxation rates (2H T1, T2 and the three relaxation rates associated with the rank 2 coherences) acquired at 11.7 T, 5 °C, for the depicted methyl groups. The global motion correlation time used was τm = 8.01 ns.12,43 The quadrupole interaction was 167 kHz, and the rCH = rCD distance 1.115Å. Note that all the values are negative.
| Methyl | RC | τ0, ps | |||
|---|---|---|---|---|---|
| L8δ1 | 1.49 | –0.20 | 0.014 | 112 | 0.13 |
| L38δ1 | 1.51 | –0.33 | 0.012 | 96 | 0.22 |
| L56δ2 | 1.54 | –0.22 | 0.015 | 120 | 0.14 |
| T55 | 1.19 | –2.66 | 0.004 | 32 | 2.24 |
| T23 | 1.53 | –0.28 | 0.016 | 128 | 0.18 |
| A18 | 1.37 | –1.66 | 0.013 | 104 | 1.21 |
| A50 | 1.51 | –0.77 | 0.010 | 80 | 0.51 |
| L38δ2 | 1.50 | –0.51 | 0.014 | 112 | 0.34 |
| T37 | 1.51 | –0.65 | 0.027 | 216 | 0.43 |
| T17 | 1.92 | –1.69 | 0.019 | 152 | 0.88 |
| V49γ2 | 1.62 | –0.85 | 0.012 | 96 | 0.59 |
| A6 | 1.55 | –0.92 | 0.020 | 160 | 0.59 |
| A61 | 1.51 | –0.95 | 0.013 | 104 | 0.63 |
| V47γ1 | 1.51 | –0.61 | 0.016 | 128 | 0.40 |
| V2γ2 | 1.56 | –0.82 | 0.012 | 100 | 0.53 |
| I9γ | 1.56 | –0.80 | 0.012 | 96 | 0.50 |
Table 5.
Best-fit parameters obtained with combined fitting of 5 relaxation rates (2H T1, T2 and the three relaxation rates associated with the rank 2 coherences) acquired at 11.7 T, 25 °C, for the depicted methyl groups. The global motion correlation time used was τm = 4.05 ns.12,43 Note that all the values are negative except for of L8δ1, marked with an asterisk.
| Methyl | RC | τ0, ps | |||
|---|---|---|---|---|---|
| L8δ1 | –0.29 | –0.50 | 0.013 | 53 | 1.72* |
| L38δ1 | 1.68 | –0.74 | 0.015 | 61 | 0.44 |
| L56δ2 | 1.60 | –0.68 | 0.016 | 65 | 0.43 |
| T55 | 1.83 | –0.95 | 0.028 | 113 | 0.52 |
| T23 | 1.84 | –0.94 | 0.020 | 81 | 0.51 |
| A18 | 1.90 | –1.0 | 0.027 | 109 | 0.53 |
| A50 | 1.81 | –0.93 | 0.013 | 53 | 0.51 |
| L38δ2 | 1.44 | –0.46 | 0.017 | 70 | 0.32 |
| T37 | 1.89 | –0.92 | 0.024 | 97 | 0.49 |
| T17 | 1.56 | –0.82 | 0.029 | 118 | 0.53 |
| T3 | 1.78 | –0.89 | 0.021 | 113 | 0.50 |
| V49γ2 | 1.68 | –0.82 | 0.015 | 61 | 0.52 |
| A6 | 1.98 | –1.02 | 0.033 | 134 | 0.52 |
| A61 | 1.68 | –0.73 | 0.019 | 77 | 0.43 |
| V47γ1 | 1.51 | –0.58 | 0.023 | 93 | 0.38 |
| V2γ2 | 1.80 | –0.79 | 0.018 | 73 | 0.44 |
| I9γ | 1.69 | –0.78 | 0.015 | 61 | 0.46 |
| A31 | 1.94 | –1.15 | 0.033 | 134 | 0.59 |
Figure 5.
Schematic representing trends in SRLS and Saxis2 MF at 5 °C (a) and 25 °C (b). The and Saxis2 values have been classified into three groups according to their magnitude, as outlined in the text. These categories are denoted as 1, 2 and 3 on the ordinate. As pointed out above, the average error in c20 has been estimated at 10%. A conservative estimate of the average error in Saxis2, based on ref. 12, is 4%. These figures translate into an average error margin of ±1.5 times the black symbol size, and ±1.0 times the red symbol size.
Hence, care is to be exerted in MF analyses in interpreting squared order parameters and local motion correlation times in terms of physical or biological properties. Saxis2 has been used extensively to derive residual configurational entropy and heat capacity, with far-fetched implications.30 Recently a new term called, “polar dynamics”, based on relative Saxis2 values, was set forth.49 Small differences in Saxis2 and τe (which is actually a composite depending on both S2 and a bare rate of local motion) have been used to elucidate communication pathways in proteins.50 Such inferences require accurate best-fit parameters.
Data fitting
Five relaxation rates (2H T1 and T2, and relaxation rates associated with two-quantum, two-spin order and antiphase rank 2 coherences) acquired at 5, 15, 25, 35 and 45°C, 11.7 T, have been measured for the methyl groups L8δ1, L38δ1, L56δ2, T55, T23, A18, A50, L38δ2, T37, T17, V49γ , A6, A61, V47γ1, V2γ2, and I9γ. These data were fit with SRLS allowing , and RC to vary while keeping βMQ fixed at 69.5°. The best-fit parameters are shown in Tables 3–7.
Table 7.
Best-fit parameters obtained with combined fitting of 5 relaxation rates (2H T1, T2 and the three relaxation rates associated with the rank 2 coherences) acquired at 11.7 T, 45 °C, for the depicted methyl groups. The global motion correlation time used was τm = 1.74 ns.12,43 Note that all the values are negative.
| Methyl | RC | τ0, ps | |||
|---|---|---|---|---|---|
| L8δ1 | 0.87 | –0.32 | 0.018 | 31 | 0.37 |
| L38δ1 | 1.39 | –0.50 | 0.020 | 35 | 0.36 |
| L56δ2 | 1.93 | –0.58 | 0.007 | 12 | 0.30 |
| T55 | 3.94 | –1.18 | 0.063 | 110 | 0.30 |
| T23 | 4.10 | –0.66 | 0.067 | 117 | 0.16 |
| A18 | 2.68 | –0.70 | 0.052 | 91 | 0.26 |
| A50 | 3.05 | –0.54 | 0.034 | 59 | 0.18 |
| L38δ2 | 1.49 | –0.50 | 0.015 | 26 | 0.34 |
| T37 | 2.51 | –0.57 | 0.044 | 77 | 0.23 |
| T17 | 2.15 | –0.48 | 0.022 | 38 | 0.22 |
| T3 | 3.06 | –0.59 | 0.049 | 125 | 0.19 |
| V49γ2 | 2.03 | –0.49 | 0.024 | 42 | 0.24 |
| A6 | 2.48 | –0.74 | 0.058 | 101 | 0.30 |
| A61 | 2.36 | –0.44 | 0.028 | 49 | 0.19 |
| V47γ1 | 1.96 | –0.49 | 0.034 | 59 | 0.25 |
| V2γ2 | 2.20 | –0.47 | 0.030 | 52 | 0.21 |
| I9γ | 2.42 | –0.74 | 0.061 | 106 | 0.31 |
| A31 | 2.03 | –1.00 | 0.050 | 87 | 0.49 |
The results of fitting the data acquired at 5 °C, shown in Table 3, feature best-fit RC values of 0.01-0.02 (with the exception of methyl T55). The strength of the local potential, given by , is approximately 1.5 in units of kBT. The highest potential asymmetry (rhombicity), as given by , is on the order of 0.6 (with the exception of methyl groups T55, A18 and T17). Inspection of the data shown in Tables 4-7, obtained at the higher temperatures, points to a diversified picture. We present in Table 8 results obtained by averaging over the methyl groups analyzed at any given temperature. It can be seen that on average RC increases, τ0 decreases, increases, decreases and decreases with increasing temperature. Similar trends were observed with the alternative fitting strategy where βMQ was also allowed to vary.
Table 4.
Best-fit parameters obtained with combined fitting of 5 relaxation rates (2H T1, T2 and the three relaxation rates associated with the rank 2 coherences) acquired at 11.7 T, 15 °C, for the depicted methyl groups. The global motion correlation time used was τm = 5.36 ns.12,43 Note that the values are negative, except for those marked with asterisks.
| Methyl | RC | τ0, ps | |||
|---|---|---|---|---|---|
| L8δ1 | 1.63 | –0.67 | 0.012 | 64 | 0.41 |
| L38δ1 | –0.11 | –0.51 | 0.011 | 59 | 4.64* |
| L56δ2 | 1.58 | –0.80 | 0.015 | 80 | 0.51 |
| T55 | 1.66 | –0.77 | 0.024 | 129 | 0.46 |
| T23 | 1.65 | –0.77 | 0.024 | 129 | 0.47 |
| A18 | 1.68 | –0.82 | 0.018 | 97 | 0.49 |
| A50 | 1.64 | –0.80 | 0.013 | 70 | 0.49 |
| L38δ2 | 1.44 | +0.35 | 0.016 | 86 | 0.24* |
| T37 | 1.77 | –0.88 | 0.022 | 118 | 0.50 |
| T17 | 1.66 | –0.80 | 0.019 | 102 | 0.48 |
| T3 | 1.58 | –0.78 | 0.025 | 200 | 0.49 |
| V49γ2 | 1.61 | –0.71 | 0.018 | 97 | 0.44 |
| A6 | 1.64 | –0.81 | 0.019 | 102 | 0.49 |
| A61 | 1.64 | –0.81 | 0.013 | 70 | 0.49 |
| V47γ1 | 1.54 | –0.68 | 0.016 | 85 | 0.44 |
| V2γ2 | 0.99 | –0.99 | 0.024 | 129 | 1.00 |
| I9γ | 1.54 | –0.61 | 0.015 | 80 | 0.40 |
Table 8.
Average best-fit values, rhombicity ratios , local motion correlation times, τ0, and time scale separations, RC, obtained for the methyl groups in Tables 3 – 7. The global motion correlation times, τm, are also given.
Decrease in the local motional correlation time, τ0, with increasing temperature is expected. An activation energy of 2 ± 0.2 kcal/mol has been derived from the data of Table 8 based on the Arrhenius relation for the rate 1/6τ0. Large site-specific variations in local motional correlation times of methyl groups in proteins have been predicted theoretically.27a,51 The value of 2 kcal/mol pertains to the theoretically predicted range, and the tenfold lower SRLS rates are in significantly better agreement with the theoretical predictions than the MF rates.27a,51
Based on the Arrhenius relation for the rate 1/6τm (with τm determined previously12,43) we obtained an activation energy of 6.72 ± 0.36 kcal/mol for the global motion of protein L, in agreement with similar values obtained for other proteins in aqueous solution (see ref. 22 and relevant papers cited therein).
Mode-coupling, as expressed by the parameter <RC> = <τ0/τm>, increases with increasing temperature, in accordance with the activation energies for global and local motion. This is an interesting result further documented below by outlining the mode-composition at various temperatures.
The asymmetry of the local potential, as expressed by , decreases with increasing temperature. This is also interesting new information indicating that the local spatial restrictions at the site of the motion of the methyl group become more axially symmetric as the temperature is raised.
When the local potential is axially symmetric and βMQ = 110.5° (with rCH = rCD = 1.115 Å) the local spatial restrictions at the methyl site are characterized solely by c20, which evaluates the potential strength. Clearly c20 is expected to decrease with increasing temperature. When the potential is rhombic and βMQ is also allowed to vary, as done (in a controlled manner – see above) in the present study, the set of parameters c20, c22 and βMQ, rather than c20 alone, evaluates the local restrictions. Changes in c20 with temperature represent in this case apparent changes in potential strength.
Inter-site comparison of the local restrictions based on parameter sets is not straightforward. We have been looking for simplified models, which evaluate this important property in a more direct manner. The combination where c20 is fixed at its Woessner-model-compatible value and βMQ at 110.5°, with c22 and the local motion correlation time, τ, allowed to vary, was found to be appropriate. It yields results, which are similar at lower temperatures, and very close at higher temperatures, to those of the complete model. The parameter τ decreases consistently with increasing temperature, as required by physical viability, and |c22| decreases with increasing temperature in most cases. The typical decrease (exceptional increase) in structural rhombicity at most (specific) methyl sites with increasing temperature constitutes interesting new information. These developments will be reported shortly elsewhere.
Temperature-dependent mode composition
We illustrate the mode-coupling concept inherent to the SRLS model. The “pure”, i.e., unrestricted by a potential local motional mode has an eigenvalue of 6, and the “pure” global motion mode has an eigenvalue of 6×RC, both in units of RL.24 In the original MF formula (eq 1), which is a limiting case of the two coupled rotator model,20,21,24,45 the weighting factors of the global and local motional modes correspond to and , respectively.
When the mode-decoupling limit is exceeded a larger number of modes contribute to the spectral density (eq 8). We show in Table 9 the dynamic modes associated with methyl I9γ at 5, 25 and 45 °C which contribute to the time correlation functions C0(t), C1(t) and C2(t) (the labels 0, 1 and 2 are abridged versions of KK’ = (0,0), (1,1) = (–1,–1) and (2,2) = (–2,–2), eq 6). We focus first on C0(t). For the largest time scale separations of RC = 0.012, and a rhombic potential with and obtained at 5 °C, three major local motional modes with eigenvalues in the vicinity of 6 make a fractional contribution of 0.778 to C0(t). The eigenvalue of the global motion mode (shown in bold face letters) is equal to the “pure” eigenvalue of 0.072 = 6×0.012, indicating that the overall tumbling mode of the protein is, as expected, distinct from the local motional modes. The fractional contribution of the global motion mode is 0.092. Additional modes with eigenvalues ranging from 4.87 to 9.17, with various individual weights, contribute 0.098. The rest (0.052) is contributed by a large number of mixed modes with small individual weights (not shown). In can be seen that the two-mode limit is exceeded even though the time scale separation is relatively low (RC = 0.012) and the potential relatively weak ( in units of kBT).
Table 9.
Eigenvalues, 1/τi (in units of RL), and weighting factors, cK.i, of the SRLS solution for C0(t), C1(t) and C2(t) obtained using (a) and RC = 0.012, (b) and RC = 0.015, and (c) and RC = 0.061. These values represent the best-fit parameters obtained with rhombic potential fitting using the data acquired at a magnetic field of 11.7 T, and at 5, 25 and 45 °C, respectively for methyl I9γ. In all the cases shown βMQ = 69.5°. The global motion mode contribution is marked in bold face letters.
| C0(t) | |||||
|---|---|---|---|---|---|
| a | b | c | |||
| 1/τi | cK.i | 1/τi | cK,i | 1/τi | cK,i |
| 5.30 | 0.297 | 5.40 | 0.309 | 6.29 | 0.396 |
| 5.94 | 0.264 | 6.06 | 0.270 | 9.46 | 0.130 |
| 7.92 | 0.194 | 8.08 | 0.188 | 7.41 | 0.123 |
| 8.08 | 0.057 | 8.23 | 0.050 | 19.83 | 0.023 |
| 4.87 | 0.031 | 4.83 | 0.024 | 9.70 | 0.021 |
| 5.94 | 0.023 | 21.7 | 0.009 | 23.03 | 0.016 |
| 9.17 | 0.010 | 9.37 | 0.008 | 14.84 | 0.006 |
| 0.072 | 0.092 | 0.090 | 0.121 | 0.355 | 0.263 |
| C1(t) | |||||
|---|---|---|---|---|---|
| a | b | c | |||
| 1/τi | cK.i | 1/τi | cK,i | 1/τi | cK,i |
| 6.14 | 0.321 | 6.09 | 0.299 | 6.41 | 0.223 |
| 5.92 | 0.309 | 6.49 | 0.290 | 1.69 | 0.221 |
| 6.43 | 0.313 | 6.85 | 0.265 | 1.51 | 0.141 |
| 1.77 | 0.051 | 1.61 | 0.130 | 7.80 | 0.140 |
| 19.75 | 0.003 | 19.80 | 0.009 | 9.17 | 0.111 |
| 9.00 | 0.068 | ||||
| 7.55 | 0.048 | ||||
| 19.85 | 0.013 | ||||
| 22.06 | 0.010 | ||||
| C2(t) | |||||
|---|---|---|---|---|---|
| a | b | c | |||
| 1/τi | cK.i | 1/τi | cK,i | 1/τi | cK,i |
| 5.42 | 0.506 | 5.11 | 0.573 | 4.70 | 0.365 |
| 6.45 | 0.380 | 6.89 | 0.335 | 4.65 | 0.242 |
| 6.80 | 0.111 | 7.47 | 0.086 | 4.82 | 0.149 |
| 20.02 | 0.002 | 20.26 | 0.005 | 9.62 | 0.097 |
| 9.70 | 0.080 | ||||
| 6.29 | 0.026 | ||||
| 7.41 | 0.008 | ||||
For RC = 0.015, and , obtained for I9γ at 25 °C, three local motional modes with eigenvalues close to 6 make a combined fractional contribution of 0.767. The global motion eigenvalue is given by 0.090 = 6×0.016, which is again equal to the “pure” eigenvalue. Its contribution has increased to 0.121. Additional modes with eigenvalues ranging from 4.83 to 9.37 contribute 0.083. The rest (0.05) is contributed by a large number of mixed modes with small individual weighting factors. Note the presence of a mixed mode with an eigenvalue of 1.77, which contributes 0.051.
At 45 °C, where RC = 0.061, and was determined for I9γ, only two modes with eigenvalues relatively close to 6 (6.29 and 7.41), with a combined contribution of 0.519, are present. The global motion eigenvalue is given by the “pure” eigenvalue of 0.366 = 6×0.061, and its contribution is 0.263. Mixed or coupled modes contribute 0.218 to C0(t). Mode-coupling is clearly important at 45 °C.
Dynamic modes with eigenvalues close to 6 contribute to C1(t) (C2(t)) 0.943, 0.854 and 0.411 (0.997, 0.994 and 0.791) at 5, 25 and 45 °C, respectively. For C1(t) mode-coupling is significant at 35 °C and dominant at 45 °C. For C2(t) mode-coupling is important at 45 °C.
The trends in the various parameters as a function of temperature have been discussed above.
Residual configurational entropy
Side-chain Saxis2 values, which exhibit significantly larger variations than backbone S2 values, have been used extensively in recent years to calculate residual configurational entropy.30,52-54 This requires the equilibrium probability distribution function, Peq(ΩC'M), of the M frame relative to the local director, C’. Peq(ΩC'M) is calculated automatically in SRLS using a potential form as general as justified by the quality of the experimental data. The coefficients of this potential, and , are determined by fitting the experimental 2H relaxation data. Model-free does not feature potential energy functions (hence equilibrium probability functions) explicitly. Equation 1 features a single order parameter consistent with axial potential/axial ordering. Hence one has to adopt a simple form of the potential after fitting, assuming that it is axially symmetric, and then use S2, which is typically inaccurate because of force-fitting, to calculate the coefficient of this (axial) potential. Hence the residual configurational entropy derived from S2 is often inaccurate.
Determining the form of the local potential compatible with data integrity, and accounting for potential rhombicity, are clear advantages of SRLS over MF. Currently the orientation of the spin-bearing bond vectors does not depend explicitly on the other degrees of freedom implying over-estimation of the partition function.52 Significant improvement on this important aspect is expected to be achieved within the scope of the “integrated approach” discussed below.
Side-chain rotamer inter-conversion
SRLS is a many-body mode-coupling approach.24 In principle it can handle any number of local motions coupled to one another and to the (asymmetric) global diffusion. The local potential is expanded in the complete basis set of the Wigner rotation matrix elements. The number of terms one may preserve is determined by the nature of the experimental data. We found that the sensitivity of the 2H relaxation data set (including in the current paper rank two coherences and relaxation rates acquired at four magnetic fields) does not justify preserving terms beyond the axial and (indispensable) rhombic L = 2 components. The local diffusion tensor is axially symmetric, accounting for diffusion about and of the C-CH3 axis.23
This scenario captures many of the major features of methyl dynamics as they emerged from early55-57 and recent (ref. 23 and the current paper) studies. The asymmetry of the local spatial restrictions is represented by the rhombicity of the SRSL potential. The dynamical coupling between the local and global motions (which may occur with arbitrary rates) is intrinsic to the SRLS model. General features of local geometry (e.g., the tilt between the magnetic and local ordering/local diffusion frames) are allowed for automatically by the SRLS formalism. All of the relevant physical quantities can be determined as best-fit parameters.
With regard to rotamer jumps − the SRLS model can include potential minima involving motion within the latter, with less frequent jumps between the minima. This is illustrated in Figure 4 of reference 25 and discussed at length in that paper. However, terms of higher rank, L, and order, K, are required to generate such potentials. As pointed out above, data sensitivity does not justify including these terms in the SRLS potential.
Fast rotamer jumps and local librations can be treated separately and combined with SRLS. We have used this strategy (using the Stochastic Liouville Equation approach) in the context of ESR of a nitroxide label tethered to a helix mimicking a protein environment.58,59 In a most general manner one can combine SRLS with molecular dynamics (MD) simulations, which account for any local motion including rotamer jumps in significantly populated conformations. Moreover, quantum chemical calculations can be used to determine magnetic tensors, and hydrodynamic methods to determine the global diffusion tensor.
One of us currently promoting such an “integrated approach”, applied so far to small molecules.60,61 Our present and recently published23 SRLS-based methyl dynamics papers constitute a critical step toward the application of the “integrated approach” to bio-macromolecules. This is currently probably the most advanced attempt to treat methyl dynamics in proteins. The recent study of Hu et al.39 uses model-free (MF) (in particular, the squared generalized order parameter, S2) combined with molecular mechanics. Unlike SRLS, the MF method does not account for local structural asymmetry, mode-coupling and general features of local geometry.21 Only the torsional potential term associated with the C-C bond preceding the CCH3 axis is considered in reference 39. Conformational multiplicity and additional possible local motions are not accounted for.
Finally, let us point out that methyl dynamics is currently the leading method for studying with NMR mega-Dalton protein systems.62,63 Therefore efforts to improve the analysis of the experimental data are timely and important.
IV. Conclusions
By applying SRLS to an extensive set of 2H spin relaxation data we have shown that appropriate analysis of methyl dynamics requires rhombic local potentials/local ordering. The model-free Saxis2 -based “amplitude of motion” picture, implying extensive excursions of the CCH3 axis in tightly packed protein cores, has been replaced with site-specific potential rhombicity derived with SRLS. The form of the local potential is an important structural property not determined so far with NMR spin relaxation. Potential rhombicity was found to decrease with increasing temperature. The rates for local motions increase on average with increasing temperature. They are approximately 10 times lower than their MF counterparts. For the first time activation energies for methyl motion in proteins are estimated with NMR spin relaxation at 2 ± 0.2 kcal/mol. These findings are consistent with theoretical predictions derived with molecular dynamics and molecular mechanics methods. The dynamical coupling between the global and the local motions increases with increasing temperature. The two-mode approximation is not borne out by our results. Rather, methyl dynamics is given by the superposition of quite a few modes. The intrinsic ill-definition of the measurable spectral density is reduced considerably using SRLS. The accuracy of the results can be improved by including in the experimental data set rank 2 coherences. Research prospects include elucidation of highly accurate site-specific information, the calculation of residual configurational entropy from experimentally determined rhombic potentials, and enhancements of the model to include rotamer jumps.
Table 6.
Best-fit parameters obtained with combined fitting of 5 relaxation rates (2H T1, T2 and the three relaxation rates associated with the rank 2 coherences) acquired at 11.7 T, 35 °C, for the depicted methyl groups. The global motion correlation time used was τm = 2.55 ns.12,43 Note that all the values are negative except for of L38δ1, marked with an asterisk.
| Methyl | RC | τ0, ps | |||
|---|---|---|---|---|---|
| L8δ1 | 1.49 | –0.50 | 0.017 | 43 | 0.36 |
| L38δ1 | –0.54 | –0.56 | 0.015 | 38 | 1.04* |
| L56δ2 | 1.46 | –0.50 | 0.020 | 51 | 0.34 |
| T55 | 1.99 | –0.66 | 0.037 | 94 | 0.33 |
| T23 | 2.17 | –0.63 | 0.029 | 71 | 0.29 |
| A18 | 2.09 | –0.74 | 0.039 | 100 | 0.35 |
| A50 | 2.41 | –0.55 | 0.026 | 66 | 0.23 |
| L38δ2 | 1.19 | –0.52 | 0.016 | 41 | 0.44 |
| T37 | 1.89 | –0.58 | 0.032 | 82 | 0.31 |
| T17 | 1.61 | –0.54 | 0.040 | 102 | 0.30 |
| T3 | 2.12 | –0.62 | 0.030 | 122 | 0.29 |
| V49γ2 | 1.83 | –0.51 | 0.021 | 54 | 0.28 |
| A6 | 2.06 | –0.80 | 0.042 | 107 | 0.39 |
| A61 | 1.67 | –0.51 | 0.024 | 61 | 0.31 |
| V47γ1 | 1.59 | –0.51 | 0.029 | 74 | 0.32 |
| V2γ2 | 1.75 | –0.94 | 0.058 | 147 | 0.54 |
| I9g | 1.88 | –0.90 | 0.044 | 112 | 0.41 |
| A31 | 1.91 | –0.92 | 0.044 | 112 | 0.48 |
Acknowledgments
This work was supported by the Israel Science Foundation (Grant No. 279/03 to E.M.), the Binational Science Foundation (Grant No. 2000399 to E.M. and J.H.F.), and the Damadian Center for Magnetic Resonance at Bar-Ilan University, Israel. This work was also supported by the National Center for Research Resources of the National Institutes of Health (Grant No. P41-RR016292 to J.H.F.). A.P. acknowledges support of the Italian Ministry for Universities and Scientific and Technological Research projects FIRB and PRIN ex-40%.
We thank Prof. Lewis E. Kay of the University of Toronto for providing the experimental data used in this study.
References
- 1.Ishima R, Torchia DA. Nat. Struct. Biol. 2000;7:740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- 2.Case D. Acc. Chem. Res. 2002;35:325, 325–33. doi: 10.1021/ar010020l. [DOI] [PubMed] [Google Scholar]
- 3.Bruschweiler R. Curr. Opin. Struct. Biol. 2003;13:175–183. doi: 10.1016/s0959-440x(03)00036-8. [DOI] [PubMed] [Google Scholar]
- 4.Palmer AG., III Chem. Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 5.Mittermaier A, Kay LE. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 6.Igumenova TI, Frederick KK, Wand JA. Chem. Rev. 2006;106:1672–1699. doi: 10.1021/cr040422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitao A, Wagner G. Mag. Reson. Chem. 2006;44:S130–S142. doi: 10.1002/mrc.1839. [DOI] [PubMed] [Google Scholar]
- 8.Jarymowycz VA, Stone MJ. Chem. Rev. 2006;106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- 9.Palmer AG, Massi F. Chem. Rev. 2006;2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 10.Muhandiram DR, Yamazaki T, Sykes BD, Kay LE. J. Am. Chem. Soc. 1995;117:11536–11544. [Google Scholar]
- 11.Millet O, Muhandiram DR, Skrynnikov NR, Kay LE. J. Am. Chem. Soc. 2002;124:6439–6448. doi: 10.1021/ja012497y. [DOI] [PubMed] [Google Scholar]
- 12.Srynnikov NR, Millet O, Kay LE. J. Am. Chem. Soc. 2002;124:6449–6460. doi: 10.1021/ja012498q. [DOI] [PubMed] [Google Scholar]
- 13.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 14.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 15.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. J. Am. Chem. Soc. 1990;112:4989–4991. [Google Scholar]
- 16.Tugarinov V, Liang Z, Shapiro Yu. E., Freed JH, Meirovitch E. J. Am. Chem. Soc. 2001;123:3055–3063. doi: 10.1021/ja003803v. [DOI] [PubMed] [Google Scholar]
- 17.Tugarinov V, Shapiro Yu. E., Liang Z, Freed JH, Meirovitch E. J. Mol. Biol. 2002;315:171–186. doi: 10.1006/jmbi.2001.5231. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro Yu. E., Kahana E, Tugarinov V, Liang Z, Freed JH, Meirovitch E. Biochemistry. 2002;41:6271–6281. doi: 10.1021/bi012132q. [DOI] [PubMed] [Google Scholar]
- 19.Meirovitch E, Shapiro Yu. E., Tugarinov V, Liang Z, Freed JH. J. Phys. Chem.B. 2003;107:9883–9897. [Google Scholar]
- 20.Meirovitch E, Shapiro Yu. E., Liang Z, Freed JH. J.Phys. Chem. B. 2003;107:9898–9904. [Google Scholar]
- 21.Meirovitch E, Shapiro Yu. E., Polimeno A, Freed JH. J.Phys. Chem. A. 2006;110:8366–8396. doi: 10.1021/jp056975t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro Yu. E., Meirovitch E. J. Phys. Chem. B. 110:11519–11524. doi: 10.1021/jp060282a. [DOI] [PubMed] [Google Scholar]
- 23.Meirovitch E, Polimeno A, Freed JH. J. Phys. Chem. B. 2006;110:20615–20628. doi: 10.1021/jp061403+. [DOI] [PubMed] [Google Scholar]
- 24.Polimeno A, Freed JH. Adv. in Chem. Phys. 1993;83:89–210. [Google Scholar]
- 25.Polimeno A, Freed JH. J. Phys. Chem. 1995;99:10995–11006. [Google Scholar]
- 26.Liang Z, Freed JH. J. Phys. Chem. B. 1999;103:6384–6396. [Google Scholar]
- 27.a Chatfield DC, Szabo A, Brooks BR. J. Am. Chem. Soc. 1998;120:5301–5311. [Google Scholar]; b Brainard JR, Szabo A. Biochemistry. 1981;20:4618–4628. doi: 10.1021/bi00519a016. [DOI] [PubMed] [Google Scholar]; c Wallach D. J. Chem. Phys. 1967;47:5258–5268. [Google Scholar]; d Woessner DE. J. Chem. Phys. 1962;36:1–4. [Google Scholar]
- 28.Lee AL, Flynn PE, Wand AJ. J. Am. Chem. Soc. 1999;121:2891–2902. [Google Scholar]
- 29.Lee AL, Sharp KA, Kranz JK, Song X-J, Wand A. J. Biochemistry. 2002;41:13814–13825. doi: 10.1021/bi026380d. [DOI] [PubMed] [Google Scholar]
- 30.Lee AL, Wand AJ. Nature. 2001;411:501–504. doi: 10.1038/35078119. [DOI] [PubMed] [Google Scholar]
- 31.Prabhu NV, Lee AL, Wand H, Sharp KA. Biochemistry. 2003;42:562–570. doi: 10.1021/bi026544q. [DOI] [PubMed] [Google Scholar]
- 32.Bruschweiler R, Wright PE. J. Am. Chem. Soc. 1994;116:8426–8427. [Google Scholar]
- 33.Bremi T, Bruschweiler R. J. Am. Chem. Soc. 1997;119:6672–6673. [Google Scholar]
- 34.Lienin SF, Bremi T, Brutscher B, Bruschweiler R, Ernst RR. J. Am. Chem. Soc. 1998;120:9870–9879. [Google Scholar]
- 35.Mittermaier A, Kay LE. J. Am. Chem. Soc. 2001;123:6892–6903. doi: 10.1021/ja010595d. [DOI] [PubMed] [Google Scholar]
- 36.Chou JJ, Case DA, Bax A. J. Am. Chem. Soc. 2003;125:8959–8966. doi: 10.1021/ja029972s. [DOI] [PubMed] [Google Scholar]
- 37.Best RB, Clarke J, Karplus M. J. Am. Chem. Soc. 2004;126:7734–7735. doi: 10.1021/ja049078w. [DOI] [PubMed] [Google Scholar]
- 38.Best RB, Clarke J, Karplus M. J. Mol. Biol. 2005;349:185–203. doi: 10.1016/j.jmb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Hu A, Hermans J, Lee AL. J. Biomol. NMR. 2005;32:151–162. doi: 10.1007/s10858-005-5366-0. [DOI] [PubMed] [Google Scholar]
- 40.Freed HJ, Nayeem A, Rananavare SB. In: The Molecular Dynamics of Liquid Crystals. Luckhurst GR, Veracini CA, editors. Kluwer Academic Publishers; The Netherlands: 1994. pp. 271–312. Chapter 12. [Google Scholar]
- 41.Abragam A. Principles of Nuclear Magnetism. Oxford University Press (Clarendon); London: 1961. [Google Scholar]
- 42.Peng JW, Wagner G. In: Methods in Enzymology. James TL, Oppenheimer NJ, editors. Vol. 239. Academic Press; New York: 1994. pp. 563–595. [DOI] [PubMed] [Google Scholar]
- 43.Tugarinov V, Kay LE. J. Biomol. NMR. 2004;29:369–376. doi: 10.1023/B:JNMR.0000032562.07475.7f. [DOI] [PubMed] [Google Scholar]
- 44.Kay LE, Torchia DA, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 45.Freed JH. J. Chem. Phys. 1977;66:4183–4199. [Google Scholar]
- 46.a Polimeno A, Moro GJ, Freed JH. J. Chem. Phys. 1995;104:1090–1104. [Google Scholar]; b Polnaszek CF, Bruno GV, Freed JH. J. Chem. Phys. 1973;58:3185–3199. [Google Scholar]; c Polnaszek CF, Freed JH. J. Phys. Chem. 1975;79:2283–2306. [Google Scholar]
- 47.Choy W-Y, Kay LE. J. Biomol. NMR. 2003;25:325–333. doi: 10.1023/a:1023065310430. [DOI] [PubMed] [Google Scholar]
- 48.Mandel AM, Akke M, Palmer AG., III J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 49.Johnson E, Chazin WJ, Rance M. J. Mol. Biol. 2006;357:1237–1252. doi: 10.1016/j.jmb.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 50.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chatfield DC, Augusten A, D'Cunha C. J. Biomol. NMR. 2004;29:377–385. doi: 10.1023/B:JNMR.0000032553.13686.0b. [DOI] [PubMed] [Google Scholar]
- 52.Akke M, Bruschweiler R, Palmer AG., III J. Am. Chem. Soc. 1993;115:9832–9833. [Google Scholar]
- 53.Yang D, Kay LE. J. Mol. Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- 54.Lee AL, Kinnear SA, Wand A. J. Nat. Struct. Biol. 2000;7:72–77. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- 55.Wittebort RJ, Szabo A. J. Chem. Phys. 1978;69:1722–1736. [Google Scholar]
- 56.Avitable J, London RE. J. Am. Chem. Soc. 1978;100:7159–7165. [Google Scholar]
- 57.Brainard JR, Szabo A. Biochemistry. 1981;20:4618–4628. doi: 10.1021/bi00519a016. [DOI] [PubMed] [Google Scholar]
- 58.Tombolato F, Ferrarini A, Freed JH. J. Phys. Chem. B. 2006;110:26248–26259. doi: 10.1021/jp0629487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tombolato F, Ferrarini A, Freed JH. J. Phys. Chem. B. 2006;110:26260–26271. doi: 10.1021/jp062949z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barone V, Polimeno A. PhysChemChemPhys. 2006;8:4609–4628. [Google Scholar]
- 61.Barone V, Brustolon M, Cimino P, Polimeno A, Zerbetto M, Zoleo A. J. Am. Chem. Soc. 2006;128:15865–25873. doi: 10.1021/ja065475q. [DOI] [PubMed] [Google Scholar]
- 62.Tugarinov V, Sprangers R, Kay LE. J. Am. Chem. Soc. 2007;129:1743–1750. doi: 10.1021/ja067827z. [DOI] [PubMed] [Google Scholar]
- 63.Sprangers R, Kay LE. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]