Abstract
Nesprins are located at the outer and inner membranes of the nuclear envelope and help link the cytoskeleton to the nucleoskeleton. Nesprin-1α, located at the inner nuclear membrane, binds to A-type lamins and emerin and has homology to spectrin-repeat proteins. However, the mechanical and thermodynamic properties of the spectrin-like repeats (SLRs) of nesprin-1α and the potential structural contributions of the unique central domain were untested. In other spectrin superfamily proteins, tandem spectrin-repeat domains undergo cooperatively coupled folding and unfolding. We hypothesized that the large central domain, which interrupts SLRs and is conserved in other nesprin isoforms, might confer unique structural properties. To test this model we measured the thermal unfolding of nesprin-1α fragments using circular dichroism and dynamic light scattering. The SLRs in nesprin-1α were found to have structural and thermodynamic properties typical of spectrins. The central domain had relatively little secondary structure as an isolated fragment, but significantly stabilized larger SLR-containing molecules by increasing their overall helicity, thermal stability and cooperativity of folding. We suggest this domain, now termed the ‘adaptive’ domain (AD), also strengthens dimerization and inhibits unfolding. Further engineering of the isolated AD, and AD-containing nesprin molecules, may yield new information about the higher-order association of cooperative protein motifs.
Keywords: Nuclear envelope, spectrin repeat, laminopathy, emerin, LINC complex, nuclear mechanics
INTRODUCTION
In eukaryotic cells, chromosomes are enclosed by the nuclear envelope, which consists of inner and outer nuclear membranes (INM and ONM, respectively) and an underlying lamina network formed by A- and B-type lamins.1 The A-type lamins (e.g. lamins A and C), encoded by one gene (LMNA), are particularly important for the shape and mechanical properties of the nucleus and nuclear functions.2,3 Many INM proteins, including members of the nesprin family, are proposed to directly influence the A-type lamina network.4–10 The human genome encodes four nesprin genes, each of which is alternatively transcribed and spliced to yield an array of protein isoforms with different sizes, locations and functions.11–12 The giant isoforms of nesprin-1, -2 and -3 bind either actin or plectin in the cytoplasm, and are embedded in the ONM as key components of a proposed nuclear envelope-spanning LINC complex (linker of nucleoskeleton and cytoskeleton).1,13–15 In contrast to these larger ONM-localized isoforms of nesprin, other isoforms including nesprin-1α are smaller (e.g., 120 kDa) and localize to the INM.7,8 An open question in the field is whether nesprin-1α and other INM-localized nesprins participate in LINC complexes. Furthermore, INM-localized nesprins, whether attached to LINC complexes or not, might have unique structural roles. Nesprin-1α is of particular interest as a mechanical ‘connector’ because it can bind directly to itself (forming antiparallel homodimers), A-type lamins and another INM protein named emerin.16
The mechanical connections of nesprin-1α may play a role in Emery-Dreifuss muscular dystrophy, whichcan be caused by dominant mutations in LMNA or recessive mutations in EMD, the gene encoding emerin.18,19 Interestingly, mutations in the genes encoding either nesprin-1α or nesprin-2β can independently cause Emery-Dreifuss muscular dystrophy20,21, and mice with a C-terminal deletion in nesprin-1 have a phenotype similar to Emery-Dreifuss muscular dystrophy.22 Nesprin-1α is expressed at high levels in heart and skeletal muscle, the tissues most affected in this disease.8,23
By homology, nesprins belong to the spectrin superfamily, which includes spectrin, dystrophin, α-actinin and others.23,24 This superfamily is characterized by multiple repeats of a structural unit, the spectrin repeat (SR), which consists of a bundle of three antiparallel ~106-residue α-helices.24 Canonical SRs can resist unfolding induced by force, temperature or chemical denaturants.25–32 Thermal unfolding studies of tandem SRs from spectrins reveal multiple transition states and cooperatively coupled unfolding.28,29 Consistent with thermal studies, SR domains also undergo stepwise stretching and cooperative unfolding when subjected to pulling forces by Atomic Force Microscopy.32
Nesprin-1α was originally predicted to have seven spectrin-like repeats (SLRs), a central domain of unknown function located between SLR5 and SLR6, and a C-terminal transmembrane KASH domain.7,33 SLR3 and SLR5 interact mutually and mediate dimerization of nesprin-1α.16 The most commonly expressed nesprin-1α isoform lacks SLR1, and new sub-isoforms of nesprin-1α (such as nesprin-1α1 and nesprin-1α2) have been reported.16,33,34 We studied this abundant isoform but retained the original SLR numbering for consistency. The mysterious central domain of nesprin-1α was previously thought to include a ‘split’ LEM-domain based on limited amino acid sequence homology to the LAP2-emerin-MAN1 domain8. However a subsequent study detected no binding of the isolated central domain to the canonical LEM-domain partner, Barrier to Autointegration Factor (BAF), in vitro.16 We noted this central domain is rich in disordered loops and coils, suggesting its conformation might change upon binding to partners. Since the dynamic stability of canonical SRs involves their highly repetitive nature,35 we hypothesized the 230-residue central domain might confer unique properties to nesprin-1α and other nesprin isoforms in which it is conserved. This hypothesis is supported by the biophysical results reported here, and we therefore named this region the ‘adaptive domain’ (AD). Also unknown was whether the SLR domains in nesprin-1α actually behave as SRs, since there were no previous molecular-level biophysical studies of nesprins.
We studied the secondary structure and thermal unfolding of nesprin-1α polypeptides. Our results suggest the SLR domains have spectrin-like unfolding, but are collectively and uniquely stabilized by the AD. Like other SR proteins, nesprin-1α also appears to have long-range cooperative folding of multiple SLRs. One consequence of this property is that studies based on subfragments of nesprin-1α, and potentially other nesprins, may lead to false negatives (e.g., failure to bind biologically relevant partners) because these molecules lack properties or functions conferred by the AD.
MATERIALS AND METHODS
Nesprin-1α constructs
All nesprin-1α fragments were derivatives of the nesprin-1α cDNA specified by GenBank accession number AF444779. The full nesprin-1α cDNA encodes SLR2 through the C-terminal transmembrane and KASH domain. The conformational dynamics of the adaptive and SLR domains were characterized using five purified recombinant subfragments of nesprin-1α (Fig. 1). Fragment SLR2-7 consisted of full-length nesprin-1α minus the transmembrane domain. The SLR2-5 fragment consisted of the first four SLR domains. Fragment SLR5-7 consisted of the three C-terminal SLRs with the AD between SLR5 and 6. To determine whether and how the AD might influence full-length nesprin-1α, we expressed the AD domain alone and deleted the AD from SLR2-7 (designated SLR2-7ΔAD). The cDNAs encoding domains SLR2-5, SLR2-7 and SLR5-7 in the pCR-T7 vector were described previously8 and were generously donated by Elizabeth McNally (University of Chicago). To make the SLR2-7ΔAD construct, we used a described method36: we PCR-amplified the entire SLR2-7 containing vector without the adaptive domain, then self-ligated this linear DNA to regenerate the vector plus cDNA, minus the AD (SLR2-7ΔAD). The backward and forward primers were 5′-CAGGCGGATCAGTTTCTTATG-3′ and 5′-CAGAAGTGGCAGCAGTTTAAC-3′, each flanking the adaptive domain, and corresponding to nucleotides 1731–1741 and 2430–2450 respectively, of AF444779. To express AD alone, we PCR-amplified using primers 5′-ATAAGAAACTGATCCGCCTGC-3′ and 5′-GTT AAACTGCTGCCACTTCTG-3′. All five cDNAs were verified by DNA sequencing (data not shown) and then subcloned into pET22b(+) using BamHI and HindIII and finally transformed into E. coli Rosetta2(DE3) (Novogen, Madison Wl, USA) for protein expression.
Figure 1.

(A) Schematic representation of recombinant fragments of human nesprin-1α used in this work. Each black box indicates a spectrin-like repeat (SLR). The gray box indicates the adaptive domain (AD). Short grey box indicates the KASH domain. Our nesprin-1α polypeptide starts from residue 125 in Genbank accession no. AF444779. The SLRs were predicted by SMART, and the adaptive domain (AD) was named by virtue of its disorder predicted by DisEMBL but ordered structure measured in the full length protein. (B) SDS-PAGE analysis of purified recombinant SLR2-7, SLR2-5, SLR5-7 SLR2-7ΔAD and AD polypeptides.
Preparation of recombinant proteins
To obtain soluble recombinant proteins, transformed bacteria were cultured at 28°C in Terrific Broth with 100 μg/ml ampicillin and 34 μg/ml Chloramphenicol to an A600 of 2.0 and expression was induced by 0.4 mM isopropyl-β-D-thioglactopyranoside (IPTG) for 6 h. Cells were then collected, resuspended in His buffer (0.3 M NaCl, 50 mM sodium phosphate pH 8.0, 20 mM imidazole, 1 mM DMSF, 50 μg/ml pepstatin A, 20 μg/ml aprotinin, and 10 μg/ml leupeptin) and sonicated (all chemicals from Fisher Scientific). The soluble fraction was applied to Ni-NTA resin to purify 6-His-tagged proteins, and further purified by size exclusion chromatography (HiPrep™ 16/60 Sephacryl S-200 HR column) using an AKTA prime plus system (GE Life Sciences). Expression and purification were monitored by SDS-PAGE (10% gels) and protein dimers were observed by non-reducing PAGE. Protein concentration was determined by A280 (GeneQuant™ 1300 Spectrophotometer) with extinction coefficients calculated from the amino acid composition. To minimize error from any single method, we also used the Bradford Method in the same spectrophotometer, with bovine serum albumin as standard. Protein concentrations assayed by Bradford were within 8% of those measured by A280. Recombinant proteins were over 95% pure, as estimated by Coomassie stained SDS-PAGE gels (see figure 1B).
Sequence analysis and structure prediction
SLR and AD structures were predicted by SMART software (http://smart.embl-heidelberg.de/). SLR and SR domains in nesprin-1α and other spectrin superfamily proteins were compared by BLASTP programs at NCBI. To assess evolutionary conservation, adaptive amino acid sequences were analyzed by T-Coffee (http://www.tcoffee.org/). Disordered regions within nesprin-1α were predicted by DisEMBL 1.5 (http://dis.embl.de/) and the secondary structure of AD was predicted by PSIPRED (http://www.psipred.net/psiform.html).
Circular Dichroism measurement and data analysis
Protein samples were analyzed within 24 hours after purification, usually within 4 hours. Samples were centrifuged (13,000 × g, 30 min, 4°C; Beckman J-26 XP centrifuge) to remove potential aggregates. Since our 24-hour data and 4-hour data agreed, and because protein concentrations used for CD were very low (25 μg/ml), we suggest our CD data were not significantly affected by aggregation. Far-UV CD spectra were recorded using a Jasco 810 spectropolarimeter equipped with thermostated cell housing and a 1 cm path-length cell. To determine unfolding profiles, the temperature was increased from 15°C to 109°C at a rate of 0.4°C/min, and ellipticity was recorded at 2°C intervals. Thermal induced unfolding curves were calculated by ellipticity in 222 nm versus temperature. Secondary structure percentage was calculated by software CD-pro using three different programs (SELCON3, CDSSTR, CONTINLL) and averaged. The first-order derivatives of the fraction unfolded were calculated by Origin (OriginLab) using 2nd order smoothing differentiation. The first order derivative curve mathematically exaggerates any small noise in the melting curve. To distinguish signals from noise, only transition peaks with 5–12 repeatable datapoints in the first derivative curves were considered transitions.
Dynamic light scattering measurements
Protein sizes were assayed by dynamic light scattering (DLS) using a Malvern zetasizer nano system. Immediately prior to DLS, samples were centrifuged 1 hour at 250,000xg (4°C), since some SLR2-5 and SLR5-7 aggregates were seen at the high protein concentrations (~0.5 mg/mL) required for DLS. Each protein was measured 10 independent times, with 15 runs; at least two independently produced protein samples were measured for each point. The reported diameter is the Z-average hydrodynamic diameter of the sample; error bars indicate the standard deviation between runs. Polypeptides measured as a function of temperature, SLR2-7 and SLR2-7ΔAD, were equilibrated for 20 min at each measured temperature prior to measurement.
RESULTS
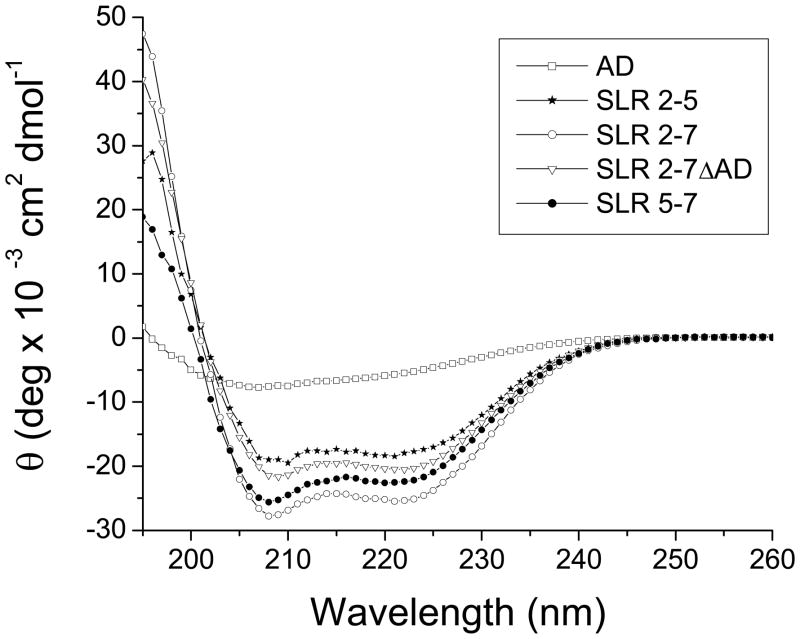
All five nesprin-1α polypeptides were predicted to have high α-helical content, and could therefore be compared by circular dichroism (CD), as shown previously for SR proteins.28–29 CD measurements of each fragment at 37°C, except the AD, revealed typical α-helical signatures with minima at about 208 and 222 nm (Fig. 2, Table 1). The four SRL-containing polypeptides displayed high α-helicity (74–88%) and little β-sheet structure (0.4–2.75), consistent with the predicted SLR-rich structure (Table 1). The AD alone showed moderate levels of both α-helix (32.7%) and β-sheet (18.3%; Table 1). These differences in the calculated secondary structure properties were significant by the student t-test (p<0.001). The standard deviation of the secondary structure reflected sample variance to a minor degree; the largest contributing factors were the biases of the three different computational programs in CD-pro (see Methods). The helicities of “pure SLR” fragments SLR2-5 and SLR2-7ΔAD averaged 74.5% and 78.4%, respectively, well within the reported helicity range of typical canonical SRs (60% to 80%).26–29 In contrast the helicity of SLR5-7, which includes the adaptive domain, was significantly higher (averaging 83.1%), suggesting the AD was more helical within the larger molecule, than either a canonical SR or the AD alone. The other AD-containing polypeptide, SLR2-7 (the largest polypeptide tested), had the highest average helicity (88.2%). This was unexpected since non-helical linking regions in larger proteins mathematically “dilute” the percentage of ordered regions. Furthermore the helical percentage of SLR2-7 was higher than any sub-fragment thereof. These results suggest full length nesprin-1α might undergo strong cooperatively-coupled folding in which the whole protein is stabilized into a highly helical structure. Supporting this conclusion, SLR2-7 had a significantly lower percentage of ‘turn’ structure than the other four polypeptides (0.9% compared to 3.8%, 4.7%, 7.7% and 20.2%; Table 1). Transitions from ‘turn’ to ‘helix’ structure can be induced by strongly cooperative coupled folding.37–39 The 88.2% helicity of full-length SLR2-7 was most closely matched by SLR5-7 (83.1%), whereas SLR2-7ΔAD and SLR2-5 had significantly less helicity (78.4% and 74.5%), suggesting the AD strongly influences the secondary structure and potentially also the tertiary structure of nesprin-1α. Notably the AD alone had only moderate (32.7%) helicity and more β-sheet structure (18.3%) than any larger AD-containing polypeptide. We therefore suggest the AD changes conformation (‘adapts’) to adjacent SLRs and thereby increases the thermodynamic stability of the native nesprin molecule.
Figure 2.
Representative CD spectra of nesprin-1α polypeptides SLR2-7, SLR2-5, SLR5-7 SLR2-7ΔAD and AD. Spectra were recorded in PBS buffer (pH 7.4) at 37°C and scaled to 10 μg/ml for comparison.
Table 1.
Structural properties of nesprin-1α fragments calculated from the CD spectra in Figure 2, and protein size measured by DLS at 25°C unless otherwise noted.
| Circular Dichroism analysis | DLS | |||||
|---|---|---|---|---|---|---|
| Protein fragments | %α-helix | %β-sheet | %Turn | θ222/θ208 | Size(nm) | monomer (nm) |
| SLR2-5 | 74.5±4.5 | 2.4±0.7 | 7.7±2.0 | 0.95 | 35.4±5.4 | |
| SLR2-7 | 88.2±4.1 | 0.4±0.2 | 0.9±0.5 | 0.91 | 99.0±14.9 | 57.4±13.1 (58°C) |
| SLR5-7 | 83.1±3.9 | 2.7±0.4 | 3.8±2.2 | 0.88 | 36.1±6.1 | |
| SLR2-7ΔAD | 78.4±5.0 | 1.6±0.3 | 4.7±1.3 | 0.96 | 76.1±7.8 | 53.6±4.45 (42°C) |
| AD | 32.7±4.3 | 18.3±2.1 | 20.2±1.3 | NTa | NT | NT |
NT indicates not tested. The θ222/θ208 ratio is useful for high helix proteins, not for AD alone. Similarly, isolated AD is in its disordered state, so size analysis is not appropriate.
The ellipticity ratio θ222/θ208 of CD signals was used as an index of coiled-coil interactions.40 The 222 nm CD signal reflects the n-π* transition and is responsive to α-helical content, whereas the 208 nm CD signal reflects the π-π* transition and is sensitive to whether the helix is monomeric or has tertiary interactions with other helices, because close apposition of rigid α-helices decreases the π-π* transition signal.7 Typical α-helical monomer structures have θ222/θ208 ratios of around 0.85, whereas α-helical bundles or dimers have a ratio close to 1.41–43 We found that SLR2-5 and SLR 2-7ΔAD, both of which lack the AD, had θ222/θ208 ratios of 0.95 and 0.96, respectively (Table 1), similar to canonical SRs and other helical bundles.27,44 However, the AD-containing polypeptides SLR5-7 and SLR2-7 had lower θ222/θ208 ratios (0.88 and 0.91, respectively), suggesting the AD does not behave as a helical bundle.
Nesprin 1α can form homodimers16, in contrast to other SR proteins such as α-spectrins, which heterodimerize with a β-spectrin partner. To investigate potential dimerization or oligomerization of nesprin-1α at the biophysical level, we used Dynamic Light Scattering (DLS) to measure the sizes of two nesprin-1α fragments, in the same buffer used for CD (10 mM sodium phosphate, pH 7.4; Table 1). Ionic strength does not affect the secondary structure of canonical SR proteins measured by CD, but can vary the DLS-measured radius of gyration by 2–3 fold.45,46 Thus, the DLS-based sizes reported here are probably larger than the native state, but provide useful information when comparing related protein fragments. The tested nesprin-1α polypeptides had DLS-measured sizes that ranged from 35 to 99 nm (Table 1), much larger than globular proteins of similar mass (such as BSA, MW. 69.3 kDa, Size 9.5 nm in DLS manufacturer standard), suggesting nesprin-1α has a long rod-shaped structure (see Discussion).
Thermally-induced transitions of nesprin-1α fragments
For most tested spectrin superfamily proteins, thermal unfolding behavior correlates with the mechanical extensibility of their spectrin repeats.25,35,44 We monitored thermal unfolding of four nesprin-1α polypeptides by CD at 222 nm, to assay loss of helicity in solution. (The 222 nm signal was not used to track the AD alone, because its CD spectrum was significantly different and had low signal; Fig. 2). Melting curves are shown in Figs. 3A–3D. The corresponding first-order derivative of each melting curve (Figs. 3E–3H) was used to precisely characterize the number of transition stages and their transition middle points (Tm). Thermal unfolding of the AD was revealed by comparing its CD spectra at low (25°C; folded) and high (101°C; unfolded) temperatures (Fig. 3I). Polypeptides SLR2-5 (three SLRs) and SLR5-7 (three SLRs plus AD) each had multiple transition stages with two obvious peaks in the first order derivative curve (Figs. 3E & 3F), characteristic of polypeptides with low numbers of tandem SRs. We defined the obvious major transition peak at lower temperature as Tm1 and the peak at higher temperature as Tm2. Tm values reflect the energy required for loss of local secondary structure, and correlates with the extension of individual SLR helix bundles. For example, SLR 2-5 displayed two transition stages peaks with similar height and width with Tm1 at 45°C and Tm2 at 61°C, suggesting SLR2-5 has distinct transitional states. Related behavior was seen for SLR5-7: one small peak (Tm1 at 59°C) and a higher and broader Tm2 peak at 75°C, suggesting a small domain of SLR5-7 unfolded around 59°C, whereas a large or multiple domains were more stable, unfolding around 75°C. Interestingly, SLR2-7ΔAD showed only one broad peak (Tm at 63°C) (Fig. 3G), similar to the cooperatively coupled unfolding property of canonical SRs: spectrin itself has only one distinguished Tm (49.5°C for erythroid spectrin and 58°C for non-erythroid spectrin); the Tm of isolated SRs can vary from 21°C to 82°C and some tandem SRs show multiple Tms with the high-Tm SR stabilizing other lower-Tm SRs.28,29 SLR2-7ΔAD resembled spectrin protein both in Tm value and in the cooperatively coupled unfolding behavior. The largest fragment tested, SLR2-7, identical to SLR2-7ΔAD except for the presence of the 230-residue AD, also showed one major peak but at Tm 73°C (Fig. 3H), significantly higher than that of SLR2-7ΔAD (63°C) or spectrins (49–58°C). The AD alone was not highly helical either before or after thermal unfolding (Fig. 3I). We concluded that the AD, by interacting with SLRs, contributes significantly to the high Tm2 of both SLR5-7 (75°C) and SLR2-7 (73°C). These findings suggest the AD is essential for the high structural stability of nesprin-1α. Given the high Tm of SLR2-7 (73°C), relative to canonical SRs of spectrin (Tm~ 55°C),29 we propose that the SLRs of nesprin-1α are “over-stabilized” by the AD as a mechanism to resist unfolding. Unfolding was partially reversible when temperature was reduced at a rate of 20°C/h. For example, 70% of SLR2-7 refolded when reduced back to the original temperature (30°C to 70°C to 30°C), and continued refolding upon further decreases (to 20°C; data not shown). Due to instrumentation limitations, cooling is more difficult to control and temperatures below 20°C were not obtainable.
Figure 3.
Profiles of the thermal unfolding of nesprin-1α fragments recorded at 222 nm (A–D) and the first order derivatives of the unfolding curves, respectively (E–H), as well as the AD spectra before and after thermal unfolding (I). A and E represent the unfolding of SLR2-5; B and F represent the unfolding of SLR5-7; C and G represent the unfolding of SLR2-7ΔAD; D and H represent the unfolding of SLR2-7. SLR2-5 and SLR2-7ΔAD are shown as open and closed circles, respectively (n = 2); SLR5-7 and SLR2-7 are shown as open and closed circles, respectively (n = 3, but 2 representative samples are shown). The apparent sample to sample variation for fragments SLR5-7 and SLR2-7, compared to other two pure SLR fragments, may be due to the effects of the adaptive domain. The unfolding of AD was maintained in 101 °C for 1 hour to reach equilibrium and the spectra are repeated twice and the average value is plotted. Each trial (n) includes the repeat of protein expression, purification and unfolding; independent repeats from the same preparation were indistinguishable.
The above CD measurements of temperature-dependent secondary structure suggested AD-containing nesprin-1α fragments were unusually stable. Models of SR unfolding suggest secondary and tertiary structures are lost simultaneously.35 Since nesprin polypeptides had the added complication of dimerization (quaternary association), which might influence transition temperatures, we used DLS to measure size as a function of temperature for SLR2-7 and SLR2-7ΔAD. At 25°C their sizes were ~99 nm and ~77 nm respectively (Fig. 4), as expected (Table 1). At 37°C SLR2-7ΔAD showed unresolved dual peaks at around 51 nm and 72 nm (asterisk at 37°C, Fig. 4), which we interpret as a population of mixed dimers and monomers. At 42°C, SLR2-7ΔAD had completely transitioned to a single population of monomers (single peak at 53 nm), which then gradually but significantly increased in size as temperature increased further, potentially reflecting extension of the SLRs (Fig. 4). SLR2-7 was significantly more stable: the unresolved dual peaks (~60 nm and 100 nm) were not detected until 50°C and 54°C, with complete dissociation (single peak, 54.1 nm) at 58°C, followed by gradual increase in size up to 66°C (Fig. 4). These results show the molecular size of the SLR2-7 dimer is significantly more thermodynamically stable than SLR2-7ΔAD, consistent with our CD results. For both polypeptides this process was reversible; when temperature was decreased slowly before reaching melting temperature, about 70% went through the dual-peak transition and then regained their original (dimer) size (not shown).
Figure 4.
Protein size measured by DLS for SLR2-7 and SLR2-7ΔAD as a function of increasing temperature. Protein size was measured at each indicated temperature, and average peak size was plotted; bars indicate standard deviation. Asterisks indicate that the plotted size is the average of two unresolved peaks, interpreted as a mixture of monomers and dimers at that temperature. With increasing temperature each construct changed from single-peak dimers, to presumed mixtures of dimers and monomers (asterisk), to single-peak monomers. SLR2-7ΔAD completely dissociates at near 42°C; SLR2-7 completely dissociates at near 58°C. Dissociated proteins then extended further, prior to denaturing above 54°C (SLR2-7ΔAD) or above 66°C (SLR2-7). (n=2 independent runs).
Properties of the adaptive domain in nesprin-1α
The AD of nesprin-1α was previously deemed a ‘rod’ domain to distinguish this unique region from surrounding SLRs.33 As shown above, the AD of nesprin-1α in SLR2-7 and SLR5-7 has a thermally stable α-helical secondary structure and helps neighboring SLR domains resist both thermal unfolding and size extension. Thus, the AD appears to stabilize nesprin-1α dimers in a manner that may distinguish nesprin-1α from canonical SR proteins. We suggest these results also apply to all other AD-containing nesprin isoforms including nesprin-2β at the INM and the nesprin-1 and -2 “giants” at the ONM. The nesprin AD is highly conserved among vertebrates, including zebrafish (Fig. 5A). One known missense mutation that causes EDMD (V729L in AF444779) affects conserved residue V729 (Fig. 5A). The AD is conserved in nesprin-2 (Fig. 5A), but appears to be missing from nesprin-3 and nesprin-4. Our conservation comparison is in agreement with Simpson et al.’s evolutionary conservation analysis of nesprins, which reported that an unstructured region and a region called the ΔSR region in AD are among the most highly conserved34. We therefore propose that the AD confers unique structural and functional properties to all AD-containing nesprin-1 and nesprin-2 isoforms.
Figure 5.
Evolutionary conservation of the Adaptive Domain in nesprin-1 and nesprin-2. (A) Amino acid sequence of the AD of nesprin-1 and nesprin-2 from human, mouse, Bos taurus, chicken and Zebrafish (residues 558-786 in human nesprin-1 isoform, AF444779). Overlines indicate left and right halves of the putative ‘divided LEM-like domain’.8 Underlines in human nesprin-1 indicate ‘Hot loops’ (high mobility) predicted by DisEMBL. The V729L mutation (boxed) causes Emery-Dreifuss muscular dystrophy.21 (B) Schematic diagrams to scale, showing the relative positions of the ‘divided LEM-like domain in nesprin-1α8, disordered regions predicted by the ‘loops/coils’ definition (DisEMBL), disordered regions predicted by the ‘hot-loops’ definition (DisEMBL), and secondary possible structure (predicted by PSIPRED). Star highlights residue V729.
The isolated AD polypeptide had only moderate levels of α-helix (32.7%) and β-sheet (18.3%) as well as moderate level of turns (20.2%), which might seem to contradict the high measured helicity and very low β-sheet and turns in SLR2-7 and SLR5-7. Prediction of the AD domain, such as by the secondary predictor PSIPRED, show even less secondary structure of α-helix (18.3%) and β-sheet (1.3%) but high coil (80.4%) in the AD (Fig. 5B). This suggests that there may be longer-range forces required to stabilize secondary structures within the domain. The protein disorder prediction software DisEMBL, predicts the AD is the most disordered region in nesprin-1α.47 Over half (56%) of nesprin-1α AD residues are predicted to be disordered by the loops/coils definition, and 29% are predicted to be disordered by the “hot loops” definition (meaning high degree of mobility) (Fig. 5B). Disordered coils are thought to become ordered only when bound to other molecules,48,49 demonstrating ‘adaptive’ features. The longest predicted “hot loops” region is found within the most highly-conserved region of the AD (Fig. 5A, shaded; corresponds to human nesprin-1α residues 610–629), and is also highly conserved in nesprin-2. Our measurements clearly demonstrate the AD is less helical when expressed on its own, but highly helical and stable within larger SLR-containing molecules, which our DLS measurements suggest are dimers. We therefore propose the AD uniquely stabilizes native nesprin molecules by reinforcing nesprin homodimer structures, by interacting with flanking SLR domains, or by both mechanisms.
DISCUSSION
These results show that the predicted SLRs in nesprin-1α have biophysical properties typical of canonical SRs, as compared and summarized in Table 2. Both have high α-helical content, large dynamic size and multiple transition stages during thermal unfolding. Our analysis of SLR2-7ΔAD, which lacks the AD, allowed us to compare the SLRs in nesprin, to canonical SR proteins. SLR2-7ΔAD had a helical content and θ222/208 ratio consistent with helical bundles. A helical bundle is a group of helices packed nearly parallel or antiparallel to one another. To form such bundles, multiple non-helical loops of significant length are needed to transition and allow tight parallel association between helices. During thermal unfolding, SLR2-7ΔAD had temperature transitions in its secondary structure similar in magnitude to other spectrin proteins. SLR2-7ΔAD also showed quaternary dissociation slightly above 37°C, followed by increases in protein size, presumably from changes in tertiary and secondary structure. These findings all suggest that nesprin SLRs are similar to, or the same as, typical SRs in other SR proteins. Conversely SLR2-7 has helical and θ222/208 values inconsistent with spectrin, and transition temperatures significantly higher than canonical spectrins, suggesting the AD contributes uniquely to the thermal properties of nesprin-1.
Table 2.
Biophysical properties of SLRs, canonical SRs, nesprin-1α and spectrin.
| Properties | SLR | SR | Nesprin-1α | Spectrin |
|---|---|---|---|---|
| Constitution | 99~108 aa | ~106 aa | SLRs+AD | SRs |
| Helicity | 74~78% | 60%~80% | 83% | 60%~80% |
| Tm | 45~75°C | 21~82°C | 73°C | a49.5°C, b58°C |
| Multiple transitions | + | + | − | − |
| Cooperative folding | + | + | + | + |
erythroid spectrin;
non-erythroid spectrin.
Hydrodynamic diameters measured by DLS generally fall between protein length and width, probably closer to length for nesprins given their extended conformation.45,46 For bovine serum albumin (molecular dimensions ~5.5 × 5.6 × 12 nm50), our DLS-measured hydrodynamic diameter was 9.6 nm, close to the reference value of 9.5 nm (manufacturer standard). From the DLS data (Table 1 and Figure 4) we extracted information about the dimerization of nesprin-1α fragments in these buffer conditions. Our DLS results do not indicate actual size, since nesprin fragments may have a skewed hydrodynamic diameter making them appear larger; however the sizes and single- or bi-modal distributions of different nesprin molecules can be compared. Nesprin-1α were reported to form antiparellel homodimers, via SLR3 and SLR5.16 Our DLS results support this idea, since the measured size differences between SLR2-7ΔAD monomers (54 nm) and dimers (76 nm) and SLR2-7 monomers (58 nm) and dimers (99 nm) can be explained by anti-parallel homodimerization via SLR3 and SLR5. The SLR2-7 dimer (99 nm) is larger than the SLR2-7ΔAD dimer (76 nm), whereas the corresponding monomers of SLR2-7 (58 nm, 58°C) and SLR2-7ΔAD (54nm, 42°C) are roughly the same size. This similarity suggests the AD, in the context of SLR2-7 monomers, is disordered and less anisotropic, consistent with our other data suggesting the AD is mostly disordered by itself, but interacts within full-length SLR2-7 dimers to stabilize the entire cooperatively-coupled molecule.
The importance of structural integrity for experiments involving nesprin-1α
Our evidence shows that domain folding is strongly cooperative in nesprin-1α, typical of spectrin superfamily proteins, and that the AD confers an additional, unexpected level of stability to larger nesprin fragments that may be crucial for the functional integrity of native nesprins. Our results appear to reconcile apparently-conflicting results from two previous studies, which tested either single domains of nesprin-1α20 or larger fragments.16 Mislow et al. reported that the full SLR1-7 molecule bound emerin very tightly (equilibrium affinity of 4 nM), and that SLR2-5 and SLR5-7 also bound emerin but with slightly weaker equilibrium affinities (53 nM and 250 nM, respectively); they concluded that optimal emerin binding required full-length nesprin-1α.16 Wheeler et al. tested emerin binding to the AD alone, SLRs 2–3, and individual SLR4, SLR5, SLR6 or SLR7 domains, and detected binding only to the AD.20 They also tested emerin binding to the AD alone or single SLRs of nesprin-2β, and detected binding only to one tested SLR.20 These results are all consistent with, and explained by, the strongly cooperative coupled folding behavior of nesprin-1α, which we conclude provides the optimal binding surface for emerin. Our findings strongly suggest that single SLR domains and AD alone lack the cooperatively coupled folding seen in polypeptides with tandem SLRs, and also lack stabilizing interactions between the AD and SLRs. A major prediction from our study is that cooperative folding may do three things: promote nesprin-1α dimerization, help nesprin-1α bind emerin (and potentially other partners) stably and with high affinity, and increase resistance to imposed force, including thermal and (we propose) mechanical stretching forces.
Nesprins are a newly-discovered family of proteins with emerging links to human disease. Our findings reveal the importance of maintaining the structural integrity of the whole molecule, or large fragments thereof, in future studies. This is particularly important given our thermal unfolding results for nesprin-1α: nesprin-1α appears to be much more thermodynamically stable and (we propose) much stiffer than both erythroid spectrin (Tm 49.5°C) and non-erythroid spectrin (αII and βII spectrin; Tm 58°C),28,29 which confer elasticity and deformability properties to the red blood cell membrane and other types of cells. The significantly higher Tm (73°C) and thermodynamically stable dimer structure of full length nesprin-1α (SLR2-7) suggests the AD confers unusual stiffness to nesprin-1α; its conservation during evolution suggests this stiffness is essential for its molecular functions.
Implications for ‘giant’ nesprins at the outer nuclear membrane
Nesprin-1α was previously reported to form anti-parallel homodimers, through binding between SLR3 and SLR5.16 Our results suggest these dimer interactions are strongly reinforced by the AD. Since this entire region including the AD is identical in the ONM-localized nesprin-1 giant isoform, and highly conserved in the nesprin-2 giant, we propose that ONM-localized nesprin-1 and nesprin-2 giants form similarly stable, potentially ‘knotted’, dimers as depicted schematically in Figure 6. Each nesprin giant has an N-terminal actin-binding domain and a C-terminal KASH domain that binds a SUN-domain protein in the lumenal space of the nuclear envelope.13,15 SUN-domain proteins are known to dimerize.51 Our ‘knotted dimer’ model for nesprins, suggests a nano-structural mechanism to mechanically balance their interactions with SUN-domain dimers, which would significantly enhance the mechanical stability of these NE-spanning LINC structures. We speculate that these AD-mediated ‘knots’ might be positively or negatively regulated by adaptive binding to alternative partners, such as emerin at the INM (for nesprin-1α) or unknown proteins that might contact nesprin giants at the ONM. Further protein engineering studies of this interesting domain, and its effects on SLRs, are strongly warranted.
Figure 6.
Speculative model extending our results for INM-localized nesprin-1α to the ONM-localized nesprin-1 giant isoform. Adaptive domain (AD)-mediated self-folding and stabilization of dimers is predicted to form INM- and ONM-localized “knots” that mechanically secure and balance the formation of NE-spanning LINC complexes with SUN-domain protein dimers.
Implications for nuclear mechanotransduction and disease
Spectrin superfamily proteins have two major roles in cells: as mechanical and elastic crosslinkers between cytoskeletal elements, and as linear scaffolds for protein complexes.5 Despite relatively weak homology (21–28%) between nesprin-1α, human α- and β-spectrins and dystrophin,33 our blast searches reveal that the ‘pure’ SLR-containing nesprin-1α polypeptide (SLR2-7ΔAD) is 42–45% similar to spectrins and other spectrin superfamily proteins, including dystrophin and Microtubule-Actin Crosslinking Factor 1 (MACF1). We therefore propose nesprin-1α has a scaffolding and crosslinking role, like other spectrin superfamily proteins. Nesprin-1α, located at the inner nuclear membrane, can directly bind many nuclear proteins including lamins, emerin, SUN1 and mAKAP.8,13,16,51,52 It is possible that during evolution, nesprins retained both SR-like functions (e.g. actin binding and scaffolding), and unique (e.g. adaptive domain) reinforcement mechanisms to help connect the cytoskeleton and nucleoskeleton. Compared to the ONM-localized nesprin-1 giant isoform, which has more SLRs and is probably more spectrin-like, we predict that shorter INM nesprins are much stiffer since they are more dominated by the adaptive domain. Indeed nuclei are stiffer than most cytoskeletal elements.53 We suggest nesprin-1α and other ‘short’ nesprins contribute significantly to this stiffness. Nesprin-1α’s potential role in force transduction in concert with lamins and/or emerin and contribution to Emery-Dreifuss Muscular Dystrophy are important topics for future investigation.
Acknowledgments
We gratefully acknowledge E. McNally (Univ. Chicago) for the nesprin-1α constructs, and colleagues at Carnegie Mellon: S.D. Chapman for help with recombinant protein production, T. Przybycien for advice and CD equipment, L. Walker for help with DLS, and A. M. Laurent and V. Lam for advice. This research was supported by funding from NIH (GM48646 to K.L.W.), a National Research Service Award Fellowship (F32 GM074502 to K.N.D.) and a Liang Jidian Fellowship (to Z. Z.).
References
- 1.Stewart CL, Roux KJ, Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- 2.Moir RD, Spann TP, Herrmann H, Goldman RD. Disruption of Nuclear Lamin Organization Blocks the Elongation Phase of DNA Replication. J Cell Biol. 2000;149:1179–1192. doi: 10.1083/jcb.149.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- 5.Young KG, Kothary R. Spectrin repeat proteins in the nucleus. BioEssays. 2005;27:144–152. doi: 10.1002/bies.20177. [DOI] [PubMed] [Google Scholar]
- 6.Holaska JM. Emerin and the Nuclear Lamina in Muscle and Cardiac Disease. Circ Res. 2008;103:36. doi: 10.1161/CIRCRESAHA.108.172197. [DOI] [PubMed] [Google Scholar]
- 7.Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J Biol Chem. 2000;275:31986–31995. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- 8.Mislow JM, Kim MS, Davis DB, McNally EM. Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J Cell Sci. 2002;115:61–70. doi: 10.1242/jcs.115.1.61. [DOI] [PubMed] [Google Scholar]
- 9.Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 10.Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, Karakesisoglou I, Korenbaum E. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp Cell Res. 2004;295:330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Crisp M, Burke B. The nuclear envelope as an integrator of nuclear and cytoplasmic architecture. FEBS Lett. 2008;582:2023–2032. doi: 10.1016/j.febslet.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Ragnauth C, Greener MJ, Shanahan CM, Roberts RG. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 2002;80:473–481. [PubMed] [Google Scholar]
- 15.Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 16.Mislow JM, Holaska JM, Kim MS, Lee KK, Segura-Totten M, Wilson KL, McNally EM. Nesprin-1α self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 2002;525:135–140. doi: 10.1016/s0014-5793(02)03105-8. [DOI] [PubMed] [Google Scholar]
- 17.Broers JLV, Ramaekers FCS, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- 18.Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau1 M, Toniolo D, Schwartz1 K. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nat Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 19.Fairley EA, Kendrick-Jones J, Ellis JA. The Emery-Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J Cell Sci. 1999;112:2571–2582. doi: 10.1242/jcs.112.15.2571. [DOI] [PubMed] [Google Scholar]
- 20.Wheeler MA, Davies JD, Zhang Q, Emerson LJ, Hunt J, Shanahan CM, Ellis JA. Distinct functional domains in nesprin-1α and nesprin-2β bind directly to emerin and both interactions are disrupted in X-linked Emery-Dreifuss muscular dystrophy. Exp Cell Res. 2007;313:2845–2857. doi: 10.1016/j.yexcr.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth C, Yi Q, Mellad JA, Warren DT, Wheeler MA, Ellis JA, Skepper JN, Vorgerd M, Schlotter-Weigel B, Weissberg PL, Roberts RG, Wehnert M, Shanahan CM. Nesprin-1 and -2 are involved in the pathogenesis of Emery-Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- 22.Puckelwartz MJ, Kessler E, Zhang YD, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, Pytel P, McNally EM. Disruption of nesprin-1 produces an Emery-Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci USA. 2005;102:4359–4364. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513:119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald RI, Pozharski EV. Free energies of urea and of thermal unfolding show that two tandem repeats of spectrin are thermodynamically more stable than a single repeat. Biochemistry. 2001;40:3974–3984. doi: 10.1021/bi0025159. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald RI, Cummings JA. Stabilities of folding of clustered, two-repeat fragments of spectrin reveal a potential hinge in the human erythroid spectrin tetramer. Proc Natl Acad Sci USA. 2004;101:1502–1507. doi: 10.1073/pnas.0308059100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhasin N, Law R, Liao G, Safer D, Ellmer J, Discher BM, Sweeney HL, Discher DE. Molecular Extensibility of Mini-dystrophins and a Dystrophin Adaptive Construct. J Mol Biol. 2005;352:795–806. doi: 10.1016/j.jmb.2005.07.064. [DOI] [PubMed] [Google Scholar]
- 28.An X, Guo X, Zhang X, Baines AJ, Debnath G, Moyo D, Salomao M, Bhasin N, Johnson C, Discher D, Gratzer WB, Mohandas N. Conformational stabilities of the structural repeats of erythroid spectrin and their functional implications. J Biol Chem. 2006;281:10527–32. doi: 10.1074/jbc.M513725200. [DOI] [PubMed] [Google Scholar]
- 29.An X, Zhang M, Salomao X, Guo X, Yang Y, Wu Y, Gratzer W, Baines AJ, Mohandas N. Thermal Stabilities of Brain Spectrin and the Constituent Repeats of Subunits. Biochemistry. 2006;45:13670–13676. doi: 10.1021/bi061368x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kusunoki H, Minasov G, MacDonald RI, Mondragon A. Independent movement, dimerization and stability of tandem repeats of chicken brain α-spectrin. J Mol Biol. 2004;344:495–511. doi: 10.1016/j.jmb.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 31.Batey S, Randles LG, Steward A, Clarke J. Cooperative folding in a multi-domain protein. J Mol Biol. 2005;349:1045–1059. doi: 10.1016/j.jmb.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 32.Law R, Carl P, Harper S, Dalhaimer P, Speicher DW, Disher DE. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys J. 2003;84:533–544. doi: 10.1016/S0006-3495(03)74872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114:4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- 34.Simpson JG, Roberts RG. Patterns of evolutionary conservation in the nesprin genes highlight probable functionally important protein domains and isoforms. Biochem Soc Trans. 2008;36:1359–1367. doi: 10.1042/BST0361359. [DOI] [PubMed] [Google Scholar]
- 35.Ortiz V, Nielsen SO, Klein ML, Discher DE. Unfolding a Linker between Helical Repeat. J Mol Biol. 2005;349:638–647. doi: 10.1016/j.jmb.2005.03.086. [DOI] [PubMed] [Google Scholar]
- 36.Imai Y, Matsushima Y, Sugimura T, Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahr SJ, Engel DE, Stayrook SE, Maglio O, North B, Geremiad S, Lombardic A, DeGradoa WF. Analysis and Design of Turns in α-Helical Hairpins. J Mol Biol. 2005;346:1441–1454. doi: 10.1016/j.jmb.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 38.Suenaga A, Okimoto N, Ebisuzaki T. Refolding molecular dynamics simulations of small- and middle-sized proteins in an explicit solvent. Molecular Simulation. 2002;28:337–357. [Google Scholar]
- 39.Street TO, Fitzkee NC, Perskie LL, Rose GD. Physical-chemical determinants of turn conformations in globular proteins. Protein Science. 2007;16:1720–1727. doi: 10.1110/ps.072898507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou NE, Zhu B, Kay CM, Hodges RS. The two-stranded α-helical coiled-coil is an ideal model for studying protein stability and subunit interactions. Biopolymers. 1992;32:419–426. doi: 10.1002/bip.360320419. [DOI] [PubMed] [Google Scholar]
- 41.Woolley GA, Wallace BA. Temperature dependant of the interaction of alamethicin helicies in membranes Biochemistry. Biochemistry. 1993;32:9819–9825. doi: 10.1021/bi00088a037. [DOI] [PubMed] [Google Scholar]
- 42.Brown OJ, Lopez SA, Fuller AO, Goodson T. Formation and Reversible Dissociation of Coiled Coil of Peptide to the C-Terminus of the HSV B5 Protein: A Time-Resolved Spectroscopic Analysis. Biophys J. 2007;93:1068–1078. doi: 10.1529/biophysj.106.100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obataya I, Sakamoto S, Ueno A, Mihara H. Design and synthesis of 3-helix peptides forming a cavity for a fluorescent ligand. Biopolymers. 2001;59:65–71. doi: 10.1002/1097-0282(200108)59:2<65::AID-BIP1006>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 44.Law R, Liao G, Harper S, Yang GL, Speicher DW, Discher DE. Pathway shifts and thermal softening in temperature-coupled forced unfolding of spectrin domains. Biophys J. 2003;85:3286–3293. doi: 10.1016/S0006-3495(03)74747-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Budzynski DM, Benight AS, LaBrake CC, Fung LWM. Dynamic light scattering investigations of human erythrocyte spectrin. Biochemistry. 1992;31:3653–3660. doi: 10.1021/bi00129a014. [DOI] [PubMed] [Google Scholar]
- 46.DeSilva TM, Harper SL, Kotula L, Hensley P, Curtis PJ, Otvos L, Speicher DW. Physical Properties of a Single-Motif Erythrocyte Spectrin Peptide: A Highly Stable Independently Folding Unit. Biochemistry. 1997;36:3991–3997. doi: 10.1021/bi962412j. [DOI] [PubMed] [Google Scholar]
- 47.Linding R, Jensen L, Diella F, Bork P, Gibson T, Russell R. Protein disorder prediction: Implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Radhakrishnan I, Perez-Alvarado G, Parker D, Dyson H, Montminy M, Wright P. Solution structure of the KIX domain of CBP bound to the trans-activation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 49.Wright P, Dyson H. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 50.Carter DC, Ho JX. Structure of serum albumin. Adv Protein Chem. 1994;45:153–203. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- 51.Pare GC, Easlick JL, Mislow JM, McNally EM, Kapiloff MS. Nesprin-1α contributes to the targeting of mAKAP to the cardiac myocyte nuclear envelope. Exp Cell Res. 2005;303:388–399. doi: 10.1016/j.yexcr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 52.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear Shape, Mechanics, and Mechanotransduction. Circ Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]