Abstract
In a companion paper (Tombolato, F.; Ferrarini, A.; Freed, J.H., ... ) a study of the conformational dynamics of methanethiosulfonate spin probes linked at a surface exposed α-helix has been presented. Here, on the basis of this analysis, X-band ESR spectra of these spin labels are simulated, within the framework of the Stochastic Liouville Equation methodology. Slow reorientations of the whole protein are superimposed on fast chain motions, which have been identified with conformational jumps and fluctuations in the minima of the chain torsional potential. Fast chain motions are introduced in the SLE for the protein reorientations through partially averaged magnetic tensors and relaxation times calculated according to the motional narrowing theory. The 72R1 and 72R2 mutants of T4 lysozyme, which bear the spin label at a solvent-exposed helix site, have been taken as test systems. For the side-chain of the R2 spin label, only a few non-interconverting conformers are possible, whose mobility is limited to torsional fluctuations, yielding almost identical spectra, typical of slightly mobile nitroxides. In the case of R1, more complex spectra result from the simultaneous presence of constrained and mobile chain conformers, with relative weights which can depend on the local environment. The model provides an explanation for the experimentally observed dependence of the spectral lineshapes on temperature, solvent, and pattern of substituents in the pyrroline ring. The relatively simple methodology presented here allows the introduction of realistic features of the spin probe dynamics into the simulation of ESR spectra of spin labeled proteins; moreover, it provides suggestions for a proper account of such dynamics in more sophisticated approaches.
I. INTRODUCTION
Different kinds of ESR experiments have been devised to probe the structural features of proteins, as well as the properties of different protein environments and the dynamical processes occurring in these systems, which are characterized by a wide range of time-scales.1,2 The X-band (9GHz) spectra have typical shapes, usually three broadened lines, indicating the absence of enough fast and large amplitude motion to fully average out the anisotropy of the g and A hyperfine tensors. Simple inspection of the spectra can provide an insight into the mobility of the spin probe, at least at a qualitative level; so, for example, differences in line broadening observed for spin labels at different sites are taken as indicators of low or high mobility at the local site of the spin label. On the other hand, accurate lineshape analysis, which is capable of providing quantitative information, is not a simple task in the presence of complex dynamics. Namely, a variety of processes can produce nitroxide reorientation: they range from overall protein tumbling and refolding processes, to backbone fluctuations and motions of the tether.
The analysis of ESR lineshapes can be carried out by the Stochastic Liouville Equation (SLE) formalism, to describe the time evolution of the density matrix for the spin degrees of freedom, treated quantum-mechanically, coupled to the probability density for the orientational degrees of freedom, described as classical stochastic variables.3–5 Efficient implementations make use of the expansion on a complete basis set, which is the direct product of basis sets for the spin and the classical variables. This methodology has been widely used and software packages are available for the simulation of ESR lineshapes of spin probes undergoing Brownian rotational motion.6 In principle there is no restriction on the number of classical stochastic variables, and multivariate Fokker-Planck equations for various kinds of motional models can be formulated. In practice, however, the rapidly growing dimension of the stochastic Liouville matrix needed to represent many stochastic variables, and the increase in computational time, pose substantial limits to the complexity of motions which can be treated. So, only simplified or approximate treatments of the variety of processes affecting the spin relaxation of nitroxide spin labels attached to proteins are currently feasible. Thus some approaches have been proposed which include some key features of the systems and still circumvent the computational limits.
When the overall motions are so slow that their dynamic effects can be neglected, the so called Microscopic Order Macroscopic Disorder model (MOMD) can be used.7 Here the spectrum is calculated for a static isotropic distribution of rotors, undergoing Brownian motion in an ordered environment. The nitroxide spin probe is identified with the rotor and the constraints deriving from the chain geometry and from the steric hindrance of the environment are taken into account through an orienting potential. The MOMD approach has been used for proteins in high viscosity media and/or low temperature,8 as well as for very high frequency ESR,9 but of course it is no longer suitable when the rigid limit cannot be assumed for the protein motions.
The overall protein reorientation can be taken into account by a more sophisticated approach, known as the Slowly Relaxing Local Structure (SRLS) model, wherein the rotational diffusion of a rotor, identified with the spin probe, is superimposed onto protein rotations.10–12 Even in this case local constraints are introduced through an orienting potential. This model has been used for the simultaneous simulations of 9GHz and 250 GHz spectra of T4 lysozyme (T4L) in water.13 A large number of parameters enter the model: the diffusion tensor of the protein and that of the nitroxide in the protein frame, in addition to the coefficients specifying the orienting potential experienced by the spin probe. It follows that a large set of experimental data are needed.
A valuable feature of the SLE methodology is that it provides a relatively simple description of a complex system, in terms of a few physical properties, that are introduced in a parametric way. These parameters can then be optimized by simulating experimental spectra with best fitting procedures. The drastic reduction in the dimension of the problem deriving from a suitable selection of the variables deemed relevant, which characterizes standard SLE methods, has the drawback that mapping the results into precise information on the protein is not necessarily straightforward. So, for instance, the interpretation of diffusion coefficients and order parameters for the spin probe in terms of specific molecular motions and local constraints can remain rather vague, especially given the limited sensitivity to the details of the dynamics provided by conventional ESR.9,13,14
Alternative approaches have been proposed to obtain a more detailed modeling of the systems. The dynamics of the classical degrees of freedom can be obtained in the form of trajectories by Molecular Dynamics (MD) or Brownian Dynamics simulations.15–19 A detailed picture of the local environment probed by the spin label and of the various processes modulating its orientations can be attained at a computational cost which can be very high, due to the huge number of degrees of freedom. Conformational transitions in the side-chain occur on the nanosecond time-scale; therefore long trajectories are needed, even to just sample a few conformational sites. The computational demand can be significantly reduced if strong constraints are introduced, as in MD simulations where all degrees of freedom but those of the spin label are frozen.20
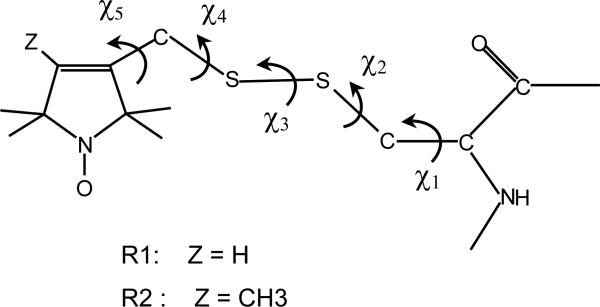
In a companion paper,21 henceforth denoted as I, we have examined the internal motions of methanethiosulfonate (MTSSL), a typical spin label whose structure is shown in Figure 1, attached to a model α-helix. We have found that the system can be described in terms of a limited number of conformers. They undergo fast, small amplitude torsional oscillations and side-chain isomerizations. In water at room temperature the former are estimated to have characteristic times shorter than a nanosecond. The latter occur on a range of time-scales which can be of the order of one nanosecond if rotations of the terminal chain bonds are involved, but are significantly slower for the bonds closer to the protein backbone. Thus, on the timescale of ESR spectra, we can distinguish two kinds of conformers: ones that are connected to other conformers by fast rotations around the terminal chain bonds, while the others are linked only by very slow transitions involving internal bonds, so they can be considered as isolated.
Figure 1.
Structure of the spin labels considered in this study. They are obtained by reaction of the sulfhydryl group of a cysteine with 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-(methyl)methanethiosulfonate (R1) and 1-oxyl-2,2,4,5,5-pentamethyl-3-pyrroline-3-(methyl)methanethiosulfonate (R2). The five dihedral angles defining the chain conformation are shown.
Here, the information derived from this analysis is used to investigate the effects of conformational dynamics on X-band (9GHz) ESR lineshapes. A simplified approach has been used, exploiting the time-scale separation between the various processes producing reorientation of nitroxide. Fast motions, identified with torsional oscillations and isomerizations of the terminal side-chain bonds, are treated according to the fast motional ESR relaxation theory.22,23 Spectral lineshapes are then obtained by introducing partially averaged magnetic tensors and T2 relaxation times, obtained in this way, into the SLE for slow protein tumblings.3,24–26 The 72R1 and 72R2 mutants of T4L, for which experimental data are available in the literature,8,9,13 are taken as test cases; the effects of side-chain dynamics on the spectral lineshape have been analyzed as a function of temperature and solvent. This approach is shown to be satisfactory for 9 GHz ESR spectra, and the generalizations needed to interpret high frequency spectra (250 GHz) are considered in the Discussion section below.
The paper is organized in the following way. In the next Section an approximate treatment for the ESR spectrum of a nitroxide spin probe in the presence of fast side-chain motions and slow overall protein reorientations is presented. Then the effects of the chain dynamics on ESR spectra of the 72R1 and the 72R2 mutants of T4L are presented. In the final Section the results of our investigation are discussed, the conclusions of this work are summarized and future outlooks are sketched.
II. ESR SPECTRUM OF A NITROXIDE SPIN LABEL
According to linear response theory, the absorbed power in an ESR experiment is the imaginary part of the Fourier-Laplace transform of the time dependent complex magnetization, M+ = MX + iMY:
| (1) |
where Tr indicates the trace over the spin variables and the bar implies an orientational average; is an electronic spin operator and ρ(Ω,t) is the spin density matrix, whose time evolution is described by the SLE:3–5
| (2) |
Here L(Ω) is the Liouville superoperator associated with the spin Hamiltonian and Γ(Ω) is a stochastic operator accounting for the orientational dynamics of the spin label. It describes the time dependence of the Euler angles Ω = (α, β, γ), specifying the orientation of the magnetic frame, MF, in the laboratory frame, LF.
The spin Hamiltonian of a nitroxide radical has the form:
| (3) |
with the Zeeman and hyperfine terms, which contain the g tensor and the hyperfine tensor A, respectively. Here and are the electronic and nuclear spin operators, and B0 represents the static magnetic field, whose orientation is chosen as the Z-axis of the laboratory frame (LF). The spin Hamiltonian can be expressed as the sum of a contribution, Hiso, independent of the orientation of the spin probe, and an orientation dependent part, δH, which fluctuates in time, due to rotations of the nitroxide:
| (4) |
It is convenient to make use of the irreducible spherical tensor formalism:27
| (5) |
| (6) |
where μ = g, A is used to denote the Zeeman and hyperfine terms, and are components of the magnetic tensors and the electronic or nuclear spin operators, respectively, all expressed in the laboratory frame: . It is convenient to express the magnetic tensors in the magnetic frame, MF; then, the orientation-dependent part of the Hamiltonian can be rewritten as
| (7) |
where are Wigner rotation matrices having as their argument the Euler angles for the rotation from the laboratory to the magnetic frame.27
A. Superposition of fast and slow motions
The orientation with respect to the laboratory frame of a spin label attached to a protein varies in time under the effect of several motions: dynamics of the chain linking the probe to the protein, fluctuations of the backbone, overall protein tumbling, domain motions,... Thus, the stochastic operator Γ in eq 2 should depend on a large number of variables, which would render the SLE too complex to treat. A simplified approach can be adopted, under the condition of time-scale separation between motions, if some of these can be assumed to be fast on the ESR time-scale.24–26 The dynamics of a given variable is assumed to be fast if its characteristic frequency, 1/τ, is larger than the magnetic anisotropy it modulates, Δω, i.e. 1/τ >> Δω. In the case of X-band (9GHz) ESR experiments, such a condition is satisfied by motions with correlation times shorter than about one nanosecond. If the fast-motional regime can be assumed for the side-chain motion, the ESR spectrum of a nitroxide spin label, in a protein at the orientation ΩD in the laboratory frame, is given by three lines whose positions are determined by the spin Hamiltonian eq 4, partially averaged by the side-chain dynamics:
| (8) |
where the upper bar denotes the average over the fast motions.
The spectral linewidths can be calculated according to the motional narrowing theory:22,23
| (9) |
with M, M′ = 0, ±1 indicating nuclear spin states, for the electron spin states connected by electronic spin transitions. The coefficients A(ΩD), B(ΩD), C(ΩD) are defined as:
| (10) |
| (11) |
| (12) |
where is the zero-frequency spectral density:
| (13) |
Here represents fluctuations of tensor components with respect to their partially averaged values, . If fast side-chain dynamics is superimposed to slow overall motions, the former can be included in the modified SLE:25
| (14) |
where L̄(ΩD) is the Liouville superoperator associated to the Hamiltonian partially averaged by the chain dynamics, eq 8, is the orientation dependent linewidth defined in eq 9 and Ω(ΩD) is the stochastic operator describing the slow protein tumbling.
B. Fast side-chain motions: average magnetic tensors and spectral linewidths
Explicit expressions for the partially averaged quantities entering the spin Hamiltonian, eq 8, and the coefficients A, B, C, eqs 10-12, can be obtained on the basis of a model for the side-chain motions. Considering the case of the MTSSL spin label linked to an α-helix, we have shown in ref.21 that a restricted number of possible conformers can be identified. All undergo fast and small-amplitude torsional oscillations, and only some of them can be connected by conformational jumps. These are characterized by transition frequencies occurring on a range of time-scales; the fastest, of the order of 1 ns−1 in water, involve rotations around the Sδ-C bond (χ4 jumps), whereas rotations around the Cβ-Sγ bond (χ2 jumps) are about an order of magnitude slower, and those involving the χ1 dihedral and the disulfide bond (χ3 transitions) are even slower. For the sake of simplicity, the approximation will be adopted that, according to their rate, conformational transitions can be assumed to be either fast or frozen. So, a spectrum will be calculated as a sum of contributions, each due to a single conformer, or to a set of conformers whose interconversion is fast on the time-scale of the experiment under examination. A contribution is obtained by solving a modified SLE, eq 14, with partially averaged Hamiltonian and spectral linewidth calculated for a given conformer or set of conformers. In the following, expressions for the quantities appearing in the SLE will be derived.
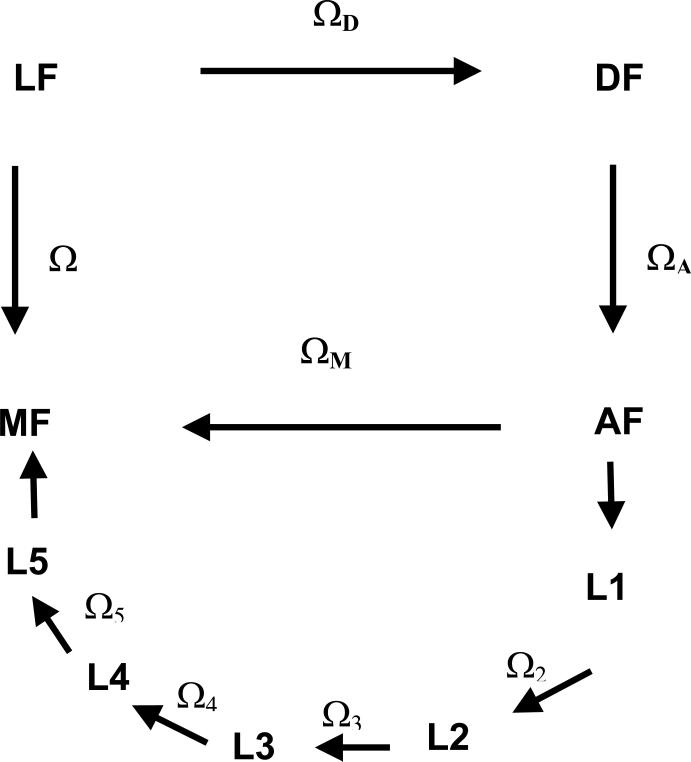
For this purpose, the rotation from the Laboratory (LF) to the Molecular (MF) frame is conveniently decomposed into a series of transformations, as shown in Figure 2. The DF frame is the Principal Axis System (PAS) of the protein diffusion tensor, AF is a frame fixed on the spin labeled amino acid residue, with the origin on the Cα carbon. The reference systems designed as Li are local frames with the origin on the side-chain carbons, whose orientation changes with the configuration of the side-chain. Finally, the MF frame is fixed in the nitroxide moiety, with the origin on the N nucleus, the zM-axis along the N-pz orbital and the xM-axis parallel to the N-O bond. Here we are focussing on the case of labeled sites in α-helices; neglecting the possibility of significant changes in the protein shape, as could be brought about by domain reorientation, and that of fluctuations of the protein backbone, the protein diffusion tensor and the orientation of AF with respect to DF will be assumed to be time independent. The validity of this assumption will be assessed by comparing theoretical and experimental results.
Figure 2.
Reference frames considered in this work.
LF: laboratory frame, with the ZL axis parallel to the magnetic field B0.
DF: PAS of the rotational diffusion tensor of the protein.
AF: frame of the amino acid residue, with the origin on the Cα bringing the nitroxide side-chain, the zA-axis perpendicular to the plane of the N-Cα-CO atoms and the xA-axis on the plane containing the zA-axis and Cα-Cβ bond.
Li: local frame, with the zLi axis parallel to the i-th chain bond and xLi in the plane of the preceding chain bond for the eclipsed configuration with χi−1 = 0° .
MF: magnetic frame, with the origin on the nitroxide N nucleus, the zM-axis along the N-pz orbital and the xM-axis parallel to the N-O bond.
The averaged Wigner rotation matrices appearing in eq 8 can be expressed as:
| (15) |
where the addition theorem of Wigner rotation matrices27 has been used to made explicit the dependence on variables which are assumed to be independent of the motions in the side-chain, i.e. ΩD and ΩA, and the average has been taken only over the fast chain motions. The matrix elements , which will be designated as order parameters, reflect the orientational distribution of the magnetic frame in the frame of the amino acid residue. They are related to the magnetic tensors partially averaged by chain motions:
| (16) |
Even for the evaluation of the spectral densities, eq 13, it is convenient to make explicit the dependence upon the variables which are modulated by chain motions; the following form is obtained:
| (17) |
where jq,k,q′,k′ is the reduced zero-frequency spectral density:
| (18) |
with .
Partially averaged magnetic tensors and reduced spectral densities for torsional oscillations and conformational jumps can be calculated as explained in I; the expressions to be used will be summarized in the following.
Torsional oscillations The order parameters , accounting for nitroxide reorientation produced by bond fluctuations about the configuration of the Jth chain conformer, are obtained by integrating over the probability distribution p(δχJ), with δχJ = χ − χJ being the displacements from the equilibrium dihedral angles, χJ. Under the assumption of independent bond contributions, the torsional probability can be factorized into single bond contributions; using the harmonic approximation for the torsional potential about a minimum, these can be expressed as:
| (19) |
where is the curvature of the torsional potential for the χi dihedral, calculated at . Then, the order parameters can be calculated as:21
| (20) |
where are the Euler angles for the local transformation Li → Li+1. Under the same conditions, the following form is obtained for the reduced spectral density eq 18:
| (21) |
The torsional angle fluctuations can be described by a Fokker-Planck-Smoluchowski equation, which is conveniently solved by the normal mode transformation: δχJ = Uy.28–30 The transformation matrix U is defined by , where DJ is the diffusion tensor for the side-chain in the Jth conformation. It is related to the friction matrix opposing bond rotations, ξJ, by the Stokes-Einstein relation . The presence of this matrix, which couples all torsional angles, makes it impossible to factorize the correlation function appearing in the last row of eq 21 into single bond contributions. However the correlation function can be easily calculated in terms of the normal modes:
| (22) |
where , . By truncating the Taylor series expansion of the exponentials at the first terms, the following expression is obtained for the correlation functions appearing in the product:
| (23) |
with , , and . Thus the reduced spectral density eq 21 can be easily calculated using the expression:
| (24) |
where the prime indicates that the term with all i values equal to zero is excluded from the summations.
Conformational jumps For conformers which, in addition to torsional oscillations, undergo also conformational jumps, the order parameters in eq 15 can be approximated as21
| (25) |
where the sum is extended to all the interconverting conformers, are order parameters calculated by averaging over the torsional fluctuations of the Jth conformer, eq 20, and PJ is the conformer probability. This is defined as , where VJ and are the potential energy and the matrix of the second derivatives of the potential energy for the Jth conformer, respectively.21
Spectral densities can be obtained as31
| (26) |
where is the symmetrized transition matrix for conformational jumps. Its elements are defined as , where −WJ J′ is the J′→J transition rate.21 The vector v in eq 26 has the Jth element defined as . Conformer probabilities can be evaluated in terms of the chain energetics, while transition probabilities have been determined on the basis of torsional potentials and friction opposing bond rotations,21,31 within the framework of the multidimensional Kramers theory.32,33
III. RESULTS
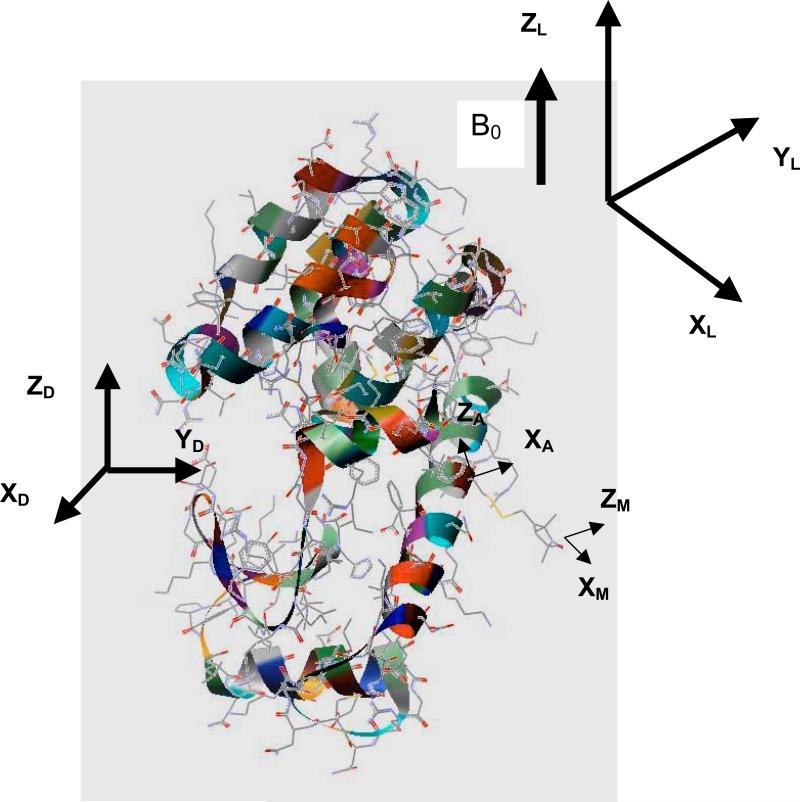
With the information derived from the conformational dynamics of the side-chain, we have calculated the ESR spectra of the R1 and R2 mutants of T4L, with the spin probe linked at site 72, which is solvent-exposed and located in the middle of a 5-turn α-helix, as shown in Figure 3. The theoretical results are compared with experimental spectra recorded at different temperatures and in different solvents, which are available in the literature.8,13
Figure 3.
Structure of T4L (3LZM.pdb)35 with the nitroxide probe linked at site 72. Some of the reference frames defined in this study are shown: laboratory frame (LF), diffusion frame (DF), frame of the amino acid residue (AF), magnetic frame (MF).
A spectrum has been obtained as the sum of contributions, each due to an independent set of conformers, with suitable weights. Each spectral component is calculated as explained in the previous Section. Partially averaged magnetic tensors and spectral densities modulated by chain dynamics, are introduced into the slow-motion simulation programs of the NLLS software package,6 which has been modified to adapt to the present model (see Appendix). The Brownian diffusion model has been used for overall protein reorientation.
The input parameters for the calculation of each spectral contribution can be summarized:
g and A tensors of the spin-label;
intrinsic linewidth;
rotational diffusion tensor D0 of the protein;
Euler angles ΩA, defining the orientation of the amino acid residue frame (AF) with respect to the diffusion frame (DF).
All parameters are obtained either by modeling or from experimental data. Magnetic tensors, expressed in the MF frame for the 72R1 mutant of T4L, are taken from the literature;13 they are reported in Table 1. The diffusion tensor D0 has been calculated with a hydrodynamic model,34 on the basis of the crystal structure of wild type T4L (3LZM.pdb).35 Also the values of the Euler angles ΩA have been obtained from the X-ray structure. Principal values of the diffusion tensor in water solution at T=298 K (η= 0.9 mPa · s) are reported in Table 1, along with the ΩA Euler angles. A slightly anisotropic diffusion tensor is obtained, in agreement with the protein shape. An intrinsic Lorentzian linewidth of 1 Gauss has been used in all lineshape calculations,13 in addition to the linewidth contribution deriving from chain motions, calculated as explained in the previous Sections.
TABLE I.
Magnetic tensors for the nitroxide in the 72R1 mutant of T4L;13 components of the rotational diffusion tensor calculated for T4L in water at T=298 K (η = 0.9 mPa · s) and angles from the DF to the AF frame.34
| gxx=2.00803 | gyy=2.00582 | gzz=2.00218 |
| Axx=6.42 G | Ayy=5.95 G | Azz=35.83 G |
| D0,∥ = 1.2 · 107s–1 | D0,⊥ = 1.9 · 107s–1 | |
| αA = –68° | βA = 166° | γA = –89° |
A. R2 spin probe
For the R2 spin label, large amplitude χ4 and χ5 rotations are hampered by the steric hindrance deriving from the methyl substituent at the 4-position in the pyrroline ring. So, within our approximation only isolated conformers are possible, which undergo fast and small amplitude torsional fluctuations.21 Ten conformers can be identified, which are listed in Table 2. A spectrum is calculated as the sum of ten independent contributions, each due to a single conformer, with a weight, PJ, estimated on the basis of the torsional potential.21 For a given conformer, reorientation of the nitroxide with respect to the magnetic field is produced by rotational diffusion of the whole protein and librations in the minima of the torsional potential.
TABLE II.
Torsional angles and probabilities for the sterically allowed conformers of the nitroxide side-chain of the R1 spin label, at a site located in the middle of a poly-Ala helix. Statistical weights were calculated at T=298 K on the basis of the single bond torsional potential, V′ in eq 1. Conformers allowed for the R2 spin probe are denoted by an asterisk; probability and χ5 values for these are reported in parentheses.
| conformer | χ1(°) | χ2(°) | χ3(°) | χ4(°) | χ5(°) | PJ |
|---|---|---|---|---|---|---|
| C1* | -60 | -75 | -90 | 180 | +77 (+85) | 0.143 (0.201) |
| C2* | -60 | -75 | -90 | 180 | -77 (-85) | 0.143 (0.201) |
| C3 | -60 | -75 | -90 | -75 | +100 | 0.045 |
| C4 | -60 | -75 | -90 | -75 | -8 | 0.074 |
| C5* | -60 | 180 | 90 | 180 | +77 (+85) | 0.187 (0.263) |
| C6* | -60 | 180 | 90 | 180 | -77 (-85) | 0.187 (0.263) |
| C7 | -60 | 180 | 90 | +75 | -100 | 0.059 |
| C8 | -60 | 180 | 90 | +75 | +8 | 0.098 |
| C9* | 180 | 180 | -90 | 180 | +77 (+85) | 0.009 (0.013) |
| C10* | 180 | 180 | -90 | 180 | -77 (-85) | 0.009 (0.013) |
| C11 | 180 | 180 | -90 | -75 | +100 | 0.003 |
| C12 | 180 | 180 | -90 | -75 | -8 | 0.005 |
| C13* | 180 | 180 | 90 | 180 | +77 (+85) | 0.009 (0.013) |
| C14* | 180 | 180 | 90 | 180 | -77 (-85) | 0.009 (0.013) |
| C15* | 180 | 75 | 90 | 180 | +77 (+85) | 0.007 (0.010) |
| C16* | 180 | 75 | 90 | 180 | -77 (-85) | 0.007 (0.010) |
| C17 | 180 | 75 | 90 | +75 | -100 | 0.002 |
| C18 | 180 | 75 | 90 | +75 | +8 | 0.004 |
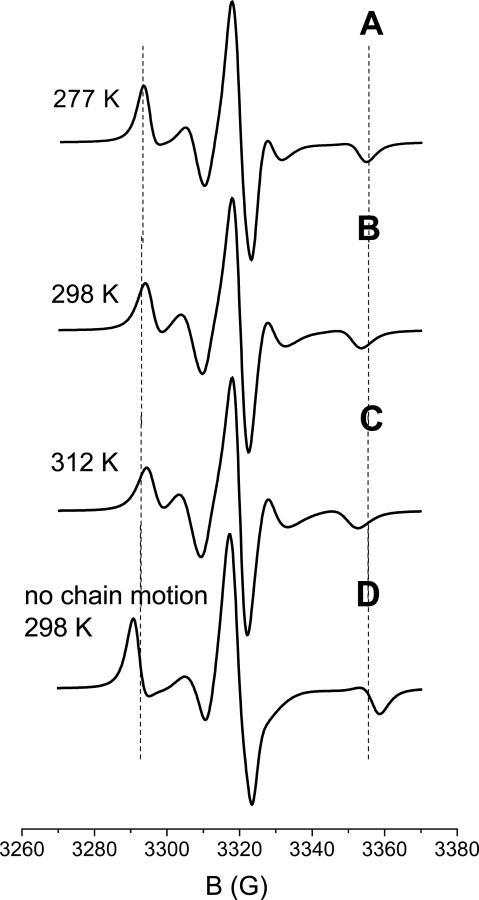
Figures 4-A to 4-C show the spectra calculated for 72R2-T4L in a 30% wt sucrose solution at the temperatures of 277 K, 298 K and 312 K. The overall diffusion tensor of the protein has been estimated scaling the values reported in Table 1 according to the change in viscosity of the medium.36 Thus, the following values have been assumed: D0,⊥ = 0.3 · 107 s-1 and D0,∥ = 0.47 · 107s−1 (T=277 K), D0,⊥ = 0.6 · 107 s-1 and D0,∥ = 0.95 · 107s−1 (T=298 K), D0,⊥ = 0.8 · 107 s-1 and D0,∥ = 1.35 · 107 s−1 (T=312 K). Due to the limited amplitude (about 10° for each bond)21 and the short correlation times of such motions, the reduced spectral densities, defined in eq 18, are small, of the order of 10−11s. This yields very small spectral densities, given by eq. 17, corresponding to a 10 mG linewidth contribution at X-band. Even at frequencies as high as 250 GHz, such values remain smaller than the inverse of the magnetic anisotropies that they average out, thereby justifying the fast-motional approximation for the torsional motions. The rotational diffusion coefficients of the protein in the viscous medium are quite small; the spectra of the ten conformers are similar, and not very different from the spectrum of a powder sample of a nitroxide spin probe with the g and A tensors partially averaged by torsional oscillations.
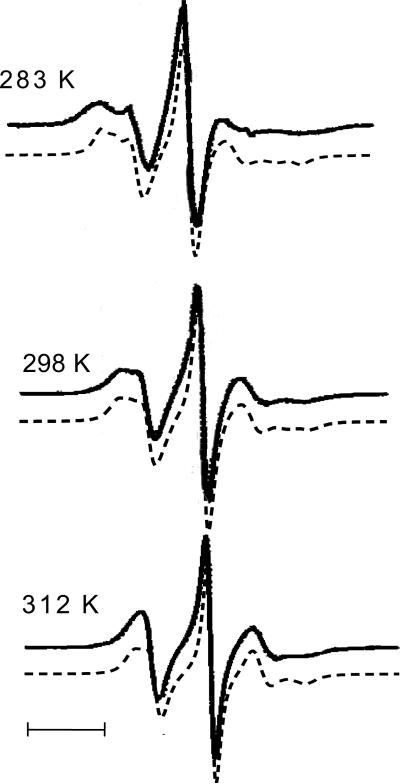
Figure 4.
Spectra calculated for the 72R2 mutant of T4L in a 30% wt sucrose solution at different temperatures (A-C). The spectrum calculated at T=298 K in the absence of chain motions is shown in (D). Experimental spectra are reported in Fig. 5(b) of ref.8. The dashed vertical lines are drawn as a guide to the eye for comparison.
Although of small amplitude, the torsional oscillations are sufficient to produce sizeable effects on the spectral lineshapes via partial averaging of the magnetic tensors. This clearly appears from comparison of the spectrum calculated at T=298 K, in Figure 4-B, with that obtained without any averaging, which is shown in Figure 4-D. The differences between the two lineshapes match those experimentally found between the spectra recorded for 72R2-T4L and for another mutant, having an MTSSL derivative with a bulky substituent at the 4-position of the pyrroline ring, which is likely to freeze almost completely the chain motions (see Figure 6(g) in ref.8). The small differences experimentally observed between the spectra of 72R2 at various temperatures, (see Figure 5(a) in ref.8), are also reflected by the lineshapes reported in Figures 4-A to 4-C. In our model, changes result from: (i) the decrease of viscosity, which brings about a twofold increase in the overall diffusion rate of the protein and the reduction, by a factor of two, of the spectral densities for chain oscillations (actually, the latter effect is scarcely detectable, in view of the small magnitude of such spectral densities); (ii) the increase in amplitude of torsional oscillations, which for each angle is of the order of 1° for a 20° temperature increase. We can see that the main features of the experimental spectra are well predicted, without requiring a significant role of the other processes, which we have not taken into account in our model. In addition to transitions involving χ2 to χ5 rotations in the side-chain, which we found to be very slow,21 these could include local backbone fluctuations and other interesting dynamical modes of the protein. Thus, our results suggest that backbone motions at sites which, like 72 in T4L, are located at the center of a well-structured α-helix will not be major contributors to the 9GHz ESR spectra.8,13 In the future, the approach presented here could be extended to include local fluctuations and protein dynamics, e.g. in the form of normal modes,37,38 for a more general analysis of ESR spectra. Further investigation, combining theoretical and experimental work, possibly with the use of multifrequency experiments and analysis, could provide new insights into the dynamics of proteins.
Figure 6.
Spectra calculated for the 72R1 mutant of T4L in 30% wt sucrose solution at different temperatures (dashed) and the corresponding experimental spectra taken from Figure 5(a) in ref.8 (solid). A small vertical offset between calculated and experimental spectra is introduced to help their comparison. The bar corresponds to a field scan of 20 Gauss.
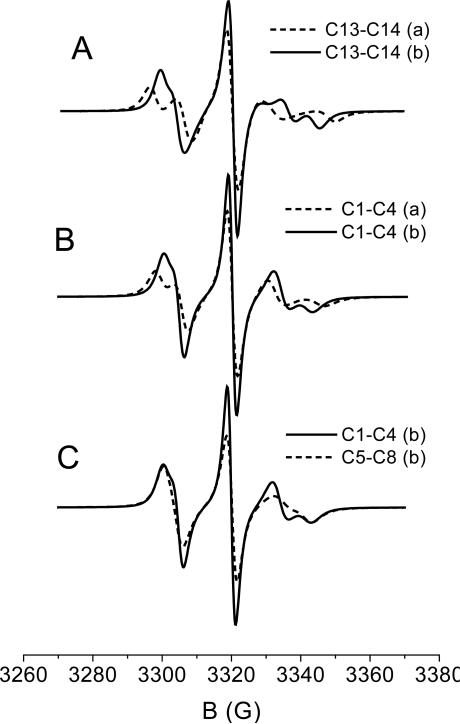
Figure 5.
Contributions to the spectrum of R1 spin label deriving from different sets of conformers, as indicated in the labels, and under different conditions for the χ5|χ4=t distribution. (a): Gaussian distribution about the minima, with root mean square amplitude ; (b): distribution derived from the calculated torsional potential (ROHF/6-31G**),21 in the range 0° to ±120°. Parameters suitable for the 72R1-T4L mutant in 30% wt sucrose solution at T=298 K are used.
B. R1 spin probe
The side-chain of the R1 spin probe is less sterically hindered than that of R2, and its dynamics becomes more complex, due to the possibility of conformational transitions.21 For the sake of simplicity, the fast-motional approximation has been assumed for torsional librations and χ4 and χ5 jumps, whereas conformational transitions involving the χ1, χ2 and χ3 dihedrals are considered to be slow enough that they could be been neglected. This choice can be justified by the observation that conformational jumps occurring through rotations of internal bonds are slower, by virtue of the higher friction opposing them; moreover, since they involve the displacement of larger chain portions, they are more likely to be hampered by neighboring chains. Thus, five independent blocks of conformers can be identified, which will be designated according to the notation reported in Table 2. The two conformers in the block C13-C14 interconvert through χ5 jumps, whereas the four conformers in each of the blocks C1-C4, C5-C8, C9-C2, C14-C18 are connected by χ4 and χ5 rotations. In 30% wt sucrose solutions at T=298 K, reduced spectral densities are of the order of 5 · 10−11s. Thus, given the magnitude of the magnetic anisotropies averaged by the two kinds of side-chain motions, the motional narrowing approximation can be adopted for them, in the case of 9GHz ESR spectra;21 however, at higher frequency such an approximation is likely inappropriate.
An important difference with the case of R2 is that the independent blocks of conformers of R1 yield significantly different spectra. The situation is made even more complicated by the possible effects of local interactions on the conformer distribution and the uncertainty in the torsional potential V (χ5) when χ4 is in the trans state, (χ5|χ4=t). Actually, a barrier of the order of kBT is predicted from ab initio calculations at the ROHF/6-31G** level;21 however such a value is sensitive to the level of the calculations, and also the possibility exists that it can be affected by interactions involving the pyrroline ring.39 Examples of the spectra which can be obtained are shown in Figure 5. In all cases the values D0,⊥ = 0.6 · 107 s-1 and D0,∥ = 0.95 · 107 s−1, appropriate for a 30% wt sucrose solution at T=298 K, are used. Given the uncertainty in the torsional profile for χ5|χ4=t and the fact that adoption of Gaussian distributions centered at the minima is not fully justified if barriers are not sufficiently high, we have considered two different torsional distributions for the χ5|χ4=t dihedral angle. In case (a) Gaussian distributions with oscillations of root mean square amplitude about the minima at ±77° are assumed; this value has been derived from the curvature of the torsional potential in the minima. In case (b) a wider distribution has been assumed, obtained from the calculated V(χ5) torsional potential in the range 0° to ±120°. The exact form of the distribution is not relevant; analogous results would be obtained with a Gaussian distribution centered in the minima, of root mean square amplitude . Figure 5-A shows the spectra calculated for the C13-C14 pair of conformers with the two choices; they look quite different, with case (b) exhibiting a higher degree of averaging, as expected. Figure 5-B shows the lineshapes obtained for the block of conformers C1 to C4, with the same choices, (a) and (b), for the χ5|χ4=t distribution. A similarity to the spectra obtained for the C13-C14 pair clearly appears, though the presence of χ4 jumps produces here somewhat more averaging. Finally, Figure 5-C displays the spectra calculated for the C1-C4 and the C5-C8 blocks; the main reason for the differences between the two lineshapes is the difference in chain isomerization rates,21 which reflects different friction opposing bond rotations, as a consequence of differences in the chain geometry. The spectra obtained for the C15-18 and the C9-C12 blocks are very similar to those shown in Figure 5-C for the C1-C4 and the C5-C8 blocks, respectively.
The examples reported in Figure 5 clearly show that the chain conformational dynamics is sufficient to produce a variety of spectra for the R1 spin label. The weight of the various contributions to the lineshape is likely to depend on the location of the spin label and on the nature of the neighboring amino acid residues. This consideration is confirmed by ESR experiments on T4L mutants with R1 linked at different solvent exposed α-helix sites, or on mutants having R1 at a given site and different nearby residues.41 Such spectra have a common structure, with minor differences which can be ascribed to the different weight of the same contributing components. Having distinguished the possible spectral components, we have tried to make the spectral analysis more manageable, retaining a minimal number of independent of contributions. Taking as a reference the spectrum of 72R1-T4L recorded at T=298 in 30% wt sucrose solution,8 these have been identified as:
a constrained component, deriving from pairs of conformers with χ4 in the t state, in which the χ5 dihedral undergoes jumps and torsional oscillations of root mean square amplitude (case (a) in Figure 5-A);
a mobile component, originating from groups of four conformers experiencing χ4 and χ5 transitions, with a wide χ5|χ4=t distribution (case (b) in Figure 5-B);
an intermediate contribution, which differs from the latter only in the assumption of a more confined χ5|χ4=t distribution around the minima of the torsional potential, with root mean square amplitude (case (a) in Figure 5-B).
Calculations have been performed giving the three components the weights 0.4, 0.3 and 0.3, respectively, which have been adjusted to reproduce the experimental lineshape of 72R1-T4L in 30% wt sucrose solution at T=298 K.8 The effect of temperature on fluctuation amplitudes and rates of conformational transitions have been taken into account. Dynamic parameters, like the overall diffusion tensor of the protein and rates of internal motions have been scaled according to the change in solution viscosity.36 The diffusion tensor components used are: D0,⊥ = 0.4 · 107 s-1 and D0,∥ = .63 · 10 s−1 (T=283 K), D0,⊥ = 0.6 · 107 s-1 and D0,∥ = 0.95 · 107 s−1 (T=298 K), D0,⊥ = 0.8 · 107 s-1 and D0,∥ = 1.35 · 10 s−1 (T=312 K). The temperature dependence of the rates of internal motion are found to be due to both the activated processes (33%) and the decreased viscosity (67%) as the temperature is raised. Figure 6 shows the calculated spectra at temperatures of 283 K, 298 K and 312 K, superimposed on the experimental spectra, taken from Figure 5(a) in ref.8. We can see that the main features of the spectra and their temperature dependence are well reproduced by the simulation. Some discrepancies are observed at the lowest temperature (T=283 K); they probably reflect the inadequacy of the fast motional assumption for the chain dynamics in the highly viscous sucrose solution at this low temperature.
We have also calculated the 9GHz spectra of 72R1 in water at different temperatures, giving again the weights 0.4, 0.3 and 0.3 to the three spectral components. At a given temperature, differences from the spectra in sucrose solution only arise from the change in viscosity, which affects both transition rates and the protein diffusion tensor. For the latter, the values reported in Table 1 have been used for T=298 K, while the values at the other temperatures have been scaled according to the change in solution viscosity.36 The following values have been used: D0,⊥ = 0.8 · 107 s-1 and D0,∥ = 1.35 · 107s−1 (T=283 K), D0,⊥ = 1.2 107 s-1 and D0,∥ = 1.9 · 107 s−1 (T=312 K). Calculated and experimental lineshapes are shown in Figure 7; the good quality of the simulations as compared to experiment can be recognized. Interestingly, the observed disappearance, at higher temperatures, of a less mobile component, clearly recognizable at low temperature, naturally emerges from the calculations.
Figure 7.
Experimental13 (solid) and calculated (dashed) spectra of the 72R1 mutant of T4L in water at different temperatures.
Summarizing, we have seen that the spectra of 72R1-T4L can be simulated as the superposition of three components, obtained from modeling of the side-chain dynamics. The only adjustable parameters are the weights of such components; the choice of all the other parameters is justified not by spectral fitting, but rather by independent experimental findings and theoretical considerations. In addition to torsional oscillations involving all the dihedral angles, the majority of conformers can undergo both χ4 and χ5 transitions; for a smaller number of conformers, t → g transitions of the χ4 dihedral are forbidden for sterical reasons, and only χ5 jumps and remain possible. The value of 40% obtained for the constrained component from spectral analysis is much greater than the value of the calculated probability for just the C13-C14 pair, which are the only conformers which do not permit rotations of the χ4 dihedral (see table 3 in ref.21). We thus infer that in the real system the number of conformers which only undergo χ5 jumps is actually larger than just the C13-C14 pair. A plausible explanation for such behavior could be the hampering of χ4 transitions by the neighboring side-chains in the real system; effects of this kind, which depend on the structure and conformation of the nearby residues, were only partially taken into account in our model of a poly-Ala α-helix used to derive the conformer distribution. If we take, for example, the C1-C4 block, it is likely that, due to the constraints of the the specific protein environment, χ4 interconversion between some of the four conformers becomes highly hindered; so, the four conformers of the C1-C4 block would in practice behave as two blocks of conformers connected by χ5 jumps, whose contribution to the ESR spectra would be similar to that of the C13-C14 pair. Similar comments could apply to other blocks. Moreover, the physical significance of the fitted weights of the spectral components should be taken with care, because even small variations in the contributing lineshapes can bring about significant changes in the relative weights estimated from spectral fitting. This matter is exacerbated when, as in the present case, just a few components are taken as representative of a more extensive collection of states. Thus, for example, the 30% weights estimated for the (ii) and (iii) components should be better interpreted as an indication of the simultaneous presence of conformers experiencing oscillations of different amplitudes about the χ5|χ4=t stable states.
TABLE III.
Order parameters for the magnetic frame of MTSSL in the 72R1 mutant of T4L in water at T=10° C, as obtained from this work and from the simultaneous fit of multifrequency spectra.13 Order parameters for the constrained and the mobile spectral components are reported. The x, y, z axes define the principal frame of the ordering matrix SZZ;43 z is parallel to the N-pz orbital.
In view of our analysis, we now wish to reconsider the results of simulation of the multi-frequency spectra of 72R1-T4L in water.13 The uncertainty in the molecular interpretation of the best fit parameters of Liang et al.,13 and the differences in the present approach from that previously used by them13 limit somewhat the comparison that can be made.
The presence of two components with different mobility emerged from fitting of multi-frequency ESR experiments on 72R1-T4L, at least at the lower temperatures.13 Dominance of the less mobile contribution was inferred, in agreement with our result. A quantitative comparison of the weights obtained in the two cases is not really meaningful, for the reason mentioned above: the separation of sharp and broad spectral components is not straightforward, and can be strongly affected by the assumptions of the underlying model. The sensitivity of spectral weights to model assumptions was also pointed out in ref.13, where different values were obtained from separate fitting of spectra at different frequencies.
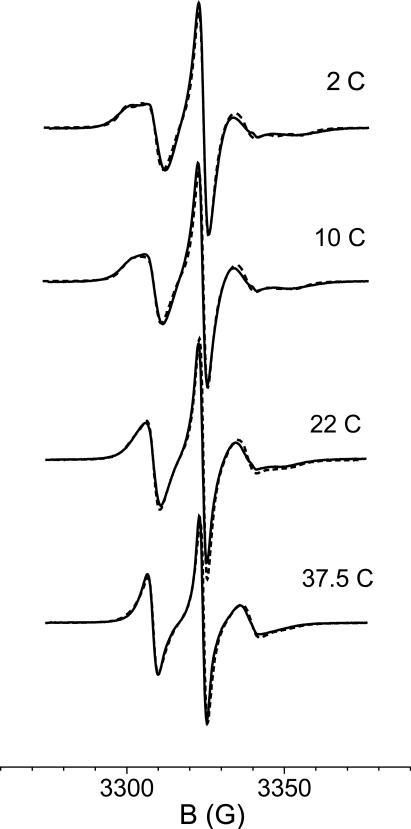
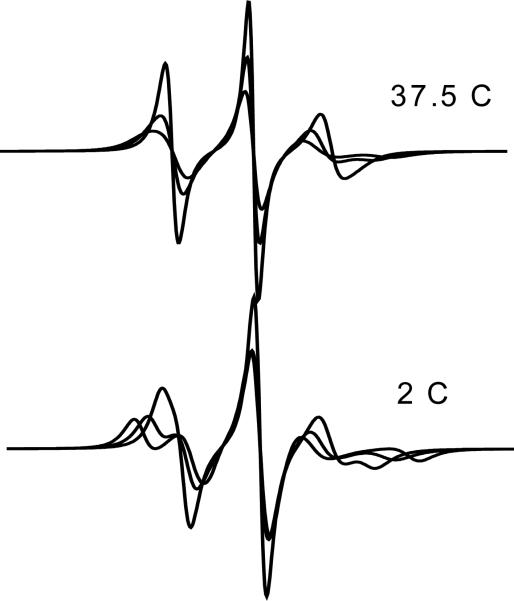
In ref.13 two spectral components were detected at low temperatures, but only one at higher temperatures; thus, the possibility of dynamic exchange between two conformers was suggested. Here, from the modeling of the side-chain dynamics, we have shown that merging of the two components naturally results with increasing temperature, from the increase in overall protein mobility and in amplitude of torsional fluctuations. The change of spectral components with temperature is shown in Figure 8. The possibility of exchange narrowing cannot be ruled out, but this is not necessary to explain the observed behaviour.
Figure 8.
Constrained and mobile contributions to the spectra of 72R1-T4L in water at T=2° C and T=37.5° C.
Table 3 reports the order parameters calculated in this work and those obtained in ref.13. The values obtained for the more mobile component are similar, although in the present case higher biaxiality of order is predicted, i.e. a larger difference in the propensity of the x and y principal axes of the magnetic frame to align along the Z director. The order parameters derived in ref.13 for the more constrained component are roughly between those obtained here for the two constrained contributions. In ref.13 the alignment axes were assumed to be parallel to the principal axes of the magnetic A and g tensors; from our modeling of the side-chain motions, only the z axis comes out to be roughly parallel to the N-pz orbital.
Let us now consider the parameters characterizing the chain dynamics. In ref.13 a higher rotational mobility perpendicular than along the N-pZ orbital was found for both components; a particularly high value of the perpendicular diffusion coefficient was obtained for the mobile component. The physical interpretation of the motional parameters derived from fitting of spectra with more or less sophisticated models of rotational diffusion has been a matter of concern.9 Our modeling of the side-chain motions o ers this interpretation: the fastest motions are χ5 rotations, which produce reorientations of the magnetic tensors roughly perpendicular to the N-pZ orbital. Such motions are expected to be particularly fast in conformers with wider χ5|χ4=t distribution around the stable states, i.e. lower barrier for the χ5 dihedral.
IV. DISCUSSION AND CONCLUSIONS
The main objective of this work has been the identification of the general features introduced into the ESR lineshape of spin-labeled proteins by the structure and intrinsic dynamics of the nitroxide side-chain. For this purpose, a methodology for the simulation of spectra of MTSSL spin labels linked at an ’ideal’ α-helix has been developed. It has been assumed that the reorientation of the magnetic tensors with respect to the static magnetic field is modulated by the conformational dynamics of the nitroxide side-chain, superimposed on overall reorientations of the protein. The latter, which fall in the slow time-scale for the experiments under examination, have been treated in the framework of the SLE. A detailed description of the chain dynamics has been adopted, on the basis of the results of the conformational analysis.21 For the sake of simplicity, a separation has been taken, between sufficiently fast chain motions, whose effect is approximated by the fast motional theory,22,23 and slower bond rotations, which have been assumed to be frozen on the ESR time-scale. The fast motions are described as jumps between stable conformers and librations about the minima of the chain torsional potential. Our approach, although approximate, contains key realistic features, which have led to some rather general results on the geometry and kinetics of such motions.
This methodology has been used to analyze X-band (9GHz) spectra of T4L with two variants of the MTSSL spin probe at the 72 residue. The spectra calculated with our modeling approach, utilizing a limited number of parameters (either derived from experiments or from theory), are in very good agreement with experiment. Details of protein dynamics are not included in the model at this stage, and from our results it can be inferred that they probably do not significantly affect the lineshape of a spin probe located on a rather rigid α-helix.
The case of the 72R2 mutants was first considered. It is actually simpler, because the presence of the methyl substituent at the 4-position of the pyrroline ring freezes out almost completely the MTSSL side-chain dynamics. Actually, 10 non-interconverting conformers turn out to be possible, which only undergo small amplitude torsional oscillations. We have shown that the main features of the spectra recorded in sucrose solution at different temperatures can be explained as the sum of contributions calculated for each of these conformers, with magnetic tensors partially averaged by oscillations about the minima of the torsional potential and an overall diffusion tensor estimated on the basis of the protein structure. Spectral changes with temperature are simply accounted for through the thermal change in oscillation amplitude and the solution viscosity, i.e. protein diffusion tensor.
The 72R1 mutants have greater torsional freedom and present a more complex dynamics. All conformers undergo, in addition to small amplitude torsional oscillations of the χ1 − χ4 dihedrals, also conformational jumps and wider amplitude fluctuations involving the χ5 dihedral. Moreover, some of the conformers can also experience χ4 conformational transitions. As a consequence, the dynamics of the side-chain is expected to yield complex spectra, resulting from the superposition of a variety of contributions. A key result of our analysis is the identification of the simultaneous presence of ’constrained’ and ’mobile’ components in the spectra. From the modeling of the chain dynamics, the former is ascribed to conformers which, in addition to rather confined fluctuations about all the minima of the torsional potential, can only undergo χ5 jumps. On the other hand, the most mobile component is attributed to sets of conformers experiencing χ4 and χ5 conformational jumps, together with wider amplitude χ5 fluctuations about the minima. To make the spectral analysis simpler, three components are taken as a minimal number sufficient to account for the experimental behavior; in this way good simulations of the 9GHz ESR spectra at different temperatures in different solvents have been obtained, with only the spectral weights as free parameters.
The presence of different components is a feature displayed in many cases by the spectra of spin labeled proteins, and several reasons for it have been proposed.8,9,13 The simultaneous presence of different conformers has been hypothesized,39,40 as well as that of different local environments,8 and the possibility of a temperature dependent dynamic exchange mechanism;13 the two-state nature of the disulfide bond has also be indicated as a possible explanation.18,39 On the basis of our model, we suggest here that the multicomponent character is an intrinsic feature of the ESR spectra of the R1 spin label, which derives from the simultaneous presence of conformers with different mobilities. Depending on the experimental conditions and the labeling site, the spectral components can sometimes be more or less well resolved, and sometimes only a single component can be inferred from the analysis of a single spectrum.
It is of interest to compare the spectra of the 72R1 mutant of T4L with those obtained for 131R1, which are also reported in refs.8 and13. Despite a little larger anisotropy of the magnetic tensors,13 less anisotropic lineshapes are obtained for 131R1 than for 72R1, which cannot be simply obtained by adjusting the weights of the components considered for 72R1. Due to its location, the spin probe in 131R1 is more solvent exposed than in 72R1; this hypothesis is supported by the larger hyperfine tensor components. Accordingly, less hindrance to conformational jumps is expected, together with larger amplitude χ5 motions, even for isolated conformers. But most likely this is not the main reason behind the narrowing of the spectra; actually, the presence of a constrained component can be detected for 131R1 in 30% wt sucrose solution, at sufficiently low temperatures. This suggests that the backbone dynamics is likely to be the main reason for the observed differences between the spectra for 72R1 and 131R1.8,13 In fact, we notice that 131R1 is located on a small two and a half turn helix, which is expected to provide less rigidity to the motion.13
A clear distinction between the R1 and R2 spin labels emerges from our analysis. In the latter case the chain dynamics is very restricted. Therefore it has small effects on the lineshapes, and can be easily accounted for. Moreover it can be treated under the fast-motional approximation up to higher frequencies, and different conformers of the side-chain yield very similar spectral components. These features turn out to be very advantageous for the interpretation of experiments, and make R2 a very useful spin probe for the investigation of protein dynamics. Namely, the possibility of accounting in a simple way for the side-chain dynamics opens up the prospect of introducing into the Stochastic Liouville Equation the protein dynamics, e.g. by relatively simple approaches like the Gaussian (or elastic network) model.37,38 The situation is less straightforward in the case of the R1 spin label, which is more mobile and has an intrinsic multicomponent nature; for these reasons spectral analysis and disentanglement of the various motions become harder. On the other hand, the possibility of exploring more extended regions make R1 more suitable to distinguish different local environments, through their effects on the probe mobility.
The methodology presented here can be used as long as the fast motional regime can be assumed for the side-chain motions; a more general description, which would also be suitable for high field experiments, would require relaxing the motional narrowing approximation for the nitroxide side-chain dynamics. (Our preliminary analysis of 72R-T4L spectra obtained at 250 GHz13 has indicated that the fast motional approximation is no longer appropriate.) To this purpose, the spectral contribution of a set of interconverting conformers could be described by explicitly including in the SLE the dynamics of the χ4 and χ5 dihedral angles, characterized by their torsional potentials and their frictional effects, in addition to the overall protein reorientation. Solution of the problem should remain feasible at a resonable computational cost, even after the inclusion of these two further degrees of freedom. The description of the conformational dynamics would not require the introduction of new parameters, since the relevant quantities, i.e. torsional potentials and friction opposing bond rotations, could be calculated as we have shown in this report. Only the height of the χ5 torsional barrier, which is likely to be strongly dependent on the specific environment experienced by the spin probe, could be taken as an adjustable parameter. This could be obtained through the best fit of experimental lineshapes, together with the weights of the spectral components and the diffusion tensor of the whole protein. The advantage of such an approach over other approaches would be the clear physical interpretation of the parameters, and, as a consequence, a better characterization of structural and dynamic features of the protein.
An alternative methodology for the introduction of the side-chain dynamics of R1 in the SLE framework could be based on the use of the SRLS model,12 together with MD simulations. From these, the parameters for internal motions and orienting potential of the spin probe in its specific environment could be derived. As shown here and in the companion paper,21 the presently available version of the SRLS model could be conveniently improved by introducing a more flexible form of the orienting potential, suitable to account for the orientational distribution of the spin probe. Of course, an important requirement of trajectories is a representative sampling of the conformational space explored by the spin probe. We have seen that the constraints imposed by a generic α-helix environment lead to sets of non-interconverting conformers. This means that in a given trajectory the spin probe could remain trapped in a subspace, which depends on the starting condition; this point should be carefully considered in order to obtain reliable information from MD.
Aknowledgments
J.H.F. acknowledges financial support by a grant from NIH/NCRR. A.F. and F.T have been supported by MIUR (PRIN-2005) and Università di Padova (CPDA057391). They gratefully acknowledge Anna Lisa Maniero, Lorenzo Franco and Antonio Toffoletti for stimulating discussions.
Abbreviations
- PAS
Principal Axis System
- SLE
Stochastic Liouville Equation
- MTSSL
methanethiosulphonate
- T4L
T4 lysozyme
- SRLS
Slowly Relaxing Local Structure
- MD
Molecular Dynamics
Appendix. Introduction of fast internal motions in the SLE
Extension of the slow-motional ESR theory42 to include fast internal motions has been performed in the following way.
The partially averaged magnetic tensors, which are used in place of the nitroxide magnetic tensors, are obtained from these and from the calculated order parameters, according to eq 8.
An orientation dependent linewidth is assumed, defined by eqs 9-13 and eq 17. Eq 9 is expressed in the form:
| (27) |
where with cM,M′m,m′,μ,μ′ being suitable coefficients. Thus, additional elements of the following form have to be introduced into the matrix representation of the stochastic Liouville operator:42
| (28) |
where .
The introduction of the side-chain fast motions does not appreciably increase the computing time required for lineshape simulations.
References and Notes
- 1.Hubbell WL, Gross A, Langen R, Lietzow M. Curr. Opin. Struct. Biol. 1998;8:649. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 2.Borbat PP, Costa-Filho AJ, Earle KA, Moscicki JK, Freed JH. Science. 2001;291:266. doi: 10.1126/science.291.5502.266. [DOI] [PubMed] [Google Scholar]
- 3.Freed JH. In: Spin Labeling. Theory and Application. Berliner LJ, editor. Academic Press; New York: 1976. p. 53. [Google Scholar]
- 4.Schwartz LJ, Stillman AE, Freed JH. J. Chem. Phys. 1982;77:5410. [Google Scholar]
- 5.Schneider DJ, Freed JH. Adv. Chem. Phys. 1989;73:387. [Google Scholar]
- 6.Budil DE, Lee S, Saxena S, Freed JH. J. Magn. Res. 1996;120:155. programs available at www.acert.cornell.edu.
- 7.Meirovitch E, Nayeem A, Freed JH. J. Phys. Chem. 1984;88:3454. [Google Scholar]
- 8.Columbus L, Kámás T, Jekö J, Hideg K, Hubbell WL. Biochemistry. 2001;40:3828. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 9.Barnes JP, Liang Z, Mchaourab HS, Freed JH, Hubbell WL. Biophys. J. 1999;76:3298. doi: 10.1016/S0006-3495(99)77482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polnaszek CF, Freed JH. J. Phys. Chem. 1975;79:2283. [Google Scholar]
- 11.Freed JH. J. Chem. Phys. 1977;66:4183. [Google Scholar]
- 12.Polimeno A, Freed JH. J. Phys. Chem. 1995;99:10995. [Google Scholar]
- 13.Liang Z, Lou Y, Freed JH, Columbus L, Hubbell WL. J. Phys. Chem. B. 2004;108:17649. [Google Scholar]
- 14.Jacobsen K, Oga S, Hubbell WL, Risse T. Biophys. J. 2005;88:4351. doi: 10.1529/biophysj.105.059725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinhoff H-J, Hubbell WL. Biophys. J. 1996;71:2201. doi: 10.1016/S0006-3495(96)79421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson BH, Slutsky LJ, Auteri FP. J. Chem. Phys. 1992;96:2609. [Google Scholar]
- 17.Stoica I. J. Phys. Chem. B. 2004;108:1771. [Google Scholar]
- 18.Murzyn K, Rg T, Blicharski W, Dutka M, Pyka J, Szytula S, Froncisz W. Proteins. 2006;62:1088. doi: 10.1002/prot.20838. [DOI] [PubMed] [Google Scholar]
- 19.Budil DE, Sale KL, Khairy KA, Fajer PJ. J. Phys. Chem. B. 2006;110:3703. doi: 10.1021/jp054738k. [DOI] [PubMed] [Google Scholar]
- 20.Timofeev VP, Nikolsky DO. J. Biomol. Struct. Dyn. 2003;21:367. doi: 10.1080/07391102.2003.10506932. [DOI] [PubMed] [Google Scholar]
- 21.Tombolato F, Ferrarini A, Freed JH. J. Phys. Chem. B submitted, companion paper. Hereafter referred to as I.
- 22.Redfield AG. Adv. Magn. Reson. 1965:1. [Google Scholar]
- 23.Freed JH, Fraenkel GK. J. Chem. Phys. 1963;39:326. [Google Scholar]
- 24.Cassol R, Ferrarini A, Nordio PL. J. Phys. Chem. 1993;97:2933. [Google Scholar]
- 25.Cassol R, Ge M-T, Ferrarini A, Freed JH. J. Phys. Chem. B. 1997;101:8782. [Google Scholar]
- 26.Liang Z, Freed JH. J. Phys. Chem. B. 1999;103:6384. [Google Scholar]
- 27.Zare RN. Angular Momentum. Wiley; New York: 1987. [Google Scholar]
- 28.Shore JE, Zwanzig R. J. Chem. Phys. 1975;63:5445. [Google Scholar]
- 29.Zientara GP, Freed JH. J. Chem. Phys. 1983;69:3077. [Google Scholar]
- 30.Risken H. The Fokker-Planck Equation. Springer-Verlag; Berlin: 1984. [Google Scholar]
- 31.Ferrarini A, Moro G, Nordio PL. Mol. Phys. 1988;63:225. [Google Scholar]
- 32.Kramers HA. Physica. 1940;7:284. [Google Scholar]
- 33.Langer JS. Ann. Phys. 1969;54:258. [Google Scholar]
- 34.Garcia de la Torre J, Huertas ML, Carrasco B. J. Magn. Res. 2000;147:138. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 35.Matsumura M, WozniaK JA, Dao-pin S, Mattews BW. J. Biol. Chem. 1989;264:16059. [PubMed] [Google Scholar]
- 36.Weast RC. Handbook of Chemistry and Physics. CRC Press; Cleveland: 1975. [Google Scholar]
- 37.Tirion MM. Phys. Rev. Lett. 1996;77:1905. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 38.Bahar I, Atilgan AR, Erman B. Fold. Des. 1997;2:173. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 39.Langen R, Joon Oh K., Cascio D, Hubbell WL. Biochemistry. 2000;39:8396. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]
- 40.Pyka J, Iinicki J, Altenbach C, Hubbell WL, Froncisz W. Biophys. J. 2005;89:2059. doi: 10.1529/biophysj.105.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mchaourab MS, Lietzow MA, Hideg K, Hubbell WL. Biochemistry. 1996;35:7692. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 42.Meirovitch E, Igner E, Igner D, Moro G, Freed JH. J. Chem. Phys. 1982;77:3915. [Google Scholar]
- 43.Zannoni C. In: The Molecular Physics of Liquid Crystals. Luckhurst GR, Gray GW, editors. Academic Press; New York: 1979. p. 51. [Google Scholar]