Abstract
Iron−sulfur (Fe−S) proteins contain prosthetic groups consisting of two or more iron atoms bridged by sulfur ligands, which facilitate multiple functions, including redox activity, enzymatic function, and maintenance of structural integrity. More than 20 proteins are involved in the biosynthesis of iron−sulfur clusters in eukaryotes. Defective Fe−S cluster synthesis not only affects activities of many iron−sulfur enzymes, such as aconitase and succinate dehydrogenase, but also alters the regulation of cellular iron homeostasis, causing both mitochondrial iron overload and cytosolic iron deficiency. In this work, we review human Fe−S cluster biogenesis and human diseases that are caused by defective Fe−S cluster biogenesis. Fe−S cluster biogenesis takes place essentially in every tissue of humans, and products of human disease genes, including frataxin, GLRX5, ISCU, and ABCB7, have important roles in the process. However, the human diseases, Friedreich ataxia, glutaredoxin 5-deficient sideroblastic anemia, ISCU myopathy, and ABCB7 sideroblastic anemia/ataxia syndrome, affect specific tissues, while sparing others. Here we discuss the phenotypes caused by mutations in these different disease genes, and we compare the underlying pathophysiology and discuss the possible explanations for tissue-specific pathology in these diseases caused by defective Fe−S cluster biogenesis.
Human Cellular Iron Homeostasis
Iron is essential for all eukaryotes and most prokaryotes, where it is used in the synthesis of heme, iron−sulfur (Fe−S),1 and other cofactors. Fe−S proteins are involved in catalysis, redox reactions, respiration, DNA replication, and transcription. Hemes are found in hemoglobin, myoglobin, and cytochromes, and recent studies have implicated heme proteins in the regulation of circadian rhythmicity (1) and microRNA processing (2).
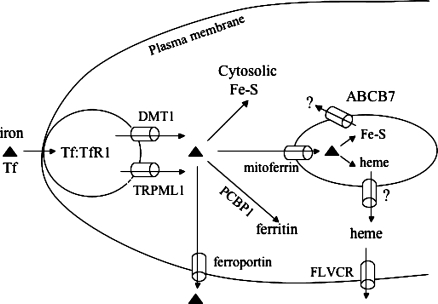
Iron homeostasis is tightly regulated to avoid iron toxicity or iron deficiency. In human systemic iron metabolism, iron uptake, trafficking, export, and utilization are highly regulated, and those processes have been reviewed recently (3−5). Here, for background, we briefly describe some of the main proteins involved in intracellular iron trafficking (Figure 1). Iron-laden transferrin (Tf) in the mammalian circulation binds to the transferrin receptor (TfR1) on the plasma membrane, inducing endocytosis of the Tf−TfR1 complex. In the endosome, iron is released from Tf upon acidification of the endosome. Upon reduction to the ferrous form, iron is transported from the endosome to the cytoplasm primarily by the iron importer, DMT1. However, there is also a DMT1-independent endosomal ferrous iron permeable channel, TRPML1, in late endosomes and lysosomes (6). Mutations in the TRPML1 gene in humans cause mucolipidosis type IV disease, a syndrome characterized by motor impairment, mental retardation, retinal degeneration, and iron-deficiency anemia. Upon its entry into the cytosol, iron can be assembled into Fe−S clusters for the maturation of cytosolic and nucleic proteins (7), incorporated into enzymes such as ribonucleotide reductase (8), or stored in ferritin. The loading of iron into ferritin is facilitated by PCBP1 protein, a recently described cytosolic chaperone that trafficks iron into the storage protein, ferritin (9). Mitochondria are the major iron-consuming subcellular organelles in cells. Iron is imported into mitochondria by mitoferrin (10) and is subsequently assembled into Fe−S clusters and heme. Some of the synthesized heme must be exported out of mitochondrion to the cytosolic/nuclear compartment to provide heme cofactors to nonmitochondrial proteins such as guanylate cyclase, but a mitochondrial heme exporter has not yet been identified.
Figure 1.
Subcellular iron trafficking in human cells. Iron is bound to transferrin Tf in serum, which interacts with TfR1 on the cell membrane. Formation of the Tf−TfR1 complex induces endocytosis. Upon acidification of the endosome, iron is released and then exported into the cytosol by DMT1 or TRPML1. Iron in cytosol has four main fates. It can be used to assemble Fe−S clusters and other iron proteins in cytosol, undergo transport into mitochondria via the mitochondrial iron importer, mitoferrin, be loaded into ferritin with the coordination of the PCBP1 chaperone, or be exported by ferroportin. Upon its import into mitochondria, iron is used to synthesize Fe−S clusters and heme. An unknown compound that represents the mitochondrial iron status and is a product of Fe−S cluster synthesis appears to be exported by ABCB7, which can affect cytosolic Fe−S cluster biogenesis. Heme is exported from the mitochondria by an unknown mechanism and is exported out of some cells by a heme transporter known as FLVCR.
In mammalian cells, Fe−S clusters are synthesized in both mitochondria and the cytosol (11), in contrast to yeast, in which it is asserted that de novo Fe−S cluster biogenesis occurs solely in mitochondria (12). Export of an unknown compound from mitochondria by a transporter known as ABCB7 in mammals, and as Atm1 in yeast, appears to be needed for some aspect of cytosolic Fe−S cluster biogenesis (12). Excess iron may be exported from cytosol by ferroportin on the plasma membrane, whereas excess cytosolic heme is thought to be exported by the putative heme transporter, FLVCR (13,14). At the cellular level, iron homeostasis is regulated by two iron regulatory proteins, IRP1 and IRP2 (15,16). IRP1 senses the iron status via an Fe−S switch mechanism: by losing its Fe−S cluster, IRP1 is activated to become an IRE-binding protein, which increases the rate of cellular iron uptake and decreases the rate of iron sequestration by stabilizing TfR1 mRNA and repressing ferritin synthesis. Fe−S cluster biogenesis is therefore important for the IRP1-mediated regulation of human iron homeostasis.
Human Fe−S Cluster Biosynthesis
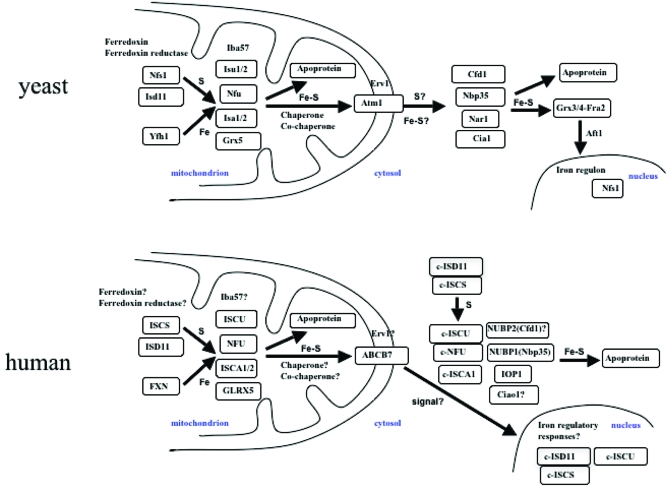
Much of what is known about eukaryotic Fe−S cluster biogenesis was learned from bacterial Fe−S proteins, which were identified initially in nitrogen-fixing bacteria, and subsequently in Escherichia coli and other bacteria (17). More than 20 proteins involved in Fe−S cluster biosynthesis have been identified in yeast (12,18), and both the bacterial and yeast model systems have revealed much about Fe−S cluster biogenesis. In the central mitochondrial Fe−S cluster synthesis machinery of yeast (Figure 2), the Nfs1−Isd11 complex functions as a cysteine desulfurase, providing sulfur. Frataxin is thought to provide iron by binding iron with low affinity to an extended acidic ridge in the protein (19). Isu1, its homologue Isu2, Nfu, Isa1, and Isa2 are all proposed to function as scaffold proteins upon which Fe−S clusters are transiently assembled. Ferredoxin (Yah1) and ferredoxin reductase (Arh1) are redox proteins that are thought to provide electrons for later steps of Fe−S cluster assembly (12,18). A chaperone and cochaperone pair, known as HscA and HscB in bacteria or Ssq1 and Jac1 in yeast (20), respectively, along with a glutaredoxin, Grx5, are thought to facilitate the transfer of Fe−S clusters from scaffolds to target apoproteins (12,18). Recently, interaction of a new component, Iba57, with Isa1 and Isa2 was observed and shown to have a specific role in the maturation of mitochondrial aconitase-type and radical SAM Fe−S proteins (21). It has been proposed that an Fe−S compound formed in the yeast mitochondrial matrix is transported to the cytosol via an export machinery in the mitochondrial inner membrane and intermembrane space, involving the transporter Atm1, a sulfhydryl oxidase, Erv1, and glutathione (GSH). The exported compound has been proposed to provide the Fe−S core for assembly of Fe−S clusters in cytosol and nuclei. Two cytosolic P-loop NTPases, Cfd1 and Nbp35, are proposed to act as scaffolds for cytosolic Fe−S cluster biogenesis. The ATPase proteins, Nar1 and Cia1, may function in cytosolic cluster transfer (12,22), and a newly identified protein, Dre2 (23), may also contribute to cytosolic Fe−S cluster biogenesis in yeast (Figure 2, top panel).
Figure 2.
Comparison of Fe−S cluster biogenesis pathways in eukaryotes. In yeast mitochondria, Nfs1 and Isd11 form a complex of cysteine desulfurase, which provides sulfur to scaffold proteins for Fe−S cluster assembly. The pair of ferredoxin (Yah1) and ferredoxin reductase (Arh1) perhaps provides reducing equivalents needed for cluster assembly. The yeast frataxin homologue, Yfh1, provides iron. Iron−sulfur clusters are assembled on scaffolds, and there are several alternative scaffolds, including Isu1 and -2, Nfu, Isa1 and -2, and Grx5. Iba57 is thought to function with the Isa1 and -2 scaffold proteins. Facilitated by chaperone (Ssq1) and cochaperone (Jac1) activities, the preassembled Fe−S clusters of Isu1 and -2 are delivered to target apoproteins in mitochondria. The mitochondrial inner membrane transporter Atm1 has been proposed to export either Fe−S cluster or sulfur to the cytosol, with assistance from Erv1 in the intermembrane space of mitochondria, and this compound “X” could then be used to assemble Fe−S clusters in the cytosol by scaffold proteins, including Cfd1, Nbp35, Nar1, and Cia1. The assembled Fe−S clusters are delivered to apoproteins, including the Grx3/4−Fra2 complex, which regulates the nucleocytoplasmic translocation of Aft1 by an unknown mechanism. As a transcription factor, Aft1 regulates transcription of the iron regulon in the yeast nucleus. Interestingly, there is a fraction of Nfs1 proteins present in yeast nucleus (109). In human mitochondria, ISCS and ISD11 form a complex that provides sulfur to scaffold proteins for Fe−S cluster biogenesis, while FXN is thought to provide iron. Potential scaffold proteins include ISCU, NFU, ISCA1/2, and GLRX5. The assembled Fe−S clusters are delivered to apoproteins for maturation. The roles of human ferredoxin, ferredoxin reductase, Iba57, chaperone, and cochaperone homologues in mitochondrial Fe−S cluster biogenesis remain to be confirmed. An unknown molecule that depends on mitochondrial Fe−S cluster biogenesis for function is exported by ABCB7. We propose that the ABCB7-exported molecule may serve as a signal that induces iron transcriptional remodeling in the nucleus to appropriately regulate mitochondrial iron homeostasis in response to a signal received from the mitochondria. In the cytosol, the cytosolic forms of ISCS and ISD11, c-ISCS and c-ISD11, respectively, provide sulfur, while iron can be directly acquired from cytosol, though its exact molecular source is not known. Fe−S clusters are likely synthesized de novo on c-ISCU, c-NFU, c-ISCA1, or NUBP1 (Nbp35) scaffolds. IOP1 may function in the delivery of the cluster to apoproteins. The involvement of the human Cfd1 homologue (NUBP2) and Cia1 homologue (Ciao1) in cytosolic Fe−S cluster biogenesis remains to be studied.
Compared to the yeast model system, human Fe−S cluster biogenesis shares many features, but it also differs in several important ways (7,24). In the mitochondrial machinery (Figure 2), ISCS is a cysteine desulfurase that provides sulfur (25) in association with ISD11 protein, which may stabilize the conformation of ISCS (26). Frataxin is a proposed iron donor (27); ISCU (28,29) and NFU are scaffold proteins (30), and ISCA is likely an alternative scaffold (31). GLRX5 may also function as a scaffold and may facilitate the transfer of Fe−S clusters to target apoproteins (32). In the export machinery, the Atm1-like transporter ABCB7 delivers an unknown signal to the cytosol that perhaps reflects the mitochondrial iron status. Components of the mammalian cytosolic Fe−S machinery include c-ISCS (25), c-ISD11 (26), c-NFU (30), and c-ISCU (28,29). Interestingly, the mitochondrial and cytosolic isoforms of ISCU are generated from a single gene through alternative splicing, and similarly, the mitochondrial and cytosolic isoforms of NFU are also generated from a single gene by alternative splicing products. Importantly, the cytosolic isoform of ISCU has been shown to have a role similar to that of its mitochondrial counterpart in Fe−S cluster formation in the cytosol (reviewed in refs (7) and (11)). Recently, a new cytosolic component IOP1, the Nar1 homologue in humans, was observed to be required for human cytosolic Fe−S cluster biogenesis (33) and to regulate hypoxia inducible factor (HIF) activity in the hypoxia response (34). Human Nbp35 forms a complex with its close homologue human Cfd1 in vivo, suggesting that a heteromeric P-loop NTPase complex is required for cytosolic Fe−S cluster assembly and cellular iron homeostasis (35). A specific candidate for the role of the iron donor for cytosolic Fe−S cluster biogenesis remains to be characterized; interestingly, some frataxin has been detected in the cytosol (36,37).
Although the human and yeast Fe−S cluster biogenesis pathways share many components, the mechanisms by which cytosolic and nuclear Fe−S clusters are assembled appear to differ significantly. In yeast, it has been proposed that the Atm1 transporter provides either an Fe−S cluster or a type of modified sufur for use in cytosolic Fe−S cluster assembly. Cytosolic Fe−S cluster biogenesis in yeast is thought to depend on the mitochondrial Fe−S machinery for this vital ingredient, which has not been molecularly characterized. Moreover, the transcriptional regulation of the iron regulon in yeast nuclei also depends on mitochondrial Fe−S cluster biogenesis (38−40). Using a mitochondrial substrate, an Fe−S cluster is thought to be assembled in the cytosol and delivered to a complex of Grx3, Grx4, and Fra2 proteins. The complex of Grx3, Grx4, and Fra2 constitutes a crucial part of a signaling pathway that regulates expression of the iron regulon by regulating nucleocytoplasmic translocation of Aft1, the major factor that regulates transcription of genes of the yeast iron regulon (38,40). Although the mechanism is not known, the Grx3/4−Fra2 complex may acquire a [2Fe-2S] cluster in iron-replete cells, and this complex may cause Aft1 to multimerize and exit the nucleus; on the other hand, Aft1 remains in the nucleus of iron-depleted cells and increases the level of transcription of genes of the iron regulon. In contrast, in human cells, there are cytosolic isoforms of the cysteine desulfurase and other basic components, and it is thought that Fe−S clusters can be synthesized de novo in the cytosol. However, problems with mitochondrial Fe−S cluster biogenesis adversely affect cytosolic iron status by inducing the cell to remodel the iron status. Dysfunctional mitochondrial Fe−S cluster biogenesis leads to mitochondrial iron overload and cytosolic iron deficiency. This iron deficiency directly impairs cytosolic Fe−S cluster biogenesis, even though all protein components of cytosolic Fe−S assembly machinery are readily available and the cytosolic Fe−S cluster biogenesis machinery is independent.
Fe−S Cluster Biosynthesis and Human Diseases
In the past two decades, several diseases have been found to be able to be attributed to mutations in genes that encode proteins involved in human Fe−S cluster biogenesis. This subject was summarized previously (11), but with new progress in the field, here we revisit the role of compromised Fe−S cluster synthesis in the pathogenesis of human diseases.
Frataxin and Friedreich's Ataxia
Frataxin (FXN) is a mitochondrial iron binding protein involved in Fe−S cluster biosynthesis. Human frataxin is transcribed into a primary mRNA 1.3 kb in size (41). Human FXN protein is synthesized as a 210-amino acid precursor protein, but multiple smaller intermediate forms are found which do not conform to the sizes expected after traditional mitochondrial import, cleavage, and processing. The different sizes of frataxin maturation may arise from tissue-specific differences in processing (42). Under normal conditions in adult tissues, FXN is localized to the mitochondria, while under oxidative stress, a cytosolic FXN has been observed which may facilitate the regeneration of Fe−S clusters damaged by reactive oxygen species (ROS) (43). Various studies suggest that FXN functions as either an iron chaperone (44), an iron storage protein (45), an iron donor for heme biosynthesis (46), an iron donor for Fe−S cluster repair, or a regulator of iron export (44,47). However, it is widely believed that the main role of FXN is to supply iron in a bioavailable form for mitochondrial Fe−S cluster synthesis.
Mutations in frataxin cause Friedreich's ataxia (FRDA), a recessive neurodegenerative disease that is the most common form of inherited ataxia in humans. FRDA is a degenerative disease that involves the central and peripheral nervous systems and the heart (48), which is characterized by a progressive gait and limb ataxia, a lack of tendon reflexes in the legs, loss of position sense, dysarthria, and leg weakness. Neurons in the dorsal root ganglia are adversely affected, and hypertrophic cardiomyopathy is found in almost all patients (49). The age of onset is usually around puberty, and always before age 25. In the affected tissues, excess iron accumulation is observed in mitochondria of cardiac myocytes and neurons (50−52). Molecular analysis reveals that activities of Fe−S proteins such as aconitase and succinate dehydrogenase are reduced in FRDA patient cells. Excess iron is imported and accumulated in mitochondria, which leads to iron deficiency in the cytosol. IRP proteins are activated, and cellular iron homeostasis is impaired (53,54).
Approximately 2% of FRDA patients are found to have point mutations in the frataxin gene, whereas 98% of patients have frataxin genes that contain an unstable GAA trinucleotide expansion in the first intron (48). Normal alleles contain 6−34 of these GAA repeats, but the expanded alleles of patients contain 66−1700 repeats. The expansion of the GAA repeat has been proposed to lead to gene silencing by allowing heterochromatin formation to silence transcription (55) or to inhibit transcription of the frataxin gene by facilitating formation of a persistent DNA−RNA hybrid. The age of onset and the severity of the disease are inversely correlated with the length of the GAA repeat sequence, and there is evidence that the GAA repeats may expand as patients age. Therapies for FRDA are aimed at alleviating transcriptional inhibition and reversing heterochromatin formation using histone deacetylase inhibitors (55). Additionally, a coenzyme Q homologue antioxidant, idebenone, was reported to have a positive effect on the cardiomyopathy in most patients, but it did not have noticeable effects on the ataxia (56,57). Other therapies focus on removing excess iron from mitochondria and redistributing it to the cytosol (58). Thus far, there has been little success in the treatment of FRDA.
GLRX5 and Sideroblastic Anemia
The glutaredoxins (Grx) are a group of thioltransferases that reduce disulfide bonds or catalyze reversible protein de- or glutathionylation (59). The Grx5 genes, including human GLRX5, encode a highly conserved mitochondrial monothiol glutaredoxin. Grx5 homologues in yeast and zebrafish are required for Fe−S cluster biogenesis (60,61), although their precise functions in the process remain unknown. Recently, two plant chloroplastic monothiol glutaredoxins were shown to ligate a [2Fe-2S] cluster upon reconstitution in vitro, leading to the suggestion that Grx5-like proteins may function as scaffolds (62). In grx5 knockout zebrafish, the extent of Fe−S cluster assembly is substantially decreased. By losing its Fe−S cluster, IRP1 of zebrafish is activated to an IRE-binding protein, which blocks the expression of ALAS2, the first enzyme of heme biosynthesis in erythroid cells, because the ALAS2 transcript contains an iron responsive element (IRE) in its 5′ UTR. The deletion of grx5 in fish results in severe hypochromic anemia and is embryonically lethal (61). A complementation experiment has demonstrated that human GLRX5 is able to rescue Fe−S cluster synthesis in grx5 mutant yeast, suggesting that the function of GLRX5 has been conserved during evolution (63).
In humans, the GLRX5 mutation causes microcytic sideroblastic anemia (SA), as shown by a recently identified male patient from Italy (64). Sideroblastic anemias are anemias characterized by the presence of ring sideroblasts, which represent erythroid precursor cells that contain iron-overloaded mitochondria that congregate in a ring around the nucleus and are detected by the Prussian blue stain for iron. The patient has a homozygous A294G mutation in the third nucleotide of the last codon of GLRX5 exon 1. This substitution does not change the encoded amino acid (glutamine), but it interferes with the correct splicing and removal of intron 1 and drastically reduces grx5 mRNA (64). Western blotting confirms that the GLRX5 protein is absent in multiple tissues of the patient, including hematopoietic cells (e.g., lymphoblast) and nonhematopoietic cells (e.g., fibroblast) (32). Like counterparts in other eukaryotes, human GLRX5 is also an Fe−S cluster synthetic protein in mitochondria, which likely functions as a scaffold protein. The GLRX5 protein deficiency results in a defect in Fe−S cluster biogenesis in multiple tissues, but not all tissues develop a pathologic phenotype. Activation of IRP proteins suppresses the translation of 5′ IRE-containing transcripts (mostly iron-utilizing genes) but promotes the stability and translation of 3′ IRE-containing transcripts involved in iron uptake such as TfR1. The imbalance of mitochondrial iron homeostasis induced by GLRX5 knockdown somehow induces mitochondrial iron overload. An ultimate consequence is the accumulation of iron in mitochondria and iron deficiency in the cytosol (32,64).
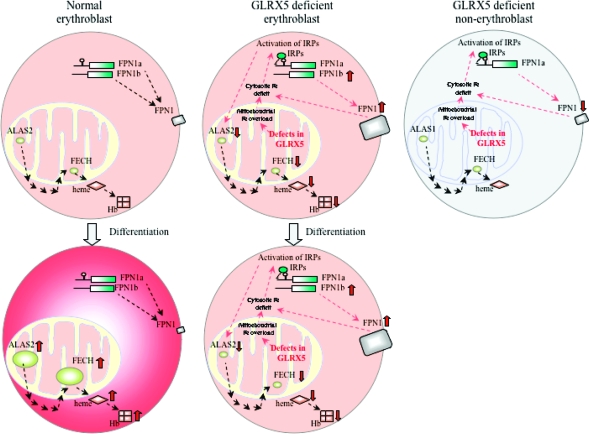
Microarray analysis of fibroblasts from the GLRX5-deficient patient indicates that there is significant transcriptional remodeling in the patient cells. In particular, the iron exporter gene ferroportin (FPN1) is significantly upregulated, potentially exacerbating the cytosolic iron deficiency. Although GLRX5 knockdown affects both Fe−S cluster biogenesis and heme synthesis pathways and affects almost all tissues, most tissues do not manifest significant pathology, perhaps because other proteins can compensate for the defect of Fe−S cluster biogenesis, and cells other than erythroid cells do not need to make large amounts of heme. However, the situation is totally different in erythroid cells, which express a heme synthetic ALAS2 isoform that contains a 5′ IRE and is subject to IRP repression and a specific FPN1b transcript that does not have a 5′ IRE and can therefore evade repression by IRPs when there is cytosolic iron deficiency (65). The GLRX5 knockdown activates IRPs and thereby decreases the level of ALAS2 expression and interrupts heme biosynthesis, but activated IRPs cannot repress FPN1b expression, which is induced during the stress response to mitochondrial iron overload and worsens cytosolic iron deficiency in eythroid cells (Figure 3). The combined effect is that the red blood cell function is highly compromised by a failure to synthesize sufficient heme and by iron deficiency (32).
Figure 3.
GLRX5 deficiency causes anemia but does not significantly affect non-erythroid tissues. In normal erythroblasts, ALAS2 and FECH in mitochondria contribute to heme synthesis, which is incorporated into hemoglobin (Hb). Erythroblasts are the only cells that express ALAS2, which contains a IRE in its 5′ UTR. All other cells express ALAS1, which is not regulated by the IRE−IRP system. Erythroblasts encode two forms of the ferroportin (FPN1) transcript, IRE-FPN1a and non-IRE-FPN1b. Both encode an identical FPN1 protein that functions as the iron exporter. Upon erythroid differentiation, both ALAS2 expression and FECH expression are upregulated, thus increasing the level of synthesis of heme and hemoglobin, while FPN1 expression is downregulated (65). In contrast, Fe−S cluster biogenesis is defective in GLRX5-deficient erythroblasts, iron accumulates in mitochondria, and associated iron deficiency in cytosol activates IRP proteins. The level of ALAS2 expression is decreased by IRP repression of its 5′ IRE. The level of FECH expression is also decreased, most likely because FECH does not acquire the Fe−S cluster it needs for stabilization (69). Together, heme synthesis and subsequent hemoglobinization are inhibited. Levels of expression of both FPN1 transcripts, including both FPN1a and FPN1b, are increased, perhaps in response to the stress caused by mitochondrial iron overload. Because FPN1b lacks the IRE, the expression of FPN1b evades the IRP repression and increases the level of expression of FPN1, which may exacerbate cytosolic iron deficiency. As a result, the GLRX5-deficient erythroblasts fail to produce enough heme for hemoglobinization upon differentiation. Although GLRX5-deficient non-erythroblasts also demonstrate iron overload in mitochondria and cytosolic iron deficiency, they express ALAS1, which does not have IRE and is not repressed by IRPs. Thus, heme synthesis is not significantly impaired in non-erythroid cell types. In addition, non-erythroblast cells express only FPN1a, which can be repressed by IRPs as cells develop cytosolic iron depletion.
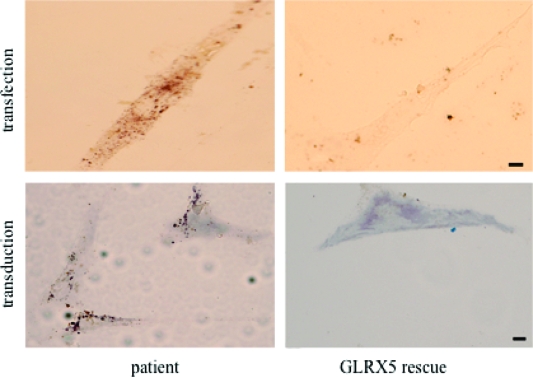
In the clinic, the personal history of the unique GLRX5-deficient patient was unremarkable until age 44, when mild anemia and type 2 diabetes were diagnosed. Laboratory tests showed high levels of blood Tf saturation, serum ferritin, bilirubin, and liver transaminases. At age 60, the patient manifested severe sideroblastic anemia, insulin-dependent diabetes, and cirrhosis. XLSA with ataxia was ruled out by the absence of neurologic symptoms. Laboratory tests showed increased serum ferritin, urinary iron, and liver iron concentrations. Bone marrow biopsy showed erythroid hyperplasia and abundant iron. Blood transfusion made the preexisting iron overload worse. Instead, iron chelation treatment was started at age 60 with subcutaneous deferoxamine (Dfo) at 30 mg/kg. Clinical improvement was noticed within 6 months. Serum ferritin and liver iron levels were decreased, and hemoglobin levels were increased (64). A recent rescue experiment of GLRX5 patient fibroblasts using a lentiviral vector that carries the wild-type GLRX5 gene sequence demonstrated that restored GLRX5 expression could improve cell growth, reverse mitochondrial iron overload, and increase aconitase activity (Figure 4) (32). These results indicate that gene therapy of the patient’s bone marrow cells could offer a potential treatment for congenital GLRX5-deficient patients in the future.
Figure 4.
Example of mitochondrial iron overload and its reversal when iron−sulfur cluster biogenesis is restored. Shown in the left panels are several fibroblasts that are stained with the Perls’ DAB iron staining technique, which detects ferric iron and enhances the signal with precipitation of DAB. Iron overload is present in a reticular pattern consistent with the mitochondrial network of patient fibroblast cells (left), but the iron deposits disappeared in patient cells rescued by the wild-type GLRX5 gene (right), introduced either by transfection (top) or by viral transduction (bottom) (32). The scale bars are 10 μm.
ISCU and Myopathy
ISCU is the major scaffold protein upon which the transient Fe−S clusters are initially assembled and from which they are then delivered to recipient proteins. The human ISCU gene encodes two forms of protein by alternative splicing. The 14 kDa protein is mitochondrial (m-ISCU) and the 15 kDa protein cytosolic (c-ISCU), and m-ISCU is much more abundant (28). In vitro reconstitution assays indicate that both forms of ISCU protein are active in Fe−S cluster assembly (66). Silencing of c-ISCU by siRNA impairs Fe−S cluster assembly in cytosol, suggesting that c-ISCU is fully functional in vivo (29). In contrast, silencing of m-ISCU decreases Fe−S protein activity in both mitochondria and cytosol, perhaps because it disturbs mitochondrial and cytosolic iron homeostasis.
A point mutation of ISCU was found to be responsible for hereditary myopathy with severe exercise intolerance in a group of Swedish patients described in the past four decades (67,68). Nineteen patients from nine families have been identified in Sweden, and all but one could be connected to a common ancestor. These patients have a homozygous intronic G7044C mutation at a potential acceptor splice site between exons 4 and 5 of the ISCU gene. The substitution creates a polypyrimidine tract that promotes spliceosome assembly and leads to the inclusion of an additional exon, exon 4A, in the transcript. ISCU protein levels are drastically reduced in patient muscle samples, perhaps because the mutation introduces a premature stop codon that destabilizes the protein. The ISCU depletion leads to defective Fe−S cluster biogenesis and to mitochondrial iron overload in skeletal muscle. Without sufficient Fe−S cluster biogenesis, the heme synthetic enzyme ferrochelatase (FECH) is unstable and is significantly degraded in patient muscle (69). More recently, a new ISCU mutation was identified in two male siblings (70), who are heterozygous for the intronic G7044C mutation commonly seen in all other myopathy patients. Additionally, the brothers have a novel G149A missense mutation in exon 3 that changes a highly conserved glycine residue to glutamate. ISCU proteins in these patients are expressed at a normal level, but they are nonfunctional. The two brothers demonstrate more severe muscle pathology than other ISCU mutation patients. Furthermore, in contrast to others, these brothers show the additional phenotype of cardiac hypertrophic myopathy, which represents a new manifestation for diseases caused by ISCU mutations.
Clinically, the patients have lifelong severe exercise intolerance that begins in childhood. Minor exertion causes fatigue of active muscles, shortness of breath, and cardiac palpitations in association with lactic acidosis (67,68). Patients also experience episodes of rhabdomyolysis associated with muscle swelling and pain, weakness, and myoglobinuria. At present, there is no effective therapy available for ISCU-deficient myopathy, but a potential strategy is to manipulate gene expression by using antisense oligonucleotides (71), in which the antisense treatment induces skipping of the pseudoexons in patients and results in restoration of wild-type mRNA expression.
The Mitochondrial ATP Binding Cassette Protein, ABCB7, X-Linked Sideroblastic Anemia and Ataxia Syndrome (XLSA/A), and a Proposed Connection to Iron−Sulfur Cluster Biogenesis
ABCB7 proteins, including human ABCB7, are ABC half-transporters in the mitochondrial inner membrane (72,73). As shown in the yeast ABCB7-like homologue, Atm1, the transporter exposes a C-terminal ATP binding domain to the mitochondrial matrix and spans the membrane with six transmembrane segments. Atm1 has been proposed to export an unknown form of iron or Fe−S compound from the mitochondrion to cytosol for the maturation of cytosolic Fe−S proteins (74−77) and is proposed to be a key component of the putative Fe−S cluster export machinery. However, an extensive analysis of ATM3 gene mutant plants has suggested that the Atm1-like proteins in plants do not export any iron but rather export one or two compounds required for both Fe−S cluster and molybdenum cofactor (Moco) assembly in the cytosol (78). As a Moco synthesis intermediate, cPMP, accumulated in mitochondria of ATM3 mutant plants (79), the ATM3 protein was proposed to be a cPMP transporter. On the basis of the fact that mutations of SLC25A38, SLC19A2, ABCB7, and heme synthetic ALAS2 all cause a similar sideroblastic anemia phenotype (80) and the prediction that SLC25A38 may provide the glycine substrate of heme biosynthesis and SLC19A2 may provide thiamine for pyruvate dehydrogenase, it is possible that ABCB7 exports a heme or heme precursor molecule to the cytosol that conveys a signal about mitochondrial iron status to the cytosolic/nuclear compartment. If levels of this putative signaling molecule decrease, the cell imports more iron from the cytosol into mitochondria, resulting in mitochondrial iron overload and cytosolic iron deficiency, which slows cytosolic Fe−S cluster biogenesis. These molecular events could explain the sideroblastic and anemic phenotypes. However, since human patients with mutations of FECH, which catalyzes the last step in heme synthesis, do not usually show mitochondrial iron overload, we hypothesize that the ABCB7 substrate could be a product of an earlier step in heme synthesis. One such candidate is aminolevulinic acid (δ-ALA), the product of condensation of glycine and succinyl-CoA catalyzed by ALAS2. The mitochondrial carrier protein SLC25A38 is the putative glycine importer (81). The high-affinity thiamine transporter SLC19A2 may provide the thiamine cofactor for pyruvate dehydrogenase which is required for the production of succinyl-CoA (82). Notably, mutations in SLC25A38, SLC19A2, and ALAS2 all cause sideroblastic anemia.
Human ABCB7 has an additional role in heme synthesis and hematopoiesis as revealed by a putative interaction between ABCB7 and FECH which has been demonstrated in pull-down assays, and anemia caused by ABCB7 depletion (83,84). In contrast to non-hematopoietic tissues that do not demonstrate an observable phenotype, the bone marrow is markedly hypocellular in ABCB7 knockout mice (84) and the the levels of hemoglobin, absolute reticulocytes, white blood cells, and platelets are all decreased. Mutations of ABCB7 in humans cause hereditary X-linked sideroblastic anemia and ataxia (XLSA/A) (73). The mutated ABCB7 gene is on the X chromosome; in males (XY), one copy of the mutated gene is sufficient to cause the disorder, while in females (XX), a mutation must be present in both copies to cause the condition. Men are therefore affected more frequently than women. Another well-known XLSA is caused by mutations in the ALAS2 gene which encodes the first enzyme of heme synthesis in erythroid cells. Three families of patients with ABCB7-caused XLSA/A have been studied so far, and all affected patients are males, consistent with the X-linked feature. In the first family, the nucleotide T at position 1200 is mutated to G (T1200G), which changes the amino acid of Ile (ATT) to Met (ATG) at position 400 (I400M) in the protein. This residue is within the predicted fifth transmembrane domain of ABCB7 (85). In the second family, a single missense mutation is found in exon 10 of the ABCB7 gene in two affected brothers, and the mutation is a G-to-A transition at nucleotide 1305, resulting in a Glu-to-Lys substitution at residue 433 (E433K) in the putative sixth transmembrane domain of ABCB7 (72). In the third family, the affected brothers have a G-to-C transition at position 1299, which predicts a V411L substitution in the sixth putative transmembrane domain (86). All these disease-causing mutations are in transmembrane segments, indicating the importance of transmembrane segments to ABCB7 function.
In animal models, ABCB7 depletion decreases the rate of cytosolic Fe−S protein maturation. IRP1 converts to its IRE-binding form and inhibits the expression of ALAS2 in heme synthesis. Cellular iron homeostasis is impaired, and iron accumulates in mitochondria (77). This observation provides a clue for understanding the human disease. Although the level of protoporphyrin IX, the substrate of ferrochelatase (FECH), was found to be slightly increased (1.33-fold) in ABCB7 knockdown HeLa cells (87), there is no evidence that free protoporphyrin IX accumulates in ABCB7-deficient human patients and mouse models (77,84,86). Rather, the level of zinc protoporphyrin was increased in some patients and mouse models (84,86). The increase in the levels of zinc protoporphyrin indicates that FECH is functional, but ferrous iron is not available for production of the iron protoporphyrin, heme (84). In addition to its importance in erythropoiesis, ABCB7 may be required for mitochondrial iron homeostasis in neural cells, as suggested by ataxia symptoms in patients.
ABCB7-caused XLSA/A is characterized by nonprogressive cerebellar ataxia (with an infantile to early childhood onset) and mild hypochromic microcytic anemia (85). In some patients, the level of protoporphyrin IX is increased in erythrocytes despite normal iron stores. Bone marrow biopsy confirms the presence of ring sideroblasts (86), and some patients show retarded development (72). There is no cure for XLSA/A.
Other Genes and Possible Diseases
Human ISCA is likely an alternative scaffold, since it complements the growth of the yeast ISA1 mutant, which has been demonstrated to function in Fe−S cluster assembly in yeast (88). ISCA is present as an autoantigen in a patient with Sjogren’s syndrome, an autoimmune disease (31), but how and why ISCA is associated with the disease remain unknown. The molecular chaperone/cochaperone (HscA/HscB) system required for Fe−S cluster assembly in bacteria and yeast may be in the same pathway as frataxin in Fe−S cluster synthesis, because the phylogenetic distributions of hscA/hscB and frataxin genes overlap significantly (89). It was further hypothesized that the mutation of human hscA/hscB may cause hereditary ataxia syndromes similar to that caused by frataxin mutations (90), based on the observations that the expression pattern of human HscA/HscB in mitochondria-rich tissues is similar to that of frataxin and human HscB and frataxin both have conserved regions that probably play a role in protein−protein interactions (20,91). However, patients with mutations in HscA or -B have not been identified. Finally, because FECH deficiency causes erythropoietic protoporphyria (EPP), manifesting painful skin photosensitivity due to protoporphyrin accumulation (92), and deficient Fe−S cluster biogenesis severely diminishes FECH protein levels (69), there might be some cases of EPP in human patients caused by deficient Fe−S cluster assembly.
Concluding Remarks and Perspectives
Spatial Separation of Fe−S Cluster Biogenesis Machinery
In spite of the significant role of mitochondria in cellular Fe−S cluster biogenesis, it is apparent now that Fe−S clusters are assembled separately in distinct subcellular compartments in mammalian cells (11) and in other organisms. Even in microsporidia, primitive intracellular parasites of other eukaryotes that have tiny mitochondrial remnants called mitosomes, some essential components of Fe−S biogenesis are not located in the mitosome (93). Whereas microsporidial Hsp70 and Nfs1 are located in mitosomes, the main pools of Isu1 and frataxin are cytosolic. These observations are most compatible with the notion that the full Fe−S synthetic machinery is represented both inside and outside the mitosomes of microspordia, and Fe−S cluster biogenesis can occur independently in both compartments. The existence of multiple distinct Fe−S assembly systems is well-recognized in photosynthetic eukaryotes, which contain Fe−S assembly systems in mitochondria, plastids, and the cytosol (94−97). To date, it is thought that Fe−S cluster biosynthesis in plastids is independent of mitochondria. Unlike that in yeast, the essential cytosolic Fe−S protein Nbp35 acts without the Cfd1 partner in plants (98). Photosynthetic eukaryotes express Cfd1 proteins; one Cfd1 homologue called HCF101 is located in chloroplasts, whereas the other localizes to mitochondria rather than the cytosol (97). It is likely that the spatially separated Fe−S synthetic machinery of the mitochondria and plastids of a single cell needs to be coordinated and regulated, to optimize cellular iron homeostasis and compartmental distribution.
Disease Mechanisms
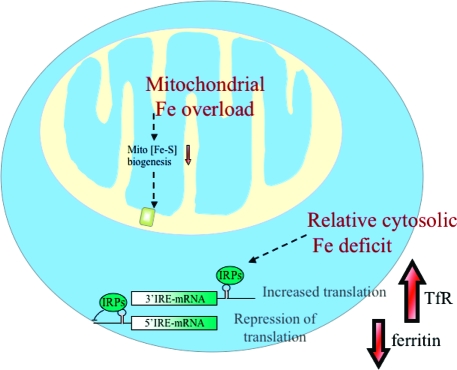
In each of the diseases caused by defective Fe−S cluster biogenesis, the failure of mitochondrial Fe−S cluster biogenesis leads to diminished activity of mitochondrial Fe−S proteins, mitochondrial iron overload, and cytosolic iron depletion (Figure 5). The cytosolic iron sensors IRP1, which senses iron levels through an iron−sulfur cluster, and IRP2, which senses iron levels through iron-dependent degradation by the hemerythrin-like ubiquitin ligase, FBXL5 (99−101), are activated, and they repress synthesis of proteins such as ferritin and ferroportin, which are encoded by transcripts that contain an IRP binding site in the 5′ UTR and increase the rate of synthesis of iron uptake proteins such as TfR1. However, these disorders produce strong tissue specificity; in Friedreich's ataxia, cardiomyocytes and a subset of neurons, especially sensory neurons, are adversely affected, whereas other tissues are spared. In ISCU myopathy, only skeletal muscles and cardiac cells are affected, and in glutaredoxin 5 deficiency, the only disease manifestation is anemia (Table 1). Thus, Friedreich ataxia patients have sensory impairments and heart problems; ISCU myopathy patients are weak, and the glutaredoxin 5-deficient patient is anemic. The phenotypes of these diseases differ significantly, even though each disease gene adversely affects the fundamental process of Fe−S cluster biogenesis. Pathology is manifested as sensory problems in Friedreich's ataxia, sideroblastic anemia in GLRX5 deficiency, and myopathy in ISCU deficiency, but it is unlikely that the fundamental defect is limited to these specific tissues, at least for some gene mutations. For instance, although the ABCB7 missense mutations affect all tissues, the major phenotype is sideroblastic anemia and ataxia, perhaps suggesting the ABCB7 protein has specific roles in neural and erythroid cells. In the case of GLRX5 deficiency, although it has been shown that asymptomatic tissues also lack the GLRX5 protein, the primary phenotype is limited to erythroid cells. Recent studies have suggested that the tissue specificity is due to the special combination of ALAS2, which contains a 5′ IRE, and FPN1b, which lacks a 5′ IRE (32); these two transcripts are simultaneously expressed only in erythroid cells, where activation of IRPs would be expected to cause unique effects.
Figure 5.
Deficiency in mitochondrial Fe−S cluster biogenesis causes mitochondrial iron overload, relative cytosolic iron deficiency, and activation of the cytosolic iron sensor proteins, IRP1 and IRP2. Deficiency in many of the major proteins involved in mitochondrial Fe−S cluster biogenesis results in defective mitochondrial Fe−S cluster biogenesis, which in turn results in transcriptional remodeling, mitochondrial iron overload, and cytosolic iron deficiency. IRP proteins are activated to be RNA regulatory proteins, which increase the rate of translation of 3′ IRE-mRNA, such as TfR1, whereas they inhibit the translation of mRNAs that contain IREs in the 5′ UTR, such as ferritin and ferroportin.
Table 1. Primarily Affected Tissues in Fe−S Cluster Synthesis Diseasesa.
| FXN | GLRX5 | ISCU | ABCB7 | |
|---|---|---|---|---|
| erythroblasts | + | + | ||
| sensory and cerebellar neurons | + | + | ||
| cardiomyocytes | + | + | ||
| skeletal myocytes | + |
The primarily affected cell types are sensory neurons and cardiac myocytes for frataxin-deficient Friedreich's ataxia, erythroblasts and cerebellar neurons for ABCB7-deficient XLSA/A, skeletal and cardiac myocytes for ISCU-deficient myopathy, and erythroblasts for GLRX5-deficient sideroblastic anemia.
It has been found that mitochondrial dysfunction leads to nuclear genome instability and that a reduced level of Fe−S cluster biogenesis especially in the cytosol (involving NAR1) is sufficient to induce genome instability (102). These results suggest that Fe−S cluster biogenesis is involved in a form of mitochondrion-to-genome communication. Another piece of evidence is that the cytosolic Fe−S synthetic protein, IOP1 (the NAR1 homologue in humans), was found to regulate the expression of HIF-1α (34), an important transcription factor. We propose that mammalian cells use a product of Fe−S cluster biogenesis to gauge and regulate mitochondrial iron homeostasis, similar to the role of cytosolic Fe−S cluster biogenesis in the regulation of the cytosolic regulatory protein, IRP1.
Elusive Mechanism That Links the Failure of Mitochondrial Fe−S Clsuter Biogenesis to Mitochondrial Iron Overload and Cytosolic Iron Deficiency
As noted before in the human diseases described, and also in tissue culture and knockdown studies of several Fe−S proteins, including ISCU (29), frataxin (53), and ISD11 (26), defective mitochondrial Fe−S cluster assembly appears to trigger a chain of events. First, mitochondrial Fe−S proteins, including aconitase and succinate dehydrogenase, fail to acquire their Fe−S prosthetic groups. Then there is a change in mitochondrial iron homeostasis that results in profound mitochondrial iron overload. This mitochondrial iron overload likely results from an increased level of iron import by the dedicated iron transporter, mitoferrin, and also from a diminished level of export by uncharacterized mitochondrial iron exporters. It is likely that mitochondrial iron overload contributes significantly to disease progression by oxidatively damaging the mitochondria, which further impairs function. Since mitochondrial iron overload appears to be caused by a failure of Fe−S cluster biogenesis and to contribute significantly to disease, it is increasingly important to understand the molecular events that contribute to mitochondrial iron overload. In the yeast model system, it was initially postulated that iron overload occurred because mitochondria lacked a dedicated iron exporter, and iron could exit the mitochondria mainly in the form of fully formed Fe−S clusters that would load clusters onto cytosolic and nuclear apoproteins. The loss of function of the yeast counterpart of ABCB7, Atm1, resulted in a loss of activity of the cytosolic Fe−S protein, Leu1 (103,104), and this information was interpreted to mean that Atm1 exported Fe−S clusters for insertion into cytosolic Fe−S proteins such as Leu1p and that its homologue ABCB7 exported Fe−S clusters for use in the cytosol of higher eukaryotes.
However, the notion that the mitochondria were the sole site of Fe−S cluster biogenesis in eukaryotes was undermined by the discovery that isoforms of the cysteine desulfurase, ISCS, the scaffold, ISCU, and the alternative scaffold, NFU, not only were present in the cytosol but also were shown to be important for cytosolic Fe−S cluster biogenesis in mammalian cells (29). Just as mammalian cytosolic iron homeostasis is regulated in part by a cytosolic iron−sulfur protein, IRP1, which functions either as a cytosolic aconitase or as an RNA regulatory protein depending on iron status, it is very likely that mitochondrial iron homeostasis is also highly regulated. Perhaps an iron−sulfur protein or a product of an iron−sulfur protein is exported from mitochondria and programs transcription of genes involved in mitochondrial iron uptake and export to increase or decrease the level of mitochondrial iron in response to this proposed exported signaling molecule. When synthesis of this mitochondrial iron status signaling molecule is defective, the cell responds to misperceived mitochondrial iron depletion by increasing the rate of mitochondrial iron uptake and decreasing the rate of export. Indeed, the level of transcriptional expression of the mitochondrial importer, mitoferrin, is increased in a Friedreich's ataxia model (54). Moreover, in a yeast model of frataxin deficiency, iron is mobilized from the iron overloaded mitochondria within minutes after normal frataxin expression is restored (105).
The phenomenon of mitochondrial iron overload is not limited to defects in iron−sulfur cluster biogenesis; it also occurs in other diseases, such as the sideroblastic anemia caused by ALAS2 mutations, even though ALAS2 mediates the first condensation of heme biosynthesis and is not directly involved in Fe−S cluster biogenesis. It is also interesting that in fibroblasts and lymphoblasts from patients with Friedreich's ataxia (53) and glutaredoxin 5 deficiency (32), impaired mitochondrial iron−sulfur cluster biogenesis is accompanied by cytosolic iron depletion, which may result in part from the fact that mitochondrial iron uptake occurs at the expense of cytosolic iron. However, recently another contributing factor has also been identified, because it appears that mitochondrial iron overload causes oxidative stress which leads to activation of genes that combat oxidative stress, including expression of the iron exporter ferroportin (32). The fact that cytosolic iron depletion occurs in conjunction with mitochondrial iron overload may explain why cytosolic iron−sulfur proteins have been known to be deficient in function. With insufficient iron for de novo cytosolic iron−sulfur cluster synthesis, activities of iron−sulfur proteins decrease, with significant implications for activities of known cytosolic iron−sulfur proteins such as IRP1 (15), the DNA repair enzymes XPD and FancJ (106), and PRI2, which is involved in DNA replication (107). Interestingly, array studies in yeast have revealed that interruption of mitochondrial iron−sulfur cluster biogenesis results in remodeling of iron metabolism, and loss of frataxin and loss of Atm1 both result in a markedly reduced level of transcription of Leu1 (108), the cytosolic iron−sulfur protein that was originally the basis for the hypothesis that mitochondria were the sole source of iron−sulfur clusters. Clearly, if the level of transcription of Leu1 is diminished, expression of the protein and enzymatic activity will be decreased, which was observed, though this loss of Leu1p activity was attributed previously to weakened acquisition of the iron−sulfur prosthetic group by the intact and abundant apoprotein.
On the basis of the problems with iron−sulfur cluster biogenesis in three human diseases, and the problems that are engendered in mitochondrial and cytosolic iron homeostasis, we suggest that the next frontier will consist of molecular characterization of a molecule synthesized in mitochondria that is exported to the cytosolic/nuclear compartment, perhaps by ABCB7, and which represents a gauge of mitochondrial iron status by possessing or depending on an iron−sulfur cluster for function. Regulation of mitochondrial iron homeostasis likely depends on mitochondrial signaling and transcriptional remodeling, and elucidation of the molecular details of this regulatory pathway may reveal important insights into an emerging class of human diseases.
Acknowledgments
We thank Wing-Hang Tong for help with making the model pictures. We apologize to colleagues whose work is not cited due to space limitations.
The work is funded by the National Institute of Child and Health and Human Development intramural program.
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: Fe−S, iron−sulfur; GLRX5, glutaredoxin 5; Grx, glutaredoxin; ABCB7, ATP-binding cassette, subfamily B, member 7; ISCU, iron−sulfur cluster scaffold homologue to NifU; FXN, frataxin; GSH, glutathione; Tf, transferrin; TfR1, transferrin receptor 1; DMT1, divalent metal transporter 1; FPN1, ferroportin 1; IRP, iron regulatory protein; ROS, reactive oxygen species; FRDA, Friedreich's ataxia; IRE, iron responsive element; ALAS2, aminolevulinate, delta, synthase 2; FECH, ferrochelatase; SA, sideroblastic anemia; XLSA/A, X-linked sideroblastic anemia and ataxia; Dfo, deferoxamine; Moco, molybdenum cofactor; δ-ALA, δ-aminolevulinic acid; EPP, erythropoietic protoporphyria; UTR, untranslated region; Atm1, mitochondrial ABC transporter 1 of Saccharomycas cerevisiae; Leu1, isopropylmalate isomerase, which catalyzes the second step in the leucine biosynthesis pathway of S. cerevisiae; Hb, hemoglobin.
References
- Yin L.; Wu N.; Curtin J. C.; Qatanani M.; Szwergold N. R.; Reid R. A.; Waitt G. M.; Parks D. J.; Pearce K. H.; Wisely G. B.; Lazar M. A. (2007) Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789. [DOI] [PubMed] [Google Scholar]
- Faller M.; Matsunaga M.; Yin S.; Loo J. A.; Guo F. (2007) Heme is involved in microRNA processing. Nat. Struct. Mol. Biol. 14, 23–29. [DOI] [PubMed] [Google Scholar]
- Hentze M. W.; Muckenthaler M. U.; Andrews N. C. (2004) Balancing acts: Molecular control of mammalian iron metabolism. Cell 117, 285–297. [DOI] [PubMed] [Google Scholar]
- De Domenico I.; McVey Ward D.; Kaplan J. (2008) Regulation of iron acquisition and storage: Consequences for iron-linked disorders. Nat. Rev. Mol. Cell Biol. 9, 72–81. [DOI] [PubMed] [Google Scholar]
- Ganz T. (2008) Iron homeostasis: Fitting the puzzle pieces together. Cell Metab. 7, 288–290. [DOI] [PubMed] [Google Scholar]
- Dong X. P.; Cheng X.; Mills E.; Delling M.; Wang F.; Kurz T.; Xu H. (2008) The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 455, 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A.; Tong W. H. (2005) Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell Biol. 6, 345–351. [DOI] [PubMed] [Google Scholar]
- Liu A.; Graslund A. (2000) Electron paramagnetic resonance evidence for a novel interconversion of [3Fe-4S]+ and [4Fe-4S]+ clusters with endogenous iron and sulfide in anaerobic ribonucleotide reductase activase in vitro. J. Biol. Chem. 275, 12367–12373. [DOI] [PubMed] [Google Scholar]
- Shi H.; Bencze K. Z.; Stemmler T. L.; Philpott C. C. (2008) A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G. C.; Cope J. J.; Li L.; Corson K.; Hersey C.; Ackermann G. E.; Gwynn B.; Lambert A. J.; Wingert R. A.; Traver D.; Trede N. S.; Barut B. A.; Zhou Y.; Minet E.; Donovan A.; Brownlie A.; Balzan R.; Weiss M. J.; Peters L. L.; Kaplan J.; Zon L. I.; Paw B. H. (2006) Mitoferrin is essential for erythroid iron assimilation. Nature 440, 96–100. [DOI] [PubMed] [Google Scholar]
- Rouault T. A.; Tong W. H. (2008) Iron-sulfur cluster biogenesis and human disease. Trends Genet. 24, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R.; Muhlenhoff U. (2008) Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77, 669–700. [DOI] [PubMed] [Google Scholar]
- Keel S. B.; Doty R. T.; Yang Z.; Quigley J. G.; Chen J.; Knoblaugh S.; Kingsley P. D.; De Domenico I.; Vaughn M. B.; Kaplan J.; Palis J.; Abkowitz J. L. (2008) A heme export protein is required for red blood cell differentiation and iron homeostasis. Science 319, 825–828. [DOI] [PubMed] [Google Scholar]
- Quigley J. G.; Yang Z.; Worthington M. T.; Phillips J. D.; Sabo K. M.; Sabath D. E.; Berg C. L.; Sassa S.; Wood B. L.; Abkowitz J. L. (2004) Identification of a human heme exporter that is essential for erythropoiesis. Cell 118, 757–766. [DOI] [PubMed] [Google Scholar]
- Rouault T. A. (2006) The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2, 406–414. [DOI] [PubMed] [Google Scholar]
- Wallander M. L.; Leibold E. A.; Eisenstein R. S. (2006) Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta 1763, 668–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. C.; Dean D. R.; Smith A. D.; Johnson M. K. (2005) Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281. [DOI] [PubMed] [Google Scholar]
- Lill R.; Muhlenhoff U. (2005) Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30, 133–141. [DOI] [PubMed] [Google Scholar]
- Pandolfo M.; Pastore A. (2009) The pathogenesis of Friedreich ataxia and the structure and function of frataxin. J. Neurol. 256, Suppl. 19–17. [DOI] [PubMed] [Google Scholar]
- Vickery L. E.; Cupp-Vickery J. R. (2007) Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit. Rev. Biochem. Mol. Biol. 42, 95–111. [DOI] [PubMed] [Google Scholar]
- Gelling C.; Dawes I. W.; Richhardt N.; Lill R.; Muhlenhoff U. (2008) Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol. Cell. Biol. 28, 1851–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J.; Pierik A. J.; Netz D. J.; Muhlenhoff U.; Lill R. (2004) The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23, 2105–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Lyver E. R.; Nakamaru-Ogiso E.; Yoon H.; Amutha B.; Lee D. W.; Bi E.; Ohnishi T.; Daldal F.; Pain D.; Dancis A. (2008) Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 28, 5569–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. H.; Rouault T. A. (2007) Metabolic regulation of citrate and iron by aconitases: Role of iron-sulfur cluster biogenesis. BioMetals 20, 549–564. [DOI] [PubMed] [Google Scholar]
- Land T.; Rouault T. A. (1998) Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell 2, 807–815. [DOI] [PubMed] [Google Scholar]
- Shi Y.; Ghosh M. C.; Tong W. H.; Rouault T. A. (2009) Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet. 18, 3014–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon T.; Cowan J. A. (2003) Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125, 6078–6084. [DOI] [PubMed] [Google Scholar]
- Tong W. H.; Rouault T. (2000) Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 19, 5692–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. H.; Rouault T. A. (2006) Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 3, 199–210. [DOI] [PubMed] [Google Scholar]
- Tong W. H.; Jameson G. N.; Huynh B. H.; Rouault T. A. (2003) Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. U.S.A. 100, 9762–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozar-Castellano I.; del Valle Machargo M.; Trujillo E.; Arteaga M. F.; Gonzalez T.; Martin-Vasallo P.; Avila J. (2004) hIscA: A protein implicated in the biogenesis of iron-sulfur clusters. Biochim. Biophys. Acta 1700, 179–188. [DOI] [PubMed] [Google Scholar]
- Ye H.; Jeong S. Y.; Ghosh M. C.; Kovtunovych G.; Silvestri L.; Ortillo D.; Uchida N.; Tisdale J.; Camaschella C.; Rouault T. A. (2010) Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J. Clin. Invest. 120, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.; Lee F. S. (2008) A role for IOP1 in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 283, 9231–9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; Song D.; Flores A.; Zhao Q.; Mooney S. M.; Shaw L. M.; Lee F. S. (2007) IOP1, a novel hydrogenase-like protein that modulates hypoxia-inducible factor-1α activity. Biochem. J. 401, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O.; Netz D. J.; Niggemeyer B.; Rosser R.; Eisenstein R. S.; Puccio H.; Pierik A. J.; Lill R. (2008) Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Mol. Cell. Biol. 28, 5517–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquaviva F.; De Biase I.; Nezi L.; Ruggiero G.; Tatangelo F.; Pisano C.; Monticelli A.; Garbi C.; Acquaviva A. M.; Cocozza S. (2005) Extra-mitochondrial localisation of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2. J. Cell Sci. 118, 3917–3924. [DOI] [PubMed] [Google Scholar]
- Condo I.; Ventura N.; Malisan F.; Tomassini B.; Testi R. (2006) A pool of extramitochondrial frataxin that promotes cell survival. J. Biol. Chem. 281, 16750–16756. [DOI] [PubMed] [Google Scholar]
- Ojeda L.; Keller G.; Muhlenhoff U.; Rutherford J. C.; Lill R.; Winge D. R. (2006) Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 281, 17661–17669. [DOI] [PubMed] [Google Scholar]
- Li H.; Mapolelo D. T.; Dingra N. N.; Naik S. G.; Lees N. S.; Hoffman B. M.; Riggs-Gelasco P. J.; Huynh B. H.; Johnson M. K.; Outten C. E. (2009) The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48, 9569–9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumanovics A.; Chen O. S.; Li L.; Bagley D.; Adkins E. M.; Lin H.; Dingra N. N.; Outten C. E.; Keller G.; Winge D.; Ward D. M.; Kaplan J. (2008) Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 283, 10276–10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano V.; Montermini L.; Lutz Y.; Cova L.; Hindelang C.; Jiralerspong S.; Trottier Y.; Kish S. J.; Faucheux B.; Trouillas P.; Authier F. J.; Durr A.; Mandel J. L.; Vescovi A.; Pandolfo M.; Koenig M. (1997) Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 6, 1771–1780. [DOI] [PubMed] [Google Scholar]
- Condo I.; Ventura N.; Malisan F.; Rufini A.; Tomassini B.; Testi R. (2007) In vivo maturation of human frataxin. Hum. Mol. Genet. 16, 1534–1540. [DOI] [PubMed] [Google Scholar]
- Martelli A.; Wattenhofer-Donze M.; Schmucker S.; Bouvet S.; Reutenauer L.; Puccio H. (2007) Frataxin is essential for extramitochondrial Fe-S cluster proteins in mammalian tissues. Hum. Mol. Genet. 16, 2651–2658. [DOI] [PubMed] [Google Scholar]
- Bulteau A. L.; O’Neill H. A.; Kennedy M. C.; Ikeda-Saito M.; Isaya G.; Szweda L. I. (2004) Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305, 242–245. [DOI] [PubMed] [Google Scholar]
- Gakh O.; Adamec J.; Gacy A. M.; Twesten R. D.; Owen W. G.; Isaya G. (2002) Physical evidence that yeast frataxin is an iron storage protein. Biochemistry 41, 6798–6804. [DOI] [PubMed] [Google Scholar]
- Yoon T.; Cowan J. A. (2004) Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J. Biol. Chem. 279, 25943–25946. [DOI] [PubMed] [Google Scholar]
- Radisky D. C.; Babcock M. C.; Kaplan J. (1999) The yeast frataxin homologue mediates mitochondrial iron efflux. Evidence for a mitochondrial iron cycle. J. Biol. Chem. 274, 4497–4499. [DOI] [PubMed] [Google Scholar]
- Campuzano V.; Montermini L.; Molto M. D.; Pianese L.; Cossee M.; Cavalcanti F.; Monros E.; Rodius F.; Duclos F.; Monticelli A.; Zara F.; Canizares J.; Koutnikova H.; Bidichandani S. I.; Gellera C.; Brice A.; Trouillas P.; De Michele G.; Filla A.; De Frutos R.; Palau F.; Patel P. I.; Di Donato S.; Mandel J. L.; Cocozza S.; Koenig M.; Pandolfo M. (1996) Friedreich's ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427. [DOI] [PubMed] [Google Scholar]
- De Biase I.; Rasmussen A.; Endres D.; Al-Mahdawi S.; Monticelli A.; Cocozza S.; Pook M.; Bidichandani S. I. (2007) Progressive GAA expansions in dorsal root ganglia of Friedreich's ataxia patients. Ann. Neurol. 61, 55–60. [DOI] [PubMed] [Google Scholar]
- Seznec H.; Simon D.; Bouton C.; Reutenauer L.; Hertzog A.; Golik P.; Procaccio V.; Patel M.; Drapier J. C.; Koenig M.; Puccio H. (2005) Friedreich ataxia: The oxidative stress paradox. Hum. Mol. Genet. 14, 463–474. [DOI] [PubMed] [Google Scholar]
- Puccio H.; Koenig M. (2002) Friedreich ataxia: A paradigm for mitochondrial diseases. Curr. Opin. Genet. Dev. 12, 272–277. [DOI] [PubMed] [Google Scholar]
- Rotig A.; Sidi D.; Munnich A.; Rustin P. (2002) Molecular insights into Friedreich's ataxia and antioxidant-based therapies. Trends Mol. Med. 8, 221–224. [DOI] [PubMed] [Google Scholar]
- Li K.; Besse E. K.; Ha D.; Kovtunovych G.; Rouault T. A. (2008) Iron-dependent regulation of frataxin expression: Implications for treatment of Friedreich ataxia. Hum. Mol. Genet. 17, 2265–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. L.; Becker E. M.; Whitnall M.; Rahmanto Y. S.; Ponka P.; Richardson D. R. (2009) Elucidation of the mechanism of mitochondrial iron loading in Friedreich's ataxia by analysis of a mouse mutant. Proc. Natl. Acad. Sci. U.S.A. 106, 16381–16386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. D. (2008) DNA triplexes and Friedreich ataxia. FASEB J. 22, 1625–1634. [DOI] [PubMed] [Google Scholar]
- Rustin P.; von Kleist-Retzow J. C.; Chantrel-Groussard K.; Sidi D.; Munnich A.; Rotig A. (1999) Effect of idebenone on cardiomyopathy in Friedreich's ataxia: A preliminary study. Lancet 354, 477–479. [DOI] [PubMed] [Google Scholar]
- Rustin P.; Rotig A.; Munnich A.; Sidi D. (2002) Heart hypertrophy and function are improved by idebenone in Friedreich's ataxia. Free Radical Res. 36, 467–469. [DOI] [PubMed] [Google Scholar]
- Kakhlon O.; Manning H.; Breuer W.; Melamed-Book N.; Lu C.; Cortopassi G.; Munnich A.; Cabantchik Z. I. (2008) Cell functions impaired by frataxin deficiency are restored by drug-mediated iron relocation. Blood 112, 5219–5227. [DOI] [PubMed] [Google Scholar]
- Herrero E.; de la Torre-Ruiz M. A. (2007) Monothiol glutaredoxins: A common domain for multiple functions. Cell. Mol. Life Sci. 64, 1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Manzaneque M. T.; Tamarit J.; Belli G.; Ros J.; Herrero E. (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 13, 1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert R. A.; Galloway J. L.; Barut B.; Foott H.; Fraenkel P.; Axe J. L.; Weber G. J.; Dooley K.; Davidson A. J.; Schmid B.; Paw B. H.; Shaw G. C.; Kingsley P.; Palis J.; Schubert H.; Chen O.; Kaplan J.; Zon L. I. (2005) Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature 436, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S.; Gama F.; Molina-Navarro M. M.; Gualberto J. M.; Claxton R.; Naik S. G.; Huynh B. H.; Herrero E.; Jacquot J. P.; Johnson M. K.; Rouhier N. (2008) Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 27, 1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina M. M.; Belli G.; de la Torre M. A.; Rodriguez-Manzaneque M. T.; Herrero E. (2004) Nuclear monothiol glutaredoxins of Saccharomyces cerevisiae can function as mitochondrial glutaredoxins. J. Biol. Chem. 279, 51923–51930. [DOI] [PubMed] [Google Scholar]
- Camaschella C.; Campanella A.; De Falco L.; Boschetto L.; Merlini R.; Silvestri L.; Levi S.; Iolascon A. (2007) The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood 110, 1353–1358. [DOI] [PubMed] [Google Scholar]
- Zhang D. L.; Hughes R. M.; Ollivierre-Wilson H.; Ghosh M. C.; Rouault T. A. (2009) A ferroportin transcript that lacks an iron-responsive element enables duodenal and erythroid precursor cells to evade translational repression. Cell Metab. 9, 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.; Tong W. H.; Hughes R. M.; Rouault T. A. (2006) Roles of the mammalian cytosolic cysteine desulfurase, ISCS, and scaffold protein, ISCU, in iron-sulfur cluster assembly. J. Biol. Chem. 281, 12344–12351. [DOI] [PubMed] [Google Scholar]
- Mochel F.; Knight M. A.; Tong W. H.; Hernandez D.; Ayyad K.; Taivassalo T.; Andersen P. M.; Singleton A.; Rouault T. A.; Fischbeck K. H.; Haller R. G. (2008) Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am. J. Hum. Genet. 82, 652–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A.; Lind L.; Thornell L. E.; Holmberg M. (2008) Myopathy with lactic acidosis is linked to chromosome 12q23.3−24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum. Mol. Genet. 17, 1666–1672. [DOI] [PubMed] [Google Scholar]
- Crooks D. R.; Ghosh M. C.; Haller R. G.; Tong W. H.; Rouault T. A. (2010) Post-translational stability of the heme biosynthetic enzyme ferrochelatase is dependent on iron availability and intact iron-sulfur cluster assembly machinery. Blood 115, 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollberg G.; Tulinius M.; Melberg A.; Darin N.; Andersen O.; Holmgren D.; Oldfors A.; Holme E. (2009) Clinical manifestation and a new ISCU mutation in iron-sulphur cluster deficiency myopathy. Brain 132, 2170–2179. [DOI] [PubMed] [Google Scholar]
- Kollberg G.; Holme E. (2009) Antisense oligonucleotide therapeutics for iron-sulphur cluster deficiency myopathy. Neuromuscular Disord. 19, 833–836. [DOI] [PubMed] [Google Scholar]
- Bekri S.; Kispal G.; Lange H.; Fitzsimons E.; Tolmie J.; Lill R.; Bishop D. F. (2000) Human ABC7 transporter: Gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron-sulfur protein maturation. Blood 96, 3256–3264. [PubMed] [Google Scholar]
- Lill R.; Kispal G. (2001) Mitochondrial ABC transporters. Res. Microbiol. 152, 331–340. [DOI] [PubMed] [Google Scholar]
- Csere P.; Lill R.; Kispal G. (1998) Identification of a human mitochondrial ABC transporter, the functional orthologue of yeast Atm1p. FEBS Lett. 441, 266–270. [DOI] [PubMed] [Google Scholar]
- Savary S.; Allikmets R.; Denizot F.; Luciani M. F.; Mattei M. G.; Dean M.; Chimini G. (1997) Isolation and chromosomal mapping of a novel ATP-binding cassette transporter conserved in mouse and human. Genomics 41, 275–278. [DOI] [PubMed] [Google Scholar]
- Shimada Y.; Okuno S.; Kawai A.; Shinomiya H.; Saito A.; Suzuki M.; Omori Y.; Nishino N.; Kanemoto N.; Fujiwara T.; Horie M.; Takahashi E. (1998) Cloning and chromosomal mapping of a novel ABC transporter gene (hABC7), a candidate for X-linked sideroblastic anemia with spinocerebellar ataxia. J. Hum. Genet. 43, 115–122. [DOI] [PubMed] [Google Scholar]
- Pondarre C.; Antiochos B. B.; Campagna D. R.; Clarke S. L.; Greer E. L.; Deck K. M.; McDonald A.; Han A. P.; Medlock A.; Kutok J. L.; Anderson S. A.; Eisenstein R. S.; Fleming M. D. (2006) The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum. Mol. Genet. 15, 953–964. [DOI] [PubMed] [Google Scholar]
- Bernard D. G.; Cheng Y.; Zhao Y.; Balk J. (2009) An allelic mutant series of ATM3 reveals its key role in the biogenesis of cytosolic iron-sulfur proteins in Arabidopsis. Plant Physiol. 151, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschner J.; Lachmann N.; Schulze J.; Geisler M.; Selbach K.; Santamaria-Araujo J.; Balk J.; Mendel R. R.; Bittner F. (2010) A Novel Role for Arabidopsis Mitochondrial ABC Transporter ATM3 in Molybdenum Cofactor Biosynthesis. Plant Cell 22, 468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A. K.; Campagna D. R.; McLoughlin E. M.; Agarwal S.; Fleming M. D.; Bottomley S. S.; Neufeld E. J. (2010) Systematic molecular genetic analysis of congenital sideroblastic anemia: Evidence for genetic heterogeneity and identification of novel mutations. Pediatr. Blood Cancer 54, 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey D. L.; Jiang H.; Campagna D. R.; Evans S. C.; Ferguson M.; Kellogg M. D.; Lachance M.; Matsuoka M.; Nightingale M.; Rideout A.; Saint-Amant L.; Schmidt P. J.; Orr A.; Bottomley S. S.; Fleming M. D.; Ludman M.; Dyack S.; Fernandez C. V.; Samuels M. E. (2009) Mutations in mitochondrial carrier family gene SLC25A38 cause nonsyndromic autosomal recessive congenital sideroblastic anemia. Nat. Genet. 41, 651–653. [DOI] [PubMed] [Google Scholar]
- Abboud M. R.; Alexander D.; Najjar S. S. (1985) Diabetes mellitus, thiamine-dependent megaloblastic anemia, and sensorineural deafness associated with deficient α-ketoglutarate dehydrogenase activity. J. Pediatr. 107, 537–541. [DOI] [PubMed] [Google Scholar]
- Taketani S.; Kakimoto K.; Ueta H.; Masaki R.; Furukawa T. (2003) Involvement of ABC7 in the biosynthesis of heme in erythroid cells: Interaction of ABC7 with ferrochelatase. Blood 101, 3274–3280. [DOI] [PubMed] [Google Scholar]
- Pondarre C.; Campagna D. R.; Antiochos B.; Sikorski L.; Mulhern H.; Fleming M. D. (2007) Abcb7, the gene responsible for X-linked sideroblastic anemia with ataxia, is essential for hematopoiesis. Blood 109, 3567–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R.; Raskind W. H.; Hutchinson A.; Schueck N. D.; Dean M.; Koeller D. M. (1999) Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum. Mol. Genet. 8, 743–749. [DOI] [PubMed] [Google Scholar]
- Maguire A.; Hellier K.; Hammans S.; May A. (2001) X-linked cerebellar ataxia and sideroblastic anaemia associated with a missense mutation in the ABC7 gene predicting V411L. Br. J. Haematol. 115, 910–917. [DOI] [PubMed] [Google Scholar]
- Cavadini P.; Biasiotto G.; Poli M.; Levi S.; Verardi R.; Zanella I.; Derosas M.; Ingrassia R.; Corrado M.; Arosio P. (2007) RNA silencing of the mitochondrial ABCB7 transporter in HeLa cells causes an iron-deficient phenotype with mitochondrial iron overload. Blood 109, 3552–3559. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U.; Gerl M. J.; Flauger B.; Pirner H. M.; Balser S.; Richhardt N.; Lill R.; Stolz J. (2007) The ISC [corrected] proteins Isa1 and Isa2 are required for the function but not for the de novo synthesis of the Fe/S clusters of biotin synthase in Saccharomyces cerevisiae. Eukaryotic Cell 6, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M. A.; Snel B.; Bork P.; Gibson T. J. (2001) The phylogenetic distribution of frataxin indicates a role in iron-sulfur cluster protein assembly. Hum. Mol. Genet. 10, 2463–2468. [DOI] [PubMed] [Google Scholar]
- Sun G.; Gargus J. J.; Ta D. T.; Vickery L. E. (2003) Identification of a novel candidate gene in the iron-sulfur pathway implicated in ataxia-susceptibility: Human gene encoding HscB, a J-type co-chaperone. J. Hum. Genet. 48, 415–419. [DOI] [PubMed] [Google Scholar]
- Cupp-Vickery J. R.; Vickery L. E. (2000) Crystal structure of Hsc20, a J-type Co-chaperone from Escherichia coli. J. Mol. Biol. 304, 835–845. [DOI] [PubMed] [Google Scholar]
- Todd D. J. (1994) Erythropoietic protoporphyria. Br. J. Dermatol. 131, 751–766. [DOI] [PubMed] [Google Scholar]
- Goldberg A. V.; Molik S.; Tsaousis A. D.; Neumann K.; Kuhnke G.; Delbac F.; Vivares C. P.; Hirt R. P.; Lill R.; Embley T. M. (2008) Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature 452, 624–628. [DOI] [PubMed] [Google Scholar]
- Xu X. M.; Moller S. G. (2008) Iron-sulfur cluster biogenesis systems and their crosstalk. ChemBioChem 9, 2355–2362. [DOI] [PubMed] [Google Scholar]
- Ye H.; Pilon M.; Pilon-Smits E. A. (2006) CpNifS-dependent iron-sulfur cluster biogenesis in chloroplasts. New Phytol. 171, 285–292. [DOI] [PubMed] [Google Scholar]
- Balk J.; Lobreaux S. (2005) Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 10, 324–331. [DOI] [PubMed] [Google Scholar]
- Godman J.; Balk J. (2008) Genome analysis of Chlamydomonas reinhardtii reveals the existence of multiple, compartmentalized iron-sulfur protein assembly machineries of different evolutionary origins. Genetics 179, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bych K.; Netz D. J.; Vigani G.; Bill E.; Lill R.; Pierik A. J.; Balk J. (2008) The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J. Biol. Chem. 283, 35797–35804. [DOI] [PubMed] [Google Scholar]
- Salahudeen A. A.; Thompson J. W.; Ruiz J. C.; Ma H. W.; Kinch L. N.; Li Q.; Grishin N. V.; Bruick R. K. (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht A. A.; Zumbrennen K. B.; Huang X.; Powers D. N.; Durazo A.; Sun D.; Bhaskaran N.; Persson A.; Uhlen M.; Sangfelt O.; Spruck C.; Leibold E. A.; Wohlschlegel J. A. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A. (2009) Cell biology. An ancient gauge for iron. Science 326, 676–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch J. R.; McMurray M. A.; Nelson Z. W.; Gottschling D. E. (2009) Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 137, 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G.; Csere P.; Guiard B.; Lill R. (1997) The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 418, 346–350. [DOI] [PubMed] [Google Scholar]
- Kispal G.; Csere P.; Prohl C.; Lill R. (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen O. S.; Hemenway S.; Kaplan J. (2002) Inhibition of Fe-S cluster biosynthesis decreases mitochondrial iron export: Evidence that Yfh1p affects Fe-S cluster synthesis. Proc. Natl. Acad. Sci. U.S.A. 99, 12321–12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J.; Makrantoni V.; Ingledew W. J.; Stark M. J.; White M. F. (2006) The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23, 801–808. [DOI] [PubMed] [Google Scholar]
- Klinge S.; Hirst J.; Maman J. D.; Krude T.; Pellegrini L. (2007) An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F.; Talibi D. (2001) Mitochondrial control of iron homeostasis. A genome wide analysis of gene expression in a yeast frataxin-deficient strain. J. Biol. Chem. 276, 7762–7768. [DOI] [PubMed] [Google Scholar]
- Nakai Y.; Nakai M.; Hayashi H.; Kagamiyama H. (2001) Nuclear localization of yeast Nfs1p is required for cell survival. J. Biol. Chem. 276, 8314–8320. [DOI] [PubMed] [Google Scholar]