Summary
The incorporation of histone H3 variants has been implicated in the epigenetic memory of cellular state. Using genome editing with zinc finger nucleases to tag endogenous H3.3, we report genome-wide profiles of H3 variants in mammalian embryonic stem (ES) cells and neuronal precursor cells. Genome-wide patterns of H3.3 are dependent on amino acid sequence, and change with cellular differentiation at developmentally regulated loci. The H3.3 chaperone Hira is required for H3.3 enrichment at active and repressed genes. Strikingly, Hira is not essential for localization of H3.3 at telomeres and many transcription factor binding sites. Immunoaffinity purification and mass spectrometry reveal that the proteins Atrx and Daxx associate with H3.3 in a Hira-independent manner. Atrx is required for Hira-independent localization of H3.3 at telomeres, and for the repression of telomeric RNA. Our data demonstrate that multiple and distinct factors are responsible for H3.3 localization at specific genomic locations in mammalian cells.
Introduction
Genetic and biochemical evidence have recently converged to connect epigenetic mechanisms at the level of chromatin (Bernstein et al., 2007; Goldberg et al., 2007; Henikoff, 2008). In addition to nucleosome remodeling and covalent modifications, eukaryotic cells generate variation in chromatin by the introduction of variant histone proteins (Henikoff, 2008). Mammalian cells express three major types of non-centromeric histone H3 variants, H3.1, H3.2, and H3.3 (Hake and Allis, 2006; Hake et al., 2006). Although histone H3.3 differs from H3.2 and H3.1 at only 4 or 5 amino acids (Figure S1A), H3.3 is specifically enriched at transcriptionally active genes and regulatory elements in non-pluripotent cells (Ahmad and Henikoff, 2002; Jin et al., 2009; Mito et al., 2005, 2007).
Histone H3.3 is incorporated into chromatin in both a replication-coupled (RC) and replication-independent (RI) manner, while the incorporation of H3.2 is coupled to replication (Ahmad and Henikoff, 2002; De Koning et al., 2007). The histone chaperone CAF-1 is found in a complex with H3.1, and mediates RC nucleosome assembly (Smith and Stillman, 1989; Tagami et al., 2004). In contrast, the histone chaperone Hira has been found in a complex with H3.3, and mediates RI nucleosome assembly (Ray-Gallet et al., 2002; Tagami et al., 2004).
Hira has been implicated in H3.3-specific deposition and chromatin assembly (Loyola and Almouzni, 2007). Although Hira is required for chromatin assembly and H3.3 deposition in the male pronucleus of Drosophila, Hira is not required for global H3.3 deposition in Drosophila embryos or adult cells, suggesting that alternate pathways may mediate H3.3 nucleosome assembly (Bonnefoy et al., 2007; Loppin et al., 2005). Indeed, the chromatin remodeling factor CHD1 was shown to physically associate with Hira, and has been suggested to work with Hira to mediate H3.3 incorporation into chromatin in Drosophila (Konev et al., 2007).
In Drosophila, both Hira and H3.3 are required for fertility and for transcriptional regulation of specific genes, but not for developmental viability (Bonnefoy et al., 2007; Hodl and Basler, 2009; Nakayama et al., 2007; Sakai et al., 2009). However, in mice, targeted mutagenesis of Hira results in a more severe phenotype, with gastrulation defects leading to early embryonic lethality (Roberts et al., 2002). Given the conserved association between H3.3 and active chromatin, H3.3 has been speculated to play an important role in mammalian ES cells (Creyghton et al., 2008; Gaspar-Maia et al., 2009). However, no genome-wide studies in pluripotent cells distinguish between H3 variants, nor do they examine the genome-wide role of Hira or other histone chaperones in specifying H3.3 localization at specific genomic regions.
Here we report genome-wide profiles of histone H3 variant localization in mammalian ES cells and neuronal precursor cells (NPCs), and we establish the dependence and independence of these patterns on Hira. We find that Hira is required for genome-wide H3.3 enrichment at active and repressed genes in ES cells. Surprisingly, H3.3 enrichment at many transcription factor binding sites (TFBS) and telomeres is Hira-independent. To identify factors that might mediate specific Hira-independent localization of H3.3, we use immunoprecipitation and mass spectrometry. We identify Atrx and Daxx as proteins that specifically associate with H3.3 in both pluripotent and non-pluripotent cells, both in the presence and in the absence of Hira. Unlike Hira, Atrx is not required for H3.3 localization at genes or TFBS. However, Atrx is specifically required for enrichment of H3.3 at telomeres in ES cells, and for the repression of telomeric RNA.
Results
Genome-wide patterns of H3.3 enrichment are dependent upon endogenous amino acid sequence
To distinguish H3 variants in our study without altering endogenous levels of H3 variant expression, we used designed zinc finger nucleases (ZFNs) (Carroll, 2008) both in gene addition (Moehle et al., 2007) and correction (Urnov et al., 2005) modes to engineer a panel of heterozygous ES lines carrying one allele of wild-type H3.3B, and another allele of H3.3B with a C-terminal EYFP tag (H3.3-EYFP), HA tag (H3.3-HA), or H3.3B with an epitope tag and simultaneous mutation towards H3.2 or H3.1 (H3.2-EYFP, H3.1-EYFP, H3.2-HA, H3.1S31-HA) (Figure S1). All heterozygously tagged ES cells retain three “wild-type” copies of H3.3 genes, including one copy of unmodified H3.3B (Figure S1G), and two copies of H3.3A. Western blots demonstrate equal protein expression in the epitope-tagged H3 variant ES lines (Figure S1H).
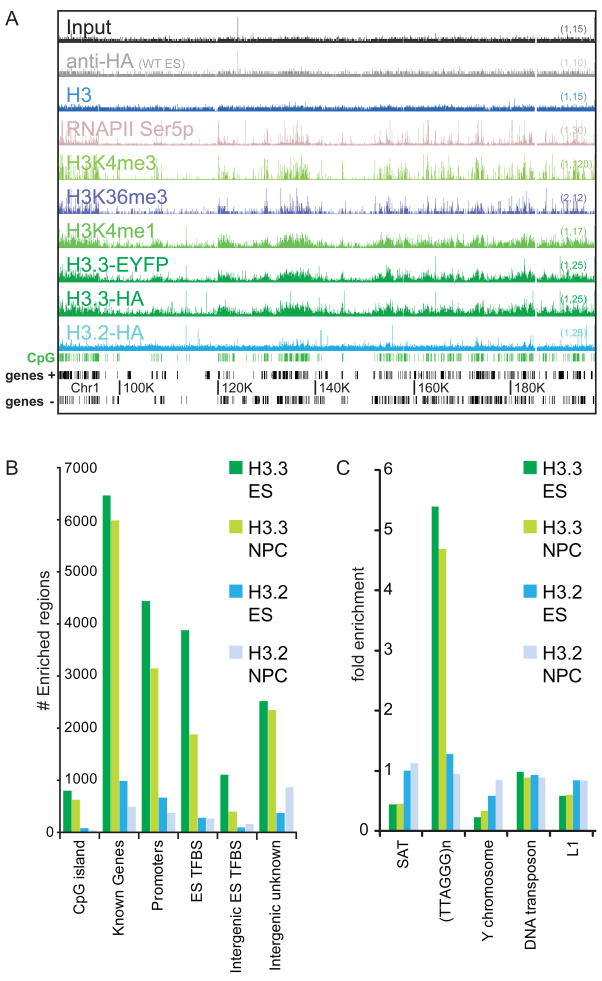
To assess the genome-wide localization of histone H3 variants at high resolution, we used chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq) (Barski et al., 2007; Mikkelsen et al., 2007). We found 10,099 total genic and intergenic regions highly enriched for H3.3 in crosslinking ChIP-seq of ES cells, in contrast to 1,442 regions enriched for H3.2, and we observe that gene-rich regions of the genome show greater enrichment of H3.3 than H3.2 (Figure 1A, B). On a chromosomal scale map, H3.3 enrichment correlates with markers of transcription, including Ser-5 phosphorylated RNA polymerase II (RNAPII), H3K4me3, H3K36me3, and H3K4me1 (Figure 1A). Unbiased clustering analyses confirm the genome-wide correlation of H3.3 with active histone modifications, particularly H3K4me1 (Table S1). Despite different epitope tags, we found extremely similar profiles of H3.3-HA and H3.3-EYFP (Figure 1A, Table S1, Figure S2A–B). Genome-wide analysis also reveals specific enrichment of H3.3 at previously identified genic and intergenic ES TFBS (Chen et al., 2008), as well as peaks of H3.3 in specific unannotated intergenic regions (Figure 1B).
Figure 1. Genomic localization of histone H3.3 is dependent on amino acid sequence.
A) ChIP-seq profiles across 120 MB of chromosome 1 in mouse ES cells, using antibodies as indicated. Y-axis represents the number of reads spanning a genomic position (scale on right, (baseline, maximum)). CpG islands (green) and genes (refseq (+) and refseq (−)) are shown below plots. Data for general H3 in ES cells is from (Mikkelsen et al., 2007).
B) Annotations of H3 variant enriched regions. Y-axis is the number of enriched regions identified for each H3 variant, while the X-axis represents different categories of gene annotation and ES TFBS (defined in Chen et al., 2008).
C) Association of H3.3 with repetitive elements. Data are presented as fold enrichment over input. SAT = satellite repeats, (TTAGGG)n = telomeric DNA repeats. See Figure S1 for details of ZFN-mediated generation and characterization of ES cell lines, and Table S1 for genome-wide correlation of ChIP-seq datasets.
To analyze H3 variant enrichment in different classes of repeats, we determined the relative enrichment of specific repeat sequences from our ChIP-seq experiments and compared it to that from control input DNA. H3.3 was reproducibly depleted in satellite repeat sequences and in Y-chromosomal repeat DNA (Figure 1C). We found the most significant enrichment of H3.3 in the (TTAGGG)n repeat that is the conserved telomeric DNA sequence among vertebrates (Meyne et al., 1989) (Figure 1C). This telomeric enrichment of H3.3 is consistent with telomeric foci of incorporation visible on the largely heterochromatic Y-chromosome (Figure S1J–O), and with a recent report of telomeric localization of transfected epitope-tagged H3.3 in ES cells (Wong et al., 2009). Mutation of H3.3B to H3.2 or H3.1S31 alters genome-wide patterns of H3.3 enrichment, demonstrating that the amino acid sequence of an endogenous H3.3 gene determines its genomic localization in mammalian cells.
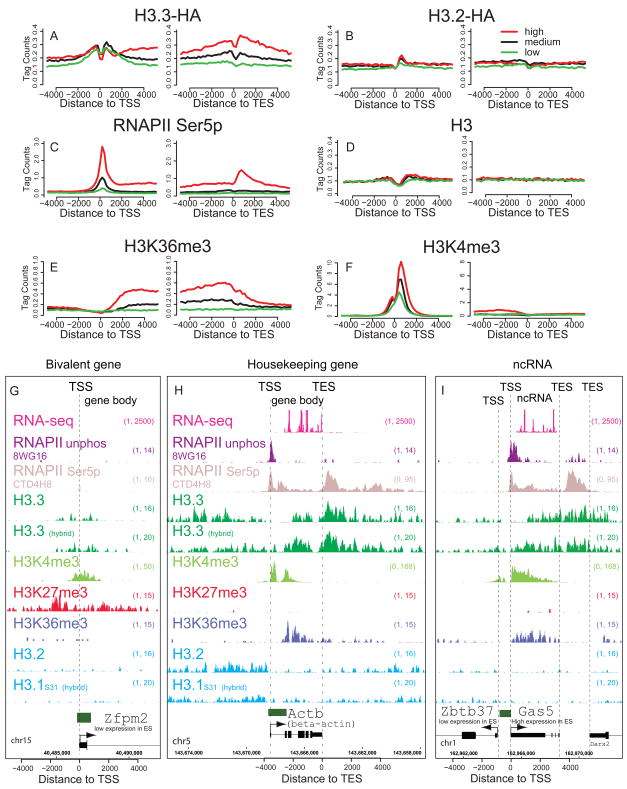
H3.3 is enriched around transcription start sites of both active and repressed genes with high CpG content promoters, and in the bodies and end sites of transcribed sequences
As there has been some disagreement regarding the patterns of H3.3 at active versus repressed genes in vertebrates (Jin and Felsenfeld, 2006; Jin et al., 2009; Sutcliffe et al., 2009; Tamura et al., 2009), we used ChIP-seq to address the genome-wide patterns of H3 variant enrichment around gene transcription start sites (TSS) in mouse ES cells. We find that H3.3 is not exclusively a marker of transcriptionally active genes in ES cells and NPCs.
Previous studies show that the majority of genes with high CpG content promoters (HCP genes) in both ES cells and differentiated cells are marked by histone H3K4me3 and the presence of RNAPII, regardless of whether the gene is active or repressed (Barski et al., 2007; Guenther et al., 2007; Mikkelsen et al., 2007). When we divide HCP genes into low, medium, and high expression in ES cells (see Methods), we find that H3.3, like H3K4me3 and RNAPII, is enriched around the TSS of both active and repressed genes (Figure 2A). In both native and crosslinking ChIP-seq, we find H3.3 less enriched at the TSS itself (Figure 2A, D, Figure S2A–D). This depletion of the −1 nucleosome at the TSS of active and inactive genes (Schones et al., 2008) has been attributed both to the instability of nucleosomes containing H3.3 and H2A.Z, and to the presence of specific sequences at CpG promoters that directly reduce nucleosome stability (Jin et al., 2009; Ramirez-Carrozzi et al., 2009).
Figure 2. Specific patterns of H3.3 and phosphorylated RNA polymerase at active and repressed genes.
A–F) Profiles of H3 variants, H3, H3 modifications, or RNAPII as indicated above each panel across the TSS and TES for highly active (red), medium expressing (black), or low expressing (green) CpG rich genes in ES cells. Y-axis represents the average number of tags per gene per 200 bp per 1 million mapped reads. H3 data is from (Mikkelsen et al., 2007). A–E is from crosslinking ChIP-seq, while F is from native ChIP-seq.
G) H3.3 is enriched around the TSS, but not into the gene body of the H3K4me3/H3K27me3 bivalent and transcriptionally repressed Zfpm2 gene in ES cells. In G-I, hybrid indicates F1 hybrid ES background.
H) H3.3 and RNAPII Ser5p are enriched in the gene body and after the TES of the highly expressed gene Actb (beta-actin) in ES cells. Data for the RNAPII unphos 8WG16 track are from (Mikkelsen et al., 2007). RNA-seq data are from (Cloonan et al., 2008).
I) H3.3 is enriched around the TSS and into the body of the non-coding RNA gene Gas5, but less enriched in the gene body of the neighboring low expressing Zbtb37 gene in ES cells. Data is represented as in Figure 1D. Gene is shown to scale below plot, with start and direction or transcription indicated by arrow. Green rectangles above the gene represent CpG islands. See also Figure S2.
More than one-fifth of HCP genes in ES cells carry both H3K4me3 and H3K27me3 in their promoters, and these transcriptionally repressed ‘bivalent’ genes are proposed to represent genes that are poised for activation following cell differentiation (Bernstein et al., 2006; Mikkelsen et al., 2007). When we analyze the pattern of H3 variants at bivalent TSS by ChIP-seq, we find that H3.3 is enriched around the TSS of bivalent genes in ES cells, while mutation of H3.3 towards H3.2 or H3.1 abolishes this enrichment (Figure 2G, S3).
Although H3.3 is incorporated around the TSS of both active and repressed genes in ES cells, H3.3 is enriched in the body of active genes, but not that of repressed genes (Figure 2A, G, H, S2A–D). Mononucleosome resolution analysis indicates that H3.3 is incorporated into the +1 nucleosomes in both active and repressed genes, but up to +3 nucleosomes and further into the coding regions of active genes (Figure S2Q–W). The level of H3.3 in gene bodies is correlated with gene expression, particularly at highly expressed genes (Spearman’s rank correlation coefficient ρ = 0.54, p < 2.2e-16; see Figure 2A, S2). Upon mutation of H3.3 to H3.2 or H3.1S31, H3.3 specific patterns of enrichment around the TSS and gene body are lost, generating patterns similar to general H3 (Figure 2B, D).
In accordance with previous studies (Henikoff et al., 2009; Jin et al., 2009; Mito et al., 2005), H3.3 enrichment often extends beyond the gene body and past the transcriptional end site (TES) at highly expressed genes in ES cells (Figure 2A, H) and in differentiated NPCs (Figure S2CC). At highly expressed genes such as beta-actin, H3.3 enrichment increases immediately after the TES, and peaks at approximately +700 bp ± 200 bp after the TES (Figure 2H, S2A–D). We observe a similar pattern of H3.3 localization in the bodies and after the TES of transcribed non-coding RNA (Guttman et al., 2009) (Figure 2I). Interestingly, the distribution of post-TES peaks of serine-5 phosphorylated RNAPII are closely co-localized with peaks of H3.3 at active genes, with the phosphorylated RNAPII peak slightly downstream (~200 bp) from the H3.3 peak. We did not observe any significant post-TES enrichment of histone modifications that are associated with active genes, such as H3K4me3, H3K4me1, and H3K36me3 (Figure 2E, F, Figure S2). Post-TES enrichment of H3.3 is also dependent on amino acid sequence (Figure 2B, D, Figure S2). From these analyses, we conclude that H3.3 is localized around the TSS of both active and repressed HCP genes, and both H3.3 and phosphorylated RNAPII are significantly enriched beyond the TES of highly expressed genes in undifferentiated and differentiated mammalian cells.
The profile of H3.3 at cell-type specific genes and regulatory elements changes with cell differentiation
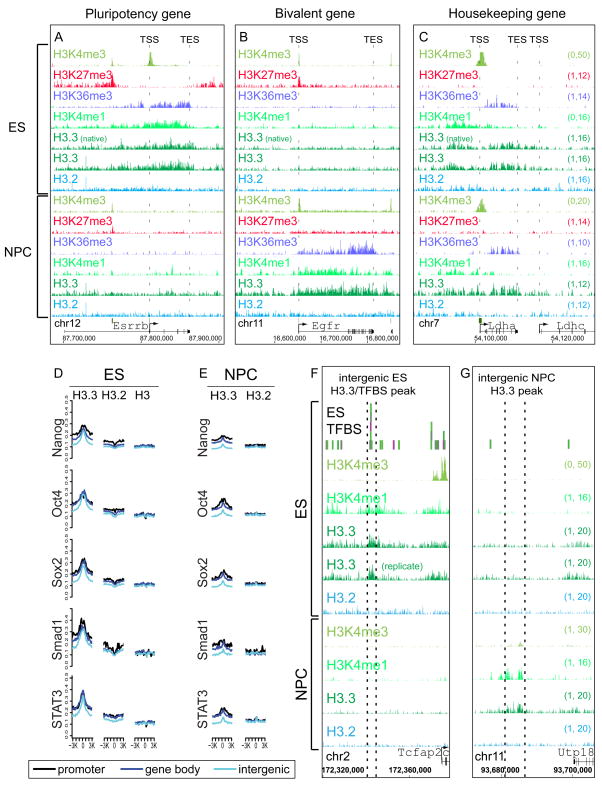
To determine how genome-wide patterns of H3.3 change with cell differentiation, we differentiated both H3.3-HA and H3.2-HA ES cells to NPCs (Figure S3A) (Conti et al., 2005). In ES cells, H3.3 is enriched in the bodies of expressed pluripotency genes such as Esrrb, Nanog, and Oct4, correlated with H3K36me3 (Figure 3A, Table S2, S3B). Upon differentiation of ES cells to NPCs, the expression of most pluripotency genes is lost (Conti et al., 2005). Accordingly, the enrichment of H3.3 and H3K36me3 in the bodies of pluripotency genes largely disappears upon cell differentiation (Figure 3A, S3B).
Figure 3. Cell-type specific enrichment of H3.3 at transcription factor binding sites and developmentally regulated genes.
A) H3.3, H3K4me1, and H3K36me3 are enriched in the gene body of the highly expressed pluripotency gene Esrrb in ES cells, and this enrichment is largely lost upon differentiation to NPCs.
B) The epidermal growth factor receptor gene Egfr is H3K4me3/H3K27me3 bivalent and transcriptionally repressed in ES cells. Upon differentiation to NPCs, H3.3, H3K4me1, and H3K36me3 are enriched in the Egfr gene body.
C) H3.3, H3K4me1, and H3K36me3 are enriched in the gene body of the housekeeping gene lactate dehydrogenase A Ldha in both ES cells and NPCs, and is not enriched in the neighboring Ldhc gene. The Y-axes in panels A-C are identical (indicated in the right side of panel C).
D–E) H3.3 is enriched genome-wide around ES cell TFBS. Crosslinking ChIP-seq genome-wide profiles of H3 variants as indicated around ES binding sites for Nanog, Oct4, Sox2, Smad1, and STAT3. TFBS in mouse ES cells were from a previous ChIP-seq study (Chen et al., 2008), and classified as promoter (black), gene body (dark blue), or intergenic (light blue). Panel D represents data from ES cells, while panel E represents data from NPCs at identical regions. Y-axis represents number of tags per binding site per 200 bp per 1 million mapped reads. Data for H3 in ES cells is from (Mikkelsen et al., 2007). In A–E, “(native)” indicates native H3.3-HA ChIP-seq, and “(replicate)” indicates biological replicate H3.3-HA ChIP-seq from F1 hybrid ES background.
F) H3.3 and H3K4me1 are enriched at an intergenic region bound by multiple transcription factors (Chen et al., 2008) specifically in ES cells.
G) H3.3 and H3K4me1 are enriched at an intergenic region specifically in NPCs. See also Figure S3.
Following differentiation of ES cells to NPCs, the profile of H3.3 changes with resolution of bivalent domains. In bivalent ES genes that resolve to H3K4me3 and become transcriptionally active in NPCs, H3.3 is maintained around the TSS and also incorporated into the gene body, in correlation with H3K36me3 and H3K4me1 (Figure 3B, S3C). For example, upon differentiation to NPCs, H3.3 extends into the gene body of the active epidermal growth factor receptor gene Egfr, correlating with H3K36me3 and H3K4me1 (Figure 3B). In contrast, for bivalent genes that remain transcriptionally repressed and resolve to either H3K27me3 or no mark in NPCs, H3.3 enrichment is reduced at the TSS upon differentiation (Figure S3C). As expected, H3.2 remains unenriched at the TSS in both ES cells and NPCs (Figure S3C).
While the pattern of H3.3 changes at cell-type-specific genes, housekeeping genes that remain highly expressed through differentiation retain similar enriched patterns of H3.3 incorporation. For example, H3.3 remains enriched around the TSS and within the gene body of the housekeeping gene lactate dehydrogenase A Ldha (Figure 3C, S3A). In both ES cells and NPCs, we find the greatest enrichment of H3.3 at highly expressed metabolic and housekeeping genes (Figure S3D). Our data show that the overall relationship between H3.3 incorporation and gene expression is retained in ES and NPCs, with H3.3 and H3K4 methylation marking the TSS of active and poised HCP genes, and H3.3 enriched in the body and TES of active HCP genes. At cell-type specific genes, H3.3 localization changes with cellular state.
Genome-scale studies have demonstrated that in addition to genes (Mito et al., 2005), H3.3 replacement marks the boundaries of regulatory elements (Jin et al., 2009; Mito et al., 2007). 36% (3,627) of H3.3 enriched regions in mouse ES cells are located outside of known annotated genes (Figure 1B, Figure 3D–F). We therefore compared our genome-wide distributions for histone H3 variants in ES cells and NPCs with established genome-wide maps for 13 distinct sequence-specific TFs in ES cells (Chen et al., 2008). We find that H3.3 is enriched genome-wide at genic and intergenic TFBS for all 13 characterized TFs in ES cells (Figure 3D, Figure S3E). Remarkably, 3,878 (38%) of the overall regions enriched for H3.3 in ES cells (Figure 1B) correspond to previously identified TFBS (Chen et al., 2008). Again, peaks of H3.3 at TFBS are dependent on H3.3 amino acid sequence (Figure 3D–G, S3E).
Many of the specific peaks of H3.3 at TFBS are cell-type specific, especially those located in intergenic regions (Figure 1B, 3D–G, S3B, data not shown). We find that H3.3 incorporation at TFBS bound by multiple TFs increases significantly with the number of bound TFs in ES cells, but these same elements do not show increased H3.3 in NPCs (Figure 3F, S3F). Genomic locations bound by more than four TFs are called multiple transcription factor-binding loci (MTL), and a subset of MTL bound by Oct4-Sox2-Nanog have been shown to serve as ES cell-specific enhanceosomes (Chen et al., 2008). For example, an intergenic region near the AP-2 transcription factor gene Tcfap2c is a target for multiple transcription factors in ES cells (Chen et al., 2008). We find H3.3 enriched at this region specifically in ES cells, with enrichment lost in NPCs (Figure 3F).
Following differentiation to NPCs, genome-wide enrichment of H3.3 at ES TFBS is reduced, but not eliminated (Figure 3E, S3E). At some specific TFBS (Figure 3F), H3.3 and H3K4me1 enrichment is lost upon differentiation to NPCs. However, at other TFBS, such as those 5′ and 3′ of Nanog, H3.3 and H3K4me1 enrichment is reduced, but partially maintained (Figure S3B). H3K4me1 and H3.3 have been separately shown to significantly co-localize with TFBS and other regulatory elements (Heintzman et al., 2007; Mito et al., 2007; Wang et al., 2008), but the patterns of H3.3 and H3K4me1 have never been directly compared. In our analysis, 26% of H3K4me1 peaks overlap with H3.3 peaks, and peaks of H3.3 are 5 times more likely to be associated with peaks of H3K4me1 than H3K4me3 (Table S1, p ≪ 0.00001). In addition to ES cell specific intergenic peaks of H3.3 and H3K4me1 (Figure 3F), we also found 724 intergenic peaks of H3.3 and H3K4me1 that are specific to differentiated NPCs and that do not correspond to any known TFBS (Figure 3G, data not shown). It is possible that these represent uncharacterized cell-type specific enhancers. Overall, our data demonstrate that the genome-wide localization of H3.3 changes with cell differentiation at cell-type specific genes and regulatory elements.
Hira is required for enrichment of H3.3 at active and repressed genes
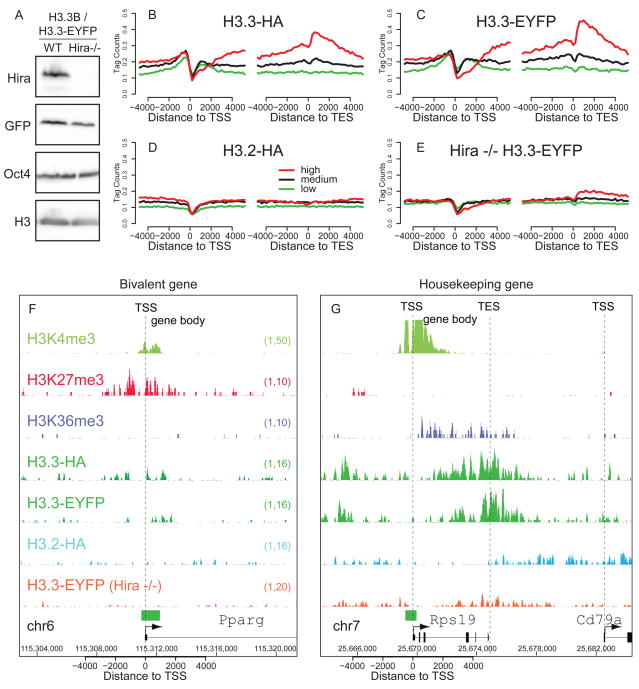
After establishing genome-wide patterns of H3.3 in undifferentiated ES cells and differentiated NPCs, we sought to determine whether these patterns were dependent on the H3.3 chaperone Hira. We therefore used ZFNs to tag one allele of the endogenous H3.3B gene in Hira −/− ES cells (Meshorer et al., 2006; Roberts et al., 2002) with a C-terminal EYFP tag (Figure 4A, Figure S4A), and compared the genome-wide localization of H3.3-EYFP in the presence and absence of Hira.
Figure 4. Enrichment of H3.3 at active and repressed genes is Hira-dependent.
A) Immunoblots showing expression of H3.3-EYFP in wild-type and Hira−/− ES cells, with antibodies as indicated.
B–E) Native ChIP-seq profiles of H3 variants, in wild-type ES cells (B-D) and Hira −/− ES cells (E) across the TSS and TES for highly active (red), medium expressing (black), or low expressing (green) CpG rich genes, with data represented as in Figure 2. See also Figure S4.
F) H3.3 enrichment at the TSS of repressed bivalent gene Pparg is Hira-dependent.
G) H3.3 enrichment at the active, ribosomal-protein coding gene Rps19 is Hira-dependent.
Using native ChIP-seq, we find that Hira is required for genome-wide H3.3 enrichment at active and repressed HCP genes (Figure 4B–G). In the absence of Hira, the pattern of H3.3 around active and repressed genes resembles H3.2, with no enrichment surrounding the TSS, and depletion of H3.3 at the TSS itself (Figure 4D, E). For example, in Hira −/− ES cells, the enrichment of H3.3 around the TSS of the repressed bivalent gene Pparg is abolished (Figure 4F). At highly expressed housekeeping genes such as the ribosomal protein-coding gene Rps19, the gene body and post-TES enrichment of H3.3 is similarly decreased in the absence of Hira (Figure 4G). Our data demonstrate that the vast majority of H3.3 enrichment at both active and repressed genes in ES cells is Hira-dependent.
One possible explanation for the loss of H3.3 localization at active and repressed genes in Hira −/− ES cells is a significant alteration in patterns of gene expression. Remarkably, microarray analysis reveals that global patterns of ES cell gene expression are maintained in Hira −/− ES cells (Figure S4B-D). Moreover, genome-wide patterns of H3K36me3 are extremely similar in wild-type and Hira −/− ES cells (Figure S4E-H). This result is similar to the recent observation that global patterns of ES cell gene expression are maintained following knock-down of CHD1 (Gaspar-Maia et al., 2009). We conclude that genome-wide enrichment of H3.3 at active and repressed genes is Hira-dependent, but is not required for maintenance of the undifferentiated ES cell transcriptome.
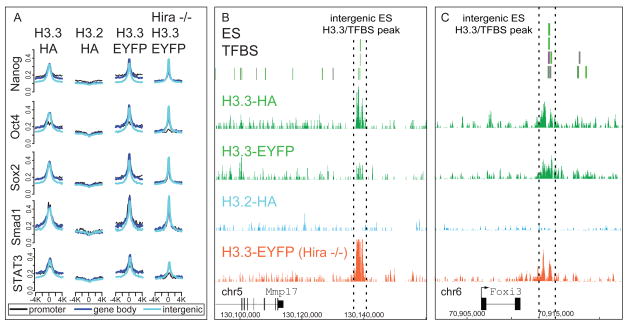
Hira-independent enrichment of H3.3 at transcription factor binding sites
We next examined the Hira-dependence of H3.3 enrichment at genic and intergenic TFBS. Surprisingly, global H3.3 profiles are largely maintained at ES cell TFBS in Hira −/− ES cells (Figure 5A, S5, Table S3), indicating that H3.3 enrichment at most known regulatory elements is Hira-independent. For example, H3.3 enrichment at an intergenic MTL approximately 30 kb from the Mmp17 gene is maintained in Hira −/− ES cells (Figure 5B), as is H3.3 enrichment at an intergenic MTL near the repressed Foxi3 gene (Figure 5C). Indeed, levels of H3.3 are increased at some TFBS in Hira −/− ES cells (Figure S5A). However, levels of H3.3 are also reduced at other TFBS in Hira −/− ES cells (Table S3, Figure S5B). Of all previously identified ES TFBS (Chen et al., 2008), 34% show greater than 2-fold more H3.3 tags in Hira +/+ than in Hira −/−, while 12% show greater than 2-fold more H3.3 tags in Hira −/−, indicating that targeting of H3.3 to the majority (54%) of known ES TFBS is Hira-independent (Table S3, Figure S5). Global profiles of H3K4me1 are very similar in wild-type and Hira −/− ES cells (Figure S4E–F, S5D), indicating that Hira is also not required to maintain the localization of H3K4me1. Overall, these data demonstrate that Hira is not essential for the localization of H3.3 at many TFBS in mammalian ES cells.
Figure 5. Hira is not essential for H3.3 enrichment at transcription factor binding sites.
A) Native ChIP-seq genome-wide profiles of H3.3 around previously described ES TFBS (Chen et al., 2008), with data analyzed as in Figure 3.
B–C) Localization of H3.3 in ES cells at intergenic enhancer elements near Mmp17 (B) and Foxi3 (C) is Hira-independent. See also Figure S5 and Table S3.
Hira-independent association of Atrx and Daxx with histone H3.3
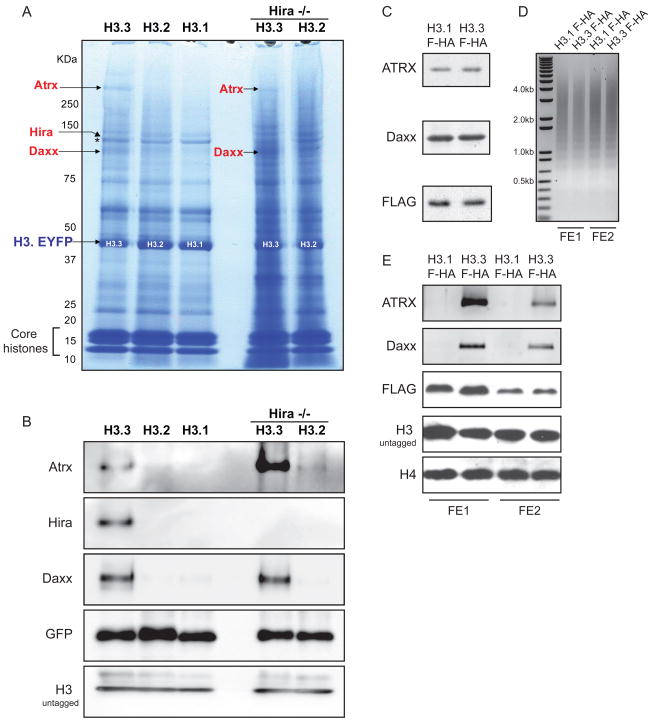
To identify candidates that might mediate Hira-independent localization of H3.3, we used immunoaffinity purification and mass spectrometry (Cristea et al., 2005) (Figure 6A, S6). We found many interacting proteins common to all H3 variants in wild-type and Hira −/− ES cells, including core histones and previously described members common to both RC and RI chromatin assembly complexes, such as Nasp, Asf1a, Asf1b, and Rbap48 (see Table S4). Notably, the previously described H3.3 chaperone Hira (Tagami et al., 2004) was identified specifically in proteins isolated with H3.3 (Figure 6A–B).
Figure 6. Atrx and Daxx association with H3.3 is specific, Hira-independent, and conserved in mouse and human cells.
A) H3.3-EYFP, H3.2-EYFP, H3.1-EYFP, and Hira −/− H3.3-EYFP associated proteins were immunopurified from heterozygous H3.3B ES cells via the EYFP tag, resolved by SDS-PAGE, and visualized by Coomassie Blue. Proteins were identified by mass spectrometry, and those listed in red are specific to H3.3. The arrow indicating Hira points to a polypeptide in the H3.3 lane running immediately above the polypeptide Nasp (*), which is common to all purified H3 variants. A full list of identified proteins is presented in Table S4.
B) Immunoblots of immunopurified H3 variant associated proteins from ES cells with antibodies as indicated. See also Figure S6.
C) Immunoblots of Atrx, Daxx and FLAG-HA-H3 from HeLa cells stably expressing H3.3-FLAG-HA (FHA) or H3.1-FHA (Tagami et al., 2004).
D) Oligonucleosomes purified from H3.3-FHA and H3.1-FHA HeLa cells. Micrococcal nuclease digested chromatin was incubated with M2 agarose beads in order to purify H3.3-FHA and H3.1-FHA oligonucleosomes and associated proteins. Two adjacent FLAG elution fractions (FE1 and FE2) from each H3.3-FHA and H3.1-FHA purification are shown in D–E. Oligonucleosomal DNA was purified and run on a 1% agarose gel, with fragment size indicated.
E) Immunoblots of eluate from two adjacent FLAG elution fractions (FE1 and FE2) following FLAG affinity purification from H3.3-FHA and H3.1-FHA oligonucleosomes (D).
In addition to Hira, we also identified Atrx and Daxx as proteins that specifically associate with H3.3 (Figure 6A–B). Atrx is a member of the SNF2 family of chromatin remodeling factors (Picketts et al., 1996). Mutations of human ATRX give rise to the ATR-X syndrome, a disorder characterized by a form of X-linked mental retardation that is frequently associated with alpha thalassemia (Gibbons et al., 2008). ATRX co-exists in a chromatin-remodeling complex with the death domain-associated protein Daxx, and these proteins localize to heterochromatin and promyelocytic leukemia (PML) nuclear bodies in human and mouse cells (Tang et al., 2004; Xue et al., 2003). As Atrx and Daxx specifically associate with H3.3 in both wild-type and Hira −/− ES cells, we conclude that this association is Hira-independent.
To determine if the association between H3.3, Atrx, and Daxx was conserved in differentiated human cells, we isolated oligonucleosomes and chromatin-associated proteins from HeLa cells stably expressing FLAG-HA tagged H3.3 or H3.1 (Tagami et al., 2004) (Figure 6C–D). Following FLAG affinity purification, immunoblots of H3.3 and H3.1-associated proteins reveal that both Daxx and Atrx are specifically associated with H3.3 but not H3.1 oligonucleosomes in human cells (Figure 6E).
Atrx is required to maintain H3.3 localization at telomeres and for repression of telomeric repeat-containing RNA (TERRA) in ES cells
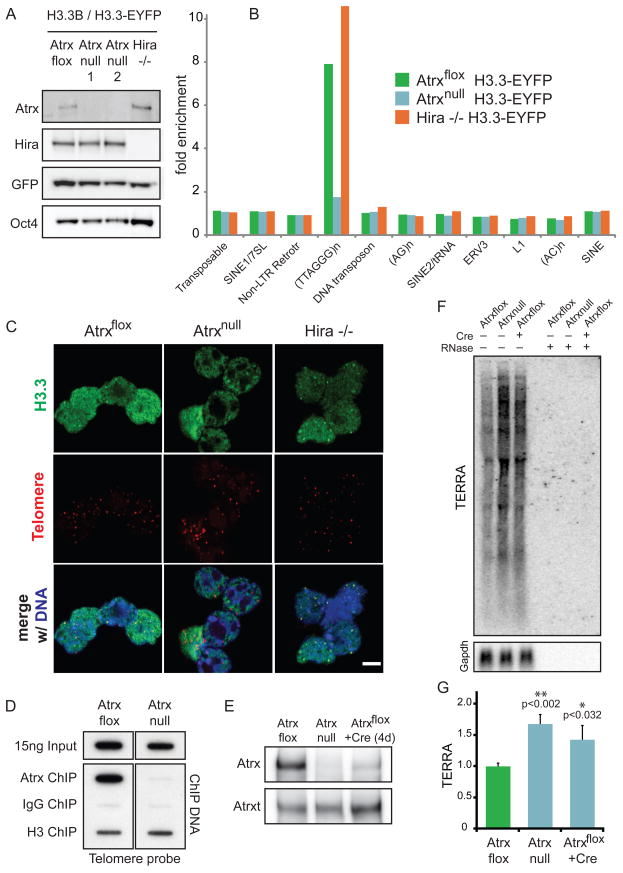
To determine if Atrx is required for H3.3 localization, we again used ZFNs to knock-in an epitope tag into the endogenous allele of H3.3B, generating heterozygous H3.3B/H3.3B-EYFP in Atrxflox and Atrxnull mouse ES cells (Garrick et al., 2006) (Figure 7A). We then used native ChIP-seq to generate genome-wide profiles of H3.3 in the presence and absence of Atrx. We find that Atrx is not required for H3.3 incorporation at active or repressed genes, or at regulatory elements (Figure S5A, S7A-E), as genome-wide profiles of H3.3 are similar at genes and TFBS in Atrxflox and Atrxnull ES cells. Strikingly, both ChIP-seq (Figure 7B) and cell imaging analysis (Figure 7C) demonstrate that Atrx, but not Hira, is specifically required for H3.3 enrichment at telomeres. In accordance with the requirement of Atrx for telomeric localization of H3.3, ChIP analysis shows that Atrx itself is physically associated with telomeres in Atrxflox ES cells (Figure 7D).
Figure 7. Atrx is required for Hira-independent enrichment of H3.3 at telomeres, and for repression of telomeric repeat-containing RNA.
A) Immunoblots showing expression of H3.3-EYFP in Atrxflox, Atrxnull, and Hira −/− ES cells, with antibodies as indicated.
B) ChIP-seq analysis of H3.3 enrichment at repetitive elements in Atrxflox, Atrxnull, and Hira −/− ES cells. Fold enrichment of repetitive elements in ChIP-seq data are plotted over input.
C) Immunofluorescence (IF) and telomere fluorescence in situ hybridization (FISH) of Atrxflox, Atrxnull, and Hira −/− ES cells expressing H3.3B/H3.3-EYFP. Representative confocal images of interphase ES cells demonstrate that H3.3 co-localization with telomeres is lost in Atrxnull ES cells (middle), but maintained in Hira −/− ES cells (right). Bar = 5μm.
D) ChIP of Atrx, control IgG, and H3 was performed in Atrxflox and Atrxnull ES cells, followed by slot blot with a telomere probe.
E) Immunoblots of Atrx in Atrxflox, Atrxnull, and Atrxflox ES cells 4 days after treatment with retroviral Cre. Atrxt refers to a previously identified alternative splicing product of Atrx that remains expressed in Atrxnull ES cells (Garrick et al., 2006).
F) Representative northern blot for TERRA (upper panel) and Gapdh (lower panel) in Atrxflox and Atrxnull ES cells in the presence and absence of Cre. To ensure that signal is from RNA, samples on the right were treated with RNase for 30 minutes as indicated.
G) Quantification of TERRA normalized to Gapdh. Error bars represent the standard deviation of three independent experiments. See also Figure S7.
To investigate the functional consequences of Atrx deletion and the loss of Atrx-dependent telomeric enrichment of H3.3, we examined the chromatin state and transcriptional output of ES cell telomeres. The chromatin of ES and induced pluripotent cell telomeres has previously been shown to have lower levels of the heterochromatin marker H3K9me3 and increased transcription of telomeric repeat-containing RNA (TERRA) in comparison to differentiated cells (Marion et al., 2009). In particular, TERRA has recently been identified as a component of telomeric heterochromatin, and levels of TERRA have been shown to be regulated by chromatin modifying enzymes (Azzalin et al., 2007; Luke and Lingner, 2009; Schoeftner and Blasco, 2008). ChIP of H3K4me3 and H3K9me3 does not show a significant difference in telomeric enrichment between Atrxflox and Atrxnull ES cells (Figure S7F). However, northern blots from Atrxflox, Atrxnull, and Atrxflox ES cells 4 days after treatment with Cre reveal reproducible (~1.7-fold) upregulation of TERRA in the absence of Atrx (Figure 7E–G). Our data demonstrate that Atrx is required for Hira-independent localization of H3.3 at telomeres and for repression of TERRA.
Discussion
In this study, we examine H3 variant localization in mammalian ES cells and differentiated NPCs. Genome-wide patterns of H3.3 are dependent on H3.3-specific amino acid sequence; the enrichment of H3.3 at cell-type specific genes and TFBS is dependent on cellular state. We describe three general categories of H3.3 enrichment in mammalian cells: 1) genes and other transcribed non-repetitive sequences, 2) TFBS, and 3) telomeres. Remarkably, we find that each of these general categories of H3.3 enrichment in ES cells is mediated by distinct mechanisms. As expected, Hira is required for genic enrichment of H3.3. Unexpectedly, localization of H3.3 at specific TFBS and telomeres is Hira-independent, and we have identified Atrx as required for H3.3 localization at telomeres. Our results demonstrate that distinct factors control H3.3 localization at specific genomic locations in mammalian cells.
We find that H3.3 is constitutively enriched around the TSS of active and repressed HCP genes in mammalian ES cells and NPCs, including the TSS of repressed bivalent genes in ES cells. Although a recent genome-wide study found that H3.3 is unenriched at the TSS of repressed genes in HeLa cells (Jin et al., 2009), these results are not necessarily in conflict with our findings. Low CpG content promoter (LCP) and HCP genes have been described to display distinct modes of regulation (Mikkelsen et al., 2007; Ramirez-Carrozzi et al., 2009; Saxonov et al., 2006). Most HCP genes show evidence of transcriptional initiation, assemble unstable nucleosomes, and do not require SWI/SNF nucleosome remodeling complexes for gene induction, while LCP genes assemble stable nucleosomes and require SWI/SNF (Guenther et al., 2007; Ramirez-Carrozzi et al., 2009). Less differentiated cells such as ES cells contain large numbers of HCP genes with characteristics of transcriptional initiation (Guenther et al., 2007; Mikkelsen et al., 2007). Indeed, nearly all (99%) of HCP genes are marked by H3K4me3 in mouse ES cells, whether they are transcriptionally active or repressed (Mikkelsen et al., 2007). In contrast to HCP genes, we do not observe any significant pattern of H3.3 enrichment at LCP genes in ES cells and NPCs (data not shown). Our results are therefore consistent with a model in which H3K4 methylation and H3.3 localization at HCP TSS are coupled to transcriptional initiation.
We find that enrichment of H3.3 in the gene body and after the TES is proportional to transcriptional activity. As with previous studies in Drosophila and human cells (Henikoff et al., 2009; Jin et al., 2009; Mito et al., 2005), we find peaks of H3.3 after the TES of highly active genes, and we observe that these peaks are closely paralleled by peaks of Ser-5 phosphorylated RNAPII itself. We demonstrate that chromatin-based “transcriptional punctuation” (Siegel et al., 2009; Talbert and Henikoff, 2009) by H3.3 and phosphorylated RNAPII marks the boundaries of highly expressed genes in both undifferentiated and differentiated mammalian cells, calling attention to a potentially more universal mechanism for histone variant utilization as a genomic “boundary marker.”
To our knowledge, our report is the first genome-wide study to compare chromatin in the presence and absence of a mammalian histone chaperone. We find that H3.3 enrichment at active and repressed genes is dependent on the histone chaperone Hira. Previous studies suggest that H3.3 deposition in actively transcribed gene bodies may be coupled to transcription, potentially mediated by factors associated with elongating polymerase (Daury et al., 2006; Janicki et al., 2004; Schwartz and Ahmad, 2005). Our data are consistent with Hira-dependent transcription-coupled deposition of H3.3 at transcribed non-repetitive sequences.
Intriguingly, we do not observe significant abnormalities in Hira −/− ES cells, despite a global lack of H3.3 enrichment at active and repressed genes, and despite the requirement of Hira for early embryonic development (Roberts et al., 2002). We speculate that Hira −/− ES cells may be rescued by the replication-coupled deposition of histones during the frequent S-phases of rapidly dividing ES cells (Burdon et al., 2002). Hira −/− ES cells divide as rapidly as wild-type ES cells, and show a similar preponderance of cells in S phase (A.C. and P.J.S., unpublished data). Overall, our data are consistent with a role for Hira in genic deposition of H3.3.
H3.3 enrichment has recently been shown at TFBS in Drosophila and human cells (Jin et al., 2009; Mito et al., 2007). Deposition of H3.3 at TFBS may serve as a mechanism for the maintenance of regulatory elements in a more accessible chromatin conformation (Henikoff, 2008). Close comparison of our data to a recent dataset of 13 different TFs in mouse ES cells (Chen et al., 2008) shows H3.3 enriched in ES cells at all known types of TFBS genome-wide, whether in gene bodies, promoters, or intergenic regions. We also find a strong positive correlation between MTL and H3.3 localization, indicating particular enrichment of H3.3 at enhancer elements. Our data demonstrate that Hira is involved in H3.3 localization at some genic and intergenic TFBS. However, we also find that genome-wide H3.3 enrichment at many regulatory elements is Hira-independent and Atrx-independent. Our data therefore suggest that H3.3 localization at TFBS may be mediated by multiple and distinct factors, including Hira, with as yet unidentified factors mediating H3.3 localization at specific regulatory elements.
We find that H3.3 is specifically enriched in the canonical (TTAGGG)n repeat that is the hallmark of telomeres in vertebrates (Meyne et al., 1989). Previous immunofluorescence studies localizing GFP-tagged Hira to telomeres suggested that Hira facilitates H3.3 deposition at this location (Wong et al., 2009). However, using genome-wide ChIP-seq and cell imaging analyses in Hira −/− ES cells, we show that localization of H3.3 at telomeres in ES cells is Hira-independent. Further, we identify Atrx and Daxx as proteins that associate with H3.3 nucleosomes in the presence and absence of Hira.
Recent studies have shown that the Drosophila homolog of Atrx, XNP, co-localizes with H3.3 at sites of nucleosome replacement on polytene chromosomes, but is not required for H3.3 localization at these sites (Schneiderman et al., 2009). We find that Atrx is required for enrichment of H3.3 at mammalian ES cell telomeres, suggesting a divergence of homolog function. Moreover, we demonstrate that in the absence of Atrx, ES cells show upregulation of TERRA. Could ATRX and Daxx serve as specific H3.3 variant deposition machinery for specialized regions of heterochromatin? The ATRX/Daxx complex has previously been shown to have chromatin remodeling activity (Tang et al., 2004; Xue et al., 2003), and we show that Atrx is physically associated with ES cell telomeres. In addition to telomeres, our preliminary studies indicate that Atrx is also required for H3.3 enrichment at ribosomal DNA (data not shown), another transcribed repetitive element with characteristics of heterochromatin (McStay and Grummt, 2008).
Our findings raise multiple questions. What is the function of H3.3 at genes, TFBS, and telomeres? Do cellular requirements for H3.3 differ in dividing versus post-mitotic cells, where replication-independent deposition might play a larger role? Is Atrx-mediated localization of H3.3 also replication-independent, like Hira, or does it occur during replication? Broadly, our study raises the prospect that distinct, region-specific chaperone and remodeling complexes may mediate the localization of a single histone variant (H3.3) to particular genomic regions. Although key factors required for region-specific H3.3 localization have now been identified, the exact deposition mechanisms at play remain an important challenge for future work.
Experimental Procedures
ES cell culture and differentiation
Mouse ES cells were cultured under standard conditions and differentiated to NPCs as previously described (Conti et al., 2005). Hira −/− ES cells, Atrxflox, and Atrxnull ES cells have been described previously (Garrick et al., 2006; Meshorer et al., 2006). For more detail, see Supplemental Experimental Procedures.
ZFN design, targeting, and verification
ZFNs directed against the mouse H3.3B gene were designed using an archive of validated two-finger modules (Doyon et al., 2008; Urnov et al., 2005). ES cells were transfected with ZFNs and donor using Amaxa nucleofection. Fluorescent ES colonies were picked and screened by genomic PCR of H3.3B alleles, sequencing of PCR products, flow cytometry, and Southern blot. For more detail, see Supplemental Experimental Procedures.
Cellular extract preparation
Whole cell extracts were prepared by resuspending cell pellets in SDS-Laemmli sample buffer, followed by brief sonication and boiling.
ChIP and ChIP-seq
Crosslinking and native ChIP were performed as described (Barski et al., 2007; Lee et al., 2006), with minor modifications detailed in Supplemental Experimental Procedures. ChIP DNA was validated by real-time PCR, prepared for Illumina/Solexa sequencing, and sequenced using the Illumina Genome Analyzer II. The CTD4H8 antibody was raised against a chemically synthesized phospho-Ser 5 peptide sequence from the CTD of the largest RPB1 subunit of RNAPII, and has been extensively characterized previously (Stock et al., 2007). Other antibodies and more detailed ChIP-seq methods are described in Supplemental Experimental Procedures. ChIP-seq assays performed are listed in Table S5. To confirm telomere enrichment, ChIP DNA from Atrxflox and Atrxnull ES cells was probed with a TTAGGG repeat probe as described (Sfeir et al., 2009).
ChIP-seq data analysis
ChIP-seq or input reads were mapped to the mouse genome (build 37, or mm9) using the ELAND alignment software within the Illumina Analysis Pipeline. Profiles in specific genomic regions were displayed in the Affymetrix Integrated Genome Browser. For analysis of repetitive elements, reads were aligned directly to a library of mouse consensus repetitive sequences (http://www.girinst.org) and enrichments were computed for ChIP against input samples. For TSS/TES profiling, we segregated reference genes (refSeq) into low, medium and high expression based on a previous microarray analysis (Mikkelsen et al., 2007). For TFBS profiling, binding sites for 13 TFs in mouse ES cells were obtained from a previous ChIP-seq analysis (Chen et al., 2008). To generate density profiles ± 5kb around TSS, TES, bivalent genes, and TFBS, we used a sliding window method to count the number of ChIP-seq reads in each 200 bp window, and the resulting counts were then normalized by the totals of genes or TFBS and the total mapped reads. For details of data analysis, see Supplemental Experimental Procedures.
Gene expression analysis of wild-type and Hira −/− ES cells
RNA expression data for W9.5 and Hira −/− ES cells was generated from polyA RNA and random primers using the GeneChip Mouse Gene 1.0 ST Array kit (Affymetrix).
Isolation of protein complexes and mass spectrometric analysis
Immunoaffinity purifications of EYFP-tagged H3.3, H3.2, H3.1, HIRA−/− H3.3, and HIRA −/− H3.2 were performed as described (Cristea et al., 2005). MALDI MS and MS/MS analyses were performed as described (Luo et al., 2009). For more detail, see Supplemental Experimental Procedures.
Immunofluorescence and telomere fluorescence in situ hybridization (FISH)
Telomere FISH of ES cells was performed using a peptide nucleic acid TAMRA-TelG telomere probe (Sfeir et al., 2009), and immunofluoresence was performed using a previously described anti-GFP antibody (Cristea et al., 2005). For more detail, see Supplemental Experimental Procedures.
TERRA analysis
RNA was isolated using RNeasy Mini Kit (Qiagen), and TERRA analysis was performed as described (Azzalin et al., 2007; Sfeir et al., 2009).
Accession Numbers
Our ChIP-seq and microarray datasets have been deposited in the GEO database with accession numbers GSE16893 and GSE19542.
Supplementary Material
Acknowledgments
We thank L Baker, EM Duncan, and GG Wang for critical reading of the manuscript, P Wu, A Sfeir, and T de Lange for telomere reagents and helpful discussion, PD Adams for Hira antibodies, R Jaenisch for F1 hybrid male 129SVJae x M. m. castaneus ES cells, G Almouzni for H3.1 and H3.3-FLAG-HA HeLa cells, K Zhao for sharing his native ChIP-seq protocol, E Moehle for drawing the gene editing schemes, N Jina of the UCL Genomics Core, KR Molloy for assistance with mass spectrometric analysis, and S Mazel and A North of the RU Flow Cytometry and Bio-Imaging Resource Centers. ADG is supported by NIH MSTP grant GM07739. LAB is a Damon Runyon CRF fellow. This work was funded by institutional support from The Rockefeller University and grants from the Tri-Institutional Stem Cell Initiative (funded by the Starr Foundation), Empire State Stem Cell fund through NYSDOH Contract #C023046, HHMI (SR), BBRC UK and BHF (PJS), startup funds from AECOM of Yeshiva University (DZ), and NIH Grants RR00862, RR022220, DP1DA026192 (IMC), GM53122 and GM53512 (CDA).
Footnotes
While this manuscript was in press, Wong et al. reported that Atrx associates with histone H3.3 and localizes to telomeres in ES cells (Wong et al., 2010). These data further support our conclusion that distinct factors control H3.3 localization at specific genomic regions.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bonnefoy E, Orsi GA, Couble P, Loppin B. The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization. PLoS Genet. 2007;3:1991–2006. doi: 10.1371/journal.pgen.0030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- Daury L, Chailleux C, Bonvallet J, Trouche D. Histone H3.3 deposition at E2F-regulated genes is linked to transcription. EMBO Rep. 2006;7:66–71. doi: 10.1038/sj.embor.7400561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Sharpe JA, Arkell R, Dobbie L, Smith AJ, Wood WG, Higgs DR, Gibbons RJ. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006;2:e58. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Wada T, Fisher CA, Malik N, Mitson MJ, Steensma DP, Fryer A, Goudie DR, Krantz ID, Traeger-Synodinos J. Mutations in the chromatin-associated protein ATRX. Hum Mutat. 2008;29:796–802. doi: 10.1002/humu.20734. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis”. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodl M, Basler K. Transcription in the Absence of Histone H3.3. Curr Biol. 2009 doi: 10.1016/j.cub.2009.05.048. [DOI] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Distribution of histone H3.3 in hematopoietic cell lineages. Proc Natl Acad Sci U S A. 2006;103:574–579. doi: 10.1073/pnas.0509974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009 doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Marking histone H3 variants: how, when and why? Trends Biochem Sci. 2007;32:425–433. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009 doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Li T, Yu F, Kramer T, Cristea IM. Resolving the Composition of Protein Complexes using a MALDI LTQ Orbitrap. Journal of the American Society for Mass Spectrometry. 2009 doi: 10.1016/j.jasms.2009.08.026. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci U S A. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Nishioka K, Dong YX, Shimojima T, Hirose S. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 2007;21:552–561. doi: 10.1101/gad.1503407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Quivy JP, Scamps C, Martini EM, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol Cell Biol. 2002;22:2318–2328. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and Developmental Functions of the H3.3 Histone Variant in Drosophila. Curr Biol. 2009 doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiderman JI, Sakai A, Goldstein S, Ahmad K. The XNP remodeler targets dynamic chromatin in Drosophila. Proc Natl Acad Sci U S A. 2009;106:14472–14477. doi: 10.1073/pnas.0905816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009 doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Sutcliffe EL, Parish IA, He YQ, Juelich T, Tierney ML, Rangasamy D, Milburn PJ, Parish CR, Tremethick DJ, Rao S. Dynamic histone variant exchange accompanies gene induction in T cells. Mol Cell Biol. 2009;29:1972–1986. doi: 10.1128/MCB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Chromatin-based transcriptional punctuation. Genes Dev. 2009;23:1037–1041. doi: 10.1101/gad.1806409. [DOI] [PubMed] [Google Scholar]
- Tamura T, Smith M, Kanno T, Dasenbrock H, Nishiyama A, Ozato K. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J Biol Chem. 2009 doi: 10.1074/jbc.M805651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010 doi: 10.1101/gr.101477.109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.