Abstract
This paper describes a SlipChip-based approach to perform bead-based heterogeneous immunoassays with multiple nanoliter-volume samples. As a potential device to analyze the output of the chemistrode, the performance of this platform was tested using low concentrations of biomolecules. Two strategies to perform the immunoassay in the SlipChip were tested: 1) a unidirectional slipping method to combine the well containing a sample with a series of wells preloaded with reagents; 2) a back-and-forth slipping method to introduce a series of reagents to a well containing the sample by reloading and slipping the well containing the reagent. The SlipChips were fabricated with hydrophilic surfaces on the interior of the wells and with hydrophobic surfaces on the face of the SlipChip to enhance filling, transferring, and maintaining aqueous solutions in shallow wells. Nanopatterning was used to increase the hydrophobic nature of the SlipChip surface. Magnetic beads containing the capture antibody were efficiently transferred between wells and washed by serial dilution. An insulin immunoenzymatic assay showed a detection of limit of ~13 pM. Forty eight droplets of nanoliter volume were analyzed in parallel, including an on-chip calibration. The design of the SlipChip is flexible to accommodate other types of immunoassays, both heterogeneous and homogeneous. This work establishes the possibility of using SlipChip-based immunoassays in small volumes for a range of possible applications, including analysis of plugs from a chemistrode, detection of molecules from single cells, and diagnostic monitoring.
Introduction
This paper describes a method of using the SlipChip1–3 to analyze many nL-volume samples in parallel by a bead-based heterogeneous immunoassay. Low volume analysis is a bottleneck for a range of approaches that produce small volumes (10−1 – 102 nL), and immunoassays are a class of widely used analytical techniques in biological research. Heterogeneous immunoassays are attractive for detecting protein markers due to their high specificity and sensitivity, but require washing steps and are therefore difficult to do on small scales. Clinical research or diagnosis often involves serially monitoring a specific small group of cells, such as monitoring a tumor in vivo over time, and requires repeated sampling and analysis of small volumes.4 Understanding dynamic biological systems requires tools to deliver, capture, and interpret molecular signals with high temporal resolution. The recently developed chemistrode5–8 addresses this need by recording molecular signals in an array containing hundreds of nanoliter-volume plugs, which are subsequently analyzed by multiple independent techniques in parallel. Achieving the full potential of the chemistrode requires methods to analyze the nanoliter-volume recording plugs with higher throughput and sensitivity than provided by homogeneous fluorescence correlation spectroscopy (FCS)-based immunoassays5. To use heterogeneous immunoassays as an efficient method of detecting and quantifying biomolecules in small volumes for these and other applications, the bottlenecks associated with processing small volumes in a high throughput manner must first be overcome.
Although microfluidic devices that perform immunoassays for multiple nL-volume samples in parallel are available,9,10 these systems require complicated microfluidic chips, control systems, and assay-specific surface modifications (protein coatings). Instead of putting an assay-specific protein coating on the surface of the device, bead-based immunoassays using pre-made beads are more attractive as they make fabrication of the microfluidic chips simpler. Nanoliter droplets present a number of attractive opportunities for serial analysis,11–18 but current devices for arranging nanoliter droplets in fixed arrays do not allow for additional manipulations of droplets such as adding reagents and handling beads.11–14 A digital microfluidic platform19,20 to perform bead-based immunoassays in droplets is also available, but requires slightly larger volumes (~0.3 μL scale) and also involves a complex electrowetting system. Devices that are easier to operate, such as flow-through devices21–29 and CD-based immunoassays,30,31 cannot deal with nL-volume samples. To meet the need for a simple, easy-to-operate device that is capable of performing bead-based heterogeneous immunoassays on many small volumes in parallel, we developed a SlipChip-based system to analyze small-volume samples.
The SlipChip is capable of robustly handling many multi-step processes on nL-volumes in parallel without using complex instruments.1–3 The SlipChip consists of two plates that can move (or “slip”) relative to one another. A program for complex manipulations of fluids can be encoded into the chip as a pattern of wells and ducts imprinted into the plates. The wells can be pre-loaded with reagents1 or configured for user-loading.2,3 Each well remains isolated until it overlaps with a well or a duct on the opposite plate. The encoded program is executed by simply slipping the two plates relative to one another. As plates move, wells in the two plates come in and out of contact in a precisely define sequence, creating and breaking up transient fluidic pathways, and bringing reagents in and out of contact. Previous experiments were performed in the context of protein crystallization, at relatively high concentrations of biomolecules. Here we wished to test the SlipChip and associated surface chemistries in the context of low (pM-nM) concentrations of biomolecules. We describe a simple approach that uses the SlipChip to perform a bead-based heterogeneous immunoassay to analyze many small-volume samples in parallel. We used an ultrasensitive immunoenzymatic insulin assay as the test model. We first illustrated and characterized the SlipChip-based system with standard samples loaded onto the chip via pipetting. We then designed a SlipChip capable of analyzing a pre-formed array of nL-plugs, similar to an array that would be collected by a chemistrode, to test the compatibility of the system with droplets. We also demonstrated that a back-and-forth slipping motion could be used to perform the immunoassay rather than the usual unidirectional slipping motion, to conserve space in future SlipChip designs.
Experimental Section
Fabrication of the SlipChip
In preliminary experiments, a thin aqueous film was observed between the two plates of the SlipChip during the slipping steps. This was caused by the solutions wetting the surface of the SlipChip, despite a silanization step during fabrication that made the surface hydrophobic. The thin aqueous film connected wells that should be separated after slipping and caused cross-contamination, and this problem was alleviated by using nanopatterns on the surface of the top plate of the SlipChip.2 We followed the fabrication procedure previously described2 with modifications given in Supporting Information. The fabrication of the bottom plate of the SlipChip with hydrophilic wells is also described in Supporting Information.
Operation of SlipChip
The SlipChip was assembled, loaded and slipped as described in Supporting Information. The step-by-step procedure of the assay in the SlipChip is presented in the text with schematic diagrams in Figure 1 and Figure 5.
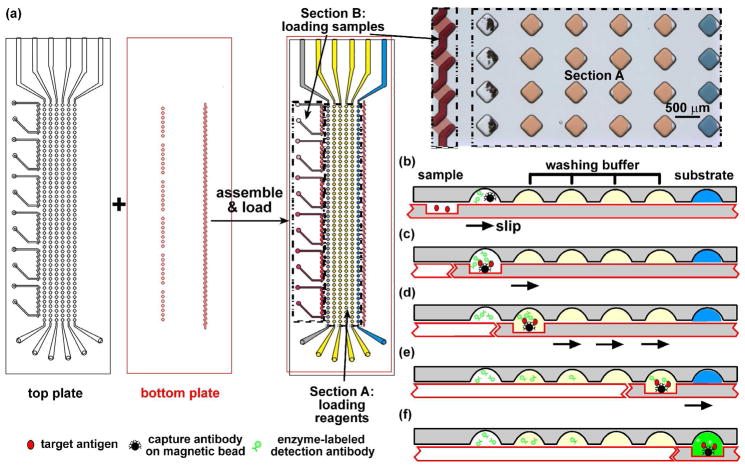
Figure 1.
Performing heterogeneous immunoassays with multiple nL samples in SlipChip. (a) A schematic of the SlipChip designed for calibration on the two plates of microfabricated glass. The top plate of this SlipChip (outlined in black) contained inlets, outlets, and wells for the various reagents (Section A), and inlets, outlets, and ducts to load the samples: six standard solutions (Section B). All wells and ducts are 80 μm deep. The bottom plate (outlined in red) of this SlipChip contained the 80 μm deep ducts to load the reagents (Section A), and 10 μm deep wells for the sample (Section B). The two plates are assembled to form the fluidic path for loading the reagents and samples. In Section A, the wells were loaded with reagents. The grey wells of row 1 were loaded with the solution containing magnetic beads coupled with the capture antibody and an enzyme-labeled detection antibody. Wells in rows 2–5 (yellow) contained the washing buffer. Wells in row 6 (blue) contained the substrate. Section B is designed to load six samples into seven wells each. A microphotograph on the right shows the wells filled with different dye solutions (rows 2–6 of section A and Section B) or a suspension of beads (row 1 of section A). (b–f) Schematics of step-by–step operation of the bead-based immunoassay in SlipChip. See details in text.
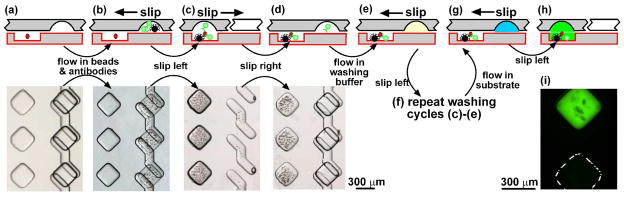
Figure 5.
Performing a bead-based immunoassay in SlipChip by back-and-forth slipping to reduce the number of wells used in the assay, and instead use introduction of reagents by flow during the assay. Top panel: Schematics of the step-by-step operation. Bottom panel: Experimental microphotographs of the SlipChip in operation. (a) Assembled SlipChip with the sample loaded. (b) The solution containing the capture-antibody coated magnetic beads and enzyme-labeled detection antibody was injected. (c) The SlipChip was slipped to combine the analyte and reagent solutions. The solutions were incubated to form the sandwich complex. (d) After the beads were pulled down on the bottom plate by a magnet, the chip was slipped back. (e) The washing buffer was injected. (f) The chip was slipped to combine the washing buffer and the beads as in step (c). This step and steps (d) and (e) were repeated multiple times to remove any unbound detection antibody. (g) The enzymatic substrate was injected. (h) The chip was slipped to combine the substrate and the beads, and the fluorescence was monitored. (i) A fluorescent microphotograph of two neighboring wells after the completion of the assay described in steps a to h. The lower well was initially loaded with blank insulin solution and the upper well was loaded with 1050 pM insulin.
Loading the reagents and samples in SlipChip
Reagents were preloaded in eight steps: (a) The SlipChip was assembled so that the wells of row 1 in Section A were connected by the reagent ducts. (b) The reagent solution (Figure 1a, grey) containing the capture-antibody coated superparamagnetic beads and enzyme-labeled detection antibody was injected into the SlipChip and the wells in row 1 of Section A were filled. (c) The chip was slipped to connect the wells in row 2 of Section A by ducts. (d) Fluorocarbon was injected through the ducts to remove any remaining solution in the ducts. (e) Washing buffer (Figure 1a, yellow) was injected to fill the wells in that row in the SlipChip. (f) The chip was slipped to connect the wells of the next row by ducts. (g) Steps (d), (e), and (f) were repeated three times to fill rows 3, 4 and 5 of Section A with washing buffer. (h) Fluorocarbon was injected through the ducts to remove any remaining solution, and the enzymatic substrate (Figure 1a, blue) was injected to fill row 6 of Section A.
Samples were loaded in two steps: (a) The SlipChip was slipped to connect the wells in Section B by the ducts. (b) Solutions of the analytes were injected by pipetting through the inlets.
Results and Discussion
Heterogeneous immunoassays have two specific features: 1) the antigen in the sample is first bound by a capture antibody linked on the surface of a solid phase, such as a magnetic bead or surface of a microfluidic channel, and a detection antibody binds to the antigen to form a sandwich complex; 2) a process of separation, or washing, is required to remove the unbound samples and detection antibody in the bulk from the sandwich complex on the surface of the solid phase. The detected signal (radioactive, colorimetric, fluorescent, or electrochemical) is generated by the label functionalized on the detection antibody, such as an enzyme. In this paper, we have constructed a SlipChip to perform a bead-based heterogeneous immunoassay (Figure 1, b to f,). Slipping of the SlipChip enables i) combination of the samples with the solution of magnetic beads and antibodies to form the sandwich complex (Figure 1, b to c); ii) introduction of the washing buffer to wash the beads by using a serial dilution (Figure 1, d to e), and iii) addition of the substrate for detection to produce a signal (Figure 1, f).
The SlipChip was designed as shown in Figure 1a. It contained two sections: Section A was used to load the reagents for the assay and Section B was used to load the samples. Section A contained six rows of 48 wells (9 nL in volume, 80 μm deep) on the top plate, and each of the 48 columns of wells were used to perform one assay on one sample. Wells in each row were used as described in Figure 1a.
The design of Section B was variable to accommodate the different loading requirements of different sources of small-volume samples. To characterize the bead-based immunoassay in SlipChip by using standard solutions to generate a calibration curve, this section was initially designed as six groups of seven wells (1 nL in volume, 10 μm deep) on the bottom plate. Each group of wells contained one standard solution (7 parallel assays for each standard solution and 42 total assays in this case). When the wells for the sample (bottom plate, Figure 1a) and ducts for the sample (top plate, Figure 1a) were aligned, six separate fluidic paths were formed, and each fluidic path was filled by pipetting. To improve filling of the 10 μm deep wells, the surfaces on the face of the device were silanized to be hydrophobic, while the 10 μm deep wells were protected during silanization to maintain a hydrophilic surface within the wells (see details in Supporting Information).
The bead-based immunoassay on SlipChip involved three general steps: (1) preloading reagents, (2) loading samples, and (3) performing the assay. The SlipChip was first assembled and then the reagents for the assay were loaded into the six rows of well in Section A (Figure 1a). Detailed step-by-step operation of preloading the reagents is described in Experimental Section. Because the six rows of wells on the top plate were loaded through only one row of ducts on the bottom plate, the ducts were slipped down to connect the following row of wells once a row of wells was filled. Therefore, the wells in a row were disconnected right after filling, minimizing potential back-flow of the solutions and minimizing incomplete filling. After loading the reagents, the SlipChip was slipped into the position to form the fluidic paths to load the samples (Figure 1a).
After loading the reagents and samples, the immunoassay was performed in five steps: (1) The SlipChip was slipped to generate 1 nL samples in the wells of the bottom plate, and to combine these samples with the solution containing the antibodies and beads. The solution was incubated in the overlaid wells (10 nL total volume) to allow the sandwich complex form (Figure 1, b to c). (2) A magnet was used to pull the beads down into the wells of the bottom plate, and the SlipChip was slipped to combine the beads and the washing buffer (Figure 1, c to d). (3) Step (2) was repeated three more times (Figure 1, d to e). (4) A magnet was used to pull the beads down into the wells of the bottom plate, and the SlipChip was slipped to combine the beads and the substrate. The fluorescence intensity was monitored using a fluorescent microscope to determine the concentration of analyte (Figure 1, e to f). To reduce the wetting of the solutions on the surface of the SlipChip and eliminate potential cross-contamination, a nanopattern2 was fabricated on the surface of the top plate (see details Experimental Section), and 0.4 mg/mL RfOEG, a surfactant, was added to the FC-40 lubricating fluid. Wetting on the surface was significantly reduced after these procedures were applied.
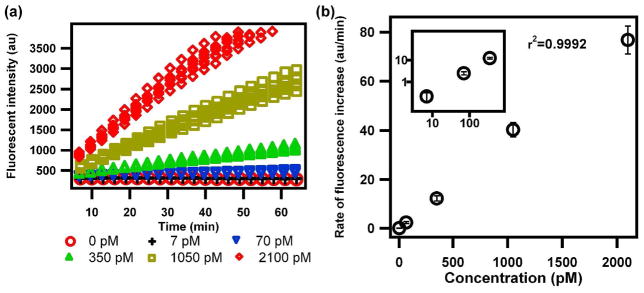
We used a bead-based immunoenzymatic assay for human insulin to demonstrate the analysis of multiple nanoliter samples in the SlipChip following the above procedures. In this assay, the superparamagnetic beads were coated with the capture antibody, and the detection antibody was labeled with alkaline phosphatase (ALP). We injected the six standard solutions of insulin (0 pM, 7 pM, 70 pM, 350 pM, 1050 pM, and 2100 pM) into the wells in section B as samples. The sample solutions of insulin were combined with a mixed solution of the antibodies and the blocking buffer to form the sandwich complex in a one-step incubation at 37 °C for 30 min (Figure 1c). To produce the signal for detection after washing, we used a fluorescent substrate for the enzyme, fluorescein diphosphate (FDP), which becomes fluorescent upon hydrolysis by alkaline phosphatase. Fluorescence intensity was measured every three minutes for each of the 42 samples at room temperature (Figure 2a). It was found that the fluorescence increased linearly within the initial ~ 30 min. A calibration curve was obtained by plotting the initial increase rates (within ~30 min) of fluorescence against the concentration (Figure 2b). The limit of detection (defined here as three times the standard deviation of the background signal) was about 13 pM (76 pg/mL) in this experiment. A detection limit in the pictogram/mililiter range is adequate for analysis of many molecular markers at physiological concentrations.32
Figure 2.
Performing bead-based insulin immunoassays in SlipChip. (a) Kinetic curves of fluorescence over time for all the 42 samples on the SlipChip. As the insulin concentration increased, the rate of the enzymatic reaction increased. (b) A graph showing a calibration curve correlating insulin concentration to rate of fluorescence intensity on SlipChip. The insert shows the region of low concentration, plotted on a logarithmic scale. (n=7).
An important step for heterogeneous immunoassays is the separation of the unbound residual antigen and detection antibody in the bulk solution from the sandwich complex that forms on the surface of the solid phase. Using magnetic beads as the solid phase provides a convenient way to do this separation. However, the quality of the results of the assays strongly depends on the efficiency of the manipulation of the magnetic beads, such retention and washing of the beads. In the SlipChip-based immunoassay, we found that ~97% or more of the beads remained in the wells on the bottom plate during slipping after using a magnet to pull down the beads (Figure 3a). No beads were observed to be trapped between the two plates of the SlipChip.
Figure 3.
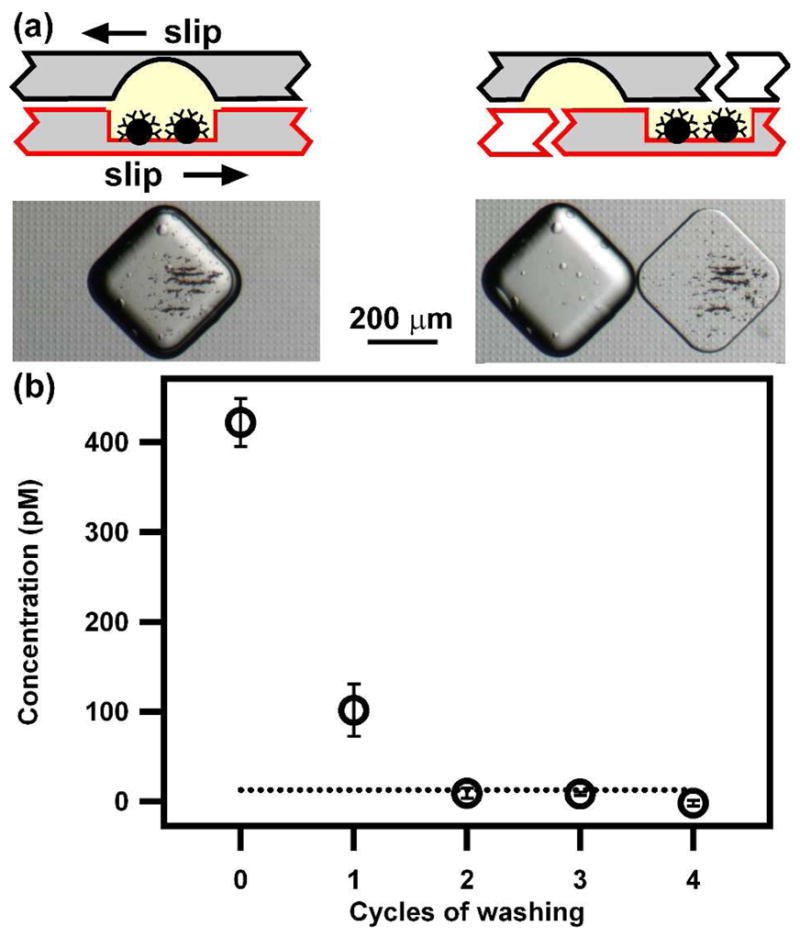
Magnetic bead retention and washing efficiency in SlipChip. (a) Images of magnetic beads being transferred during slipping. (b) A graph showing the concentration of the remaining detection antibody in the bulk of the wells with the beads in the SlipChip after with different cycles of washing (see text for details). The dotted line shows the detection limit of 13 pM.
Washing of the beads was performed by slipping the wells containing the beads to combine them with a series of wells containing washing buffer. During this series of washing steps, the residual detection antibody diffused into the washing buffer, was exponentially diluted, and eventually reached a negligible level. To maximize the washing efficiency and minimize the space and number of steps required, we fabricated the SlipChip with a high volumetric ratio between the wells on the top plate (9 nL) to the wells on the bottom plate (1 nL). Therefore, after four cycles of washing, the residual detection antibody was diluted by a factor of 104, assuming complete mixing in every washing cycle. To facilitate mixing of solutions during washing, the beads were moved using a magnet. To characterize the washing and test the washing efficiency, we performed a series of assays with a blank solution containing no insulin as the sample, and with different cycles of washing. Cycles of washing were determined by how many rows of wells on the top plate were loaded with the washing buffer. Because no insulin was added in these assays, the signals detected were produced by the residual enzyme-labeled detection antibody. After two, three, and four cycles of washing in the SlipChip, the signal from the enzyme-labeled detection antibody was below the 13 pM detection limit, corresponding to 9 pM, 8 pM, and −2 pM levels (Figure 3b).
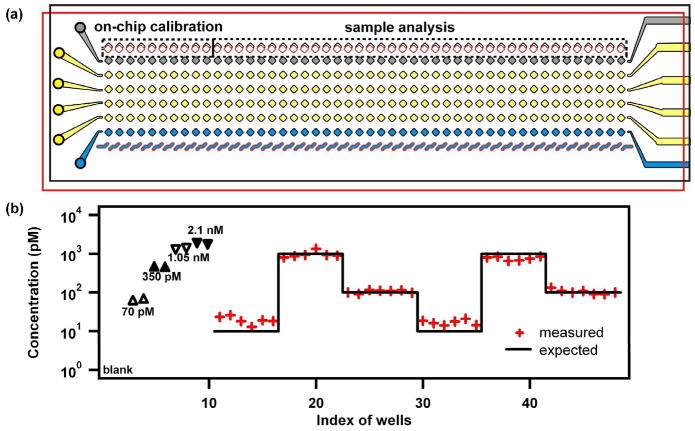
To test the suitability of this platform for analysis of plugs generated during operation of the chemistrode,5 we then modified the design of Section B of the SlipChip to allow direct loading of nL-plugs (Figure 4 and Figure S1). In this design, Section B had a row of 48 wells (9 nL) on the top plate, which was used to load the plugs, and a row of 48 wells (1 nL) on the bottom plate; Section A was the same as previously described (Figure 4a and Figure S1). We generated cartridges of plugs (~5 nL each) containing insulin of different concentrations, including five standard solutions, and then deposited the plugs in the first row of wells on the top plate under lubricating fluid. Plugs from different cartridges were deposited in a sequence as shown in Figure 4b. Ten wells were loaded with the standard solutions, two wells for each, for an on-chip calibration. For the sample solutions, we included a 10 pM solution of insulin, close to the detection limit, to test if such low concentrations could be reliably detected. We also used 100 pM and 1 nM solutions of insulin to test whether these concentrations could be reliably measured. The SlipChip was assembled by placing the bottom plate over the top plate and aligning the plates to connect the wells and ducts of Section A. After loading the reagents as described in the Experimental Section, the wells of Section B on the bottom plate were slipped to connect them with the wells on the top plate containing the deposited droplets of insulin. Surface tension drove the transfer of the droplets of insulin from the wells of the top plate to fill the wells of the bottom plate, because the wells on top plate were hydrophobic and the wells on the bottom plate were hydrophilic. Slipping apart the two plates generated a 1 nL volume of insulin solution in each of the bottom wells, and these wells were slipped through the wells in Section A to perform the insulin bead-based immunoassay as described above (Figure 1). The pattern of the concentration of insulin that was measured on the SlipChip was in good agreement with the deposited pattern, showing no biases for the wells at boundaries between regions of high and low concentrations, indicating that no significant cross-contamination occurred (Figure 4b). The 10 pM solution was robustly detected, and measured concentrations of 100 pM and 1 nM solutions were in good agreement with the expected values (Figure 4b).
Figure 4.
SlipChip is used to measure insulin in nL-plugs by bead-based immunoassay. (a) A schematic of the SlipChip designed to analyze samples in plugs. The top plate is outlined in black, and the bottom plate is outlined in red. In Section B, plugs were loaded into the wells on the top plate. Wells 1–10 were loaded with plugs of standard solutions for calibration, and wells 11–48 were loaded with plugs for analysis. (b) A graph showing the concentrations of the insulin measured on-chip. Plugs containing 10 pM insulin were deposited in wells 11–16 and 30–35, plugs containing 1 nM insulin were deposited in wells 17–22 and 36–41, and plugs containing 100 pM were deposited in wells 23–29 and 42–48.
Performing heterogeneous immunoassays in SlipChips as described above is easy to operate for users, because all reagents are preloaded into a series of wells prior to loading samples and performing the assays. However, situations that require analysis of many samples on a chip require a “space-saving” approach with fewer wells used per assay. For example, the chemistrode could generate arrays of thousands of plugs that must all be analyzed simultaneously. A space-saving design was tested by using a back-and-forth slipping motion in the SlipChip and is illustrated in Figure 5. The bottom plate of the SlipChip contained wells for loading samples and ducts for loading reagents, and the top plate contained wells for loading reagents. In this design, there was only one row of wells for the reagents, instead of six rows in the design discussed above. After loading the samples into the wells on the bottom plate and assembling the SlipChip (Figure 5a), a reagent was first injected into the wells on the top plate through the ducts (Figure 5b), and then the wells containing the reagent were slipped to combine the reagent with the wells containing samples (Figure 5c). After sufficient mixing or incubation, these wells on the top plate were slipped back to be aligned to the ducts on the bottom plate, and a solution of the second reagent was flowed to replace the former reagent in these wells (Figure 5d and e). The cycle of slipping, incubation, and slipping back was repeated as described above (Figure 5f) for each reagent of the heterogeneous immunoassay (the capture antibody on magnetic beads, the detection antibody, washing buffer, and the detection substrate). Beads were held in the wells containing samples using a magnet, and washed by serial dilution by repeating the slipping, mixing, slipping back cycle twelve times. After the substrate was added (Figure 5g), the fluorescence signal was monitored as described previously (Figure 5h). The results of this procedure are illustrated for two adjacent wells, the lower loaded with no insulin and the upper loaded with 1050 pM insulin (Figure 5i). The lower well produced low background intensity (65, background 50 ±12), indicating good washing and no cross-contamination from the well containing high concentration of insulin. The upper well produced strong signal, indicating successful capture of the insulin molecules (Figure 5i). This approach trades the ease of operation for the user for higher sample density. These are only preliminary results establishing proof of concept, but they are encouraging enough to warrant further optimization and characterization.
Conclusions
In this paper, we presented a SlipChip-based method to perform a bead-based heterogeneous immunoassay with nanoliter samples. We conclude that in addition to handling high-concentrations of biomolecules as was described previously,1,2,3 the SlipChip-based approach is suitable for analyzing biomolecules in the nM-pM range. The SlipChip is massively parallel and simple, and has less potential for user-error than manually-operated ELISA plates where reagents must be added by hand after defined time intervals. We used an insulin assay to demonstrate the method, but other bead-based immunoassays can be performed using similar procedures. The advantage of bead-based immunoassays is that the SlipChip does not require assay-specific surface modifications, and the same SlipChip can be used to perform different immunoassays by using magnetic beads coated with different capture antibodies; by loading different beads in different wells, highly multiplexed immunoassays can be conducted. Different assays may be conducted in parallel, including the ones that require different timing and reagents, using a different pattern of wells and reagents in each “track” (column of rows in the SlipChip) for each assay. A SlipChip that incorporates rotational motion may be used to create paths through which the slipping motion is occurring at different velocities.
If necessary, capture antibodies can also be immobilized on the surface of the SlipChip using surface modifications described in previous work.33–42 Control of surface chemistry using fluorous surfactants, to both prevent protein adsorption43 and to induce specific protein adsorption,44 is an attractive opportunity and should enable analysis of complex samples such as blood.16,45 Moreover, improvements in technology for fabrication can also enhance the performance of the device. Faster and more efficient washing is possible by fabricating even shallower wells. Varying the number of rows for the reagents and the distribution of reagents in these rows enables assays for samples requiring two steps to form the sandwich complex and two series of washing steps. Other types of signals can be detected by loading different substrates, and readout can be performed even without using a substrate, such as by detecting frustrated total internal reflection.24 Homogeneous immunoassays, such as HTRF® immunoassays provided by Cisbio, can be performed in a SlipChip with a much simpler design to mix multiple nL-volume samples with reagents both in the pre-loaded1 and user-loaded2, 3 formats. Performance of the assays can also be enhanced by a number of fields and effects; for example, electrical concentration using electrical fields to concentrate molecules near nanopores or channels.46–52
Technical advances in SlipChip technology presented here may find applications beyond immunoassays. For example, multi-step processing is a general problem in sample preparation and assays; multi-step serial dilution could find applications in assays with high dynamic range; and hydrophobic-to-hydrophilic transfer of solutions within the SlipChip could be used for reliable metering of a solution from a single well into one or more smaller wells. We demonstrated two types of serial dilution, used here to wash beads: by forward slipping over preloaded wells, and by back-and-forth slipping over the same well that can be refilled by flowing washing buffer through the fluidic path in the SlipChip. Preliminary experiments indicated that direct washing of beads by flow is also possible on SlipChip. Transferring magnetic beads from well to well is the basis for many laboratory operations, automated on larger scale by the Kingfisher system provided by Thermo Scientific.53 Driving flow by slipping to connect and disconnect a pattern of hydrophilic and hydrophobic wells is likely to be useful in a number of applications as well.
High sensitivity, defined as the number of molecules detected, can be achieved by analysis of small-volume samples, even without increasing sensitivity of the assay itself. In a 1 nL volume, a 13 pM concentration corresponds to ~8000 molecules per well. By using ultrasmall volumes (i.e. picoliters and below),33,54–56 e.g. combining optical fiber-based detection and SlipChip-based handling of fluids, one should be able to detect single or few molecules by a standard immunoassay on a SlipChip. In combination with the chemistrode, these capabilities would enable sampling and analysis of tissues and organisms, e.g. continuous monitoring of CSF biomarkers in small animals, currently measured via terminal sampling approaches,57 and nanovolume sampling from live tissue, e.g., a tumor or the retina.58 Single cell handling, stimulation, and analysis are critical for our understanding of complex networks, drug screening, and diagnostics. The SlipChip could complement the flow-cytometry based approaches in these areas.59–61 We expect the SlipChip to be able to handle cells without damage, although this prediction remains to be tested experimentally. The shear rate experienced by cells, even in a shallow 10 μm-deep wells, during the operation of the SlipChip, can be ~5–50 s−1, if slipping ~500 μm takes ~1–10 seconds. These values are lower, for example, than the 3000 s−1 limit62 or 40–1900 s−1 range of physiological shear in blood vessels,63 but higher than ~0.2 s−1 shear in devices designed for long-term culture.64 The shear rate can be adjusted into the desired regime by controlling the depth of wells and the rate of slipping. We believe that the SlipChip platform is sufficiently simple to support these types of assays across the spectrum of applications, settings and users. Applications may range from those commonly done on lateral flow “dipsticks” to those done by more complex instruments, such as in diagnostics, food safety testing, allergen testing, to blood characterization and blood type determination. Settings and users may range from well-equipped research laboratories, to clinical settings, to point of care and home testing, to diagnostic testing under resource-poor conditions.
Supplementary Material
Acknowledgments
This work was supported by the NSF CRC CHE-0526693 and the NIH Director’s Pioneer Award program, part of the NIH Roadmap for Medical Research (1 DP1 OD003584). We thank Heidi Park for contributions to writing and editing this manuscript.
Footnotes
Supporting Information Available: Additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Du WB, Li L, Nichols KP, Ismagilov RF. Lab Chip. 2009;9:2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Du W, Ismagilov RF. J Am Chem Soc. 2010;132:112–119. doi: 10.1021/ja908558m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L, Du W, Ismagilov RF. J Am Chem Soc. 2010;132:106–111. doi: 10.1021/ja908555n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan AC, Deb-Basu D, Orban MW, Gotlib JR, Natkunam Y, O’Neill R, Padua RA, Xu LW, Taketa D, Shirer AE, Beer S, Yee AX, Voehringer DW, Felsher DW. Nat Med. 2009;15:566–571. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D, Du WB, Liu Y, Liu WS, Kuznetsov A, Mendez FE, Philipson LH, Ismagilov RF. Proc Natl Acad Sci U S A. 2008;105:16843–16848. doi: 10.1073/pnas.0807916105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen DL, Du WB, Ismagilov RF. New J Phys. 2009:11. doi: 10.1088/1367-2630/11/7/075017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Ismagilov RF. Langmuir. 2009;25:2854–2859. doi: 10.1021/la803518b. [DOI] [PubMed] [Google Scholar]
- 8.Liu WS, Kim HJ, Lucchetta EM, Du WB, Ismagilov RF. Lab Chip. 2009;9:2153–2162. doi: 10.1039/b904958d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kartalov EP, Zhong JF, Scherer A, Quake SR, Taylor CR, Anderson WF. Biotechniques. 2006;40:85–90. doi: 10.2144/000112071. [DOI] [PubMed] [Google Scholar]
- 10.Diercks AH, Ozinsky A, Hansen CL, Spotts JM, Rodriguez DJ, Aderem A. Anal Biochem. 2009;386:30–35. doi: 10.1016/j.ab.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shim JU, Cristobal G, Link DR, Thorsen T, Jia YW, Piattelli K, Fraden S. J Am Chem Soc. 2007;129:8825–8835. doi: 10.1021/ja071820f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi WW, Qin JH, Ye NN, Lin BC. Lab Chip. 2008;8:1432–1435. doi: 10.1039/b808753a. [DOI] [PubMed] [Google Scholar]
- 13.Huebner A, Bratton D, Whyte G, Yang M, deMello AJ, Abell C, Hollfelder F. Lab Chip. 2009;9:692–698. doi: 10.1039/b813709a. [DOI] [PubMed] [Google Scholar]
- 14.Schmitz CHJ, Rowat AC, Koster S, Weitz DA. Lab Chip. 2009;9:44–49. doi: 10.1039/b809670h. [DOI] [PubMed] [Google Scholar]
- 15.Song H, Tice JD, Ismagilov RF. Angew Chem Int-Ed. 2003;42:768–772. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 16.Song H, Li HW, Munson MS, Van Ha TG, Ismagilov RF. Anal Chem. 2006;78:4839–4849. doi: 10.1021/ac0601718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song H, Ismagilov RF. J Am Chem Soc. 2003;125:14613–14619. doi: 10.1021/ja0354566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song H, Chen DL, Ismagilov RF. Angew Chem Int-Ed. 2006;45:7336–7356. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sista R, Hua ZS, Thwar P, Sudarsan A, Srinivasan V, Eckhardt A, Pollack M, Pamula V. Lab Chip. 2008;8:2091–2104. doi: 10.1039/b814922d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sista RS, Eckhardt AE, Srinivasan V, Pollack MG, Palanki S, Pamula VK. Lab Chip. 2008;8:2188–2196. doi: 10.1039/b807855f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim CT, Zhang Y. Biosens Bioelectron. 2007;22:1197–1204. doi: 10.1016/j.bios.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Sato K, Tokeshi M, Kimura H, Kitamori T. Anal Chem. 2001;73:1213–1218. doi: 10.1021/ac000991z. [DOI] [PubMed] [Google Scholar]
- 23.Hermann M, Veres T, Tabrizian M. Anal Chem. 2008;80:5160–5167. doi: 10.1021/ac800427z. [DOI] [PubMed] [Google Scholar]
- 24.Bruls DM, Evers TH, Kahlman JAH, van Lankvelt PJW, Ovsyanko M, Pelssers EGM, Schleipen J, de Theije FK, Verschuren CA, van der Wijk T, van Zon JBA, Dittmer WU, Immink AHJ, Nieuwenhuis JH, Prins MWJ. Lab Chip. 2009;9:3504–3510. doi: 10.1039/b913960e. [DOI] [PubMed] [Google Scholar]
- 25.Yang SY, Lien KY, Huang KJ, Lei HY, Lee GB. Biosens Bioelectron. 2008;24:855–862. doi: 10.1016/j.bios.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 26.Murakami Y, Endo T, Yamamura S, Nagatani N, Takamura Y, Tamiya E. Anal Biochem. 2004;334:111–116. doi: 10.1016/j.ab.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 27.Lacharme F, Vandevyver C, Gijs MAM. Microfluid Nanofluid. 2009;7:479–487. [Google Scholar]
- 28.Bronzeau S, Pamme N. Anal Chim Acta. 2008;609:105–112. doi: 10.1016/j.aca.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 29.Moser Y, Lehnert T, Gijs MAM. Lab Chip. 2009;9:3261–3267. doi: 10.1039/b907724c. [DOI] [PubMed] [Google Scholar]
- 30.Lai S, Wang SN, Luo J, Lee LJ, Yang ST, Madou MJ. Anal Chem. 2004;76:1832–1837. doi: 10.1021/ac0348322. [DOI] [PubMed] [Google Scholar]
- 31.Riegger L, Grumann M, Nann T, Riegler J, Ehlert O, Bessler W, Mittenbuehler K, Urban G, Pastewka L, Brenner T, Zengerle R, Ducree J. Sens Actuator A-Phys. 2006;126:455–462. [Google Scholar]
- 32.Rifai N, Gillette MA, Carr SA. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 33.Sasuga Y, Iwasawa T, Terada K, Oe Y, Sorimachi H, Ohara O, Harada Y. Anal Chem. 2008;80:9141–9149. doi: 10.1021/ac8016423. [DOI] [PubMed] [Google Scholar]
- 34.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. Nat Biotechnol. 2006;24:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 35.Delamarche E, Juncker D, Schmid H. Adv Mater. 2005;17:2911–2933. doi: 10.1038/nmat1435. [DOI] [PubMed] [Google Scholar]
- 36.Bange A, Halsall HB, Heineman WR. Biosens Bioelectron. 2005;20:2488–2503. doi: 10.1016/j.bios.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Linder V, Sia SK, Whitesides GM. Anal Chem. 2005;77:64–71. doi: 10.1021/ac049071x. [DOI] [PubMed] [Google Scholar]
- 38.Linder V, Verpoorte E, de Rooij NF, Sigrist H, Thormann W. Electrophoresis. 2002;23:740–749. doi: 10.1002/1522-2683(200203)23:5<740::AID-ELPS740>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Dodge A, Fluri K, Verpoorte E, de Rooij NF. Anal Chem. 2001;73:3400–3409. doi: 10.1021/ac0015366. [DOI] [PubMed] [Google Scholar]
- 40.Dong YZ, Shannon C. Anal Chem. 2000;72:2371–2376. doi: 10.1021/ac991450g. [DOI] [PubMed] [Google Scholar]
- 41.Fu E, Nelson KE, Ramsey SA, Foley JO, Helton K, Yager P. Anal Chem. 2009;81:3407–3413. doi: 10.1021/ac802672v. [DOI] [PubMed] [Google Scholar]
- 42.Hu GQ, Gao YL, Sherman PM, Li DQ. Microfluid Nanofluid. 2005;1:346–355. [Google Scholar]
- 43.Roach LS, Song H, Ismagilov RF. Anal Chem. 2005;77:785–796. doi: 10.1021/ac049061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreutz JE, Li L, Roach LS, Hatakeyama T, Ismagilov RF. J Am Chem Soc. 2009;131:6042–6043. doi: 10.1021/ja808697e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kline TR, Runyon MK, Pothiawala M, Ismagilov RF. 2008;80:6190–6197. doi: 10.1021/ac800485q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang YC, Stevens AL, Han JY. Anal Chem. 2005;77:4293–4299. doi: 10.1021/ac050321z. [DOI] [PubMed] [Google Scholar]
- 47.Dai JH, Ito T, Sun L, Crooks RM. J Am Chem Soc. 2003;125:13026–13027. doi: 10.1021/ja0374776. [DOI] [PubMed] [Google Scholar]
- 48.Dhopeshwarkar R, Li SA, Crooks RM. Lab Chip. 2005;5:1148–1154. doi: 10.1039/b509063f. [DOI] [PubMed] [Google Scholar]
- 49.Dhopeshwarkar R, Crooks RM, Hlushkou D, Tallarek U. Anal Chem. 2008;80:1039–1048. doi: 10.1021/ac7019927. [DOI] [PubMed] [Google Scholar]
- 50.Hlushkou D, Dhopeshwarkar R, Crooks RM, Tallarek U. Lab Chip. 2008;8:1153–1162. doi: 10.1039/b800549d. [DOI] [PubMed] [Google Scholar]
- 51.Kim SJ, Han JY. Anal Chem. 2008;80:3507–3511. doi: 10.1021/ac800157q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim SJ, Wang YC, Lee JH, Jang H, Han J. Phys Rev Lett. 2007;99:044501. doi: 10.1103/PhysRevLett.99.044501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makinen J, Marttila H, Viljanen MK. J Magn Magn Mater. 2001;225:134–137. [Google Scholar]
- 54.Chiu DT, Lorenz RM, Jeffries GDM. Anal Chem. 2009;81:5111–5118. doi: 10.1021/ac900306q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rissin DM, Walt DR. J Am Chem Soc. 2006;128:6286–6287. doi: 10.1021/ja058425e. [DOI] [PubMed] [Google Scholar]
- 56.Walt DR. Chem Soc Rev. 2010;39:38–50. doi: 10.1039/b809339n. [DOI] [PubMed] [Google Scholar]
- 57.Liu MC, Akle V, Zheng WR, Kitlen J, O’Steen B, Larner SF, Dave JR, Tortella FC, Hayes RL, Wang KKW. J Neurochem. 2006;98:700–712. doi: 10.1111/j.1471-4159.2006.03882.x. [DOI] [PubMed] [Google Scholar]
- 58.Lu MJ, Pulido JS, McCannel CA, Pulido JE, Hatfield RM, Dundervill RF, Shippy SA. Exp Diabetes Res. 2007 doi: 10.1155/2007/39765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krutzik PO, Crane JM, Clutter MR, Nolan GP. Nat Chem Biol. 2008;4:132–142. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 60.Danna EA, Nolan GP. Curr Opin Chem Biol. 2006;10:20–27. doi: 10.1016/j.cbpa.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 61.Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 62.Maiorella B, Dorin G, Carion A, Harano D. Biotechnol Bioeng. 1991;37:121–126. doi: 10.1002/bit.260370205. [DOI] [PubMed] [Google Scholar]
- 63.Hathcock JJ. Arterioscler Thromb Vasc Biol. 2006;26:1729–1737. doi: 10.1161/01.ATV.0000229658.76797.30. [DOI] [PubMed] [Google Scholar]
- 64.Berthier E, Warrick J, Yu H, Beebe DJ. Lab Chip. 2008;8:860–864. doi: 10.1039/b717423c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.