Abstract
The purpose of this paper is to establish single-participant white matter atlases based on diffusion tensor imaging. As one of the applications of the atlas, automated brain segmentation was performed and the accuracy was measured using Large Deformation Diffeomorphic Metric Mapping (LDDMM). High-quality diffusion tensor imaging (DTI) data from a single-participant were B0-distortion-corrected and transformed to the ICBM-152 atlas or to Talairach coordinates. The deep white matter structures, which have been previously well documented and clearly identified by DTI, were manually segmented. The superficial white matter areas beneath the cortex were defined, based on a population-averaged white matter probability map. The white matter was parcellated into 176 regions based on the anatomical labeling in the ICBM-DTI-81 atlas. The automated parcellation was achieved by warping this parcellation map to normal controls and to Alzheimer’s disease patients with severe anatomical atrophy. The parcellation accuracy was measured by a kappa analysis between the automated and manual parcellation at 11 anatomical regions. The kappa values were 0.70 for both normal controls and patients while the inter-rater reproducibility was 0.81 (controls) and 0.82 (patients), suggesting “almost perfect” agreement. A power analysis suggested that the proposed method is suitable for detecting FA and size abnormalities of the white matter in clinical studies.
Keywords: Human, White matter, Atlas, Association fiber, Magnetic resonance imaging, Diffusion tensor, Alzheimer’s disease
Introduction
The white matter (WM) of the human brain has been attracting attention as an important area affected by various neurodegenerative diseases. There is growing evidence that the neuronal degeneration associated with these diseases begins in the neuronal periphery, such as in the axons and dendrites (Chevalier-Larsen and Holzbaur, 2006; Gunawardena and Goldstein, 2001; Meyer-Luehmann et al., 2008; Pigino et al., 2003; Stokin et al., 2005; Wirths et al., 2006). In addition, some neurodegenerative diseases are characterized by alterations in the myelin (Bronge et al., 2002; Matsuo et al., 1998; Sjobeck et al., 2005). Consistently, conventional magnetic resonance imaging (MRI) studies using T1- and T2-weighted contrast and the resultant images, such as magnetization transfer measurement and T2 relaxometry, have revealed WM alterations in several neurodegenerative diseases (da Rocha et al., 2004; Hirata et al., 2005; Horimoto et al., 2002; Mascalchi and Dal Pozzo, 1992; Seppi et al., 2005; Specht et al., 2003, 2005; Tambasco et al., 2003). However, these images lack the necessary contrast to differentiate the structures in the WM, and therefore, it has been difficult to precisely identify WM structures affected by these diseases.
DTI is a method that can delineate specific WM structures based on the orientation information of axonal bundles (Catani et al., 2002; Douek et al., 1991; Makris et al., 1997; Mori et al., 2005; Nakada and Matsuzawa, 1995; Pajevic and Pierpaoli, 1999; Pierpaoli and Basser, 1996; Stieltjes et al., 2001). DTI could be an ideal method for investigating the anatomy of intra-WM structures and their status under pathological conditions, which is difficult to investigate with conventional MRI. Indeed, ROI analyses of DTI data have revealed abnormalities in specific WM tracts in many neurodegenerative diseases (Bozzali et al., 2002; Fellgiebel et al., 2004; Ringman et al., 2007; Sage et al., 2007; Shiga et al., 2005; Zhang et al., 2007b). To examine the status of the entire WM anatomy, voxel-based group analysis (VBA) is an effective initial step. In VBA, the shapes of participant brains are normalized to a template space, and then voxel-by-voxel analyses are performed. This method enables us to statistically analyze the whole brain and report abnormal areas using a common coordinate system, such as ICBM-152 space (Ashburner and Friston, 1999; Chiang et al., 2008; Good et al., 2001; Mazziotta et al., 2001; Tanabe et al., 1998; Wright et al., 1995; Oishi et al., in press). Nevertheless, the voxel-based method often suffers from low statistical power because information from each voxel can be highly noisy. To ameliorate this issue, spatial blurring is often employed. However, simple isotropic blurring often introduces partial volume effects, especially for the WM in which anatomical units usually have very sharp boundaries (tracts with very different connectivity are often adjacent to each other).
Another promising approach to ameliorate this problem is to combine the voxel-based method with a pre-segmented brain atlas. Within the accuracy level of normalization, the entire brain can be automatically segmented into many 3D structural components. Auto-mozation is needed as this would be too time-consuming to perform manually. Smith et al. proposed “skeletonization” of white matter, in which WM information is condensed to the WM skeleton (called TBSS [Tract-Based spatial statistics]) (Smith et al., 2006). Yushkevich, on the other hand, employed a method to project white matter information to the surface of pre-defined white matter tracts (Yushkevich et al., 2008). If we assume that abnormalities follow white matter anatomical units, as seen in neurodegenerative diseases, these approaches make more sense than utilizing simple isotropic filtering.
The purpose of this paper is to further extend efforts in this direction, by combining a pre-defined comprehensive white matter atlas with highly non-linear image registration methods for automated 3D white matter segmentation. This approach allows us to investigate changes in anatomical (sizes and shapes) and intensity (e.g., FA, ADC, T2, MTR, etc.) for each segmented structure. This approach can also be combined with the conventional voxel-based analysis or TBSS for interpretation of affected white matter regions and with the Yushkevich approach for surface projection. In this study, we established fully segmented white matter atlases in ICBM-152 and Talairach spaces that contain as many as 176 pre-defined 3D anatomical regions. Highly non-linear Large Deformation Diffeomorphic Metric Mapping (LDDMM) was used for normalization. The results of the automated segmentation were compared to manual definition for accuracy measurements. In order to test the segmentation accuracy in pathological conditions, the technique was also applied to the brains of Alzheimer’s disease patients with severe atrophy.
Methods
Creation of the single-participant atlas
Data acquisition
Data were obtained from a 32-year-old healthy female participant without known brain or neurological disorders. A 1.5 T MR unit (Gyroscan NT, Philips Medical Systems) was used. DTI data were acquired using single-shot echo-planar imaging sequences with sensitivity encoding (SENSE) and a parallel imaging factor of 2.5 (Pruessmann et al., 1999). The imaging matrix was 112×112, with a field of view of 246×246 mm (nominal resolution: 2.2 mm), zero-filled to 256×256 pixels. Transverse sections of 2.2 mm thickness were acquired parallel to the anterior commissure–posterior commissure line (AC–PC). A total of 60 sections covered the entire hemisphere and brainstem without gaps. Diffusion weighting was encoded along 30 independent orientations (Jones et al., 1999), and the b-value was 700 mm2/s. Five additional images with minimal diffusion weighting (b=33 mm2/s) were also acquired. The scanning time per dataset was approximately 4.6 min. To enhance the signal-to-noise ratio, imaging was repeated six times. Co-registered double-echo (40 and 100 ms), Fast Spin-Echo (FSE) images (JHU-T2w atlas), and a Magnetization-Prepared Rapid Gradient Echo (MPRAGE) image (JHU-T1w atlas) were also recorded for anatomical guidance.
Data processing
The raw diffusion-weighted images (DWIs) were first co-registered to one of the least diffusion-weighted images and corrected for participant motion and eddy current distortion using a 12-mode affine transformation of Automated Image Registration (AIR) (Woods et al., 1998), and then re-sliced to 1 mm isotropic resolution (181×217×181 matrix). B0-susceptibility distortion was corrected by non-linear warping using the least diffusion-weighted image of the FSE images (echo time: 100 ms) (Huang et al., 2008; Mori et al., 2008). For the non-linear warping, a large deformation diffeomorphic metric mapping (LDDMM) was used as briefly described in the next section. The warping was applied to all raw DWIs. The six elements of the diffusion tensor were calculated for each pixel with multivariate linear fitting using DtiStudio (H. Jiang and S. Mori, Johns Hopkins University, Kennedy Krieger Institute, lbam.med.jhmi.edu or www.MriStudio.org) (Mori et al., 2008; Pierpaoli et al., 2001). After diagonalization, three eigenvalues and eigenvectors were obtained. For the anisotropy map, fractional anisotropy (FA) was used (Pierpaoli and Basser, 1996). The eigenvector (v1) associated with the largest eigenvalue was used as an indicator of fiber orientation. A 24-bit, color-coded orientation map was created by assigning red, green, and blue channels to the x (right–left), y (anterior–posterior), and z (superior–inferior) components of the v1, where intensity was proportional to FA (Makris et al., 1997; Pajevic and Pierpaoli, 1999). These procedures resulted in a B0-distortion-corrected atlas with 1 mm isotropic resolution (JHU-DTI atlas), which consists of various types of DTI-derived contrasts, including eigenvalues, eigenvectors, FA, tensor, and color maps.
Atlas creation
The atlases (JHU-DTI, JHU-T2w, and JHU-T1w) were transformed to the ICBM-152 template and the Talairach coordinates to create a single-participant atlas based on these standard coordinate systems. To create the ICBM coordinate-based atlases (JHU-DTI-MNI, JHU-T2w-MNI, and JHU-T1w-MNI), we simply transformed the JHU atlases using a 12-parameter affine transformation of AIR since the template (ICBM-152) was also created in the same manner. The JHU-T1w atlas was normalized to the ICBM-152. Then, the transformation matrix was applied to the JHU-T2w atlas and to the calculated diffusion tensor field of the JHU-DTI atlas using the method described by Parker and Alexander (2003) and Xu et al. (2002). To create the Talairach coordinate-based atlases (JHU-DTI-Talairach, JHU-T2w-Talairach, and JHU-T1w-Talairach), highly elastic deformation is required since the Talairach atlas has a brain shape that is quite different from normal participants. Therefore, we resorted to manually placed landmark sets, in which we read the coordinates of many anatomical features in the Talairach atlas and defined the corresponding locations in our single-participant data. After placing 312 landmarks on the JHU-T1w atlas, we deformed the brain using LDDMM. This procedure ensures that the coordinates of the most prominent gray and WM structures are well-aligned between the two. The transformation matrix was then applied to the JHU-T2w atlas and the tensor field of the JHU-DTI atlas. The entire normalization process was performed with in-house software called Landmarker (X. Li, H. Jiang, and S. Mori, Johns Hopkins University, Kennedy Krieger Institute, lbam.med.jhmi.edu or www.MriStudio.org), in approximately 30 min for the entire process.
Parcellation of the gray and white matter
The boundary of the cortex and WM of our DTI atlases (JHU-DTI-MNI and JHU-DTI-Talairach) was defined by an FA threshold of 0.25. Since one of our aims was to use these atlases as a DTI template to normalize WM area correctly, we set this threshold to minimize the inclusion of the cortex, which typically has an FA less than 0.15, into the WM area. Please note that this boundary does not represent the cortex–WM boundary typically defined by T1-weighted images. Then, we switched the DTI atlases to the corresponding T1-weighted images (JHU-T1w-MNI and JHU-T1w-Talairach) to manually define each gyrus and subcortical gray matter (GM) based on T1-weighted contrast. For this procedure, we inspected all three slice orientations. The atlas provided by Talairach and Tournoux (1988) was used as the reference. For the parcellation of the deep WM (DWM), we adopted our previous WM parcellation map (www.loni.ucla.edu/Atlases,lbam.med.jhmi.edu, and mri.kennedykrieger.org), which manually defines 28 deep WM regions (Mori et al., 2008). Parcellation of the superficially located WM (SWM) was accomplished according to our previous publication (Oishi et al., 2008). Namely, we defined the SWM as the area between the cortex and the DWM, and manually parcellated the SWM into nine major structures called “blades”. The 9 blades were further sub-parcellated into 23 regions based on the relationships with 24 cerebral cortical areas and the cerebellum. These processes almost completely parcellates the GM and WM structures of the JHU-DTI-MNI and JHU-DTI-Talairach atlases. We call this parcellation map the WM parcellation map (WMPM), hereafter.
Creation of the probabilistic WM map in the ICBM space
The probabilistic WM map, in which each pixel demonstrates the probability to be the WM, was created by averaging 21 binarized WMs normalized to the ICBM space.
Data acquisition
The DTI database from the International Consortium of Brain Mapping (ICBM) was used to create the probabilistic WM map. Data from the 21 normal participants were acquired at the Montreal Neurological Institute and the University of California Los Angeles, as previously described (Mori et al., 2008; Oishi et al., 2008). The imaging protocol and data processing were largely similar to those used for the single-participant atlas, and only the differences are described here. The data were obtained on Siemens 1.5 T MR units. A parallel imaging factor of 2.0 was used. The imaging matrix was 96×96, with a field of view of 240×240 mm (nominal resolution: 2.5 mm). The b-value was 1000 s/mm2. The scanning time per dataset was approximately four min. To enhance the signal-to-noise ratio, imaging was repeated twice.
Atlas creation
WM was defined using an FA threshold of 0.25 for each participant, and was labeled in a binary approach (inside the boundary=1; outside the boundary=0), which was then normalized to the single-participant atlas (JHU-DTI-MNI) described above. The entire transformation process was performed by Landmarker. We first skull-stripped T2-weighted images (images without b0-distortion) and b0 images (images with b0-distortion) using the ROIEditor software (X. Li, H. Jiang, and S. Mori, Johns Hopkins University, Kennedy Krieger Institute, www.MriStudio.org or mri.kennedykrieger.org). To correct the b0 distortion, we transformed skull-stripped b0 images into corresponding skull-stripped T2-weighted images using LDDMM (Huang et al., 2008). The transformation matrix was applied to all DTI-derived images. After b0-distortion correction, dual-channel LDDMM (Ceritoglu, 2008) was performed to register each participant’s data to the JHU-DTI-MNI template. For the dual channel registration, we used FA and b0 images simultaneously.
The LDDMM was performed according to a previous publication (Huang et al., 2008; Miller et al., 2005). Briefly, the LDDMM algorithm computes a transformation, ϕ: Ω→Ω, where Ω⊆ℜ3 is the 3D cube on which the data are defined. The computed transformation is the end point, ϕ=ϕ1, of a flow of vector fields, vt ∈ V, t ∈ [0,1], given by the ordinary differential equation
where φ0 is the identity transformation, φ0(x)= x, ∀x ∈ Ω. As shown by Dupuis et al. (1998), enforcing a sufficient amount of smoothness on the elements in the space of allowable vector fields, V, ensures that the solution to the differential equation, φ̇t = vt(φt), t ∈ [0,1], is in the space of diffeomorphisms. Smoothness is enforced throughout by defining the norm on the space, V, of smooth velocity vector fields through a differential operator, L, which generally represents Laplacian powers such that , where |||·|||2 is the standard L2 norm for square integrable functions defined on Ω.
After LDDMM, we applied the transformation matrices to the binarized WMs of the corresponding participants to create normalized binarized WMs. Then, we averaged the normalized binarized WM to obtain a “probabilistic” WM map in each pixel in the ICBM152 coordinates.
Parcellation of the white matter
For the single-participant atlas, the white matter-cortex boundary was defined by FA>0.25. While this definition carries the detailed white matter anatomy of the participant, it also contains the participant-specific anatomical features in the SWM. The probabilistic WM map in the same atlas coordinate provides alternative criteria by which to define the boundary of the SWM based on WM probability. In this paper, we used 90% white matter probability to test this idea and compared the result with the single-participant atlas.
Measurement of registration quality
Data from five normal participants (72.8+/−8.8 years old, who scored 29.2+/−1.3 on the Mini Mental State Examination (MMSE)) and five Alzheimer’s disease (AD) patients (77.2+/−5.3 years old, who scored 22.8+/−3.6 on the MMSE), which were not included in the atlas construction, were used to measure the accuracy of automated WM parcellation. The AD cases were chosen to investigate the parcellation accuracy for brains with severe anatomical abnormalities. The average brain and ventricle volumes of the five AD cases were 1150+/−40.6 mm3 and 37.6+/−21.0 mm3, respectively, while those of the five normal controls were 1200+/−64.0 mm3 and 13.9+/−2.7 mm3, indicating the severely enlarged ventricles of the AD patients. The data were first registered to the JHU-DTI-MNI atlas using affine transformation for initial adjustment of the brain size and the orientation (“prepared” images), and then subjected to the following manual and automated WM structure delineation (Fig. 1).
Fig. 1.
Accuracy measurements for four different types of normalization methods. Participant’s images were first linearly normalized to the ICBM space to create a ‘prepared’ image. Type III WMPM was transformed to the ‘prepared’ images using affine, SPM5, and LDDMM. We used T1-weighted contrast (SPM-T1) and FA contrast (SPM-FA) to drive the non-linear normalization of SPM5. Manual delineation was considered the ‘gold standard,’ and kappa analysis between manual delineation and various automated delineation was performed.
Data acquisition
We used de-identified MRI database provided by Johns Hopkins Alzheimer’s Disease Research Center. The database contain brains from 55 to 85 years old and we arbitrary selected five data from AD database and five data from the normal control participant database. DTI was acquired using a SENSE head coil on 3 Tesla whole-body MR scanners (Philips Medical Systems) equipped with explorer gradients (40 mT/m). The imaging matrix was 96×96, with a field of view of 240×240 mm (nominal resolution: 2.5 mm). The parameters other than above mentioned and the image processing were same as that were used to create single-participant atlas. Double-echo FSE was also acquired and processed using the same method used for single-participant atlas.
Manual delineation
Eleven WM structures were delineated manually on the predetermined five 2D slices of the prepared images. The criteria for structural delineation closely followed the way the WM is parcellated in the WMPM of the probabilistic WM map (Type III WMPM), defined in the result section; namely, this procedure can be considered a manual transfer of the Type III WMPM to each image without automated normalization. The use of the Type III WMPM as a visual guide also helped to increase intra- and inter-rater reproducibility. Table 1 shows the list of these WM structures. The delineated set of structures was named “standard ROI set.” To investigate intra-rater variability of the manual delineation, a rater (K.O.) repeated this manual delineation three times with a more than two-week interval. Two other raters (A.F. and J.H.) also performed manual delineation to investigate inter-rater variability.
Table 1.
List of 11 WM structures and slice locations for manual definition.
| WM structures | |
|---|---|
| Axial # | |
| z=−18 | Inferior frontal blade, parieto-temporal blade |
| z=−8 | Superior frontal blade, anterior and posterior limb of internal capsule |
| Coronal # | |
| y=0 | Cingulum, superior longitudinal fasciculus |
| y=−31 | Superior parietal blade, posterior thalamic radiation |
| Sagittal # | |
| x=0 | Corpus callosum, fornix |
Automated delineation
JHU-DTI-MNI was transformed to the “prepared” images using LDDMM. The same procedure to create probabilistic WM atlas was applied. The “standard ROI set” was readily identified by superimposing the Type III WMPM onto the LDDMM transformed images. We also non-linearly transformed JHU-DTI-MNI to the “prepared” images using SPM5 (http://www.fil.ion.ucl.ac.uk/spm/) to compare the registration quality with that of LDDMM. For SPM5, we applied JHU-T1w-MNI and JHU-DTI-MNI as templates to obtain T1 contrast-based transformation (SPM-T1) and FA contrast-based transformation (SPM-FA). The other transformation parameters were based on the default setting: frequency cutoff, 25; iterations, 16; regularization, 1.
Accuracy measurements
The various structures defined by automated and manual delineation were saved as binary maps, in which the structure of interest was defined as “1” and the rest of the pixels were “0.” By superimposing the binary maps from the automated and manual methods, each pixel could be categorized into one of the three classes: (1) pixels that were outside the structures (“0”) in either method (nn); (2) pixels that were defined as the structure of interest (“1”) in only one of the two methods (pn, np); and (3) pixels that contained the structure in both methods (pp). After this categorization, reliability analyses were performed to estimate kappa statistics based on Landis and Koch (1977). These kappa estimations within the manual trials by three different raters provided the level of precision for manual delineation, which was treated as the gold standard. Then, the kappa value comparing the automated and manual delineation was calculated for each ROI. We did not expect that the kappa between automated vs. manual delineations would be better than the kappa between three raters’ manual delineations.
Power analysis
Using the data from the 21 normal participants (ICBM database) and the atlas-based automated quantification method, the FA and the size of each parcellated area were measured. This gave us average and standard deviation values for the normal population. Using these values, we performed power analyses to determine the number of patients and controls that would be required for future experimental designs using the proposed method. Software G*power (Faul et al., 2007) was used for the power analysis to investigate the number of participants needed to detect a 10% decrease of FA or to detect a 10% volume loss for each parcellated WM area. In this analysis, we assumed the same number of participants in each group (e.g., disease group and the control group), and the statistical significance was determined by an alpha error probability less than 0.05 and a power greater than 0.95 of a t-test.
Results
Single-participant comprehensive atlas of the gray and WM structures
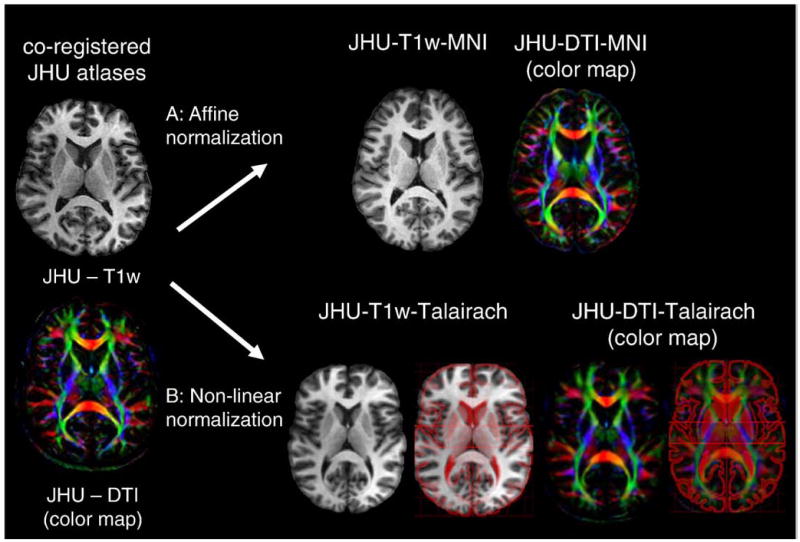
Fig. 2 shows the JHU-DTI, JHU-DTI-MNI, and the JHU-DTI-Talairach. The superimposed anatomical outline defined by Talairach (http://ric.uthscsa.edu/new/resources/talairachdaemon/talairachdaemon.html) (Lancaster et al., 2000) indicates the quality of the agreement. After the normalization, the cortex was manually parcellated based on annotations in the Talairach atlas. Each GM and WM structure is identified in this participant and annotated (Fig. 3). Fig. 4 shows the 3D reconstructed cortex (Fig. 4A), the SWM (Fig. 4B), and the DWM (Fig. 4D) of the JHU-DTI-MNI.
Fig. 2.
Transformation of single-participant MPRAGE (JHU-T1w) and DTI data (JHU-DTI) into the ICBM152 space and Talairach space. (A) JHU-DTI was transformed to the ICBM152 space using an affine transformation of AIR. The transformation matrix was obtained by normalizing JHU-T1w to the ICBM152 template. (B) JHU-DTI was transformed to the Talairach atlas using 312 anatomical landmarks. The superimposition of the Talairach drawing confirms the near-perfect transformation for both gray and white matter structures. The superimposed Talairach atlas was obtained from Talairach Daemon software (http://ric.uthscsa.edu/new/resources/talairachdaemon/talairachdaemon.html) (Lancaster et al., 2000).
Fig. 3.
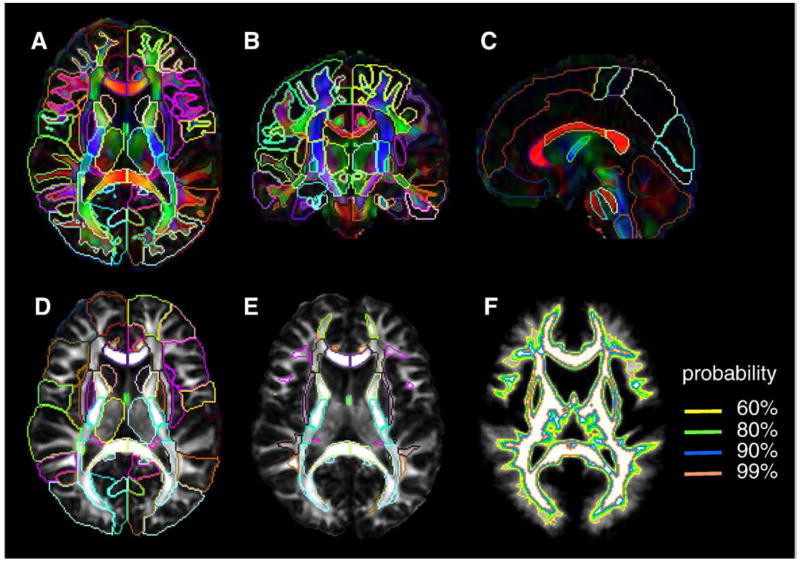
(A–C) Two-dimensional views of the JHU-DTI-MNI (color-coded orientation map), in which the entire GM and the WM are parcellated by Type I WMPM. (D) JHU-DTI-MNI (FA map) overlaid with Type II WMPM, in which the cortex and the SWM was surrounded by the same contour. (E) JHU-DTI-MNI (FA map) overlaid with Type III WMPM. (F) WM area defined by probability. To create the Type III WMPM, we adopted 90% probability as the boundary of the SWM.
Fig. 4.
Three-dimensional views of the cortical (A), SWM (B), and the DWM (D) parcellation of the Type I WMPM, and SWM parcellation of the Type III WMPM (C). The SWM (B and C) was parcellated to nine blade structures according to our previous publication (Oishi et al., 2008) as follows: purple, superior frontal blade; light green, middle frontal blade; deep green, inferior frontal blade; yellow, pre-central blade; blue, post-central blade; brown, superior parietal blade; red, parieto-temporal blade; ochre, temporal blade; and light blue, Occipital blade.
Three types of SWM definition
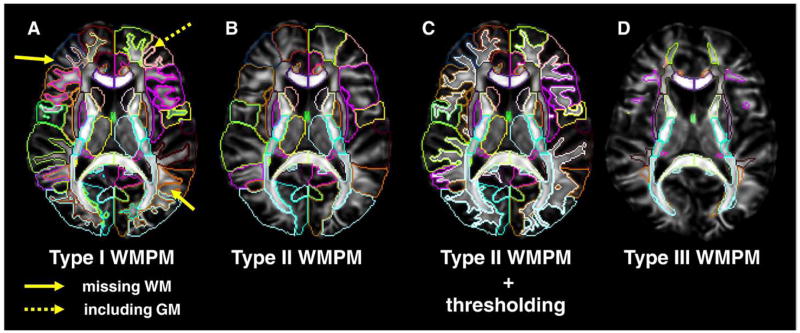
For the JHU-DTI-MNI, three types of the SWM definition were created, as shown in Figs. 3A–E. In Figs. 3A–C, the boundary between the SWM and the cortex was defined by simple FA threshold (FA>0.25) (denoted Type I hereafter). This reveals detailed SWM anatomy (Fig. 4B), but contains the individual-specific anatomy of these highly population-variable anatomical areas. In the version shown in Fig. 3D, the SWM is not defined and is combined with associated cortical parcellation (Type II). In Fig. 3E, the outer boundaries of the SWM are defined by the WM probabilistic map (Figs. 3F and 4C; Type III). For this study, a WM probability higher than 0.9 was used, meaning the defined regions were highly likely be the WM, with minimum contamination from the GM and CSF. Fig. 4 illustrates 3D views of types I and III SWM, as well as the cortical and DWM segmentations.
In Fig. 5, applications of these three types of the WMPMs are demonstrated. Data from a normal participant, which were not included in the atlas making, were transformed to the JHU-DTI-MNI template using LDDMM. When the Type I WMPM is overlaid, the segmentation of the DWM regions is well registered, but not for the SWM (Fig. 5A); the segmentation sometimes misses the WM or includes the GM. This is because of the high degree of cross-participant anatomical variability, which cannot be fully removed by LDDMM. There are two approaches to deal with this issue. First, the Type II WMPM is applied to parcellate the cortex and associated SWM together (Fig. 5B), followed by participant-by-participant delineation of the SWM (Fig. 5C). Using an FA threshold, this can be readily performed. The second approach is to use the Type III WMPM, which automatically defines core areas of the SWM (Fig. 5D).
Fig. 5.
Normalized FA map overlaid with Type I-III WMPMs. (A) Type I WMPM cannot accurately delineate the boundary of the SWM because of excessive anatomical differences between the atlas and the participant. Yellow solid arrows indicate the WM areas, which were misclassified as “cortex” based on Type I WMPM. A yellow dotted arrow indicates the cortex, which was misclassified as “WM” based on Type I WMPM. (B) Type II WMPM parcellates the cortex and associated SWM together. (C) After the type II parcellation, SWM structures can be individually parcellated using the FA threshold of 0.25. (D) Type III WMPM can label the core part of SWM, defined by the white matter probability (90% in the presented case).
Accuracy of normalization and automated segmentation
The accuracy of normalization and automated WM segmentation, using the Type III WMPM, is demonstrated for an AD patient in Fig. 6. The original image (Fig. 6A) was first affine transformed (Fig. 6B) to adjust the size of the brain to ICBM space, and then subjected to various non-linear normalizations (Fig. 6B: the atlas was non-linearly transformed to the affine-transformed patient image, or Fig. 6C: the patient image was non-linearly transformed to the atlas). To compare the result of LDDMM with that of other non-linear normalizations, we also normalized the image using SPM5. Type III WMPM was overlaid on each image. Some of the mis-registrations found in the WM area of conventional T1-contrast-based SPM transformation (SPM-T1) could be improved by using an FA contrast-based transformation (SPM-FA) (Fig. 6C, dotted arrows 5–7), but there were still WM mis-registrations (Fig. 6C, solid arrows 1–4). These mis-registrations were noticeably improved by using LDDMM.
Fig. 6.
Three types of non-linear normalization methods applied to an AD patient. Images are overlaid with the type III WMPM. (A) Original AD patient image; (B) ‘prepared’ AD image; (C) non-linearly transformed images. Upper row: SPM5 using T1-weighted contrast to drive transformation (SPM-T1). Middle row: SPM5 using FA contrast to drive transformation (SPM-FA). Lower row: LDDMM. Mis-registered structures seen by the SPM-T1 registration are indicated by yellow arrows. Yellow dotted arrows indicate mis-registrations improved by the SPM-FA registration. All the mis-registrations were significantly improved by using the LDDMM. Please note that LDDMM can generate reverse transformation simultaneously (indicated by the two-headed arrow), while SPM requires discrete transformation for participant-to-atlas and atlas-to-participant transformation (indicated by two arrows facing different directions). Abbreviations are: ALIC, anterior limb of internal capsule; EC, external capsule; Fx, fornix; CCS, splenium of corpus callosum; PLIC, posterior limb of internal capsule; CG, cingulum; IFWM, inferior frontal white matter.
The comparison of Kappa values between manual and automated WM delineation is shown in Fig. 7. Using the LDDMM, the average kappa values for the 11 anatomical regions were 0.70 for both NC and AD. These kappa values indicate “substantial agreement.” For the DWM areas, the kappa values were 0.76 (NC) and 0.75 (AD), with accuracy levels similar between the NC and the AD participants. The average intra- and inter-rater variability of manual delineation was 0.86 (0.89 for the DWM only) and 0.81 (0.84 for the DWM only), indicating a high level of reliability for the manual-based anatomical definition as our gold standard. We did not expect kappa values of the automated method to exceed 0.81. The FA-based SPM also achieved “substantial agreement” (kappa>0.6) for the NC, but not for the AD patients. Both for the NC and AD populations, significant improvement was observed with the LDDMM compared to the SPM5. This large improvement was observed particularly in the thin ‘line-like’ (e.g., cingulum) and ‘sheet-like’ structures (e.g., posterior thalamic radiation), which cannot be registered by affine or SPM5 (see also Fig. 5 for visual clues). Within SPM5, employment of the FA contrast led to better registration compared to T1-weighted contrast.
Fig. 7.
Result of the kappa analyses of five normal controls (NC, upper row), five Alzheimer’s disease patients (AD, middle row), and inter-rater comparison of three raters (bottom row). Abbreviations are; IFB, inferior frontal blade; PTB, parieto-temporal blade; SFB, superior frontal blade; and SPB, superior parietal blade (Oishi et al, 2008). Other abbreviations are summarized in the Appendix A-1 and A-4.
Power analysis for detecting FA reduction with this method
Atlas-based measurement applied to 21 normal participants was used for the initial estimation of power analyses. Tables 2 and 3 summarize the results of power analysis to detect a 10% FA reduction and a 10% volume reduction in each WM structure defined by Type III WMPM. By using ROIEditor, the FA value and volume of each WM region was automatically measured after each image was linearly normalized to the ICBM space. Note that the measured volumes of each WM structure were normalized by the total brain size, since the images were normalized to the ICBM space by affine transformation. If volumes of the un-normalized brains are required, it is straightforward to further transform the WMPM to the native space by a reverse-affine transformation.
Table 2.
Number of participants required to detect a 10% FA reduction in each WM structure.
| Required no. | WM structures |
|---|---|
| <10 | ACR, ALIC, CGC, CGH-L, CP, CST, EC, Fx/ST GCC, ICP, IFO, IFWM-L, IOG-L, MCP, MFG-R ML, MOWM-L, MTWM, PCR-L, PCT-R, PLIC, PoCWM-L, PrCWM PTR, RLIC, SCC, SCP, SCR, SFO-L, SFWM, SLF SPWM-L, SS, STWM, UNC-R (59 structures) |
| <20 | AGWM-L, BCC, CGH-R, FuWM-R, IFWM-R, IOWM-R, ITWM LFOWM-R, MFOWM, MOWM-R, PCR-R, SFO-R, SMWM, SPWM-R, UNC-L (20 structures) |
| <30 | AGWM-R, CuWM-L, FuWM-L, Fx, LFOWM-L, MFWM-L, PCT-L, PoCWM-R PreCuWM-L, RWM, SOWM, TAP (14 structures) |
| <60 | EntWM, LWM-L, PreCuWM-R (5 structures) |
| <120 | CuWM-R, LWM-R (2 structures) |
Abbreviations are summarized in the Appendix 4.
Table 3.
Number of participants required to detect a 10% volume reduction in each DWM structure.
| Required no. | WM structures |
|---|---|
| <10 | CGC-R, EC-R, SCR-R, SS-R (4 structures) |
| <20 | ACR-R, ALIC, CGC-L, CGH, CP, EC-L, Fx/ST, GCC, IFO, MCP-L, PCR-R, PLIC-R, PTR-R, RLIC, SCC, SCP, SCR-L, SLF, SS-L, UNC (31 structures) |
| <30 | ACR-L, BCC, CST-L, ICP, MCP-R, ML-R, PCR-L, PLIC-L, PTR-L (11 structures) |
| <60 | CST-R, ML-L, PCT, SFO, TAP (8 structures) |
| <120 | Fx (2 structures) |
Abbreviations are summarized in the Appendix 4.
With an alpha error probability less than 0.05 and power greater than 0.95, the majority of the WM structures require fewer than 20 participants to detect a 10% decrease in FA or volume. All DWM structures required fewer than 30 participants to detect a 10% FA decrease. However, for small structures, like the fornix or the medial lemniscus, a larger number of participants would be needed to detect a 10% volume reduction.
Discussion
Population-averaged and single-participant WM atlases
In this study, we created single-participant atlases in two different coordinate systems: ICBM-152 and Talairach. The atlases were parcellated to as many as 56 DWM, 46 SWM, 10 subcortical GM, 52 cortical areas, and 10 other areas (see Appendix). The segmentation of the DWM and the SWM was based on our previous population-based studies (Mori et al., 2008; Oishi et al., 2008).
In the earlier paper, we introduced a population-averaged WM atlas called ICBM-DTI-81, generated by linearly averaging of DTI data from 81 normal adults. When linear (or low-order non-linear) normalization was used, the adoption of this atlas maximized the registration quality; atlas-defined anatomical units were superimposed on corresponding structures in normalized brains with maximum probability. However, if highly non-linear normalization were used, the blurred anatomical definitions in the population-averaged atlases could confuse the transformation process. This prompted us to construct the single-participant WM atlases with pre-defined WM regions.
Compared to the population-averaged atlases, the advantage of the single-participant atlas is that it contains sharp definitions of anatomical structures, especially the SWM and the cortex, which are highly variable across individuals and cannot be appreciated in population-averaged atlases. However, this can also be perceived as a disadvantage for the single-participant atlas. Namely, it contains anatomical features that exist only in the arbitrarily selected participant (disadvantage 1). Even for DWM structures common to normal individuals, the question remains about how the location, size, and shape of each structure in the atlas are different from the averages of the normal population (disadvantage 2). If the transformation method of choice is perfect, the template simply serves as the origin of coordinates to measure averages, standard deviations, and differences among participant groups, and therefore, such a coordinate origin does not have to be located at the center of all the populations. However, in reality, the transformation method is hardly perfect. In this respect, we do not recommend the use of our atlas in the Talairach coordinates for voxel-based analyses, because the anatomy is severely distorted with respect to normal brains. The atlas in the Talairach coordinates, however, could be a useful tool, if one needs to utilize the cytoarchitectural map described in the Talairach atlas and investigate the relationship with WM abnormalities.
Combination of the population-averaged and single-participant anatomical definition
To ameliorate disadvantage 1, we created a hybrid atlas in which the definition of highly variable structures in the SWM was defined by population probability. Namely, we created a WM mask in each participant, normalized the mask to the single-participant template, and created a probabilistic WM map. By using a probability threshold of 0.9, the boundaries of the SWM were defined (Type III WMPM). The circular logic in this approach should be noted. We started with a single-participant atlas as a template, which contained the participant-specific anatomical features, and the resultant probabilistic map was influenced by such features, as was the choice of transformation method. There could be room for a better approach and improvement of the atlas. We also proposed a Type II WMPM (Fig. 5B) with FA thresholding (Fig. 5C).
Study of segmentation accuracy
To evaluate the impact of the second disadvantage, it was essential to determine the accuracy of the atlas-based anatomical segmentation. We used Type III WMPM and compared the segmentation results to manual-based delineation. If the agreement measurements are restricted to the DWM, the kappa value was 0.76 (NC) and 0.75 (AD), while the reliability of the manual method was 0.84, achieving almost the same accuracy as manual delineation. Accuracy measurements for the SWM are not straightforward because the boundary was difficult to define by visual inspection. This was not the case for the DWM, which was well-demarcated by clear boundaries based on sharp transitions in fiber orientations or adjacent subcortical GM nuclei in many cases. As a result, the raters had to rely on the SWM defined in the Type III WMPM as guidance. In this respect, the results shown in Fig. 7 are, strictly speaking, not the accuracy measurements in which manual delineation is the gold standard, but rather, the reproducibility of copying the SWM depicted in the Type III WMPM to each brain. The SWM in the Type III WMPM delineate the blade-type structures at the core of several distinctive cortical structures, as described previously (Oishi et al., 2008), and its boundaries are defined by probability, not by clear anatomical boundaries. Its usefulness should be evaluated in future application studies.
Another important issue when we apply the normalization-based approach to elderly brains is the influence of the extensive T2 hyperintensity often found in the WM. In this study, we visually inspected the existence of such abnormalities and identified six participants (one normal and five AD patients) with apparent T2 hyperintensity in the WM areas. The average kappa values for the 11 WM areas were 0.70 for both groups (participants with and without T2 hyperintensity). With our limited database, the result suggests that our dual-channel normalization is robust against the typical T2 hyperintensity in AD patients and in the elderly population, although further investigation with a larger data size would be required for a firmer conclusion.
Applications of the atlases
From a procedural point of view, there are two ways to use our atlases. One is to warp the atlas to individual data and the other is to warp the individual data to the atlas. Because of the reciprocal nature of the LDDM, the transformation can be accomplished either way (Appendix 5). By applying the WMPM in the native space, we can automatically parcellate the WM without image interpolation, and it is more straightforward to perform volume measurements of each parcellated compartment. If one is interested in VBA or TBSS, images need to be warped to the atlas space. In this approach, the anatomical information, such as regional volume changes, is stored in the transformation matrix and can be retrieved by using metrics such as Jacobian determinant. After VBA or TBSS analysis, the WMPM can be superimposed to evaluate which WM regions are affected. In addition to this type of qualitative utility, quantitative values can also be extracted by applying the WMPM and averaging the values of all pixels within one compartment (atlas-based analysis or ABA).
For the VBA, statistics for each pixel are derived individually. This often leads to low statistical power necessitating the frequent use of spatial filtering for pixel averaging. The filter is usually isotropic. However, some reports have criticized the use of such a filtering procedure (Jones et al., 2005), especially for the WM structures, because of sharp transitions in structural units; tracts with very different functions could be only a few pixels away. The ABA could be considered a special kind of filtering that groups pixels belonging to a specific WM unit. This, however, assumes that abnormality follows WM anatomical boundaries. Depending on pathology, this is not always the case (e.g., vasculature-dependent diseases). Therefore, VBA and ABA should be considered complementary methods.
Issues related to normalization of tensor data
For DTI, there are unique problems related to tensor manipulations at two different steps. One step is the transformation of tensor fields and the other step is the measurements of matching quality between the data and the atlas. For the tensor transformation, the transformation is not a simple issue because there are several ways to accomplish this (including tensor reorientation and interpolation), and it is difficult to conclude which is “anatomically correct” because of the massive degeneration of anatomical information in the MRI domain. Because the anatomical information within a 2 mm pixel (typical resolution of DTI) is astronomically large, any representation by MRI can be only an extremely simplified view, and it is very difficult to conclude from such a view whether the choice of transformation methods is correct. In this study, we used Gee and Alexander’s method (Alexander et al., 2001; Xu et al., 2003), which is one of the simplest and, currently, most widely used methods. For the matching measurements, we used scalar images, such as FA. It has been postulated that vector or tensor-based methods may improve the accuracy (Cao et al., 2005; Zhang et al., 2007a; Ziyan et al., 2007). These types of advanced tensor normalization methods are still being actively investigated and, in the future, incorporation of such technologies may further improve the accuracy of the atlas-based analyses introduced in this paper.
Comparison between the atlas (WMPM)-based and tractography-based analyses
In image analysis, the identification of “corresponding” regions across subjects is of central importance. The normalization-based approach is one of the most widely used methods for this purpose. Recently, tractography-based image analyses have become a popular analysis method (Glenn et al., 2003; Pagani et al., 2005; Partridge et al., 2004; Stieltjes et al., 2001; Virta et al., 1999; Wilson et al., 2003; Xue et al., 1999). Tractography can be considered a region-growing tool with which researchers can identify pixels that belong to the same tract system across the subject, and group those pixels, thereby allowing us to compare sizes and other variables, such as FA and ADC at the corresponding anatomical structures. Usually, normalization-based methods result in pixel-by-pixel statistics, while the presented atlas (WMPM)-based method leads to a grouping of pixels within segmented tract systems. In this sense, the atlas-based method can be considered a hybrid of the normalization-based and tract-based methods. However, the atlas-based and tractography-based methods have fundamental differences. First of all, in the atlas-based method, the entire white matter is segmented to one of the white matter structural units. The tractography-based methods can group pixels only for reconstructed fibers. The tractography-based method can be applied to those fibers for which a reliable reconstruction protocol (e.g., ROI placement) is established. In addition, the way pixels are grouped is different. For example, the atlas has a segment called the “posterior limb of the internal capsule (PLIC).” Using the tractography-based method, this area could be divided into the corticospinal tract, the corticopontine tract, and the thalamic radiations. If an abnormality occurs along specific connections, the tractography-based method could be more sensitive and specific. For example, if the corticospinal tract is affected, we would expect that only a portion of the PLIC would be affected (pixel grouping within the entire PLIC leads to lower sensitivity). However, the disadvantage of the tractography-based method is that, as mentioned above, not all tracts are reconstructable and cross-subject variability of reconstruction results could be large (for example, as large as a more than 30% standard deviation among a normal population in terms of size, as reported in Wakana et al., 2007). The tractography-based approach also may not work for pathological brains in which FA of certain regions is considerably decreased, and tractography results are profoundly altered. To ameliorate these problems, population-averaged probabilistic tractography approaches have been postulated (e.g., Abe et al., 2002; Hua et al., 2008). The atlas-based and tractography-based approaches should, therefore, be considered complementary to each other.
In summary, we developed single-participant atlases, which were combined with a highly elastic non-linear transformation method to achieve automated white matter parcellation. Using this approach the entire white matter was three-dimensionally parcellated into 102 regions. This type of analysis would be prohibitively time-consuming if a manual-based approach is used. On the other hand, accuracy is often an issue for this type of automated segmentation approaches compared to manual delineation. The accuracy level of our approach was measured using Alzheimer’s disease patients and age-matched controls and was comparable to the reliability level of human raters. This tool is expected to be a suitable tool for the initial whole-brain screening to detect potential abnormalities in clinical studies. The atlases developed in this study are now available for downloading from our website (lbam.med.jhmi.edu or mri.kennedykrieger.org). The software programs used in this study are also available (www.MriStudio.org or mri.kennedykrieger.org).
Acknowledgments
This publication was made possible by grant number P41 RR015241 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH. This research was supported by NIH grants U24RR021382, PO1EB00195, RO1AG20012, and by the Johns Hopkins Alzheimer’s Disease Research Center (P50AG005146). The images acquired on Alzheimer’s patients and age matched controls were supported by a methods development grant from Glaxo-Smith-Kline. Dr. van Zijl is a paid lecturer for Philips Medical Systems and is the inventor of technology that is licensed to Philips. This arrangement has been approved by Johns Hopkins University in accordance with its conflict of interest policies.
Appendix A
-
List of abbreviations (names of white matter structures are listed in Appendix 4)
AD, Alzheimer’s disease
ADC, apparent diffusion coefficient
AIR, automated image registration
DTI, diffusion tensor imaging
DWI, diffusion-weighted image
DWM, deep white matter
FA, fractional anisotropy
FSE, fast spin-echo
GM, gray matter
ICBM, international consortium of brain mapping
LDDMM, large deformation diffeomorphic metric mapping
MPRAGE, magnetization-prepared rapid gradient echo
MTR, magnetization transfer ratio
NC, normal control
ROI, region of interest
SENSE, sensitivity encoding
SPM, statistical parametric mapping
SPM-FA, FA-contrast-based SPM transformation
SPM-T1, T1-contrast-based SPM transformation
SWM, superficially located white matter
TBSS, tract-based spatial statistics
VBA, voxel-based group analysis
WM, white matter
-
Names of the atlases created in this study
Assigned coordinate system
Native space ICBM space Talairach space T1-weighted image JHU-T1w JHU-T1w-MNI JHU-T1w-Talairach T2-weighted image JHU-T2w JHU-T2w-MNI JHU-T2w-Talairach DTI JHU-DTI atlas JHU-DTI-MNI JHU-DTI-Talairach Parcellation maps Type I-III WMPM -
Explanation of the white matter parcellation map (WMPM)
WMPM Definition No. of structures parcellated
Gyri SWM DWM Subcortical GM Others Total Type I Manual parcellation of the whole brain. 176 structures (bilateral) are parcellated. 52 46 56 10 12 176 Type II Manual parcellation of the whole brain. 22 SWM of type I WMPM are included in the corresponding gyri. 52 56 10 12 130 Type III Manual parcellation of the SWM and the DWM area. Outline of the SWM is based on 90% white matter probability. Structures in the cerebellum and the cingulum white matter are not included. 44 56 100 -
Parcellated structures for Type I WMPM and the abbreviations
Each structure has a right and a left side. In the main content, tables, and figures, R represents the right side and L represents the left side.
SPG, Superior Parietal Lobule
CingG, Cingulate Gyrus left
SFG, Superior Frontal Gyrus
MFG, Middle Frontal Gyrus
IFG, Inferior Frontal Gyrus
PrCG, Pre-Central Gyrus
PoCG, Post-Central Gyrus
AG, Angular Gyrus
PrCu, Pre-Cuneus
Cu, Cuneus
LG, Lingual Gyrus
Fu, Fusiform Gyrus
PHG, Parahippocampal Gyrus
SOG, Superior Occipital Gyrus
IOG, Inferior Occipital Gyrus
MOG, Middle Occipital Gyrus
ENT, Entorhinal Area
STG, Superior Temporal Gyrus
ITG, Inferior Temporal Gyrus
MTG, Middle Temporal Gyrus
LFOG, Lateral Fronto- Orbital Gyrus
MFOG, Middle Fronto-Orbital Gyrus
SMG, Supramarginal Gyrus
RG, Gyrus Rectus
Ins, Insular Cortex
Amyg, Amygdala
Hippo, Hippocampus
Cerebellum, Cerebellum cortex
CST, Corticospinal tract
ICP, Inferior cerebellar peduncle
ML, Medial lemniscus
SCP, Superior cerebellar peduncle
CP, Cerebral peduncle
ALIC, Anterior limb of internal capsule
PLIC, Posterior limb of internal capsule
PTR, Posterior thalamic radiation (includes optic radiation)
ACR, Anterior corona radiata
SCR, Superior corona radiata
PCR, Posterior corona radiata
CGC, Cingulum (cingulate gyrus)
CGH, Cingulum (hippocampus)
Fx/ST, Fornix (cres)/Stria terminalis (cannot be resolved with current resolution)
SLF, Superior longitudinal fasciculus
SFO, Superior fronto-occipital fasciculus (could be a part of ALIC)
IFO, Inferior fronto-occipital fasciculus
SS, Sagittal stratum (includes ILF and IFO)
EC, External capsule
UNC, Uncinate fasciculus
PCT, Pontine crossing tract (a part of MCP)
MCP, Middle cerebellar peduncle left
Fx, Fornix (column and body of fornix)
GCC, Genu of corpus callosum
BCC, Body of corpus callosum
SCC, Splenium of corpus callosum
RLIC, Retrolenticular part of internal capsule
RedNc, Red Nucleus
Snigra, Substantia Nigra
TAP, Tapatum
Caud, Caudate Nucleus
Put, Putamen
Thal, Thalamus
GP, Globus Pallidus
Midbrain
Pons
Medulla
SPWM, Superior Parietal white matter
CingWM, Cingulum white matter
SFWM, Superior Frontal white matter
MFWM, Middle Frontal white matter
IFWM, Inferior Frontal white matter
PrCWM, Pre-Central white matter
PoCWM, Post-Central white matter
AWM, Angular white matter
PrCuWM, Pre-Cuneus white matter
CuWM, Cuneus white matter
LWM, Lingual white matter
FuWM, Fusiform white matter
SOWM, Superior Occipital white matter
IOWM, Inferior Occipital white matter
MOWM, Middle Occipital white matter
STWM, Superior Temporal white matter
ITWM, Inferior Temporal white matter
MTWM, Middle Temporal white matter
LFOWM, Lateral Fronto- Orbital white matter
MFOWM, Middle Fronto-Orbital white matter
SMWM, Supramarginal white matter
RGWM, Rectus white matter
CerebelWM, Cerebellum white matter
-
Test for the reciprocal nature of the LDDMM transformation
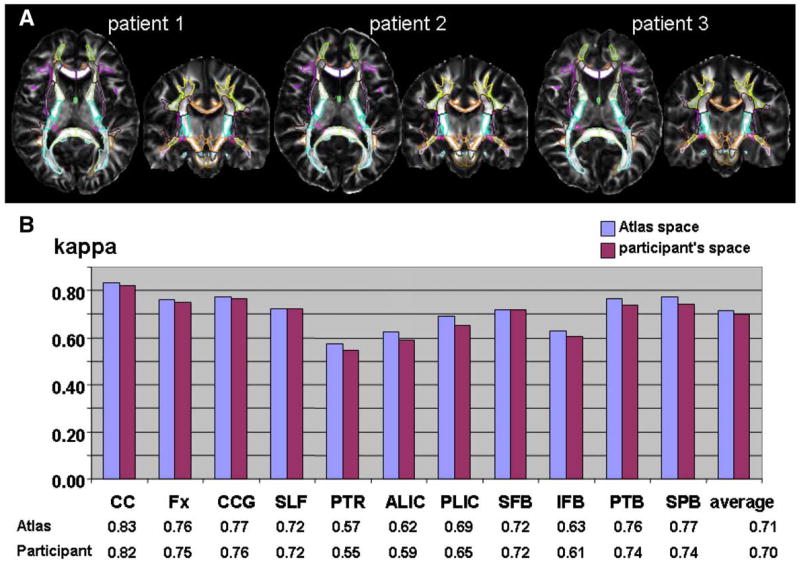
The LDDMM transformation is reciprocal: the atlas can be warped to the shape of each participant’s brain (atlas-to-data) or all participant brains can be warped to the atlas brain (data-to-atlas). The former approach can avoid interpolation of participant data, but voxel-based analyses require the latter approach. For image registration accuracy, the difference in these two approaches should remain within acceptable interpolation errors. The results reported in this paper were based on the atlas-to-data approach. Fig. 8A visually shows the normalization quality when the data-to-atlas approach is used (three randomly chosen cases). In Fig. 8B, accuracy measurement results of one arbitrary chosen patient were compared between the two approaches. The kappa values were comparable, with a slight tendency toward higher kappa values in the data-to-atlas approach. We attributed this to the fact that the manual segmentation criteria are more consistent in the data-to-atlas approach, in which all images have a very similar shape. Accordingly, the intra- and inter-rater reproducibility could also be higher in the data-to-atlas approach. We also tested FA measurements of the 100 WM structures defined by the type III WMPM using both approaches, and the correlation was Y=0.99X+0.0037, with r2=0.9941. The results support the reciprocal nature of the LDDMM-based quantitative analyses.
Fig. 8.
(A) Quality of automated white matter segmentation (Type III atlas with 100 WM regions) of 3 AD patients transformed to the atlas space. (B) Comparison of kappa values obtained by two reciprocal approaches. See Fig. 7 and Appendix A-4 for the abbreviations.
References
- Abe O, Aoki S, Hayashi N, Yamada H, Kunimatsu A, Mori H, Yoshikawa T, Okubo T, Ohtomo K. Normal aging in the central nervous system: quantitative MR diffusion-tensor analysis. Neurobiol Aging. 2002;23:433–441. doi: 10.1016/s0197-4580(01)00318-9. [DOI] [PubMed] [Google Scholar]
- Alexander DC, Pierpaoli C, Basser PJ, Gee JC. Spatial transformations of diffusion tensor magnetic resonance images. IEEE Trans Med Imag. 2001;20:1131–1139. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Hum Brain Mapp. 1999;7:254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzali M, Falini A, Franceschi M, Cercignani M, Zuffi M, Scotti G, Comi G, Filippi M. White matter damage in Alzheimer’s disease assessed in vivo using diffusion tensor magnetic resonance imaging. J Neurol Neurosurg Psychiatry. 2002;72:742–746. doi: 10.1136/jnnp.72.6.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronge L, Bogdanovic N, Wahlund LO. Postmortem MRI and histopathology of white matter changes in Alzheimer brains. A quantitative, comparative study. Dement Geriatr Cogn Disord. 2002;13:205–212. doi: 10.1159/000057698. [DOI] [PubMed] [Google Scholar]
- Cao Y, Miller MI, Winslow RL, Younes L. Large deformation diffeomorphic metric mapping of vector fields. IEEE Trans Med Imag. 2005;24:1216–1230. doi: 10.1109/tmi.2005.853923. [DOI] [PubMed] [Google Scholar]
- Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. NeuroImage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- Ceritoglu C. Multichannel Large Deformation Diffeomorphic Metric Mapping and Registration of Diffusion Tensor Images. Department of Biomedical Engineering The Johns Hopkins University; Baltimore: 2008. [Google Scholar]
- Chevalier-Larsen E, Holzbaur EL. Axonal transport and neurodegenerative disease. Biochim Biophys Acta. 2006;1762:1094–1108. doi: 10.1016/j.bbadis.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Chiang MC, Leow AD, Klunder AD, Dutton RA, Barysheva M, Rose SE, McMahon KL, de Zubicaray GI, Toga AW, Thompson PM. Fluid registration of diffusion tensor images using information theory. IEEE Trans Med Imag. 2008;27:442–456. doi: 10.1109/TMI.2007.907326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha AJ, Oliveira AS, Fonseca RB, Maia AC, Jr, Buainain RP, Lederman HM. Detection of corticospinal tract compromise in amyotrophic lateral sclerosis with brain MR imaging: relevance of the T1-weighted spin-echo magnetization transfer contrast sequence. AJNR Am J Neuroradiol. 2004;25:1509–1515. [PMC free article] [PubMed] [Google Scholar]
- Douek P, Turner R, Pekar J, Patronas N, Le Bihan D. MR color mapping of myelin fiber orientation. J Comput Assist Tomogr. 1991;15:923–929. doi: 10.1097/00004728-199111000-00003. [DOI] [PubMed] [Google Scholar]
- Dupuis P, Grenander U, Miller MI. Variational problems on flows of diffeomorphisms for image matching. Q Appl Math. 1998;56:587–600. [Google Scholar]
- Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- Fellgiebel A, Wille P, Muller MJ, Winterer G, Scheurich A, Vucurevic G, Schmidt LG, Stoeter P. Ultrastructural hippocampal and white matter alterations in mild cognitive impairment: a diffusion tensor imaging study. Dement Geriatr Cogn Disord. 2004;18:101–108. doi: 10.1159/000077817. [DOI] [PubMed] [Google Scholar]
- Glenn OA, Henry RG, Berman JI, Chang PC, Miller SP, Vigneron DB, Barkovich AJ. DTI-based three-dimensional tractography detects differences in the pyramidal tracts of infants and children with congenital hemiparesis. J Magn Reson Imaging. 2003;18:641–648. doi: 10.1002/jmri.10420. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. NeuroImage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Goldstein LS. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron. 2001;32:389–401. doi: 10.1016/s0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- Hirata Y, Matsuda H, Nemoto K, Ohnishi T, Hirao K, Yamashita F, Asada T, Iwabuchi S, Samejima H. Voxel-based morphometry to discriminate early Alzheimer’s disease from controls. Neurosci Lett. 2005;382:269–274. doi: 10.1016/j.neulet.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Horimoto Y, Aiba I, Yasuda T, Ohkawa Y, Katayama T, Yokokawa Y, Goto A, Ito Y. Longitudinal MRI study of multiple system atrophy — when do the findings appear, and what is the course? J Neurol. 2002;249:847–854. doi: 10.1007/s00415-002-0734-0. [DOI] [PubMed] [Google Scholar]
- Hua K, Zhang J, Wakana S, Jiang H, Li X, Reich DS, Calabresi PA, Pekar JJ, van Zijl PC, Mori S. Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. NeuroImage. 2008;39:336–347. doi: 10.1016/j.neuroimage.2007.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Ceritoglu C, Li X, Qiu A, Miller MI, van Zijl PC, Mori S. Correction of B0 susceptibility induced distortion in diffusion-weighted images using large-deformation diffeomorphic metric mapping. Magn Reson Imaging. 2008 doi: 10.1016/j.mri.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Horsfield MA, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magn Reson Med. 1999;42:515–525. [PubMed] [Google Scholar]
- Jones DK, Symms MR, Cercignani M, Howard RJ. The effect of filter size on VBM analyses of DT-MRI data. NeuroImage. 2005;26:546–554. doi: 10.1016/j.neuroimage.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Kochunov PV, Nickerson D, Mikiten SA, Fox PT. Automated Talairach atlas labels for functional brain mapping. Hum Brain Mapp. 2000;10:120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- Makris N, Worth AJ, Sorensen AG, Papadimitriou GM, Wu O, Reese TG, Wedeen VJ, Davis TL, Stakes JW, Caviness VS, Kaplan E, Rosen BR, Pandya DN, Kennedy DN. Morphometry of in vivo human white matter association pathways with diffusion-weighted magnetic resonance imaging. Ann Neurol. 1997;42:951–962. doi: 10.1002/ana.410420617. [DOI] [PubMed] [Google Scholar]
- Mascalchi M, Dal Pozzo G. Magnetic resonance imaging of degenerative diseases of the central nervous system. Ital J Neurol Sci. 1992;13:105–111. [PubMed] [Google Scholar]
- Matsuo A, Akiguchi I, Lee GC, McGeer EG, McGeer PL, Kimura J. Myelin degeneration in multiple system atrophy detected by unique antibodies. Am J Pathol. 1998;153:735–744. doi: 10.1016/S0002-9440(10)65617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, Koenigsknecht-Talboo J, Holtzman DM, Bacskai BJ, Hyman BT. Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer’s disease. Nature. 2008;451:720–724. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Beg MF, Ceritoglu C, Stark C. Increasing the power of functional maps of the medial temporal lobe by using large deformation diffeomorphic metric mapping. Proc Natl Acad Sci U S A. 2005;102:9685–9690. doi: 10.1073/pnas.0503892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Wakana S, Nagae-Poetscher LM, van Zijl PC. MRI Atlas of Human White Matter. Elsevier; Amsterdam, The Netherlands: 2005. [Google Scholar]
- Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziotta J. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage. 2008;40:570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada T, Matsuzawa H. Three-dimensional anisotropy contrast magnetic resonance imaging of the rat nervous system: MR axonography. Neurosci Res. 1995;22:389–398. doi: 10.1016/0168-0102(95)00917-i. [DOI] [PubMed] [Google Scholar]
- Oishi K, Zilles K, Amunts K, Faria AV, Jiang H, Li X, Akhter K, Hua K, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziotta J, Mori S. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. NeuroImage. 2008;43:447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Konishi J, Mori S, Ishihara H, Kawamitsu H, Fujii M, Kanda F. Reduced fractional anisotropy in early-stage cerebellar variant of Multiple System Atrophy. J Neuroimaging. doi: 10.1111/j.1552-6569.2008.00262.x. in press. (Electronic publication ahead of print) [DOI] [PubMed] [Google Scholar]
- Pagani E, Filippi M, Rocca MA, Horsfield MA. A method for obtaining tract-specific diffusion tensor MRI measurements in the presence of disease: application to patients with clinically isolated syndromes suggestive of multiple sclerosis. NeuroImage. 2005;26:258–265. doi: 10.1016/j.neuroimage.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pajevic S, Pierpaoli C. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn Reson Med. 1999;42:526–540. [PubMed] [Google Scholar]
- Parker GJ, Alexander DC. Probabilistic Monte Carlo based mapping of cerebral connections utilising whole-brain crossing fibre information. Inf Process Med Imag. 2003;18:684–695. doi: 10.1007/978-3-540-45087-0_57. [DOI] [PubMed] [Google Scholar]
- Partridge SC, Mukherjee P, Henry RG, Miller SP, Berman JI, Jin H, Lu Y, Glenn OA, Ferriero DM, Barkovich AJ, Vigneron DB. Diffusion tensor imaging: serial quantitation of white matter tract maturity in premature newborns. NeuroImage. 2004;22:1302–1314. doi: 10.1016/j.neuroimage.2004.02.038. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn Reson Med. 1996;36:893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Barnett A, Pajevic S, Chen R, Penix LR, Virta A, Basser P. Water diffusion changes in Wallerian degeneration and their dependence on white matter architecture. NeuroImage. 2001;13:1174–1185. doi: 10.1006/nimg.2001.0765. [DOI] [PubMed] [Google Scholar]
- Pigino G, Morfini G, Pelsman A, Mattson MP, Brady ST, Busciglio J. Alzheimer’s presenilin 1 mutations impair kinesin-based axonal transport. J Neurosci. 2003;23:4499–4508. doi: 10.1523/JNEUROSCI.23-11-04499.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. [PubMed] [Google Scholar]
- Ringman JM, O’Neill J, Geschwind D, Medina L, Apostolova LG, Rodriguez Y, Schaffer B, Varpetian A, Tseng B, Ortiz F, Fitten J, Cummings JL, Bartzokis G. Diffusion tensor imaging in preclinical and presymptomatic carriers of familial Alzheimer’s disease mutations. Brain. 2007;130:1767–1776. doi: 10.1093/brain/awm102. [DOI] [PubMed] [Google Scholar]
- Sage CA, Peeters RR, Gorner A, Robberecht W, Sunaert S. Quantitative diffusion tensor imaging in amyotrophic lateral sclerosis. NeuroImage. 2007;34:486–499. doi: 10.1016/j.neuroimage.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Seppi K, Schocke MF, Wenning GK, Poewe W. How to diagnose MSA early: the role of magnetic resonance imaging. J Neural Transm. 2005;112:1625–1634. doi: 10.1007/s00702-005-0332-2. [DOI] [PubMed] [Google Scholar]
- Shiga K, Yamada K, Yoshikawa K, Mizuno T, Nishimura T, Nakagawa M. Local tissue anisotropy decreases in cerebellopetal fibers and pyramidal tract in multiple system atrophy. J Neurol. 2005;252:589–596. doi: 10.1007/s00415-005-0708-0. [DOI] [PubMed] [Google Scholar]
- Sjobeck M, Haglund M, Englund E. Decreasing myelin density reflected increasing white matter pathology in Alzheimer’s disease—a neuropathological study. Int J Geriatr Psychiatry. 2005;20:919–926. doi: 10.1002/gps.1384. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TE. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuro-Image. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Specht K, Minnerop M, Abele M, Reul J, Wullner U, Klockgether T. In vivo voxel-based morphometry in multiple system atrophy of the cerebellar type. Arch Neurol. 2003;60:1431–1435. doi: 10.1001/archneur.60.10.1431. [DOI] [PubMed] [Google Scholar]
- Specht K, Minnerop M, Muller-Hubenthal J, Klockgether T. Voxel-based analysis of multiple-system atrophy of cerebellar type: complementary results by combining voxel-based morphometry and voxel-based relaxometry. NeuroImage. 2005;25:287–293. doi: 10.1016/j.neuroimage.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Stieltjes B, Kaufmann WE, van Zijl PC, Fredericksen K, Pearlson GD, Solaiyappan M, Mori S. Diffusion tensor imaging and axonal tracking in the human brainstem. NeuroImage. 2001;14:723–735. doi: 10.1006/nimg.2001.0861. [DOI] [PubMed] [Google Scholar]
- Stokin GB, Lillo C, Falzone TL, Brusch RG, Rockenstein E, Mount SL, Raman R, Davies P, Masliah E, Williams DS, Goldstein LS. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science. 2005;307:1282–1288. doi: 10.1126/science.1105681. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-Planar Stereotaxic Atlas of the Human Brain. Thieme; New York: 1988. [Google Scholar]
- Tambasco N, Pelliccioli GP, Chiarini P, Montanari GE, Leone F, Mancini ML, Paciaroni M, Gallai V. Magnetization transfer changes of grey and white matter in Parkinson’s disease. Neuroradiology. 2003;45:224–230. doi: 10.1007/s00234-002-0925-5. [DOI] [PubMed] [Google Scholar]
- Tanabe JL, Vermathen M, Miller R, Gelinas D, Weiner MW, Rooney WD. Reduced MTR in the corticospinal tract and normal T2 in amyotrophic lateral sclerosis. Magn Reson Imaging. 1998;16:1163–1169. doi: 10.1016/s0730-725x(98)00129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virta A, Barnett A, Pierpaoli C. Visualizing and characterizing white matter fiber structure and architecture in the human pyramidal tract using diffusion tensor MRI. Magn Reson Imaging. 1999;17:1121–1133. doi: 10.1016/s0730-725x(99)00048-x. [DOI] [PubMed] [Google Scholar]
- Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, Blitz A, van Zijl P, Mori S. Reproducibility of quantitative tractography methods applied to cerebral white matter. NeuroImage. 2007;36:630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, Tench CR, Morgan PS, Blumhardt LD. Pyramidal tract mapping by diffusion tensor magnetic resonance imaging in multiple sclerosis: improving correlations with disability. J Neurol Neurosurg Psychiatry. 2003;74:203–207. doi: 10.1136/jnnp.74.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirths O, Weis J, Szczygielski J, Multhaup G, Bayer TA. Axonopathy in an APP/PS1 transgenic mouse model of Alzheimer’s disease. Acta Neuropathol. 2006;111:312–319. doi: 10.1007/s00401-006-0041-4. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Wright IC, McGuire PK, Poline JB, Travere JM, Murray RM, Frith CD, Frackowiak RS, Friston KJ. A voxel-based method for the statistical analysis of gray and white matter density applied to schizophrenia. NeuroImage. 1995;2:244–252. doi: 10.1006/nimg.1995.1032. [DOI] [PubMed] [Google Scholar]
- Xu D, Mori S, Shen D, Davatzikos C. IEEE International Symposium on Biomedical Imaging. Macro to Nano; Washington D.C: 2002. Statisticaly-based reorientation of diffusion tensor field. [Google Scholar]
- Xu D, Mori S, Shen D, van Zijl PC, Davatzikos C. Spatial normalization of diffusion tensor fields. Magn Reson Med. 2003;50:175–182. doi: 10.1002/mrm.10489. [DOI] [PubMed] [Google Scholar]
- Xue R, van Zijl PC, Crain BJ, Solaiyappan M, Mori S. In vivo three-dimensional reconstruction of rat brain axonal projections by diffusion tensor imaging. Magn Reson Med. 1999;42:1123–1127. doi: 10.1002/(sici)1522-2594(199912)42:6<1123::aid-mrm17>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Zhang H, Simon TJ, Gee JC. Structure-specific statistical mapping of white matter tracts. NeuroImage. 2008;41:448–461. doi: 10.1016/j.neuroimage.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Avants BB, Yushkevich PA, Woo JH, Wang S, McCluskey LF, Elman LB, Melhem ER, Gee JC. High-dimensional spatial normalization of diffusion tensor images improves the detection of white matter differences: an example study using amyotrophic lateral sclerosis. IEEE Trans Med Imag. 2007a;26:1585–1597. doi: 10.1109/TMI.2007.906784. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Schuff N, Jahng GH, Bayne W, Mori S, Schad L, Mueller S, Du AT, Kramer JH, Yaffe K, Chui H, Jagust WJ, Miller BL, Weiner MW. Diffusion tensor imaging of cingulum fibers in mild cognitive impairment and Alzheimer disease. Neurology. 2007b;68:13–19. doi: 10.1212/01.wnl.0000250326.77323.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziyan U, Sabuncu MR, O’Donnell LJ, Westin CF. Nonlinear registration of diffusion MR images based on fiber bundles. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2007;10:351–358. doi: 10.1007/978-3-540-75757-3_43. [DOI] [PubMed] [Google Scholar]