Abstract
Airway hyper-responsiveness (AHR) is a critical phenotype of human asthma and animal models of asthma. Other studies have measured AHR in nine mouse strains, but only six strains have been used to identify genetic loci underlying AHR. Our goals were to increase the genetic diversity of available strains by surveying 27 additional strains, to apply haplotype association mapping to the 36-strain survey, and to identify new genetic determinants for AHR. We derived AHR from the increase in airway resistance in females subjected to increasing levels of methacholine concentrations. We used haplotype association mapping to identify associations between AHR and haplotypes on chromosomes 3, 5, 8, 12, 13, and 14. And we used bioinformatics techniques to narrow the identified region on chromosome 13, reducing the region to 29 candidate genes, with 11 of considerable interest. Our combined use of haplotype association mapping with bioinformatics tools is the first study of its kind for AHR on these 36 strains of mice. Our analyses have narrowed the possible QTL genes and will facilitate the discovery of novel genes that regulate AHR in mice.
Keywords: Mice, Genetics, Asthma
Introduction
Asthma prevalence has increased rapidly during the last 30 years, but the reason for the increase remains unknown (Moorman et al. 2007). Asthma phenotypes, such as airway inflammation, airway hyper-responsiveness (AHR), allergy, and airway remodeling are determined not only by environmental exposure, but also by the interaction of genetic determinants (Colilla et al. 2003; Hoffjan et al. 2005; Palmer and Cookson 2000). Because each phenotype has its own underlying genetic architecture, the genetic predisposition to the final disease state is complex. For a thorough understanding of this complexity, it is necessary to dissect the genetic contribution of each phenotype, a process that is dificult in humans but feasible in mice. The genetic architecture of mice is remarkably similar to that of humans, and we can readily manipulate mouse models and study them with a wide variety of genetic and bioinformatics resources. Thus, mouse models are excellent tools for narrowing the search for the genetic determinants of asthma (Peters et al. 2007; Stylianou et al. 2007).
AHR is one of the critical phenotypes of human asthma and an important phenotype in animal models of asthma (De Sanctis et al. 1999; Drazen et al. 1999; Richards 1996). AHR is defined as a significant narrowing of the airways in response to provoking stimuli, such as methacholine, that would be innocuous to an unaffected individual. There are 2 types of AHR: native AHR, which is hyper-responsiveness in the absence of allergen-induced airway inflammation, and allergen-induced AHR. Previous analysis of a limited number of inbred mouse strains for native AHR showed variation (Levitt and Mitzner 1989).
To dissect the genetic basis of complex diseases in the mouse, the traditional method has been to cross 2 strains with a phenotypic difference in a quantitative trait locus (QTL). Eight QTL studies for AHR have been conducted (Table 1); most of these studies used A/J as the hyper-responsive strain and C57BL/6J or C3H/HeJ as the hypo-responsive strain. Three of the eight QTL crosses were between the same strains, A/J and C3H/HeJ (Ackerman et al. 2005; De Sanctis et al. 1995, 1999; Ewart et al. 2000, 1996; McIntire et al. 2001; Nicolaides et al. 1997; Zhang et al. 1999). In five of these QTL studies, native AHR was determined (Ackerman et al. 2005; De Sanctis et al. 1995; De Sanctis et al. 1999; Ewart et al. 1996; Nicolaides et al. 1997) in three of the studies, allergen-induced AHR was determined in mice previously sensitized and challenged with an allergen (ovalbumin) (Ewart et al. 2000; McIntire et al. 2001; Zhang et al. 1999). Nineteen QTLs for AHR were identified: 16 (84%) are located in regions homologous to human asthma QTLs (Stylianou et al. 2007). To narrow these QTLs for AHR, additional studies must combine increased genetic diversity with improved analysis. As with successful identification of QTL genes in the mouse for other phenotypes (Pletcher et al. 2004), strategies include development of new QTL crosses with additional inbred strains and the use of novel bioinformatics tools, such as haplotype association mapping.
Table 1.
Summary of the 8 QTL studies that have been reported for AHR in the mouse
| Cross | Detection method | Pre-treatment | Challenge agent | Sex | Age (mos.) | Chr | Peak (Mb) | 95% CI (Mb) | LOD | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| (C3HxA)C3H | APTI | None | Acetylcholine | M | 5–6 | 6 | NA | 116–133 | 3.1 | Ewart et al. (1996) |
| Rrs | None | Acetylcholine | M | 5–6 | 6 | NA | 71–133 | 2.1 | ||
| (C3HxA)A | APTI | Ovalbumin | Acetylcholine | M | 4 | 2 | NA | 4.2 | ||
| APTI | Ovalbumin | Acetylcholine | M | 4 | 2 | NA | 3.7 | Ewart et al. (2000) | ||
| APTI | None | Acetylcholine | M | 4 | 7 | NA | 1.9 | |||
| (B6xD2)RI | APTI | None | Atracurium | M & f | 5–6 | 13 | 53 | 33–73 | NA | Nicolaides et al. (1997) |
| (B6xA)B6 | logED200RL | None | Methacholine | M | 5–6 | 2 | 158 | 123–163 | 3 | De Sanctis et al. (1995) |
| logED200RL | None | Methacholine | M | 5–6 | 15 | 84 | 64–103 | 3.7 | ||
| logED200RL | None | Methacholine | M | 5–6 | 17 | 11 | 4–48 | 2.8 | ||
| (C3HxA)C3H | RL | None | Methacholine | M | 5–6 | 6 | 115 | 95–135 | 3.3 | De Sanctis et al. (1999) |
| RL | None | Methacholine | M | 5–6 | 7 | 37 | 0–60 | 3.8 | ||
| RL | None | Methacholine | M | 5–6 | 17 | 43 | 23–63 | 1.7 | ||
| (B6xA)B6RC | Penh | None | Methacholine | M | 8 | 2 | NA | 142–178 | 1.1 | Ackerman et al. (2005) |
| Penh | None | Methacholine | M | 8 | 6 | NA | 4–87 | 1.4 | ||
| Penh | None | Methacholine | M | 8 | 2 | NA | 4.9 | |||
| 6 | ||||||||||
| (BiozzixBALB)F2 | Penh | Ovalbumin | Methacholine | NA | 6–8 | 9 | 37 | 28–49 | 2.5 | Zhang et al. (1999) |
| Penh | Ovalbumin | Methacholine | NA | 6–8 | 10 | 82 | 67–97 | 3.8 | ||
| Penh | Ovalbumin | Methacholine | NA | 6–8 | 11 | 90 | 70–103 | 3.7 | ||
| Penh | Ovalbumin | Methacholine | NA | 6–8 | 17 | 15 | 0–36 | 2.1 | ||
| (BALBxD2)RC | Penh | Ovalbumin | Methacholine | NA | NA | 11 | NA | 46–48 | NA | McIntire et al. (2001) |
RI, recombined inbred; RC, recombined congenic; APTI, airway pressure–time index, i.e. induced change in peak inspiratory pressure from time of challenge until baseline or plateau; Rrs, respiratory system resistance; logED200RL, effective dose required to increase RL to 200% of control values; RL, pulmonary resistance; Ova, ovalbumin; ACh, acetylcholine; MCh, methacholine; Atra, atracurium; NA, not available; Chr, chromosome; Mb, million base pairs; significant LOD scores are bolded; Ref, reference
Haplotype association mapping has become an integral alternative to QTL crosses for performing genetic studies. Haplotype association mapping is a method similar to genome-wide association studies in human populations (Cervino et al. 2007; Chesler et al. 2001; Grupe et al. 2001; McClurg et al. 2006; Payseur and Place 2007; Pletcher et al. 2004; Svenson et al. 2007). In the mouse, haplotype association mapping has been shown to successfully reproduce previously found QTL peaks, for example, for high-density lipoprotein cholesterol and gallstone susceptibility (Pletcher et al. 2004). Here, we chose haplotype association mapping because more genotype information is now available, and because we had access to comprehensive phenotypic data from 36 strains—the 9 strains used in previous studies, and the 27 strains we selected for their genetic diversity. The use of haplotype association mapping as a supplemental strategy to QTL studies allowed us to obtain more precise information about important genetic loci than with QTL studies alone.
In the present study, we detected variations of AHR in the 36 mouse strains and we applied haplotype association mapping to predict potentially new QTL regions for native AHR. We discovered several new strains that have AHR and we identified QTL regions on chromosomes (Chrs) 3, 5, 8, 12, 13, and 14 with promising novel candidate genes.
Materials and methods
Mice
We studied 6–10 female mice from each of 36 mouse strains:
| 129S1/SvlmJ | BTBR T+ tf/J | C57BR/cdJ | FVB/NJ | MRL/MpJ | PL/J |
| A/J | BUB/BnJ | C57L/J | I/LnJ | NOD/LtJ | PWD/PhJ |
| AKR/J | C3H/HeJ | CBA/J | KK/HlJ | NON/LtJ | RIIIS/J |
| BALB/cJ | C57BL/10J | CE/J | LG/J | NZO/HlLtJ | SJL/J |
| BALB/cByJ | C57BL/6J | DBA/1J | LP/J | NZW/LacJ | SM/J |
| BPN/3J | C57BLKS/J | DBA/2J | MA/MyJ | P/J | SWR/J |
All mice were raised and maintained in colonies at The Jackson Laboratory. Mice were kept on a 12 h light/12 h dark cycle in cages containing pine shaving bedding and topped with a polyester filter. They were allowed ad libitum access to water and chow diet (LabDiet® JR Rat and Mouse/Auto 6F 5K52) and were regularly monitored for (and found to be free of) 15 viruses, 17 bacterial species, Mycoplasma spp., and parasites, as described at http://jaxmice.jax.org/.
At 6–11 weeks of age, 10 mice from each strain were shipped to the University of Pittsburgh, where they were maintained under the same conditions as above for the duration of their lives. There they underwent phenotype analysis at 9–12 weeks of age, as described below, with a success rate for performing measurements of 6–10 mice per strain.
All animal protocols were reviewed and approved by the Animal Care and Use Committees at the University of Pittsburgh and The Jackson Laboratory.
Phenotype analysis
We measured airway resistance in response to methacholine (Sigma–Aldrich Inc., St. Louis, MO, USA) inhalation using a computer-controlled ventilator (flexiVent, SCIREQ, Montreal, QC, Canada). We anesthetized each mouse with 60 mg/kg pentobarbital sodium i.p. (Ovation Pharmaceuticals Inc., Deerfield, IL, USA), performed a tracheostomy, and then attached the mouse to the ventilator. Each animal then received deep lung inflation to 30 cm H2O distending pressure, after which we took baseline readings. Then we administered saline and sequentially increasing concentrations of methacholine (1, 3, 10, and 30 ml/kg) via nebulizer for 10 s through the tracheostomy.
To determine airway resistance, we fitted a constant phase model to the data obtained from the multiple frequencies simultaneously applied at the airway opening. After each of the 5 challenges, we recorded airway resistance (Raw) in 12 cycles of 30–60 s. We plotted changes in Raw as a function of the log-transformed methacholine concentration administered. We summarized AHR as the slope of the curve Raw versus log-transformed methacholine concentration.
Statistical modeling for haplotype association mapping
In recent years, haplotype association mapping has become the most commonly used technique for testing and developing the optimal method for genome-wide scans that include multiple strain genotypes and phenotypes. In addition to the strategy of scanning for associations between a phenotype and a single nucleotide polymorphism (SNP), researchers have also used a 3-SNP-window scan or a scan with a variable window size, which is determined by the local haplotype block (3, 4, 11, 14). Because previous studies showed that the local haplotype scanning method with a variable SNP-window size eliminates the false positive associations found by the 3-SNP-window method for 2 traits (high-density lipoprotein cholesterol and red blood cell count), we chose it as our method. We obtained genotype data consisting of 70,000 SNPs from the Broad Institute, Perlegen, GNF, Oxford, and Merck, selecting SNPs that were highly polymorphic and that were located approximately 40,000 base pairs apart.
We conducted computer analysis using the following as input: phenotypic trait data (slope) at strain mean level and genotype data (i.e., SNPs) across 33 inbred mouse strains (as a matrix). We excluded the wild-derived strain PWD/PhJ (because of genotype differences between it and common inbred strains) and BPN/3J and BALB/cByJ (because of missing genotype data). The SNP data set (termed 70 K SNP data set in build 36) is publically available at Gary Churchill’s website at http://cgd.jax.org. We used a hidden Markov model (HMM) to identify regions of the mouse genome that display a haplotype block structure, in order to assign individual strains to local dominant haplotypes, and to infer the genotypes of missing SNP alleles (Szatkiewicz et al. 2007). Within each inferred haplotype block, we grouped strains by the inferred local haplotype states. The number of distinct haplotype groups ranged between 2 and 5 across the genome.
We computed a regression-based test statistic and its nominal P value to measure the strength of association between the inferred haplotypes and phenotype means. All P values were transformed using −log10 (P value) in the scan plots (score). We controlled the type I error rate for multiple testing due to genome-wide searching using family-wise error rate control and the re-sampling based procedure described in Westfall and Young (1993). To generate the control population, we shuffled the phenotype data set and kept the genotype data and the inferred haplotypes intact (permutation testing). We conducted 1,000 permutation tests; the percentiles of the minimum P values (minP) distribution provided the approximate multiple test-adjusted thresholds. In each of these permutations, we recomputed test statistics and recorded the minP across all halotype blocks. The haplotype inference and missing data imputation were computed using software written in C (Szatkiewicz et al. 2007). Statistical analysis was performed using MATLAB computing environment (The Mathworks, http://www.mathworks.com).
Databases used to narrow genomic regions of interest
We narrowed genomic regions of interest using bioinformatics tools, such as haplotype analysis. Haplotype analysis compares the genotypes between strains in a region of interest. For haplotype analysis we used strain-specific genotype information (i.e., SNPs) that are publically available in the form of high-density SNP data sets provided by the Mouse Phenome Database (http://aretha.jax.org/pub-cgi/phenome/mpdcgi?rtn=docs/home). We compared SNP genotypes between the strains C57BL/6J and DBA/2J. If we found SNPs for which the genotypes differed (polymorphic SNPs), we assumed that these SNPs could explain genomic variations that may contribute to the phenotypic difference observed between the two strains. We then identified the location of a polymorphic SNP in relation to surrounding genes. Only genes that contained at least one polymorphic SNP within the gene were considered for further investigation. For identification of polymorphic SNPs in coding regions we used the SNP wizard provided by the Mouse Phenome Database (http://phenome.jax.org/pub-cgi/phenome/mpdcgi?rtn=snps/wiz1). To determine if the SNPs were located in conserved regions, we used the UCSC genome browser (http://genome.ucsc.edu/), and to determine if the SNPs were located in functional domains, we used the ExPASY browser (http://au.expasy.org/sprot/).
Gene expression experiments
The expression of the candidate genes in the lung in comparison to their expression in other tissues was tested using the Illumina Sentrix Mouse-6 BeadChip v 1.1 microarray that included 46,657 probes. Probes were annotated using the ArrayGene software as explained elsewhere (Verdugo and Medrano 2006). Total RNA was purified, amplified, and labeled from C57BL/6J animals for adrenals, brown adipose tissue, brain, colon, duodenum, heart, ileum, jejunum, kidney, liver, lung, muscle, pancreas, spleen, stomach, testis, and white adipose tissue. Five biological replicates were sampled for liver, heart, lung, kidney, adrenals, muscle, testis, gonadal adipose tissue, and brown adipose tissue. A second set of five replicates were sampled for pancreas, brain, duodenim, jejunum, ileum, colon, stomach, and spleen. Two technical replicates were hybridized for the kidney samples. Hybridizations were performed at the gene expression core facility at The Jackson Laboratory. Samples were divided into four 24-sample batches of hybridizations performed in consecutive days, i.e. 4 Mouse-6 arrays were hybridized per day. Data were extracted from tiff images with the Illumina BeadStudio v 3.0.1 software and exported as flat files. Unnormalized intensity values at probeset level were imported into R 2.7.1 (UNIX) environment for normalization and analysis. Quantile normalization was performed with the affy package for R (Bolstad et al. 2003). Quantiles normalization between samples of the same tissue was chosen at the least drastic normalization for this design. Scaling normalization was applied between samples of different tissues. Further preprocessing steps were applied to the data (e.g., local background correction, probe level values summarization, log2 data transformation). Probes in the array were filtered by detection calls and by variability across samples. Detection calls were computed from the probability of detection, which is estimated by comparing intensity values to negative probes in the same array using the BeadStudio software. A P value of 0.01 was used as a threshold to declare a target as present in a given sample. Only probes that were present in at least two samples from any tissue were kept for further analysis (McClintick and Edenberg 2006). This resulted in a dataset of 25,038 probes (14,980 unique Entrez genes).
Results
Native AHR varied among the 36 mouse strains
We surveyed 36 inbred mouse strains (6–10 mice per strain) for airway resistance (Raw) in response to increasing concentrations of methacholine aerosol challenges. We observed a dose-dependent increase in Raw values for all mice of all mouse strains. From the baseline, the percentage of Raw increased, ranging from 9% in response to 1 mg/ml methacholine inhalation to 544% in response to 30 mg/ml methacholine inhalation (Table 2).
Table 2.
Dose-dependent increase in airway resistance in mouse strains
| Strain | N | 1 mg/ml (mean ± SEM) | 3 mg/ml (mean ± SEM) | 10 mg/ml (mean ± SEM) | 30 mg/ml (mean ± SEM) |
|---|---|---|---|---|---|
| KK/HlJ | 9 | 35 ± 8 | 162 ± 24 | 489 ± 67 | 540 ± 112 |
| A/J | 10 | 39 ± 12 | 127 ± 11 | 234 ± 23 | 391 ± 35 |
| I/LnJ | 9 | 28 ± 6 | 127 ± 14 | 213 ± 33 | NA |
| MA/MyJ | 9 | 13 ± 3 | 57 ± 6 | 273 ± 39 | 442 ± 77 |
| CE/J | 9 | 30 ± 11 | 76 ± 21 | 204 ± 25 | 392 ± 47 |
| NZO/HILtJ | 9 | 31 ± 8 | 63 ± 6 | 182 ± 17 | 407 ± 30 |
| C57BR/cdJ | 7 | 48 ± 10 | 93 ± 18 | 234 ± 48 | 452 ± 103 |
| C57BLKS/J | 9 | 15 ± 2 | 49 ± 11 | 243 ± 30 | 544 ± 106 |
| PL/J | 9 | 19 ± 3 | 88 ± 14 | 207 ± 36 | 275 ± 39 |
| C57L/J | 6 | 14 ± 4 | 72 ± 15 | 202 ± 36 | 394 ± 54 |
| DBA/1 J | 9 | 22 ± 5 | 54 ± 12 | 224 ± 31 | NA |
| NOD/LtJ | 9 | 17 ± 7 | 81 ± 14 | 150 ± 6 | 407 ± 37 |
| BTBR T+tf/J | 10 | 25 ± 4 | 61 ± 11 | 225 ± 39 | 369 ± 73 |
| DBA/2 J | 10 | 12 ± 2 | 46 ± 6 | 146 ± 15 | 379 ± 64 |
| FVB/NJ | 9 | 25 ± 5 | 77 ± 10 | 154 ± 16 | 293 ± 28 |
| RIIIS/J | 6 | 45 ± 16 | 60 ± 12 | 157 ± 17 | 296 ± 38 |
| MRL/MpJ | 9 | 22 ± 4 | 53 ± 8 | 176 ± 35 | 347 ± 44 |
| PWD/PhJ | 6 | 12 ± 3 | 57 ± 8 | 181 ± 19 | 336 ± 54 |
| C57BL/10 J | 9 | 14 ± 3 | 57 ± 8 | 167 ± 16 | 244 ± 30 |
| CBA/J | 8 | 39 ± 9 | 82 ± 20 | 141 ± 36 | 241 ± 41 |
| SM/J | 10 | 17 ± 4 | 56 ± 10 | 165 ± 15 | 254 ± 23 |
| LP/J | 9 | 11 ± 2 | 40 ± 4 | 133 ± 11 | 289 ± 33 |
| NZW/LacJ | 8 | 19 ± 3 | 39 ± 6 | 121 ± 14 | 351 ± 59 |
| NON/LtJ | 9 | 21 ± 5 | 63 ± 9 | 125 ± 15 | 221 ± 20 |
| AKR/J | 9 | 14 ± 4 | 40 ± 5 | 109 ± 16 | 247 ± 29 |
| 129S1/SvImJ | 9 | 11 ± 1 | 47 ± 5 | 140 ± 10 | 250 ± 36 |
| BALB/cByJ | 8 | 23 ± 7 | 54 ± 12 | 142 ± 37 | 203 ± 23 |
| LG/J | 10 | 22 ± 6 | 38 ± 6 | 117 ± 24 | 406 ± 83 |
| BUB/BnJ | 9 | 18 ± 5 | 43 ± 10 | 123 ± 15 | 242 ± 32 |
| SWR/J | 9 | 9 ± 2 | 44 ± 4 | 144 ± 15 | 215 ± 13 |
| BALB/cJ | 8 | 34 ± 16 | 50 ± 7 | 122 ± 12 | 287 ± 36 |
| BPN/3 J | 8 | 10 ± 3 | 30 ± 3 | 105 ± 16 | 223 ± 32 |
| C3H/HeJ | 10 | 15 ± 1 | 40 ± 6 | 121 ± 9 | 233 ± 34 |
| P/J | 7 | 18 ± 3 | 42 ± 10 | 94 ± 15 | 145 ± 29 |
| SJL/J | 8 | 12 ± 6 | 30 ± 7 | 93 ± 13 | 123 ± 19 |
| C57BL/6 J | 8 | 10 ± 2 | 31 ± 5 | 65 ± 4 | 104 ± 16 |
Airway resistance was measured with flexiVent in response to methacholine challenge (1, 3, 10, and 30 mg/ml) and is expressed as % increases compared to airway resistance at saline inhalation challenge in 36 mouse strains
N number of animals per strain, NA not available, SEM standard error means
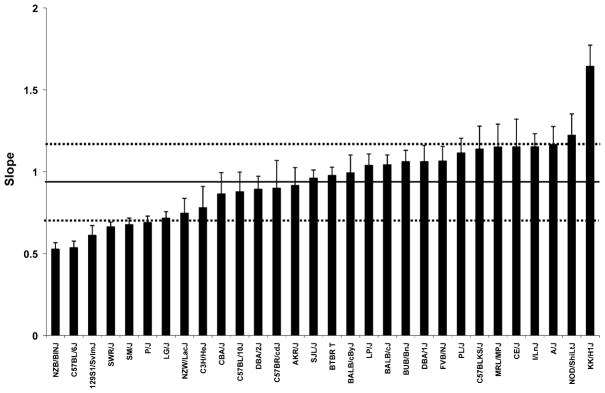
We summarized AHR as the slope of the curve of Raw versus concentration of log-transformed methacholine concentration administered. We found that considerable variation in the slope existed among the 36 mouse strains, with more than sixfold difference between the two strains with the most divergent responses, KK/HlJ and C57BL/6J (Fig. 1).
Fig. 1.
Variation in slope—a measure of airway hyper-responsiveness—among the 36 mouse strains. Bars represent strain mean ± SEM. Horizontal lines represent mean (solid line) across all 36 strains ± SD (dashed lines)
Haplotype association mapping for native AHR identified association peaks on chromosomes 3, 5, 8, 12, 13, and 14
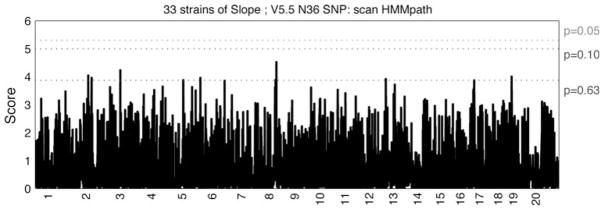
Haplotype association mapping, similar to genome-wide association studies in human populations, is a method of searching for phenotype–haplotype associations across the entire genome among several strains. In our study, we used the hidden Markov model (HMM) method to search for associations between haplotype blocks of variable SNP-window sizes and our phenotypic trait: slope. The percentiles of the minimum P value distribution, based on the 1,000 permutation tests, provided the approximate multiple test-adjusted thresholds for significant associations. For slope, the threshold score for significant associations was 4.91, while the threshold score for suggestive associations was 3.88. The genome-wide scan for slope identified 1 significant peak on Chr 14 at 115.7 Mb and 7 suggestive associations on chromosomes 3, 5, 8, 12, and 13 (Fig. 2). Only the peak on Chr 13 at 34.7 Mb was located in a QTL region found for AHR in the mouse (Table 3). The peaks on Chr 8 at 64.6 Mb, on Chr 12 at 110.3 Mb, and on Chr 13 at 34.7 Mb, also mapped to concordant regions of human asthma QTL. The association on Chr 3 at 47.7 Mb, on Chr 5 at 134.5 Mb, and on Chr 8 at 69 Mb were located less than 6, 12, and 2 Mb, respectively, from concordant regions of human asthma QTL (Table 3).
Fig. 2.
Genome-wide scan to measure the strength of association between the inferred haplotypes and the phenotype means for slope. The strength of the associations was determined by a regression-based test statistic, and its nominal P value was computed. All P values were transformed using −log10 (P value). We performed permutation testing and the minimum P values across all haplotype blocks provided the multiple test-adjusted thresholds (dotted lines). Position in the genome, divided by chromosome, is depicted along the x axis. 20 is used for the X chromosome
Table 3.
Associations between slope and haplotypes are located on mouse chromosome 3, 5, 8, 12, 13, and 14
| HAM peaks |
Mouse QTL (Mb) | Human QTLs (Location) | |
|---|---|---|---|
| Chr | Mb | ||
| 3 | 47.7 | 13q11 | |
| 5 | 48.7 | ||
| 134.5 | 13q11 | ||
| 8 | 64.6 | 4q35 | |
| 69.0 | 4q35 | ||
| 12 | 110.3 | 14q24 | |
| 13 | 34.7 | 32.9–72.9 | 6q24 |
| 14 | 115.7 | ||
HAM haplotype association mapping, Chr chromosome, Mb million base pairs
Candidate gene identification narrowed the QTL for AHR on Chr 13 to 11 genes
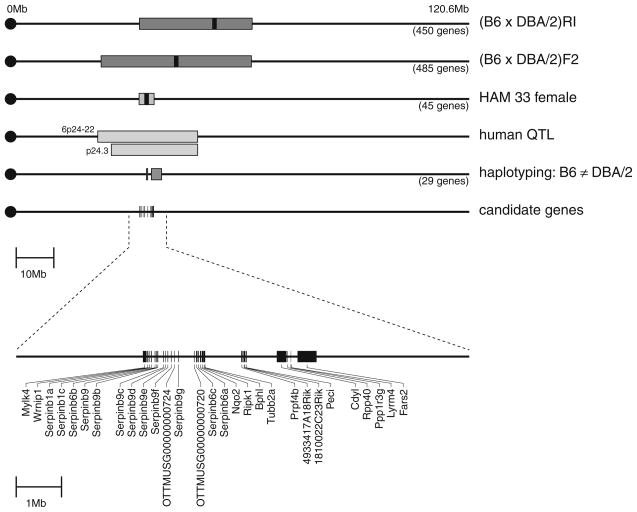
We used comparative genomics, haplotype association mapping, and haplotype analysis to narrow the region of interest on Chr 13 (Fig. 3). Previously, a QTL for AHR on Chr 13 had been found in recombinant inbred mice generated by crossing mice from the strains C57BL/6J and DBA/2J (Nicolaides et al. 1997). This QTL overlapped with a QTL for interleukin (IL-) 4 production in the lung that also had been found in an intercross between C57BL/6J and DBA/2J (Azuara and Pereira 2000). Our haplotype association peak was located in the overlapping region between these 2 previously found QTLs, which contains about 450 genes. With our haplotype association peak and a confidence interval of ± 2 Mb around the peak, we were able to narrow the region of interest from 450 to 45 genes. Twenty-nine of these 45 genes are located in polymorphic regions between C57BL/6J and DBA/2J (Fig. 3). To narrow the list of candidate genes even further, we compared expression levels between tissues of the strain C57BL/6J and gene sequences between the two cross strains, C57BL/6J and DBA/2J. Using databases we found that 6 of these 29 genes (Serpinb1a, Serpinb9c, Serpinb9e, Ripk1, Tubb2a, and an uncharacterized Riken gene [1810022C23Rik]) contain non-synonymous (Cn) SNPs, suggesting a likely change in function (Table 4). The Cn SNPs in the Serpin genes, Tubb2a, and 1810022C23Rik are located in regions of the proteins that are conserved among animals. In addition, in C57BL/6J the expression of six genes (Mylk4/EG23856, Serpinb1a, Serpinb6b, Nqo2, Cdyl, Fars2) in the lung is at a level of at least the median expression among other tissues (Table 4). Therefore, 11 of the 29 genes that are located in polymorphic regions between C57BL/6J and DBA/2J have limited evidence for being a QTL gene based either on an amino acid change or on the gene’s expression in the lung at a level of at least the median expression among other tissues.
Fig. 3.
Fine detailed map of mouse Chr 13 that explains how several sets of data were combined leading to “target regions” containing candidate genes of interest. The map shows the mouse chromosome with the previously found QTL and its 95% confidence interval, the QTL found with our haplotype association analysis, the concordant regions of human asthma QTLs, the regions that are not homologous between the parental strains of the QTL cross (C57BL/6J and DBA/2J), and the target genes
Table 4.
Genes located in polymorphic regions between C57BL/6J and DBA/2J at the association peak on chromosome 13
| Gene |
Non-synonymous SNP change |
Expression in lung of B6 | ||||
|---|---|---|---|---|---|---|
| Name | Start | End | Amino acid | Conserved region | Location | |
| Mylk4/EG238564 | 32,796 | 32,876 | Yes | |||
| Wrnip1 | 32,893 | 32,914 | ||||
| Serpinb1a | 32,933 | 32,943 | Q/L | Yes | Proteinase inhibitor domain | Yes |
| Serpinb1c | 32,973 | 32,990 | ||||
| Serpinb6b | 33,057 | 33,071 | Yes | |||
| Serpinb9 | 33,095 | 33,110 | ||||
| Serpinb9b | 33,119 | 33,134 | ||||
| Serpinb9c | 33,241 | 33,252 | P/S | Yes | Proteinase inhibitor domain | |
| Serpinb9d | 33,284 | 33,295 | ||||
| Serpinb9e | 33,341 | 33,353 | L/F | No | Proteinase inhibitor domain | |
| Serpinb9f | 33,415 | 33,427 | ||||
| OTTMUSG00000000724 | 33,487 | 33,497 | ||||
| Serpinb9g | 33,576 | 33,588 | ||||
| OTTMUSG00000000720 | 33,924 | 33,935 | ||||
| Serpinb6c | 33,971 | 33,997 | ||||
| Serpinb6a | 34,009 | 34,028 | ||||
| Nqo2 | 34,056 | 34,078 | Yes | |||
| Ripk1 | 34,094 | 34,126 | T/I | No | Proteinase inhibitor domain | No |
| Bphl | 34,129 | 34,166 | ||||
| Tubb2a | 34,166 | 34,170 | N/S | Yes | NA | |
| Prpf4b | 34,967 | 34,995 | ||||
| 4933417A18Rik | 35,016 | 35,045 | ||||
| 1810022C23Rik | 35,038 | 35,056 | No | |||
| Peci | 35,069 | 35,086 | ||||
| Cdyl | 35,751 | 35,965 | Yes | |||
| Rpp40 | 35,988 | 35,998 | ||||
| Ppp1r3 g | 36,059 | 36,062 | ||||
| Lyrm4 | 36,071 | 36,209 | ||||
| Fars2 | 36,209 | 36,629 | Yes | |||
AA amino acid, Cn non-synonymous SNP, SNP single nucleotide polymorphism, NA not available
Discussion
To identify underlying disease-causing variations of airway hyper-responsiveness (AHR) in the mouse, it is important to perform genetic studies (such as QTL studies) with a large variety of strains. Previously, Levitt and Mitzner (1989) reported a mouse strain survey of AHR in nine inbred strains. But only eight studies of QTL crosses for AHR have been published (Table 1), and these crosses included just three of the strains—A/J, C3H/HeJ, and C57BL/6J—from Levitt and Mitzner’s survey (Table 1). Most of these studies used A/J as the hyper-responsive parent and C3H/HeJ or C57BL/6J as the hypo-responsive parent (Table 1), probably because no other strains expressed widely deviating AHR phenotypes. Three of the eight QTL crosses were between the same two strains, A/J and C3H/HeJ, and for some of these crosses the marker information is no longer available. The limited number of suitable parental strains and the lack of genetic diversity and published data were serious deficiencies that precluded the use of bioinformatics and other valuable genetic tools for narrowing QTLs and identifying QTL genes related to asthma.
Our objective was to broaden the genetic diversity of strains that can be used in QTL studies for asthma, to narrow the QTLs, and to identify QTL genes. First, we quadrupled the genetic diversity by surveying 36 inbred strains (Fig. 1)—the 9 strains Levitt and Mitzner (1989) studied plus 27 additional strains that were previously untested. This new AHR data among so many genetically diverse inbred strains will allow us and others to generate novel QTL crosses, narrow QTLs at similar genetic regions using bioinformatics tools such as haplotyping and combined cross analysis (Peters et al. 2007).
We further demonstrated that our 36-mouse strain survey is a useful tool for verifying previously found QTLs for AHR and for identifying potentially novel QTL regions by haplotype association mapping. For other phenotypes, such as high-density lipoprotein (HDL) cholesterol, gallstone susceptibility and red blood cell count, Pletcher et al. (2004) used haplotype association mapping to successfully reproduce previously discovered QTL regions. This study also used haplotype association mapping to detect new QTLs for HDL and red blood cell count. Peak associations detected by haplotype association mapping for these traits were tested against known QTLs. New associations were then tested in additional crosses. When using the 3-SNP-window, 30% of the associations were false positives, but each false positive was in linkage disequilibrium with a true peak. When using the hidden Markov model (HMM), the false positives were eliminated (personnel communication Burgess-Herbert and Paigen).
Using genetic variations from multiple strains in haplotype association studies helps to identify smaller genomic regions, making identification of possible candidate genes more likely than when using only two strains in QTL cross studies, as we demonstrated by the narrowing of a Chr 13 QTL. We performed haplotype association mapping for AHR among 33 mouse strains using the HMM method, and we identified numerous novel QTL locations. Only the association on Chr 13 overlapped with a previously found QTL for AHR in the mouse (Table 3, Fig. 2). Our identification of new QTLs can be explained by two observations. First, the various studies used different methods for detecting AHR. We are unlikely to replicate QTLs that were detected with the method of whole-body plethysmography (i.e., Buxco) because this method is, in comparison to the flexiVent method, more susceptible to environmental factors (e.g., humidification and warming of the inspired air). Indeed, the lack of correlation between measures of these two methods has been published (Mitzner and Tankersley 1998; Sly et al. 2005). Also, although the method used to detect the QTL on Chr 13 in the previous study was similar to the method we used in our study, it was not identical. Second, the various studies used mice of different sexes. Whereas we used only female mice (which were also used in the study that found the QTL on Chr 13), other studies used primarily male mice (Table 1). Sex-specific differences in asthma in mice have been thoroughly discussed elsewhere (Chang and Mitzner 2007). Therefore, our results demonstrate that haplotype association mapping identifies new AHR QTL. Additional studies (e.g., QTL studies) using the same or similar methods to measure AHR will be necessary to verify the novel QTL locations as well as the QTL on Chr 13 found by our haplotype association mapping.
We further demonstrated that the combination of QTL and haplotype association studies in the mouse could accelerate the narrowing of the genomic candidate region on Chr 13 (Fig. 3). By applying comparative genomics and haplotyping, we were able to narrow a genomic region containing about 450 genes to a region containing only 29 genes. Then, with evidence on sequence and expression differences, we were able to further narrow the list of genes to 11 (Table 4). More evidence will be needed, however, to narrow this list even further. The evidence can be increased by, for example, advanced gene sequencing and expression experiments. Although dense sequence information for C57BL/6J and DBA/2J are available online, a more detailed sequence analysis will be necessary before we can argue that these SNP polymorphisms are disease-causing. Additionally, gene expression analysis must be compared to physiological data to ensure that the expression difference is involved in the regulation of AHR. Finally, one weakness of our current study was the lack of microarray gene expression results for strains other than C57BL/6J. Because C57BL/6J is a strain on the low end of the AHR scale, we might have potentially missed other relevant candidate genes. Therefore, a detailed microarray study of gene expression differences in the lung between multiple strains will be helpful to further increase the evidence for important candidate genes.
This study on airway responsiveness successfully demonstrated that our strain survey of 36 mouse strains could be utilized for verification of existing, and for identification of potentially new, QTL regions for AHR by haplotype association mapping. Now, we and others can use our strain survey to easily choose the most suitable parental strains for traditional mouse crosses in which to further verify these QTLs. Finally, by narrowing the QTL region on Chr 13, we identified potentially new candidate genes for AHR.
Acknowledgments
The authors would like to thank Jesse Hammer for assistance with the graphics, Joanne Currer for her help in preparing the manuscript, and Drs. Luanne Peters and Edward Leiter for their helpful comments on the manuscript. This work was supported by the grants HL 66611 and HL 83069 from the National Institutes of Health, U.S., and by the Cancer Core grant CA 34196 to the Jackson Laboratory.
Footnotes
Communicated by S. Hohmann.
Contributor Information
Adriana S. Leme, Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA 15261, USA
Annerose Berndt, Email: annerose.berndt@jax.org, The Jackson Laboratory, Bar Harbor, ME 04609, USA.
Laura K. Williams, Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA 15261, USA
Shirng-Wern Tsaih, The Jackson Laboratory, Bar Harbor, ME 04609, USA.
Jin P. Szatkiewicz, The Jackson Laboratory, Bar Harbor, ME 04609, USA
Ricardo Verdugo, The Jackson Laboratory, Bar Harbor, ME 04609, USA.
Beverly Paigen, The Jackson Laboratory, Bar Harbor, ME 04609, USA.
Steven D. Shapiro, Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA 15261, USA
References
- Ackerman KG, Huang H, Grasemann H, Puma C, Singer JB, Hill AE, Lander E, Nadeau JH, Churchill GA, Drazen JM, Beier DR. Interacting genetic loci cause airway hyperresponsiveness. Physiol Genomics. 2005;21:105–111. doi: 10.1152/physiolgenomics.00267.2004. [DOI] [PubMed] [Google Scholar]
- Azuara V, Pereira P. Genetic mapping of two murine loci that influence the development of IL-4-producing Thy-1dull gamma delta thymocytes. J Immunol. 2000;16:42–48. doi: 10.4049/jimmunol.165.1.42. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Cervino AC, Darvasi A, Fallahi M, Mader CC, Tsinoremas NF. An integrated in silico gene mapping strategy in inbred mice. Genetics. 2007;175:321–333. doi: 10.1534/genetics.106.065359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Mitzner W. Sex differences in mouse models of asthma. Can J Physiol Pharmacol. 2007;85:1226–1235. doi: 10.1139/Y07-116. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Rodriguez-Zas SL, Mogil JS. In silico mapping of mouse quantitative trait loci. Science. 2001;294:2423. doi: 10.1126/science.294.5551.2423a. [DOI] [PubMed] [Google Scholar]
- Colilla S, Nicolae D, Pluzhnikov A, Blumenthal MN, Beaty TH, Bleecker ER, Lange EM, Rich SS, Meyers DA, Ober C, Cox NJ. Evidence for gene-environment interactions in a linkage study of asthma and smoking exposure. J Allergy Clin Immunol. 2003;111:840–846. doi: 10.1067/mai.2003.170. [DOI] [PubMed] [Google Scholar]
- De Sanctis GT, Merchant M, Beier DR, Dredge RD, Grobholz JK, Martin TR, Lander ES, Drazen JM. Quantitative locus analysis of airway hyperresponsiveness in A/J and C57BL/6J mice. Nat Genet. 1995;11:150–154. doi: 10.1038/ng1095-150. [DOI] [PubMed] [Google Scholar]
- De Sanctis GT, Singer JB, Jiao A, Yandava CN, Lee YH, Haynes TC, Lander ES, Beier DR, Drazen JM. Quantitative trait locus mapping of airway responsiveness to chromosomes 6 and 7 in inbred mice. Am J Physiol. 1999;277:L1118–L1123. doi: 10.1152/ajplung.1999.277.6.L1118. [DOI] [PubMed] [Google Scholar]
- Drazen JM, Finn PW, De Sanctis GT. Mouse models of airway responsiveness: physiological basis of observed outcomes and analysis of selected examples using these outcome indicators. Annu Rev Physiol. 1999;61:593–625. doi: 10.1146/annurev.physiol.61.1.593. [DOI] [PubMed] [Google Scholar]
- Ewart SL, Mitzner W, DiSilvestre DA, Meyers DA, Levitt RC. Airway hyperresponsiveness to acetylcholine: segregation analysis and evidence for linkage to murine chromosome 6. Am J Respir Cell Mol Biol. 1996;14:487–495. doi: 10.1165/ajrcmb.14.5.8624254. [DOI] [PubMed] [Google Scholar]
- Ewart SL, Kuperman D, Schadt E, Tankersley C, Grupe A, Shubitowski DM, Peltz G, Wills-Karp M. Quantitative trait loci controlling allergen-induced airway hyperresponsiveness in inbred mice. Am J Respir Cell Mol Biol. 2000;23:537–545. doi: 10.1165/ajrcmb.23.4.4199. [DOI] [PubMed] [Google Scholar]
- Grupe A, Germer S, Usuka J, Aud D, Belknap JK, Klein RF, Ahluwalia MK, Higuchi R, Peltz G. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–1918. doi: 10.1126/science.1058889. [DOI] [PubMed] [Google Scholar]
- Hoffjan S, Nicolae D, Ostrovnaya I, Roberg K, Evans M, Mirel DB, Steiner L, Walker K, Shult P, Gangnon RE, Gern JE, Martinez FD, Lemanske RF, Ober C. Gene-environment interaction effects on the development of immune responses in the 1st year of life. Am J Hum Genet. 2005;76:696–704. doi: 10.1086/429418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt RC, Mitzner W. Autosomal recessive inheritance of airway hyperreactivity to 5-hydroxytryptamine. J Appl Physiol. 1989;67:1125–1132. doi: 10.1152/jappl.1989.67.3.1125. [DOI] [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClurg P, Pletcher MT, Wiltshire T, Su AI. Comparative analysis of haplotype association mapping algorithms. BMC Bioinformatics. 2006;7:61. doi: 10.1186/1471-2105-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire JJ, Umetsu SE, Akbari O, Potter M, Kuchroo VK, Barsh GS, Freeman GJ, Umetsu DT, DeKruyff RH. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol. 2001;2:1109–1116. doi: 10.1038/ni739. [DOI] [PubMed] [Google Scholar]
- Mitzner W, Tankersley C. Noninvasive measurement of airway responsiveness in allergic mice using barometric plethysmography. Am J Respir Crit Care Med. 1998;158:340–341. doi: 10.1164/ajrccm.158.1.let1. [DOI] [PubMed] [Google Scholar]
- Moorman JE, Rudd RA, Johnson CA, King M, Minor P, Bailey C, Scalia MR, Akinbami LJ. National surveillance for asthma—United States, 1980–2004. MMWR Surveill Summ. 2007;56:1–54. [PubMed] [Google Scholar]
- Nicolaides NC, Holroyd KJ, Ewart SL, Eleff SM, Kiser MB, Dragwa CR, Sullivan CD, Grasso L, Zhang LY, Messler CJ, Zhou T, Kleeberger SR, Buetow KH, Levitt RC. Interleukin 9: a candidate gene for asthma. Proc Natl Acad Sci USA. 1997;94:13175–13180. doi: 10.1073/pnas.94.24.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LJ, Cookson WO. Genomic approaches to understanding asthma. Genome Res. 2000;10:1280–1287. doi: 10.1101/gr.143400. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Place M. Prospects for association mapping in classical inbred mouse strains. Genetics. 2007;175:1999–2008. doi: 10.1534/genetics.106.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, Mural RJ, Paigen B, Wiltshire T. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards IM. Mouse models of allergic disease; how do they relate to asthma in man? Clin Exp Allergy. 1996;26:618–620. [PubMed] [Google Scholar]
- Sly PD, Turner DJ, Collins RA, Hantos Z. Penh is not a validated technique for measuring airway function in mice. Am J Respir Crit Care Med. 2005;172:256. doi: 10.1164/ajrccm.172.2.954. [DOI] [PubMed] [Google Scholar]
- Stylianou IM, Paigen B, Singh J, Schwartz DA. Comparative genomics of asthma. In: Postma DS, editor. Lung biology in health and disease. S.T.W., Informa Healthcare; New York/London: 2007. pp. 159–177. [Google Scholar]
- Svenson KL, Von Smith R, Magnani PA, Suetin HR, Paigen B, Naggert JK, Li R, Churchill GA, Peters LL. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol. 2007;102:2369–2378. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- Szatkiewicz JP, Beane G, Ding Y, Hutchins L, Pardo-Manuel de Villena F, Churchill GA. An imputed genotype resource for the laboratory mouse. Mamm Genome. 2007;19:199–208. doi: 10.1007/s00335-008-9098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdugo RA, Medrano JF. Comparison of gene coverage of mouse oligonucleotide microarray platforms. BMC Genomics. 2006;7:58. doi: 10.1186/1471-2164-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based multiple testing: examples and methods for p-value adjustment. Wiley; New York: 1993. p. 340. [Google Scholar]
- Zhang Y, Lefort J, Kearsey V, Lapa e Silva JR, Cookson WO, Vargaftig BB. A genome-wide screen for asthma-associated quantitative trait loci in a mouse model of allergic asthma. Hum Mol Genet. 1999;8:601–605. doi: 10.1093/hmg/8.4.601. [DOI] [PubMed] [Google Scholar]