Abstract
Macrophages are aptly positioned to function as the primary line of defence against invading pathogens in many organs, including the lung and peritoneum. Their ability to phagocytose and clear microorganisms has been well documented1,2. Macrophages possess several substances with which they can kill bacteria, including reactive oxygen species, nitric oxide, and antimicrobial proteins3–9. We proposed that macrophage-derived proteinases may contribute to the antimicrobial properties of macrophages. Macrophage elastase (also known as matrix metalloproteinase 12 or MMP12) is an enzyme predominantly expressed in mature tissue macrophages10 and is implicated in several disease processes, including emphysema11. Physiological functions for MMP12 have not been described. Here we show that Mmp12−/− mice exhibit impaired bacterial clearance and increased mortality when challenged with both Gram-negative and Gram-positive bacteria at macrophage-rich portals of entry, such as the peritoneum and lung. Intracellular stores of MMP12 are mobilized to macrophage phagolysosomes after the ingestion of bacterial pathogens. Once inside phagolysosomes, MMP12 adheres to bacterial cell walls where it disrupts cellular membranes resulting in bacterial death. The antimicrobial properties of MMP12 do not reside within its catalytic domain, but rather within the carboxy-terminal domain. This domain contains a unique four amino acid sequence on an exposed β loop of the protein that is required for the observed antimicrobial activity. The present study represents, to our knowledge, the first report of direct antimicrobial activity by a matrix metallopeptidase, and describes a new antimicrobial peptide that is sequentially and structurally unique in nature.
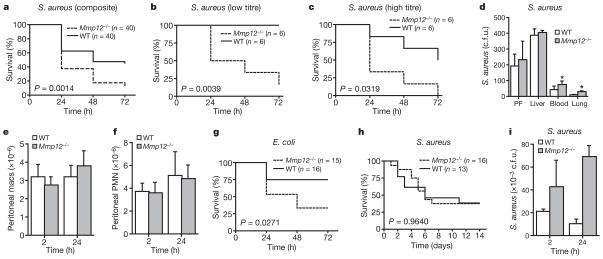
To determine whether MMP12 has a role in host defence against bacteria, we subjected Mmp12−/− mice and wild-type littermates to intraperitoneal (i.p.) injection of either Staphylococcus aureus or Escherichia coli. In each case, there was a significant survival advantage for wild-type mice as compared to Mmp12−/− mice over a 3-day period (Fig. 1a–c, g). Lower titres of i.p. S. aureus injection (1 ×107 colony-forming units (c.f.u.)), which did not result in mortality in wild-type mice, still proved >50% lethal in Mmp12−/− mice. Soon after i.p. injection of S. aureus, the local bacterial burden in the peritoneal fluid and liver was similar in both groups of mice. However, wild-type mice were significantly protected from bacterial dissemination to distal sites, such as the blood and lung (Fig. 1d). None of these observations was attributable to differences in inflammatory cell recruitment, because the macrophage and neutrophil counts were equivalent between the two groups at 2 and 24 h after i.p. S. aureus injection (Fig. 1e, f).
Figure 1. MMP12 provides survival advantage in Gram-positive and Gram-negative infection.
a, Kaplan–Meier curve for Mmp12−/− and wild-type (WT) mice after i.p. injection with 1 ×108 c.f.u. S. aureus (n = 40 for each group from six experiments); P = 0.0014, log-rank test. b, c, Kaplan–Meier curves for low (b; 1 ×107 c.f.u.) and high (c; 1 ×109 c.f.u.) S. aureus titres (n = 6 each group); P <0.05. d, Bacterial c.f.u. 24 h after i.p. S. aureus injection in peritoneal fluid (PF), blood, liver and lungs (n = 4 each group); *P <0.01. e, f, Peritoneal macrophage (macs; e) and neutrophil (f) counts 2 and 24 h after i.p. S. aureus injection (n = 6 each group). PMN, polymorphonuclear leukocytes; P >0.05. g, Kaplan–Meier curve for Mmp12−/− (n = 15) and wild-type (n = 16) mice (from three experiments) after 1 ×108 c.f.u. i.p. E. coli injection; P = 0.0271. h, Kaplan–Meier curve for intravenous S. aureus (1 ×108 c.f.u.) to wild-type (n = 13) and Mmp12−/− (n = 16) mice (from three experiments); P = 0.9640. i, Lung bacterial burden (c.f.u.) 2 h after S. aureus TVI (n = 6 each group); *P <0.005. All error bars represent s.d.
Bacteremia generated by tail vein injection (TVI) of 1 ×108 c.f.u. S. aureus in Mmp12−/− and wild-type control mice produced equivalent 2-week mortality in both genotypes (Fig. 1h). However, the bacterial burden in the lungs of Mmp12−/− mice was significantly greater than in wild-type counterparts at both 2 and 24 h after TVI (Fig. 1i). The bacterial burden was equivalent in kidney and spleen homogenates between the two groups at the same time points (data not shown). Thus, it seems that MMP12 exerts its antimicrobial properties solely in organs, such as the lung, that are fortified with macrophages.
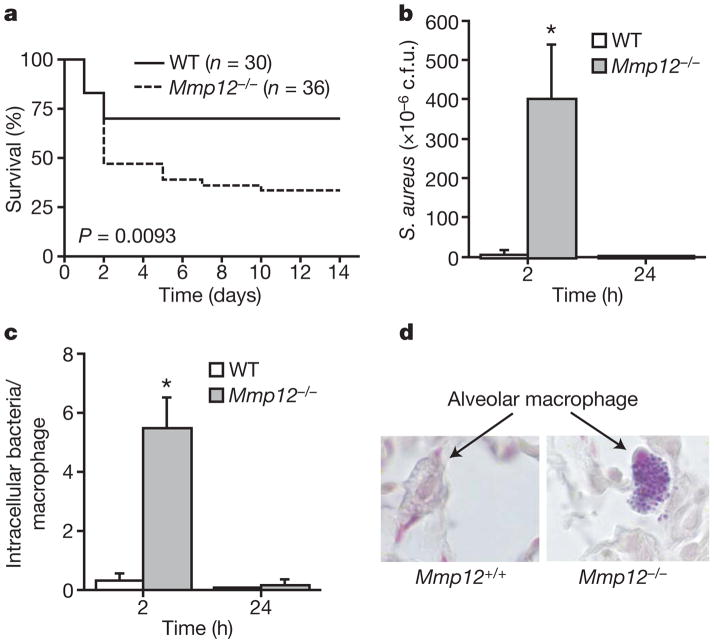
To validate this hypothesis, we subjected Mmp12−/− mice and wild-type littermates to an in vivo pneumonia model in which mice received the approximate lethal dose to 50% of animals tested (LD50 dose; 1 ×108 c.f.u.) of S. aureus intratracheally (i.t.). As with peritonitis, wild-type mice showed a decided survival advantage over Mmp12−/− mice (Fig. 2a). Repeating the experiment using a sub-lethal titre of S. aureus (1 ×106 c.f.u.) resulted in ~100-fold greater increased bacterial burden in Mmp12−/− lungs 2 h after infection (Fig. 2b). The bacteria were eliminated by 24 h in both groups, which correlated with an equivalent increase in lung neutrophils in both groups. Surprisingly, the bacterial burden in Mmp12−/− mice was predominantly located within macrophages (Fig. 2c, d).
Figure 2. MMP12 improves bacterial clearance and survival in S. aureus pneumonia.
a, Kaplan–Meier curve for Mmp12+/+(n = 30; wild-type, WT) and Mmp12−/− (n = 36) mice (from six experiments) administered with S. aureus i.t. (1 ×108 c.f.u.); P = 0.0093, log-rank test. b, Bacterial burden in the lung after sub-lethal dose of S. aureus i.t. (1 ×106 c.f.u.; n = 8 each group); *P<0.01. c, Intramacrophage bacterial particle counts on Gram stain (n =8 each group); *P <0.001. d, Mmp12+/+ and Mmp12−/− alveolar macrophages on Gram stain at the 2-h time point. Original magnification, ×100. All error bars represent s.d.
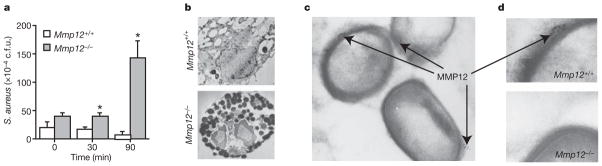
MMP12, like other MMPs, is transcriptionally regulated12. The pro-enzyme is secreted and subsequently activated in the extracellular space where MMPs are thought to perform their biological functions13. Yet, preformed intracellular pools of MMP12 exist within quiescent macrophages14, presumably for rapid secretion from the cell. To determine whether these intracellular stores of MMP12 are responsible for antimicrobial activity within macrophages, we performed macrophage intracellular bacterial killing assays. Wild-type and Mmp12−/− macrophages were co-incubated with S. aureus for 60 min in antibiotic-free medium, at which time (t =0) the medium was changed and the cells were washed with PBS to remove non-phagocytosed bacteria. The bacterial content was equal at this time point, demonstrating equal phagocytic capacity in both genotypes (Fig. 3a). Macrophages were collected after further 30- and 90-min incubations, and intracellular bacterial burden was quantified. Wild-type macrophages efficiently cleared bacteria within 90 min. In contrast, bacterial content increased over time in Mmp12−/− macrophages. Overall, the bacterial burden was tenfold greater in Mmp12−/− macrophages than in wild-type macrophages. Scanning electron microscopy of wild-type macrophages demonstrated very few intracellular bacteria after 60 min (Fig. 3b). In contrast, Mmp12−/− macrophages phagocytosed the bacteria but were unable to kill them, resulting in the presence of several intracellular bacteria even after 60 min (Fig. 3b). Immuno-gold labelling and transmission electron microscopy demonstrated the presence of MMP12 in clusters on disrupted areas of bacterial cell walls within phagolysosomes of wild-type but not Mmp12−/− macrophages (Fig. 3c, d).
Figure 3. MMP12 is required for early intracellular bacterial killing by macrophages.
Peritoneal macrophages were incubated with S. aureus in RPMI plus 10% FCS without antibiotics for 60 min (t = 0). a, Intracellular bacterial c.f.u. from macrophage lysates. Data are from an experiment in triplicate. Error bars represent s.d., *P <0.05. b, Scanning electron microscopy (original magnification, ×4,400) of macrophages (from three experiments) at t = 60 min. c, Staphylococcus aureus in phagolysosomes of wild-type macrophages. Immuno-electron-microscopy (original magnification, ×30,000) shows MMP12 (gold particles) on S. aureus cocci (c, d) within wild-type macrophages. Gold particles were not detected within Mmp12−/− macrophages (d).
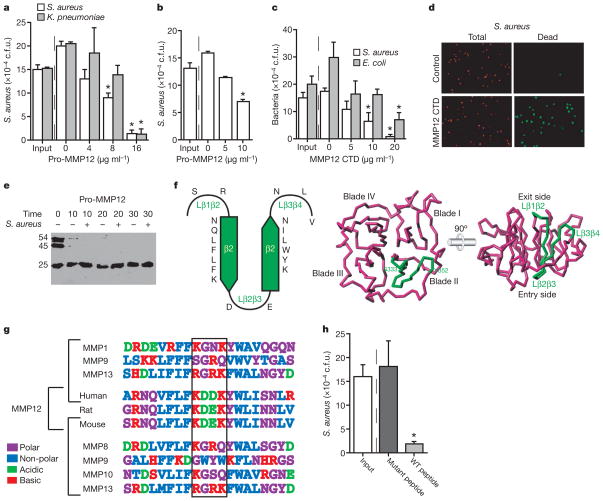
To determine whether MMP12 possesses direct antimicrobial activity, as suggested by the transmission electron microscopy studies, we incubated full-length recombinant human MMP12 with S. aureus and Klebsiella pneumoniae in tryptic soy broth (TSB) buffer (Fig. 4a). Staphylococcus aureus killing assays were repeated in RPMI with 10% FCS (Fig. 4b) in an attempt to produce a physiological environment. In both conditions we observed a dose-dependent inhibition of S. aureus growth by human MMP12. We suspect that the concentration of MMP12 within intracellular granules is much greater than the effective concentrations in these assays, although the exact concentrations are unknown. Bacterial content (c.f.u.) after incubation with >5 μg ml−1 MMP12 was less than the input amount, suggesting bactericidal activity. None of the other MMPs tested, including MMP3, 7, 8, 9 and 14, showed direct antimicrobial activity at concentrations above 100 μg ml−1 (data not shown). The antimicrobial properties of MMP12 were not dependent on catalytic activity, as determined by continued activity with the co-administration of a hydroxamic acid MMP inhibitor and the lack of activity of the catalytic domain alone. The catalytic domain may contribute to host defence by processing/degrading other proteinases or bacterial toxins. However, MMP12 does not degrade S. aureus alpha-toxin (data not shown).
Figure 4. MMP12 CTD possesses bactericidal activity.
a–c, Bacteria were incubated with human pro-MMP12 in 5% TSB (a) or RPMI plus 10% FCS (b), or with mouse MMP12 CTD (c) for 60 min at 37 °C. Data are expressed as c.f.u. (from three experiments); *P <0.005. d, Fluorescent propidium iodide exclusion assay after co-incubation of S. aureus and MMP12 CTD or control for 30 min. All bacteria stain positive for Syto 59 (red), but only those with disrupted membranes stain with S-7020 (green). e, Human pro-MMP12 incubated with and without S. aureus to study CTD processing in the presence of bacteria using a CTD antibody. The 54 kDa band represents full-length MMP12, the 45 kDa band represents shedding of pro-domain, and the 25 kDa band represents the CTD. The CTD is not processed to smaller fragments. f, Computational three-dimensional model of mouse MMP12 CTD. The SR-20 sequence is located within CTD blade II including β strands β2 and β3, as well as the connecting and flanking loops (green trace). g, SR-20 shows a high degree of homology among MMP12 orthologues but not among other MMPs. h, Staphylococcus aureus was incubated with either wild-type (WT) MMP12 SR-20 peptide or mutant peptide (Lys-Asp-Glu-Lys replaced by Ser-Gly-Arg-Gln) (both at 20 μg ml−1), in RPMI plus 10% FCS for 60 min. Data are expressed as c.f.u., *P <0.001. All error bars represent s.d.
Full-length MMP12 is a 54 kDa pro-enzyme consisting of three common MMP domains: an amino-terminal pro-domain, a zinc- and calcium-binding catalytic domain, and a haemopexin-like carboxy-terminal domain (CTD)10. CTD functions include enhancement of TIMP binding and improved catalysis of both collagen and chemokines15–18. The CTD is not required for MMP12-mediated substrate catalysis, and is commonly shed during MMP12 activation in vitro19.
To isolate region(s) of antimicrobial activity, we generated recombinant mouse MMP12 CTD (residues 280–473) in E. coli. Co-incubation of MMP12 CTD (but not MMP9 CTD) with either S. aureus or E. coli demonstrated a dose-dependent inhibition of bacterial growth equivalent to full-length MMP12 (Fig. 4c). In addition to S. aureus, K. pneumoniae and E. coli, MMP12 also kills Salmonella enteriditis. Notably, MMP12 is not active against Listeria monocytogenes, an organism that is able to survive within macrophages (data not shown).
To expand on the immuno-electron-microscopy results, which suggest that MMP12 is directly bactericidal by disrupting bacterial outer membrane integrity, we performed a propidium iodide exclusion assay. Mouse MMP12 CTD killed 98% of S. aureus in this assay (Fig. 4d). We also generated S. aureus cell-walls loaded with fluorescent dextran and performed a liposomal release assay demonstrating that MMP12 CTD can disrupt liposomal membranes (data not shown).
To more precisely locate the region of antimicrobial activity in mouse MMP12 CTD, we generated three recombinant fragments corresponding to the initial (Ser 280–Ile 342), middle (Glu 343—Glu 404), and last (Trp 405–Cys 473) third of the CTD. Only the middle fragment showed antimicrobial activity. Synthetic peptides were generated from this region of mouse MMP12 that narrowed the antimicrobial properties of the MMP12 CTD to a 20 amino acid sequence, designated SR-20 (344- SRNQLFLFKDEKYWLINNLV-363). SR-20 proved bactericidal against S. aureus in a propidium iodide exclusion assay, whereas the comparable region in mouse MMP13 (343-SRDLMFIFRGRKFWALNGYD-362) did not (data not shown). Bacterial killing assays also demonstrated that MMP12 CTD killed bacteria without further processing to smaller fragments, suggesting that SR-20 preferentially killed bacteria within the scaffold of the entire CTD (Fig. 4e).
We generated a three-dimensional homology model of mouse MMP12 CTD (Fig. 4f). The SR-20 antimicrobial peptide resides within blade II of the CTD, which is an all β structure featuring a fourfold β propeller in all experimental structures reported. Interactions with either the catalytic domain or the pro-domain are not predicted. The only exposed portions of SR-20 that could theoretically interact with bacteria are the surface accessible loops that flank and connect the two central β strands (Lβ2β3 on the entry side, and Lβ1β2 and Lβ3β4 on the exit side of the CTD disk; Fig. 4f). Lβ2β3 is of particular interest given its unique sequence of acidic amino acids flanked by basic residues. Although this amino acid sequence is homologous in rabbit, rat, mouse and human MMP12, it is not found in any other MMP (Fig. 4g). The sequence Lys-Asp-Glu-Lys is commonly found in various proteins, but a search of three-dimensional structures that contain this motif protruding from the surface of a β loop confirmed that it is sequentially and structurally unique to MMP12.
To prove that this motif is responsible for the observed antimicrobial properties of MMP12, we constructed a mutant peptide identical to SR-20 except that the Lys-Asp-Glu-Lys motif found in mouse MMP12 was replaced by the human MMP9 sequence, Ser-Gly-Arg-Gln. Incubation of S. aureus with wild-type and mutant peptide confirmed that the Lys-Asp-Glu-Lys motif is essential for the antimicrobial properties of mouse MMP12 CTD (Fig. 4h). Shorter four-amino-acid peptides (Ser-Gly-Arg-Gln, Lys-Asp-Asp-Lys and Lys-Asp-Glu-Lys) did not show antimicrobial activity (data not shown), suggesting that the loop structure of the protein is required for bacterial killing.
Previously, we, and others, have shown that neutrophil elastase is able to kill bacteria within neutrophil phagolysosomes20–22. Other serine-proteinases, including cathepsin G, proteinase 3, and the inactive azurocidin have been shown to possess antimicrobial activity, independent of catalytic activity23. However, sequence analysis of these serine-proteinases did not reveal the presence of the Lys-Asp-Glu-Lys motif (data not shown). Matrilysin (also known as MMP7), the only other MMP known to participate in bacterial killing, does so indirectly by cleaving and activating alpha-defensins within Paneth cells before their secretion into the gut24.
MMPs participate in physiological functions, including embryonic growth and development, generation and termination of the cytokine/chemokine gradients, and regulation of cell growth, differentiation and angiogenesis by cleavage of non-matrix proteins17,18,25,26. When aberrantly or excessively expressed, MMPs cause tissue destruction leading to emphysema, vascular disease and arthritis11,27,28. Although virtually all MMP functions had been thought to occur extracellularly or on the cell surface, MMPs have recently been implicated in intra-cellular functions including apoptosis and cell cycle regulation29,30. This study extends this concept to include intracellular (within the phagolysosome) antimicrobial activity.
The present study supports the participation of the macrophage in the earliest stages of the host’s defence against microorganisms, and adds to our understanding of MMP biology by demonstrating that MMP12 possesses direct antimicrobial activity. After engulfment of microorganisms by macrophages, intracellular stores of MMP12 are mobilized to phagolysosomes, where MMP12 uses a new antimicrobial peptide within the CTD to disrupt the integrity of cell wall structures in the invading microorganisms. The continued discovery of new antimicrobial mechanisms may prove essential to combat the increasing incidence of resistance to antibiotics now in use.
METHODS SUMMARY
Mmp12−/− and wild-type mice were maintained on a 129/SvJ background and housed in a sterile barrier facility. In vivo infection models were accomplished by i.p. (peritonitis), intratracheal (pneumonia) or i.v. (bacteremia) administration of bacteria. Staphylococcus aureus, S. enteritidis and K. pneumoniae (clinical isolates) and E. coli (K1 strain) were grown in TSB and isolated on Luria-Bertani broth agar (LB) plates. Organ c.f.u. were determined by plating limiting dilutions of organ homogenates on LB plates. Inflammatory cell counts were determined using a haemocytometer and Wright-stained cytospins. Macrophage killing assays were performed using thioglycollate-induced peritoneal macrophages. After 60 min incubation with bacteria, macrophages were washed with PBS and incubated for a further 30 or 90 min. Colony-forming units were determined from macrophage lysates. Recombinant protein constructs were generated in pet 20b vectors and grown in E. coli. All synthetic peptides were generated by the University of Pittsburgh protein core facility. Protein and peptide killing assays were performed in either TSB or RPMI plus 10% FCS at 37 °C for 60 min. Propidium iodide exclusion assay was performed using Syto 59 (red) to stain total bacteria and S-7020 (green) to stain bacteria with disrupted membranes (both from Molecular Probes). A three-dimensional homology model of mouse MMP12 CTD was constructed on the basis of crystal structures of related full-length MMPs. The programs CPHmodels 2.0 (www.cbs.dtu.dk/services/CPHmodels) and Swiss Model (swissmodel.expasy.org) were used for construction.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH) National Heart, Lung and Blood Institute (NHLBI) (S.D.S.) and Spanish and European public agencies (F.X.G.-R.).
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints.
Author Contributions A.M.H. performed in vivo and in vitro studies, contributed to data interpretation and mechanistic advance, prepared the manuscript and figures, and performed all revisions. W.O.H. performed in vivo and in vitro studies and contributed to study design, data analysis, and mechanistic advance. C.S.R. performed CTD processing studies. F.X.G.-R. constructed the three-dimensional homology model of MMP12 CTD. S.D.S. generated Mmp12−/− mice and all recombinant proteins, was responsible for study design, data interpretation, and mechanistic advance, and assisted with manuscript preparation.
References
- 1.Green GM, Kass EH. The role of the alveolar macrophage in the clearance of bacteria from the lung. J Exp Med. 1964;119:167–176. doi: 10.1084/jem.119.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon S. The macrophage: Past, present and future. Eur J Immunol. 2007;37:S9–S17. doi: 10.1002/eji.200737638. [DOI] [PubMed] [Google Scholar]
- 3.Shiloh MU, et al. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity. 1999;10:29–38. doi: 10.1016/s1074-7613(00)80004-7. [DOI] [PubMed] [Google Scholar]
- 4.Ganz T. Oxygen-independent microbicidal mechanisms of phagocytes. Proc Am Assoc Physicians. 1999;111:390–395. doi: 10.1111/paa.1999.111.5.390. [DOI] [PubMed] [Google Scholar]
- 5.Selsted ME, Ouellette AJ. Defensins in granules of phagocytic and non-phagocytic cells. Trends Cell Biol. 1995;5:114–119. doi: 10.1016/s0962-8924(00)88961-8. [DOI] [PubMed] [Google Scholar]
- 6.Biggar WD, Sturgess JM. Role of lysozyme in the microbicidal activity of rat alveolar macrophages. Infect Immun. 1977;16:974–982. doi: 10.1128/iai.16.3.974-982.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.del Cerro-Vadillo E, et al. Cutting edge: a novel nonoxidative phagosomal mechanism exerted by cathepsin-D controls Listeria monocytogenes intracellular growth. J Immunol. 2006;176:1321–1325. doi: 10.4049/jimmunol.176.3.1321. [DOI] [PubMed] [Google Scholar]
- 8.Hiemstra PS, van den Barselaar MT, Roest M, Nibbering PH, van Furth R. Ubiquicidin, a novel murine microbicidal protein present in the cytosolic fraction of macrophages. J Leukoc Biol. 1999;66:423–428. doi: 10.1002/jlb.66.3.423. [DOI] [PubMed] [Google Scholar]
- 9.Hiemstra PS, et al. Antimicrobial proteins of murine macrophages. Infect Immun. 1993;61:3038–3046. doi: 10.1128/iai.61.7.3038-3046.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shipley JM, Wesselschmidt RL, Kobayashi DK, Ley TJ, Shapiro SD. Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc Natl Acad Sci USA. 1996;93:3942–3946. doi: 10.1073/pnas.93.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Macrophage elastase is required for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 12.Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nature Rev Mol Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 13.Raza SL, Nehring LC, Shapiro SD, Cornelius LA. Proteinase-activated receptor-1 regulation of macrophage elastase (MMP-12) secretion by serine proteinases. J Biol Chem. 2000;275:41243–41250. doi: 10.1074/jbc.M005788200. [DOI] [PubMed] [Google Scholar]
- 14.Lijnen HR, et al. Temporal and topographic matrix metalloproteinase expression after vascular injury in mice. Thromb Haemost. 1999;81:799–807. [PubMed] [Google Scholar]
- 15.Knauper V, et al. The role of the C-terminal domain of human collagenase-3 (MMP-13) in the activation of procollagenase-3, substrate specificity, and tissue inhibitor of metalloproteinase interaction. J Biol Chem. 1997;272:7608–7616. doi: 10.1074/jbc.272.12.7608. [DOI] [PubMed] [Google Scholar]
- 16.Tam EM, Moore TR, Butler GS, Overall CM. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): the differential roles of the MMP hemopexin c domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J Biol Chem. 2004;279:43336–43344. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]
- 17.McQuibban GA, et al. Matrix metalloproteinase activity inactivates the CXC chemokine stromal cell-derived factor-1. J Biol Chem. 2001;276:43503–43508. doi: 10.1074/jbc.M107736200. [DOI] [PubMed] [Google Scholar]
- 18.McQuibban GA, et al. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science. 2000;289:1202–1206. doi: 10.1126/science.289.5482.1202. [DOI] [PubMed] [Google Scholar]
- 19.Gronski TJ, Jr, et al. Hydrolysis of a broad spectrum of extracellular matrix proteins by human macrophage elastase. J Biol Chem. 1997;272:12189–12194. doi: 10.1074/jbc.272.18.12189. [DOI] [PubMed] [Google Scholar]
- 20.Belaaouaj A, et al. Mice lacking neutrophil elastase reveal impaired host defense against gram-negative bacterial sepsis. Nature Med. 1998;4:615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 21.Belaaouaj A, Kim KS, Shapiro SD. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science. 2000;289:1185–1188. doi: 10.1126/science.289.5482.1185. [DOI] [PubMed] [Google Scholar]
- 22.Weinrauch Y, Drujan D, Shapiro S, Weiss J, Zychlinsky A. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 2002;417:91–94. doi: 10.1038/417091a. [DOI] [PubMed] [Google Scholar]
- 23.Lehrer RI, Ganz T. Antimicrobial polypeptides of human neutrophils. Blood. 1990;76:2169–2181. [PubMed] [Google Scholar]
- 24.Wilson CL, et al. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 25.Vu TH, et al. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 1998;93:411–422. doi: 10.1016/s0092-8674(00)81169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Park P, Wilson C, Parks W. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell. 2002;111:635–646. doi: 10.1016/s0092-8674(02)01079-6. [DOI] [PubMed] [Google Scholar]
- 27.Curci JA, Liao S, Huffman MD, Shapiro SD, Thompson RW. Expression and localization of macrophage elastase (matrix metalloproteinase-12) in abdominal aortic aneurysms. J Clin Invest. 1998;102:1900–1910. doi: 10.1172/JCI2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heymans S, et al. Inhibition of plasminogen activators or matrix metalloproteinases prevents cardiac rupture by impairs therapeutic angiogenesis and causes cardiac failure. Nature Med. 1999;5:1135–1142. doi: 10.1038/13459. [DOI] [PubMed] [Google Scholar]
- 29.Golubkov VS, et al. Membrane type-1 matrix metalloproteinase (MT1-MMP-exhibits an important intracellular cleavage function and causes chromosome instability. J Biol Chem. 2005;280:25079–25086. doi: 10.1074/jbc.M502779200. [DOI] [PubMed] [Google Scholar]
- 30.Limb GA, et al. Matrix metalloprteinase-1 associates with intracellular organelles and confers resistance to laminin a/c degradation during apoptosis. Am J Pathol. 2005;166:1555–1563. doi: 10.1016/S0002-9440(10)62371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.