Abstract
Background
Sensitization to house dust mite allergens is strongly correlated with asthma. Der p 7 elicits strong IgE antibody and T-cell responses in mite allergic patients. However, the structure and biological function of this important allergen are unknown. Allergen function may contribute to allergenicity as shown for the protease activity of Group 1 mite allergens and the interaction with the innate immune system by Group 2 mite allergens.
Objective
To determine the crystal structure of Der p 7 and to investigate its biological function.
Methods
X-ray crystallography was utilized to determine the Der p 7 structure. NMR analysis and biochemical assays were used to examine the binding of Der p 7 to predicted ligands.
Results
Der p 7 has an elongated structure with two 4-stranded anti-parallel β-sheets which wrap around a long C-terminal helix. The fold of Der p 7 is similar to lipopolysaccharide binding protein (LBP), which interacts with Toll-like receptors (TLRs) after binding lipopolysaccharide and other bacterially-derived lipid ligands. NMR and biochemical assays indicate that Der p 7 does not bind lipopolysaccharide but binds with weak affinity to the bacterial lipopeptide polymyxin B in the predicted binding site of Der p 7.
Conclusions
Der p 7 binds a bacterially-derived lipid product, a common feature of some allergens. The finding that the Group 7 as well as the Group 2 mite allergens are structurally similar to different proteins in the TLR pathway further strengthens the connections between dust mites, innate immunity, and allergy.
Keywords: Asthma, allergens, dust mites, Der p 7, lipopolysaccharide binding protein, TLR4, lipopeptide, innate immunity
Key Messages.
The structure of the dust mite allergen Der p 7 reveals a distant homology to a family of proteins involved in the human innate immune recognition of bacterial lipid products.
Der p 7 binds specifically to a bacterially derived lipopeptide.
Der p 7 is the second dust mite allergen identified to be structurally related to a protein in the TLR pathway.
Introduction
The development of asthma associated with allergens is known as extrinsic asthma, which differentiates it from intrinsic asthma of unknown etiology. Primarily, extrinsic asthma is associated with exposure to indoor allergens and occasionally with exposure to outdoor allergens.1 Among indoor allergens, the most commonly associated with asthma are those of the house dust mite.2 Indeed, greater than 80% of asthma patients show immediate hypersensitivity to allergens of the house dust mite.3 A significant question remains as to why dust mite allergens are so strongly associated with the etiology of asthma. Early studies to address this question focused on the discovery that many dust mite allergens are proteolytic enzymes.4 Experiments showed that these proteases can damage lung epithelia, which may help explain the chronic inflammation of the lung that characterizes asthma symptoms.5, 6 Other studies have shown that digestion of specific proteins could encourage IgE synthesis and/or skew the immune response towards a Th2 or symptomatic response instead of tolerance.7, 8
An important dust mite allergen for which there is not a known function from sequence analysis is Der p 7. The mature protein is 198 amino acids (MW= 22.2 kDa) and is estimated to be found in dust at concentrations of 2.5 μg per gram of dust.9 Natural Der p 7 has been described as a mixture of three molecules with molecular weights between 24 and 31 kDa. These different molecular forms could represent different levels of glycosylation or isoforms.10, 11 More than 50% of dust mite allergic patients react specifically to Der p 7 and some patients respond with an IgE titre as strong as that to the major allergen Der p 2.12 Previous studies suggested that Der p 7 promotes an abnormally high reactivity in T-cell proliferation assays among allergic and non-allergic patients.9, 11, 13-15 In this study, the three dimensional structure of the protein was determined in order to gain insight into the abnormal patient response and th100 e natural function of the protein.
Allergens can be enzymes, structural proteins, or ligand-binding proteins that often show specific binding affinity for lipids.16 The intrinsic adjuvant activity of lipid- binding proteins and their lipid cargo may be a general mechanism underlying allergenicity.17 Supporting this hypothesis, coexposure to allergens and lipopolysaccharide (LPS) from bacteria can drive the immune response away from benign tolerance to symptomatic response.18, 19 Interestingly, the dust mite allergen Der p 2 can bind to LPS and initiate the signaling of the TLR4 complex in MD-2-deficient mice, thus functionally substituting for the mammalian innate immune protein MD-2. This raises many interesting questions about how other allergens, especially the lipid-binding proteins, might interfere with normal host defense signaling.
Herein, we report that structural analysis reveals Der p 7 to be distantly related to another family of innate immune proteins that bind to various hydrophobic ligands. One such protein, LPS binding protein (LBP), is in the TLR4 pathway. To explore the possibility of immune protein mimicry analogous to the recently reported behavior of Der p 2 and MD-2, we examined the binding of LPS and select other hydrophobic compounds to Der p 7.
Materials and Methods
Chimeric ELISA for IgE antibodies to Der p 7
IgE binding to Der p 7 was analyzed by a chimeric ELISA as previously described.20 The plates were coated overnight at 4°C with anti-Der p 7 mAb WH9. Plates were blocked with PBS-Tween-1%BSA, followed by an incubation with 400 ng/mL of recombinant or natural Der p 7 at room temperature. The source of natural Der p 7 was a dust mite extract from Dermatophagoides pteronyssinus (HollisterStier, Spokane, WA) which contained 126 μg/ml Der p 7 (30,000 AU/ml). Sera from allergic patients was purchased from PlasmaLab International (Everett, WA). Thirty-three sera were added at 1:2 and 1:10 dilutions and bound IgE was detected using biotinylated goat anti-human IgE. Anti-Der p 2 mAb αDpX, purified natural Der p 2 at 500 ng/ml, and chimeric Ab 2B12-IgE were used for the standard curve to quantify the assay. Natural allergen (nDer p 7) and rDer p 7 expressed in Pichia pastoris21 were compared to constructs of rDer p 7 expressed in bacteria, vide infra.
Crystallographic Studies
The best results for crystallography were obtained with a customized maltose binding protein (MBP) fusion with Der p 7. Briefly, the MBP fusion was designed with surface entropy reducing mutations in MBP to enhance crystallization.22 DNA encoding residues 18-215 of Der p 7 representing the native expressed mite protein10 was cloned into a modified fixed-arm PMAL vector between the NotI and BamHI restriction sites with MBP mutations D82A/K83A/E172A/N173A/K239A.23, 24 The MBP-Der p 7 fusion protein was expressed in E. coli Rosetta2(DE3) pLacI cells (Novagen), and purified using the amylose affinity resin in batch followed by gel-filtration utilizing a 16/60 superdex 200 column and ionic exchang139 e chromatography step using Q sepharose. The purified protein was dialyzed against 25 mM HEPES pH 7.4, 75 mM NaCl, 5 mM maltose and concentrated to 27 mg/mL.
Crystals of the MBP-Der p 7 fusion protein were obtained at 4°C by mixing 1 μL of the protein solution with 1 μL of the reservoir consisting of 42.5 mM Tris pH 8.5, 12.75% PEG4000, 85 mM lithium sulfate, and 7.5% glycerol. The crystal used in this experiment reached a maximum size of 0.2×0.2×0.2mm after one week. Prior to data collection the crystal was transferred to 63.8mM Tris pH 8.5, 19.1% PEG4000, 128mM lithium sulfate, and 11.25% glycerol and then quickly transferred to 85 mM Tris pH 8.5, 25.5% PEG4000, 0.17 M lithium sulfate, and 15% glycerol. The crystal was flash frozen in liquid nitrogen and placed in a nitrogen gas stream cooled to -180°C. Data were collected to a resolution of 2.35Å, and were scaled and processed using HKL2000.25 Molecular replacement using the MBP from the MBP-RACK1 fusion structure was used to solve the position of the three molecules of MBP in the asymmetric unit.24, 26 The structures of the three Der p 7 molecules were built manually by iterative cycles of model building in O 27 as well as density modification and refinement in CNS.28 We were unable to identify a structure with sufficient homology to Der p 7 to allow solution by molecular replacement, but the coordinates from 1EWF.pdb (bactericidal/permeability increasing protein) and 3E8T.pdb (Takeout 1) were consulted for help in chain tracing and secondary structure based on structural similarity suggested by GenTHREADER.29-31 Table 1 lists important statistics for assessing the quality of the crystallographic data and the resulting structures.
Table 1.
| Crystallographic data statistics | |
|---|---|
| data set | MBP-derp7(18-215) |
| unit cell | a=193.35Å, b=117.90Å, c=92.44Å; α=γ=90°, β =114.06° |
| Space Group | C2 |
| Resolution (Å) | 50.0 -2.35 |
| # of observations | 407,861 |
| unique reflections | 76,800 |
| Rsym(%)(last shell)1 | 6.3 (35.5) |
| I/σI (last shell) | 28.4 (2.5) |
| Mosaicity range | 1.1-1.4 |
| completeness(%) (last shell) | 97.8 (97.2) |
| Refinement statistics | |
| Rcryst(%)2 | 24.9 |
| Rfree(%)3 | 29.3 |
| # of waters | 322 |
| Overall Mean B value (Å2) | 50.4 |
| Average for molecule A | 43.99 |
| molecule B | 47.49 |
| molecule C | 61.46 |
| r.m.s. deviation from ideal values | |
| bond length (Å) | 0.004 |
| bond angle (°) | 1.0 |
| dihedral angle (°) | 22.5 |
| improper angle (°) | 0.68 |
| Ramachandran Statistics4 | |
| residues in: | |
| favored (98%) regions (%) | 92.5 |
| allowed (>99.8%) regions (%) | 99.5 |
| PDB I.D. code | 3H4Z |
Rsym = Σ (| Ii - <I>|)/ Σ(Ii) where Ii is the intensity of the ith observation and <I> is the mean intensity of the reflection.
Rcryst = Σ∥ Fo| - | Fc ∥/ Σ| Fo| calculated from working data set.
Rfree was calculated from 5% of data randomly chosen not to be included in refinement.
Ramachandran results were determined by MolProbity.
NMR Constructs and Spectroscopy
Recombinant Der p 7, residues 18-215, was subcloned and expressed in the Gateway system (Invitrogen), with a cleavable 6-His tag, in order to reduce the size of the protein for NMR spectroscopic analysis. Optimal bacterial expression of this construct was obtained in the Rosetta 2 (DE3) pLacI cell line by growing the cells at 37° C to an OD600 of 0.6 and inducing overnight at 18° C. Stable isotopic labeling for NMR was done as described previously.32 The cells were lysed by sonication in 50 mM HEPES pH 7.4, 500 mM NaCl, and protein was found in the soluble fraction after centrifugation at >40,000g for 30 minutes. The rDer p 7 was purified with the Ni2+ affinity chromatography, followed by a Q-sepharose ion-exchange column. The affinity tag was cleaved with tobacco etch virus (a.k.a. TEV) protease. Subsequently, cleaved protein was passed through another Ni2+ column and further purified with size exclusion chromatography.
NMR backbone assignments were obtained on a U-2H, 13C, 15N labeled sample of 0.5 mM protein using a Varian 600 MHz spectrometer. Standard backbone assignment experiments were used to establish the connectivity of resonances through the C’, CA, and CB.32 The solution conditions for all NMR studies utilized phosphate buffered saline, with 5-10% 2H2O and 1 μM 5,5-dimethylsilapentanesulfonate (DSS) for referencing. Data were acquired at 25° C. The NMRViewJ RunAbout module was used for making the assignments.33 Aiding the analysis were assignments from the automated algorithms, MARS and PINE.34, 35 The assignments of manual inspection, MARS, and PINE agree very well.
Results
IgE antibody responses to Der p 7
The prevalence of IgE ab binding to natural allergen, rDer p 7 expressed in Pichia, and bacterially expressed rDer p 7 constructs used for crystallography or NMR were very similar (range 43-58%). There was an excellent correlation of IgE ab binding between rDer p 7 expressed in E. coli with or without MBP (r = 0.94; n = 18). IgE ab binding to the rDer p 7 with cleaved affinity tag showed an excellent correlation with IgE ab binding to natural Der p 7 from mite extracts (r = 0.99, n = 21) (data not shown). Similarly, IgE ab bound to the MBP-Der p 7 fusion protein showed a high correlation with IgE ab to Pichia-expressed Der p 7 (r = 0.94; n = 16) and to IgE bound to the natural allergen (r = 0.96, n = 21). There was an excellent correlation between IgE antibody binding to all the rDer p 7 allergens and to the natural allergen, indicating that the recombinant proteins were suitable for structural studies. Total IgE and specific IgE antibody levels to four allergens (Der f 1, Der p 1, Der p 2, Der p 7) have been reported in Table E1 (Online Repository).
Structural Analysis
rDer p 7 without an affinity tag crystallizes readily at room temperature, although no crystals diffracted at high resolution. Therefore we utilized an alternate approach in which the Der p 7 was fused to an MBP mutant protein designed for optimal crystallization characteristics. The best Der p 7 –MBP fusion protein crystals diffracted at 2.35 Å. The final model of the asymmetric unit consisted of three MBP-Der p 7 fusion proteins, shown in Figure 1a, (PDB code 3H4Z). The most extensive interactions between symmetric units are among MBP molecules, although all permutations of interactions exist, i.e. MBP-MBP, MBP-Der p 7, and Der p 7- Der p 7. The Der p 7 structure is elongated with two 4-stranded β-sheets that wrap around a long C–terminal helix (Figure 1). These two β-sheets are situated in a “head to toe” orientation appearing to form a single continuous β-sheet with a break in the middle. The N-terminal helix forms a cleft between the adjacent β-sheet and the end of the C-terminal helix, possibly for binding an unknown substrate.
Figure 1.
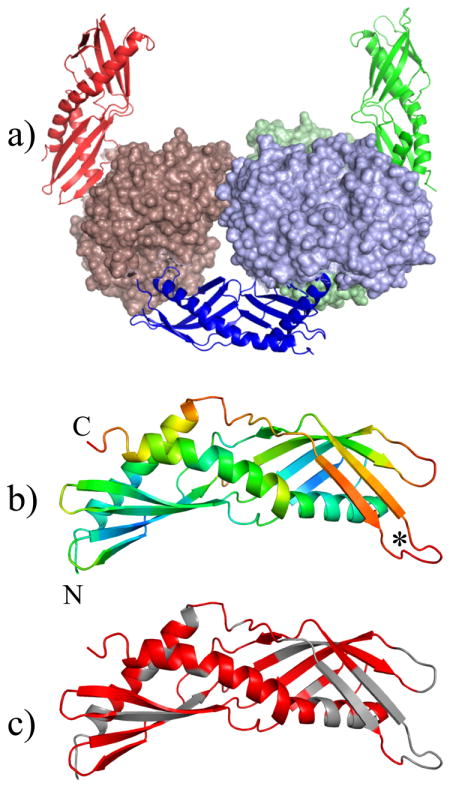
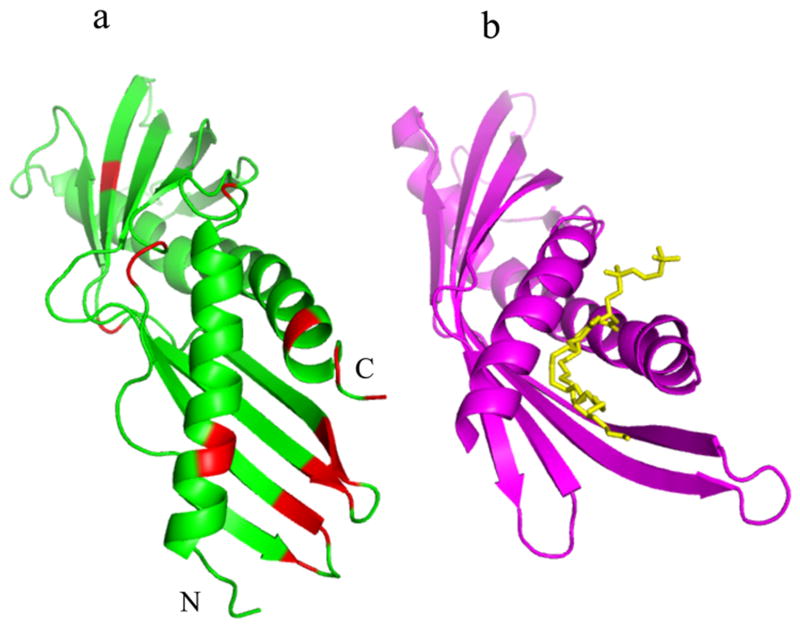
Der p 7 structural analysis. Panel a shows the three molecules in the crystallographic asymmetric unit, colored green (chain A), blue (chain B) and red (chain C). The Der p 7 sequence is rendered in ribbon style while the MBP sequence is represented with a solid surface. Panel b shows the Der p 7 molecule from chain A colored by relative B factor where red is high (greater than 80) and blue is low (less than 30). In Panel c, residues that could be assigned by NMR are colored red, and unassigned residues are grey. The N and C terminus are annotated in panel b. The asterisk indicates a loop with variable density in chains A, B, and C of the asymmetric unit.
Figure 1b shows the Der p 7 molecule from chain A colored by relative B-factor. Of note, the Der p 7 molecules generally showed higher B-factors than the attached MBP molecule with the exception of chain C, which had high B-factors for both the MBP and Der p 7 molecule (Table 1). The regions with very high B factors were difficult to trace, and in some cases density could not be found in all three chains. For example, all Der p 7 residues are traced in molecule A while molecule B and C are missing residues 63-67 and 59-67 respectively (see asterisk in Figure 1b). Additionally, the model of molecule C is missing residues 122-126 and the C terminal residue Q215. All three of the Der p 7 molecules superimpose well, with an RMSD of less than 0.51Å over 197 Cα atoms.
The NMR data acquired in solution is consistent with the structure determined by crystallography. The 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of Der p 7 is characteristic of a generally well folded protein, with well dispersed resonance intensities. The assignment of the backbone resonances was extremely challenging due to the high number of missing resonances in the HSQC and other 3D NMR experiments: 159 amide resonances appear out of a possible 192 backbone amides. Of those that appear, 134 could be confidently assigned with manual inspection of the spectra. The assigned residues are colored red on the structure in The Figure 1c. In general, the assigned residues correlate with regions of lower B-factors in the crystal structure. The correlation of missing assignments with high B-factors supports th227 e contention that most of the assignment problem results from broadening due to conformational exchange. The secondary structure prediction from the assigned shifts34 agrees very well with the secondary structure of the crystal structure, providing additional confidence in the assignments, and showing that the solution conformation of the protein is similar to the crystal structure.
Structural Homology
To investigate whether the structure of Der p 7 provided clues to the function of the protein in mites, or to its allergenic effect in humans, the software DALI36 was used to identify related protein folds in the database. The search returned over 500 structures with marginal match scores (Z-score ~ 5), and eight with very good matches (Z-score > 10). Table 2 shows the top eight results as well as a few select entries that are notable because they are also allergens. For example the 37th best score is the birch pollen allergen Bet v 1. The fold superfamily of these allergens is classified as Bet v 1-like, also termed START.37 The common feature of START domains is that they are known to bind hydrophobic ligands.
Table 2.
| No | PDB & Chain | Z-score | RMSD | Length Align | No. Residues | % Identity | Name |
|---|---|---|---|---|---|---|---|
| 1 | 2RCK-B | 12.8 | 3.7 | 175 | 208 | 13 | Juvenile Hormone Binding Protein |
| 2 | 1BP1-A | 12.8 | 3.5 | 177 | 456 | 14 | Bactericical/Permeability-Increasing Protein |
| 3 | 2RCK-A | 12.7 | 3.6 | 174 | 221 | 13 | Juvenile Hormone Binding Protein |
| 4 | 1BP1 | 12.6 | 3.6 | 174 | 456 | 14 | Bactericical/Permeability-Increasing Protein |
| 5 | 1EWF | 11.0 | 3.3 | 174 | 456 | 15 | Bactericical/Permeability-Increasing Protein |
| 6 | 3E8W | 10.7 | 4.4 | 171 | 219 | 11 | Takeout-Like Protein 1 |
| 7 | 3E8T | 10.6 | 4.5 | 172 | 296 | 10 | Takeout-Like Protein 1 |
| 8 | 2OBD-A | 10.4 | 3.5 | 169 | 427 | 12 | Cholesteryl Ester Transfer Protein |
| 37 | 1VJH-B | 4.5 | 3.7 | 99 | 120 | 11 | Bet v 1 Allergen family |
| 290 | 2VJG-D | 3.4 | 4.1 | 109 | 152 | 9 | Major Allergen Dau c 1 |
| 453 | 1H2O-A | 2.5 | 4.0 | 92 | 159 | 10 | Major Allergen Pru av 1 |
| 531 | 2BK0-A | 2.0 | 3.7 | 89 | 153 | 12 | Major Allergen Api g 1 |
The best structural matches with Der p 7 include insect proteins that have very similar folds such as juvenile hormone binding protein (JHBP) and the take-out (TO) proteins, consistent with GenTHREADER predictions.29, 31, 38 The RMSD of the superposition for Der p 7 with JHBP is 3.6 Å over 176 Cα atoms (Figure 2a). Like the TO proteins, it superimposes well over the whole molecule and shows very similar secondary structure organization. JHBP and TO proteins bind to hydrophobic ligands, similar to Bet v 1 and the other START domain-like allergens.
Figure 2.
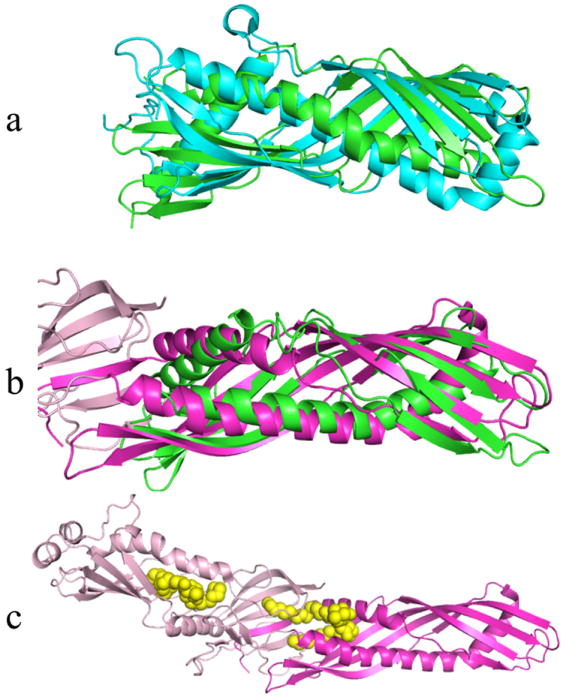
Examples of structurally similar proteins to Der p 7. Der p 7 is shown in green in all panels. Panel a is the best alignment with JHBP (cyan). Panel b shows the best alignment of Der p 7 with the N terminal domain of BPI shaded magenta, while the C terminal domain of BPI is shaded pale pink. Panel c shows BPI with two bound DSPC molecules (yellow spheres).
Another significant match to Der p 7 based on the DALI evaluation was the N terminal domain of bactericidal permeability increasing protein (BPI, pdb code 1BP130). A structural overlay is shown in Figure 2b, and has a RMSD of 3.5 Å over 174 Cα atoms. BPI contains two domains, which have a very similar fold but are distantly related sequentially. The full structure of BPI is shown in Figure 2c, with the domains shaded differently and bound ligands indicated with yellow spheres. The protein family database PFAM39 designates this fold as “BPI/LBP/CETP N terminal”, and the database CATH40 classifies the architecture as a “super-roll” based on the way the long beta strands roll around the central long helix. The fold of Der p 7 is more similar to the super-roll than to the START domains.
Lipid Binding Assays
Based on the structural similarities to other lipid binding proteins the lipid binding potential of Der p 7 was examined. Since BPI and the related LBP bind to LPS we first tested LPS binding to Der p 7. Biochemical assays indicated that Der p 7 does not bind tightly to LPS (See Figure E1 in the Online Repository). Other potential Der p 7 ligands for evaluation were identified by comparison to BPI and LBP. One type of potential Der p 7 substrate is distearoyl phosphatidyl choline (DSPC), which is found complexed to BPI in the crystal structure (Figure 2c).30 Lyso-palmitoyl PC (LPPC) was tested for interactions with Der p 7 due to its similar structure to DSPC and much higher solubility. A sample containing 50 μM U-[15N] labeled Der p 7 was titrated with increasing amounts of LPPC and two dimensional 1H-15N HSQC spectra showed very few changes. Similar negative results were obtained upon addition of SDS and N-octyl-glucoside. Figure 3a shows an example of an NMR spectrum showing minor perturbations that were obtained upon addition of LPS. In order to see if the small changes due to LPS could be reversed, polymyxin B (PB) was added, which is known to sequester LPS.41 Surprisingly, the addition of PB produced much more significant spectral changes (data not shown). To insure that the changes were due to PB, Figure 3b shows the changes in the NMR HSQC spectra of Der p 7 upon addition of PB alone. This result was followed up with a saturation transfer difference experiment42 (STD) that demonstrated that Der p 7 and PB interact. Figure 4 shows the 1H spectra of PB alone (Panel a) and the results of the STD experiment with PB and Der p 7 (Panel b). The same resonances appearing in panels a and b provide conclusive evidence of a binding interaction between PB and Der p 7.
Figure 3.
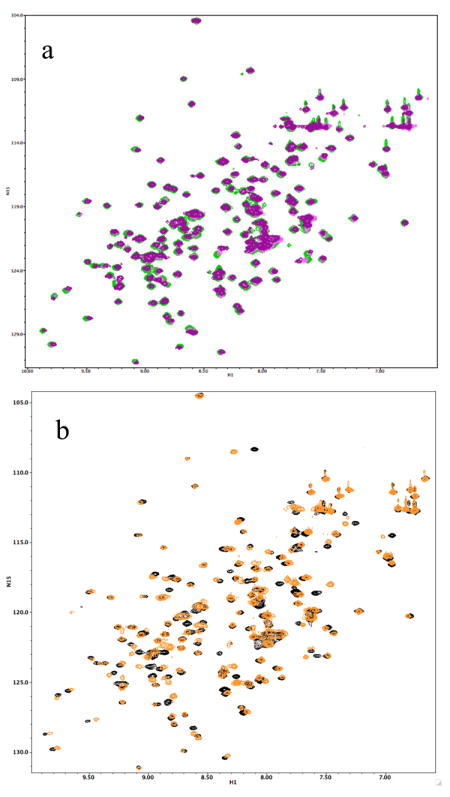
Changes in the 15N-1H HSQC spectra of Der p 7 upon addition of compounds. Panel a shows very little change in the HSQC spectra of 50 μM Der p 7 going from apo (green) to 100 uM (pink) to 200 uM (purple) LPS. In contrast, addition of polymyxin B (PB) caused dramatic changes, shown in panel b. Der p 7 apo spectra is black, and upon addition PB the spectra is colored orange.
Figure 4.
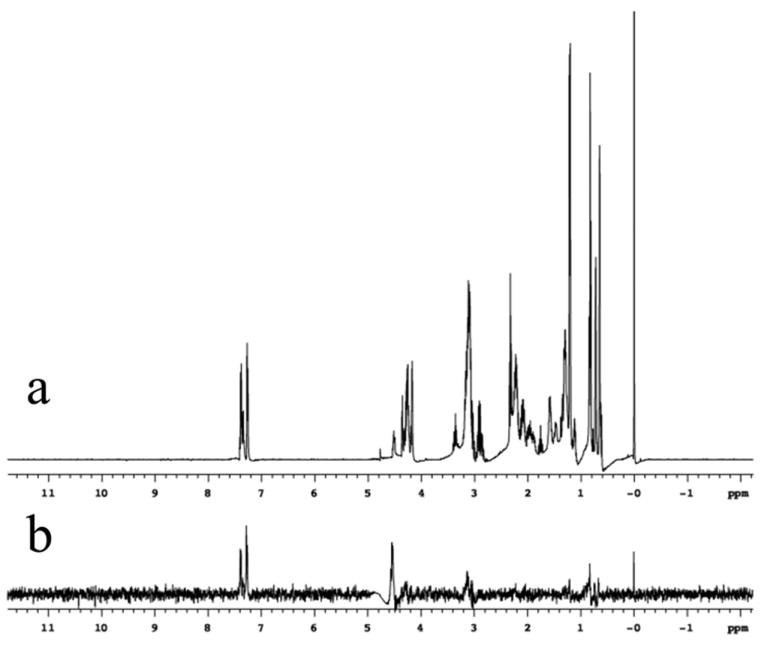
1H NMR of PB. Panel a shows the 1H NMR spectrum of 1 mM Polymyxin B, in 2H2O with 10 μM DSS (at 0 ppm) for chemical shift referencing. Panel b shows the result of the STD experiment42 with 1 mM PB and 50 μM Der p 7
To show that the binding of PB was a specific interaction, the assignments of the resonances in the HSQC spectra were determined to map the site of the interaction (See Figure E2 in the Online Repository). Figure 5a shows the residues involved in the interaction colored red. Many of the significant shift changes cluster near a cleft between the N and C terminus of Der p 7. However, because of missing assignments we are being cautious in our interpretation of the shift changes, as it is possible that additional residues are involved. Figure 5b shows the N terminal domain of the structure of BPI with the DSPC bound. BPI is aligned in a similar orientation to Der p 7. This figure shows the DSPC molecule bound in a cleft similar in location to the cleft in the Der p 7 molecule. Hence PB interactions with Der p 7 occur near the expected binding site.
Figure 5.
Proposed binding site of PB in Der p 7. The significant chemical shift changes in Figure 3b correspond to the residues colored red in Panel a. The residues map to a cleft between the N and C terminus, labeled in Panel a. This location is similar to a cleft in the N terminal domain of BPI that contains DSPC in the crystal structure, see Panel b.
Discussion
Der p 7 appears closely related to several proteins known to bind hydrophobic compounds, including the Take-out proteins and BPI and LBP. The TO proteins are involved in signaling in a wide variety of biological functions of insects.43 Since the class Insecta and the class Arachnida (mites) are both in the phylum Arthropoda this may provide some clues as to the natural function of Der p 7 in mites, but there is not a good enough match in the literature with a known natural ligand. Der p 7 does not appear to bind to LPS like BPI or LBP, but exhibits affinity for PB, which is a nine residue cyclic peptide with a lipid tail from the Gram-positive bacteria Bacillus polymyxa. The weak binding of Polymyxin B to Der p 7 does not imply that PB is the natural substrate. Future studies to identify the natural ligand(s) will require screening of a broader spectrum of potential ligands, as well as a determination of the corresponding dissociation constants. However, the mapping of the interaction site to a similar position in homologous proteins validates the conclusion that Der p 7 has affinity for a lipid substrate, in this case a bacterially-derived lipopeptide product. The importance of this is that lipid binding is a common feature of some allergens44 and natural lipid adjuvants are suspected to be important in sensitizing patients to allergens.17
LBP has been shown to interact with di- and tri-acylated lipopeptides, as well as LPS.45, 46 Bacterial and mycobacterial lipopeptides can induce the release of cytokines through interactions with TLR2 in cooperation with TLR1 or TLR6. Several TLRs may be involved in stimulating the allergic response through promotion of Th2 immunity.44 Indeed stimulation of TLR2 in the presence of allergen enhances Th2 responses,47, 48 as does TLR5 stimulation.49 We speculate that if Der p 7 can bind bacterial lipopeptides other than PB as its natural ligand, it may promote Th2 immunity (to itself) through co-stimulation of TLR2 pathways.
In addition to understanding allergen function, structural characterization of allergens can provide detailed knowledge of potential B-cell epitopes and aid in mapping antibody interactions. We previously used hydrogen exchange NMR to map monoclonal antibody binding sites on Der p 2 and NMR techniques have recently been used to define a mAb epitope on Blomia tropicalis allergen Blo t 5.50, 51 Several X-ray crystal structures of allergen/antibody complexes have also been reported including Bet v 1, β-lactoglobulin and Bla g 2.52-54 Modification of structure has been an effective way to design recombinant hypoallergenic variants of allergens, which have potential clinical application for immunotherapy in mite allergy.55, 56 Our IgE binding data confirms that Der p 7 is an important allergen and shows that the prevalence of IgE ab to rDer p 7 is comparable to that of the natural allergen.12 The purified rDer p 7 was of sufficiently high quality for crystallographic studies. These observations suggest that rDer p 7 could be used in a cocktail of recombinant mite allergens for use in subcutaneous immunotherapy and for rational design of hypoallergenic variants. A cocktail of five recombinant timothy pollen allergens was successfully used in a placebo controlled trial and resulted in a 39% reduction in symptom scores and medication use.57 Clinical improvement was associated with a strong increase in allergen-specific IgG4 antibody production, reduced levels of IgE ab and induction of IL-10 producing regulatory T-cells. Current data suggests that recombinant Der p 1, Der p 2 and Der p 7 are important immunologic targets and should be included in similar vaccine trials for mite allergen immunotherapy. The structure presented here will be useful in the rational design of hypoallergenic mutant proteins, and to understand future studies of the cross reactivity between the Group 7 mite allergens.
Knowledge of allergen function has contributed to an improved understanding of allergenicity. This was demonstrated for the protease activity of Group 1 mite allergens on the adaptive immune system7, 8 and the interaction with the innate immune system by Group 2 mit337 e allergens.17 We suggest that there may be similarities between the biological functions of the group 7 and group 2 mite allergens that involve binding of lipid substrates and that both allergens may co-opt innate immune responses into promoting allergenicity. Further studies are needed to investigate this hypothesis. The structure of Der p 7 presented here will contribute to an improved understanding of the complex interactions between the innate and adaptive immune systems in the development of allergy.
Acknowledgments
The authors wish to thank Eugene DeRose for many helpful discussions and advice on NMR spectroscopy. Additionally, we thank Thomas Kirby and Stavros Garantzlotis for critical reading of the manuscript. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences and by R01AI077653 from the National Institute of Allergy And Infectious Diseases (AP and MDC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences and by NIH grant AI077563 (AP and MDC) from the National Institute for Allergy and Infectious Diseases.
Abbreviations Used
- LBP
lipopolysaccharide binding protein
- TLR
Toll-like receptors
- PB
polymyxin B
- BPI
bactericidal/permeability increasing protein
- LPS
lipopolysaccharide (LPS)
- MBP
maltose binding protein (MBP)
- STD
saturation transfer difference
- DSPC
distearoyl phosphatidyl choline
- DSS
5,5-dimethylsilapentanesulfonate
- HSQC
heteronuclear single quantum coherence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Salo PM, Cohn RD, Zeldin DC. Exposure to mouse allergen in US homes is associated with asthma symptoms. Journal of Allergy and Clinical Immunology. 2008;121:S231–S. [Google Scholar]
- 2.Sporik R, Chapman MD, Platts-Mills TA. House dust mite exposure as a cause of asthma. Clin Exp Allergy. 1992;22:897–906. doi: 10.1111/j.1365-2222.1992.tb02062.x. [DOI] [PubMed] [Google Scholar]
- 3.Nelson RP, Jr, DiNicolo R, Fernandez-Caldas E, Seleznick MJ, Lockey RF, Good RA. Allergen-specific IgE levels and mite allergen exposure in children with acute asthma first seen in an emergency department and in nonasthmatic control subjects. J Allergy Clin Immunol. 1996;98:258–63. doi: 10.1016/s0091-6749(96)70148-3. [DOI] [PubMed] [Google Scholar]
- 4.Chapman MD, Wunschmann S, Pomes A. Proteases as Th2 adjuvants. Current Allergy and Asthma Reports. 2007;7:363–7. doi: 10.1007/s11882-007-0055-6. [DOI] [PubMed] [Google Scholar]
- 5.King C, Brennan S, Thompson PJ, Stewart GA. Dust mite proteolytic allergens induce cytokine release from cultured airway epithelium. J Immunol. 1998;161:3645–51. [PubMed] [Google Scholar]
- 6.Tomee JF, van Weissenbruch R, de Monchy JG, Kauffman HF. Interactions between inhalant allergen extracts and airway epithelial cells: effect on cytokine production and cell detachment. J Allergy Clin Immunol. 1998;102:75–85. doi: 10.1016/s0091-6749(98)70057-0. [DOI] [PubMed] [Google Scholar]
- 7.Hewitt CR, Brown AP, Hart BJ, Pritchard DI. A major house dust mite allergen disrupts the immunoglobulin E network by selectively cleaving CD23: innate protection by antiproteases. J Exp Med. 1995;182:1537–44. doi: 10.1084/jem.182.5.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz O, Sewell HF, Shakib F. Proteolytic cleavage of CD25, the alpha subunit of the human T cell interleukin 2 receptor, by Der p 1, a major mite allergen with cysteine protease activity. J Exp Med. 1998;187:271–5. doi: 10.1084/jem.187.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hales BJ, Shen HD, Thomas WR. Cytokine responses to Der p 1 and Der p 7: house dust mite allergens with different IgE-binding activities. Clinical and Experimental Allergy. 2000;30:934–43. doi: 10.1046/j.1365-2222.2000.00901.x. [DOI] [PubMed] [Google Scholar]
- 10.Shen HD, Chua KY, Lin KL, Hsieh KH, Thomas WR. Molecular cloning of a house dust mite allergen with common antibody binding specificities with multiple components in mite extracts. Clin Exp Allergy. 1993;23:934–40. doi: 10.1111/j.1365-2222.1993.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 11.Shen HD, Lin WL, Tsai LC, Tam MF, Chua KY, Chen HL, et al. Characterization of the allergen Der f 7 from house dust mite extracts by species-specific and crossreactive monoclonal antibodies. Clinical and Experimental Allergy. 1997;27:824–32. [PubMed] [Google Scholar]
- 12.Shen HD, Chua KY, Lin WL, Chen HL, Hsieh KH, Thomas WR. IgE and monoclonal antibody binding by the mite allergen Der p 7. Clin Exp Allergy. 1996;26:308–15. [PubMed] [Google Scholar]
- 13.Hales BJ, Shen HD, Thomas WR. Cross-reactivity of T-cell responses to Dermatophagoides pteronyssinus and D-farinae. Studies with group 1 and 7 allergens. Clinical and Experimental Allergy. 2000;30:927–33. doi: 10.1046/j.1365-2222.2000.00900.x. [DOI] [PubMed] [Google Scholar]
- 14.Thomas WR, Hales BJ, Shen HD, Smith W. Comparison of T-cell responses induced by major and less major allergens of the house dust mite. International Archives of Allergy and Immunology. 1999;118:214–5. doi: 10.1159/000024073. [DOI] [PubMed] [Google Scholar]
- 15.Thomas WR, Hales BJ. T and B cell responses to HDM allergens and antigens. Immunol Res. 2007;37:187–99. doi: 10.1007/BF02697369. [DOI] [PubMed] [Google Scholar]
- 16.Chapman MD, Pomes A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. Journal of Allergy and Clinical Immunology. 2007;119:414–20. doi: 10.1016/j.jaci.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Trompette A, Divanovic S, Visintin A, Blanchard C, Hegde RS, Madan R, et al. Allergenicity resulting from functional mimicry of a Toll-like receptor complex protein. Nature. 2009;457:585–8. doi: 10.1038/nature07548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J Exp Med. 2002;196:1645–51. doi: 10.1084/jem.20021340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piggott DA, Eisenbarth SC, Xu L, Constant SL, Huleatt JW, Herrick CA, et al. MyD88-dependent induction of allergic Th2 responses to intranasal antigen. J Clin Invest. 2005;115:459–67. doi: 10.1172/JCI22462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichikawa K, Iwasaki E, Baba M, Chapman MD. High prevalence of sensitization to cat allergen among Japanese children with asthma, living without cats. Clin Exp Allergy. 1999;29:754–61. doi: 10.1046/j.1365-2222.1999.00472.x. [DOI] [PubMed] [Google Scholar]
- 21.Glesner J, Chapman MD, Pomes A. Recombinant Der p 7 Constitutively Expressed in a Eukaryotic System Shows the Same Antigenic Properties as the Natural Allergen. Journal of Allergy and Clinical Immunology. 2009;123:S227–S. [Google Scholar]
- 22.Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–35. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Center RJ, Kobe B, Wilson KA, Teh T, Howlett GJ, Kemp BE, et al. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. 1998;7:1612–9. doi: 10.1002/pro.5560070715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci. 2008;17:1771–80. doi: 10.1110/ps.035121.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 27.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Hamiaux C, Stanley D, Greenwood DR, Baker EN, Newcomb RD. Crystal structure of Epiphyas postvittana takeout 1 with bound ubiquinone supports a role as ligand carriers for takeout proteins in insects. J Biol Chem. 2009;284:3496–503. doi: 10.1074/jbc.M807467200. [DOI] [PubMed] [Google Scholar]
- 30.Kleiger G, Beamer LJ, Grothe R, Mallick P, Eisenberg D. The 1.7 A crystal structure of BPI: a study of how two dissimilar amino acid sequences can adopt the same fold. J Mol Biol. 2000;299:1019–34. doi: 10.1006/jmbi.2000.3805. [DOI] [PubMed] [Google Scholar]
- 31.McGuffin LJ, Jones DT. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–81. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- 32.Mueller GA, DeRose EF, Kirby TW, London RE. NMR assignment of polymerase beta labeled with 2H, 13C, and 15N in complex with substrate DNA. Biomol NMR Assign. 2007;1:33–5. doi: 10.1007/s12104-007-9007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson BA, Blevins RA. Nmr View - a Computer-Program for the Visualization and Analysis of Nmr Data. Journal of Biomolecular Nmr. 1994;4:603–14. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 34.Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. Probabilistic Interaction Network of Evidence Algorithm and its Application to Complete Labeling of Peak Lists from Protein NMR Spectroscopy. Plos Computational Biology. 2009;5 doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung YS, Zweckstetter M. Mars - robust automatic backbone assignment of proteins. Journal of Biomolecular Nmr. 2004;30:11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- 36.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–1. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radauer C, Lackner P, Breiteneder H. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolodziejczyk R, Bujacz G, Jakob M, Ozyhar A, Jaskolski M, Kochman M. Insect juvenile hormone binding protein shows ancestral fold present in human lipid-binding proteins. J Mol Biol. 2008;377:870–81. doi: 10.1016/j.jmb.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 39.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, et al. The CATH classification revisited--architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–4. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas CJ, Surolia N, Surolia A. Surface plasmon resonance studies resolve the enigmatic endotoxin neutralizing activity of polymyxin B. J Biol Chem. 1999;274:29624–7. doi: 10.1074/jbc.274.42.29624. [DOI] [PubMed] [Google Scholar]
- 42.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angewandte Chemie-International Edition. 1999;38:1784–8. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 43.Dauwalder B, Tsujimoto S, Moss J, Mattox W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002;16:2879–92. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas WR, Hales BJ, Smith WA. Structural biology of allergens. Current Allergy and Asthma Reports. 2005;5:388–93. doi: 10.1007/s11882-005-0012-1. [DOI] [PubMed] [Google Scholar]
- 45.Schroder NW, Heine H, Alexander C, Manukyan M, Eckert J, Hamann L, et al. Lipopolysaccharide binding protein binds to triacylated and diacylated lipopeptides and mediates innate immune responses. J Immunol. 2004;173:2683–91. doi: 10.4049/jimmunol.173.4.2683. [DOI] [PubMed] [Google Scholar]
- 46.Schroder NW, Eckert J, Stubs G, Schumann RR. Immune responses induced by spirochetal outer membrane lipoproteins and glycolipids. Immunobiology. 2008;213:329–40. doi: 10.1016/j.imbio.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 47.Chisholm D, Libet L, Hayashi T, Horner AA. Airway peptidoglycan and immunostimulatory DNA exposures have divergent effects on the development of airway allergen hypersensitivities. J Allergy Clin Immunol. 2004;113:448–54. doi: 10.1016/j.jaci.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 48.Redecke V, Hacker H, Datta SK, Fermin A, Pitha PM, Broide DH, et al. Cutting edge: activation of Toll-like receptor 2 induces a Th2 immune response and promotes experimental asthma. J Immunol. 2004;172:2739–43. doi: 10.4049/jimmunol.172.5.2739. [DOI] [PubMed] [Google Scholar]
- 49.Didierlaurent A, Ferrero I, Otten LA, Dubois B, Reinhardt M, Carlsen H, et al. Flagellin promotes myeloid differentiation factor 88-dependent development of Th2-type response. J Immunol. 2004;172:6922–30. doi: 10.4049/jimmunol.172.11.6922. [DOI] [PubMed] [Google Scholar]
- 50.Mueller GA, Smith AM, Chapman MD, Rule GS, Benjamin DC. Hydrogen exchange nuclear magnetic resonance spectroscopy mapping of antibody epitopes on the house dust mite allergen Der p 2. J Biol Chem. 2001;276:9359–65. doi: 10.1074/jbc.M010812200. [DOI] [PubMed] [Google Scholar]
- 51.Naik MT, Chang CF, Kuo IC, Kung CC, Yi FC, Chua KY, et al. Roles of structure and structural dynamics in the antibody recognition of the allergen proteins: an NMR study on Blomia tropicalis major allergen. Structure. 2008;16:125–36. doi: 10.1016/j.str.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Li M, Gustchina A, Alexandratos J, Wlodawer A, Wunschmann S, Kepley CL, et al. Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody. J Biol Chem. 2008;283:22806–14. doi: 10.1074/jbc.M800937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirza O, Henriksen A, Ipsen H, Larsen JN, Wissenbach M, Spangfort MD, et al. Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J Immunol. 2000;165:331–8. doi: 10.4049/jimmunol.165.1.331. [DOI] [PubMed] [Google Scholar]
- 54.Niemi M, Jylha S, Laukkanen ML, Soderlund H, Makinen-Kiljunen S, Kallio JM, et al. Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen. Structure. 2007;15:1413–21. doi: 10.1016/j.str.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 55.Gafvelin G, Parmley S, Neimert-Andersson T, Blank U, Eriksson TL, van Hage M, et al. Hypoallergens for allergen-specific immunotherapy by directed molecular evolution of mite group 2 allergens. J Biol Chem. 2007;282:3778–87. doi: 10.1074/jbc.M607938200. [DOI] [PubMed] [Google Scholar]
- 56.Walgraffe D, Matteotti C, El Bakkoury M, Garcia L, Marchand C, Bullens D, et al. A hypoallergenic variant of Der p 1 as a candidate for mite allergy vaccines. J Allergy Clin Immunol. 2009;123:1150–6. doi: 10.1016/j.jaci.2008.11.038. [DOI] [PubMed] [Google Scholar]
- 57.Jutel M, Jaeger L, Suck R, Meyer H, Fiebig H, Cromwell O. Allergen-specific immunotherapy with recombinant grass pollen allergens. J Allergy Clin Immunol. 2005;116:608–13. doi: 10.1016/j.jaci.2005.06.004. [DOI] [PubMed] [Google Scholar]