Abstract
Embryonic stem cells (ESCs) are pluripotent stem cells from early embryos. It has been well recognized that ESC genomes are maintained in a globally transcriptional hyperactive state, which genetically poised ESCs to the high differentiation potential. However, the transcription factors regulating the global transcription activities in ESCs are not well defined. We show here that mouse and human ESCs express two transcription factors, Aire and Deaf1. Previously known to function in the thymus stromal cells and peripheral lymphoid organs respectively, Aire and Deaf1 help regulate the ectopic expression of diverse tissue-specific antigens to establish self-immune tolerance. Differentiation of ESCs greatly reduced Aire and Deaf1 expression, in a pattern similar to the pluripotent factors, Oct4 and Nanog. Knockdown of Aire in mouse ESCs resulted in significantly decreased clone-forming efficiency as well as attenuated cell cycle, suggesting Aire plays a role in ESC self-renewal. In addition, some differentiation-associated genes that are sporadically expressed in ESCs were reduced in expression upon Aire knockdown. These results suggest that transcription factors such as Aire and Deaf1, which exert global transcriptional regulatory functions, may play important roles in self-renewal of ESCs and maintaining ESC in a transcriptionally hyperactive state.
Keywords: Embryonic stem cells, Global expression, Self-renewal, Aire, Deaf1
1. Introduction
Embryonic stem (ES) cells can proliferate indefinitely in vitro while maintaining the ability to differentiate into all cell lineages [1,2]. The molecular mechanisms underlying pluripotency and self-renewal are important questions in ES cell biology. It has been well established that controlled expression of a series of transcription factors, such as Oct4, Nanog, Sox2, Klf4, is essential to maintain ES cells in a pluripotent state while keeping their self-renewal activity [3–7]. More recently, many epigenetic modifiers, including chromatin remodeling, histone modifications, and microRNAs have been shown to play important roles in stem cell pluripotency and self-renewal [8–14]. Recent studies have shown that ES cells adopt loose chromatin structures, and most genes in genome were set in an expression competent state or expressed at low levels [11,12,15–17]. In this manner, ES cells are poised for lineage-specific differentiation [12]. This contributes to pluripotency and differentiation plasticity of ES cells [17,18]. However, transcription factors responsible for the active chromosomal structure and the global gene expression are not defined.
Promiscuous expression of a large number of genes specific to peripheral tissues was first found in the thymus and has recently been demonstrated in the peripheral lymph organs [19–21]. In the thymus, the medullary thymic epithelial cells (mTECs) express a large diversity of tissue-restricted self-antigens (TRAs) at low levels, which functions to purge thymocytes strongly reactive to self-antigens, thereby establishing immunetolerance [22,23]. The promiscuous gene expression in mTECs is partially regulated by the autoimmune regulator gene, Aire [22] However, Aire is not the sole player in the ectopic gene expression of the immune system. In pancreatic lymph nodes, the transcription factor Deaf1 assumes a similar role as Aire does in the thymus [21]. Whether Aire and Deaf1 also play a role in the transcriptionally hyperactive ES cells is an interesting question to ask, an exploration of which would help us to understand how transcriptional profiles of the pluripotent ES cells are regulated.
In this study, we determined the expression of Aire and Deaf1 in mouse and human ES cells. We have found that expression of the two factors decrease dramatically during the differentiation of ES cells, similar to other pluripotent factors. Aire knockdown in mES resulted in greatly reduced clone-forming efficiency and attenuated cell cycle. In addition, expression of some tissue-specific genes in ESCs reduced upon Aire knockdown. Results showed that transcription factors such as Aire, which exert a genome-wide transcriptional regulation, may contribute to maintaining the ESCs properties.
2. Materials and methods
2.1. Cell lines and cell culture
Mouse ES (mES) cell D3 line was purchased from ATCC (Manassas, VA). ZJ1 and ZJ2 mouse ES cell lines were established in our laboratory. The cells were cultured on mitomycin C-treated mouse embryonic fibroblasts (MEFs) as previously described [24]. Human ES (hES) H9 cells were obtained from WiCell Research Institute (Madison, WI) and cultured on mitomycin C-inactivated MEFs as previously described [25].
For monolayer (MO) differentiation, ES cells were digested to single cells and then cultured on gelatin-coated tissue culture plates in differentiation medium (DMEM supplemented with 15% fetal bovine serum) for different days. For embryonic body (EB) differentiation, mES cells were cultured in suspension for 6 days in differentiation medium to form EB and then adhered to tissue culture plates for further differentiation.
2.2. Antibodies
The antibody against SSEA-1 (MAB 2155) was purchased from R&D Systems (Minneapolis, MN). The antibody against murine Aire was produced by immunizing rabbits with a peptide corresponding to the C-terminal sequence of murine Aire. The peptide was conjugated to KLH protein and emulsified in complete Freund Adjuvant. The antibodies were purified by affinity purification using antigen peptide conjugated to BSA.
Alexa Fluor 488-conjugated goat anti-rabbit IgG (A11008), Alexa555 conjugated Goat anti-rabbit IgG (A21428), Alexa Fluor 488-conjugated goat anti-mouse IgM (A21426), and Alexa Fluor 555-conjugated goat anti-mouse IgM (A21042) were purchased from Invitrogen.
2.3. Plasmids and transfection of HEK-293 cells
The full length Aire gene were amplified from mES cells by PCR and cloned into the pcDNA3.1-His plasmid (Invitrogen). The authenticity of the expression construct was confirmed by sequencing. HEK-293 cells were grown in DMEM containing 10% fetal bovine serum. These cells were transfected with the Aire-expression vector using Fugene 6 reagent (Roche, Minneapolis, MN) as previously described [26].
2.4. Lentivirus production and infection of mES cells
shRNA coding DNA sequences were obtained from The RNAi Consortium (http://www.broadinstitute.org/rnai/trc) and synthesized by Sangong (Shanghai). pLKO TRC-cloning Vector (plasmid 10878) was obtained from Addgene [27]. The vector was modified in house to replace the puromycin selection marker with EGFP. Then the shRNA-coding oligonucleotides were cloned into the modified pLKO TRC-cloning Vector. Virus particles were packaged by co-transfecting 293T cell with PsPAX (Addgene plasmid 12260), pMD2.G (Addgene plasmid 12259), and the modified pLKO plasmids bearing shRNA-coding oligonucleotides. The virus particles were enriched by ultracentrifugation to reach a tilter of approximately 106 CFU/ml. For virus infection, mES cells were seeded in 96-well tissue culture plate at a density of 3000 per well and incubated with medium containing virus for 24 h at a multiplicity of infection (MOI) of 30. Single cell clones exhibiting strong GFP fluorescent intensity were picked according to GFP fluorescence. At least three independent clones were used in the functional analysis. The pLKO-scramble (Addgene plasmid 1864) virus was used as control [28].
2.5. Clonogenicity assay
mES cells were seeded in 24-well cell culture plates on feeder cells at a density of 2000 cells per well and then allowed to grow for 4 days. ALP staining was performed with an ALP assay kit (Sigma). Macroscopic pictures were taken using a digital camera (Panasonic DMC-ZS1). The number of ALP positive clones was counted under a phase contrast microscope. The results were statistical averages of three independent wells.
2.6. MTT assay
mES cells were seeded in 96-well cell culture plate at a density of 1000 cells per well on feeder cells. After 4 days, cell growth was evaluated using an MTT assay kit (Sigma). Wells plated only with MEF were set as a control.
2.7. Immunofluorescence
Immunofluorescence was carried out as previously described [24]. The subjects were observed and images were acquired under LSM 500 Confocal Microscope (Zeiss, Germany).
2.8. RT-PCR
Total RNA was extracted from cultured ES cells using TRIzol Reagent (Invitrogen), and first strand cDNA was synthesized using reverse transcriptase. The genes of interest were then amplified by PCR. Primers corresponding to the genes of interest were designed using Primer Premier 5 software (PREMIER Biosoft International, Palo Alto CA), and are listed in Supplemental Table 1.
2.9. Real-time RT-PCR
Real-time RT-PCR was performed on an Eppendorf Mastercycler. RNA was prepared using TRIzol Reagent. Tests were carried out with SYBR Green Reagents purchased from Applied Biosystems (Foster City, CA) using primers acquired from Primer Bank (http://pga.mgh.harvard.edu/primerbank/) listed in Supplemental Table 1. All expression levels were normalized to β-actin.
2.10. Statistics analysis
The values were checked for normality and homogeneity of variance using Kolmogorov–Smirnov one-sample test and Levene’s test, respectively. When necessary, the data were transformed for normalization and to reduce heterogeneity of variance. Intergroup differences were assessed by student’s t-test using the StatView 5.0 program (SAS Institute Inc., Cary, NC, USA), *p < 0.05, **p < 0.01. The column charts were drawn by Origin 8.0 software (OriginLab, MA).
3. Results
3.1. Both Aire and Deaf1 are expressed by mES cells
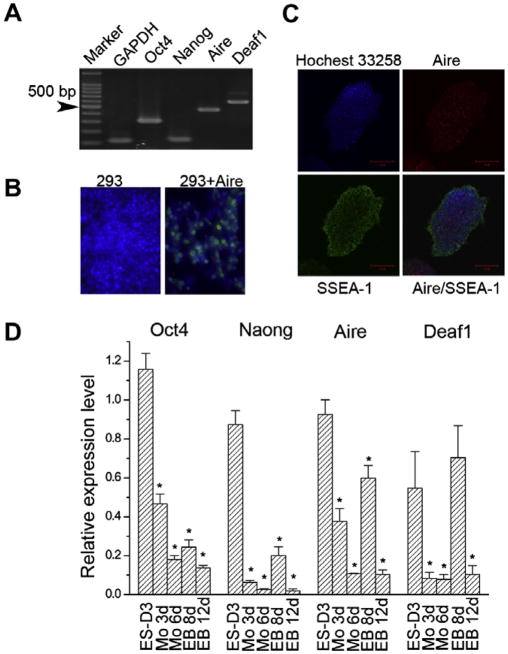
The broad expression of differentiation-associated genes suggests that ES cells exhibit promiscuous gene expression [11,17]. Such a phenomenon resembles the promiscuous expression of tissue-restricted self-antigens in mTECs and PLN stromal cells. Therefore we examined the expression of Aire and Deaf1 in mES cells. As shown in Fig. 1A transcripts of both Aire and Deaf1 were detected in undifferentiated mES cells (Fig. 1A). As expected, two ES cell markers, Oct4 and Nanog, along with the house-keeping gene GAPDH, were also expressed in mES cells from the same sample.
Fig. 1.
Expression of Aire and Deaf1 by mES cells. (A) Expression of Aire and Deaf1 on mES cells shown by RT-PCR. (B) Demonstration of the specificity of anti-Aire antibody, 293: naive 293 cells; 293 + Aire: 293 cells overexpressing Aire. (C) mES cells double stained for SSEA-1 (FITC) and Aire (Alexa555). (D) The expression of Aire and Deaf1 decreased with the differentiation of mES cells shown by Real-time RT-PCR (n = 4).
For detecting Aire protein, we prepared a polyclonal antibody against murine Aire. The specificity of the antibody was demonstrated using HEK-293 cells overexpressing Aire (Fig. 1B). Immunofluorescence was performed on mES cells using antibodies against Aire (green)2 and SSEA-1 (red). As shown in Fig. 1C, Aire co-stained with SSEA-1 in undifferentiated mES cells. The results indicate that Aire is expressed in undifferentiated mES cells.
To understand the effect of mES cell differentiation on the expression of Aire and Deaf1, we examined the expression of Aire and Deaf1 during the process of spontaneous mES cell differentiation by Real-time RT-PCR. As shown in Fig. 1D, the expression of Aire and Deaf1 both gradually attenuated upon differentiation in a time-dependent manner, showing same trend with Oct4 and Nanog. The results concerning Aire and Deaf1 expression were demonstrated in three distinct mES cell lines including D3, ZJ1, and ZJ2, although only the results obtained from D3 cells are shown here.
3.2. Aire regulates the expression of differentiation-associated genes in mES cells
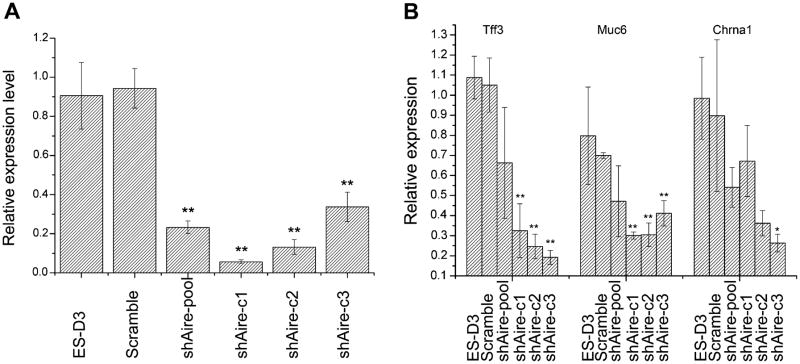
To investigate whether Aire regulates the expression of differentiation-associated genes in mES cells in a manner similar to in mTECs, we knocked down Aire expression using lentivirus-mediated RNA interference. After transfection, we established three single cell-derived clones (shAire-c1, shAire-c2, and shAire-c3) and pooled the rest of the transfected cells. As shown in Fig. 2A, Real-time RT-PCR indicated that the expression of Aire was significantly attenuated, compared to the parental cells (D3) and cells transfected with a virus encoding scramble shRNA (Scramble), both in the heterogeneous pool of transfectants (shAire-pool) and in the three single cell-derived clones. By Real-time RT-PCR, we examined the effects of Aire knockdown on the expression of three differentiation-associated genes: Tff3 (intestine), Muc6 (stomach), and Chrna1 (muscle). We chose these genes for analysis since they are transcriptionally activated by Aire in mTECs [29–31]. As shown in Fig. 2B, two of them, Tff3 (intestine), Muc6 (stomach) consistently decreased after Aire knockdown, while the third, Chrna1, showed a trend of decreased expression, although only significant in one single cell clone (shAire-c3). These results indicate that Aire regulates the expression of some differentiation-associated genes in mES cells.
Fig. 2.
Aire controlled the expression of differentiation-associated genes in mES cells. (A) RNA interference of Aire in mES cells. (B) Aire knockdown consistently inhibited the expression of three differentiation-associated genes Tff3, Muc6, while the expression of Chrna1 only changed in one shAire clone (n = 4).
3.3. Aire function in sustaining self-renewal and proliferation of mES cells
Since global gene expression has been considered one of the mechanisms for pluripotency maintenance in ES cells [15,17,18] and the expression of Aire decreases with mES cells differentiation, we analyzed the effect of Aire knockdown on self-renewal and proliferation of mES cells. Aire knockdown significantly decreased the clonogenicity of the mES cells (Fig. 3A and B). To understand the underlying mechanism we examined the effects of Aire knockdown on the expression of the two core transcription factors associated with pluripotency, Oct4 and Nanog, by Real-time RT-PCR (Fig. 3C). We found that Aire knockdown significantly attenuated the expression of both Oct4 and Nanog. Based on these observations, we propose that Aire sustained the self-renewal of mES cells by maintaining the expression of pluripotency promoting transcription factors.
Fig. 3.
Aire promoted self-renewal and proliferation of mES cells. (A) Aire knockdown inhibited colony formation. (B) Statistical analysis of colony formation (n = 3) (C) Aire knockdown inhibited the expression of Oct4 and Nanog. (D) Aire knockdown inhibited cell proliferation. (E) Aire knockdown perturbed the cell cycle distribution, HDP: hyper diploid. (F) Statistical analysis of cell cycle distributions (n = 3).
In addition to decreased clonogenicity, the cells transfected with a virus encoding Aire shRNA produced colonies that were generally smaller than Scramble mES cells under identical growth condition (Fig. 3A). Therefore, we compared the proliferation rate among the parental, Scramble, and shAire mES cells by MTT assay. As shown in Fig. 3D, the compound proliferation rate of shAire cells was significantly slower.
To delineate the mechanism underlying the attenuated proliferation in Aire knockdown cells, we examined the cell cycle distribution of Aire knockdown cells. Aire knockdown resulted in a distortion of the cell cycle distribution pattern; the fraction of cells in S phase was decreased, while the fraction of cells in G2/M phase was significantly increased (Fig. 3E and F). Furthermore, Aire knockdown resulted in significant increase of hyperdiploid cells (HDP). Although only results from a single clone (shAire-c1) were presented as a representative, similar changes were also seen with other two clones. These findings suggest that Aire functions to sustain self-renewal and proliferation of mES cells by facilitating the normal cell cycle progression.
3.4. Aire and global gene expression in human ES (hES) cells
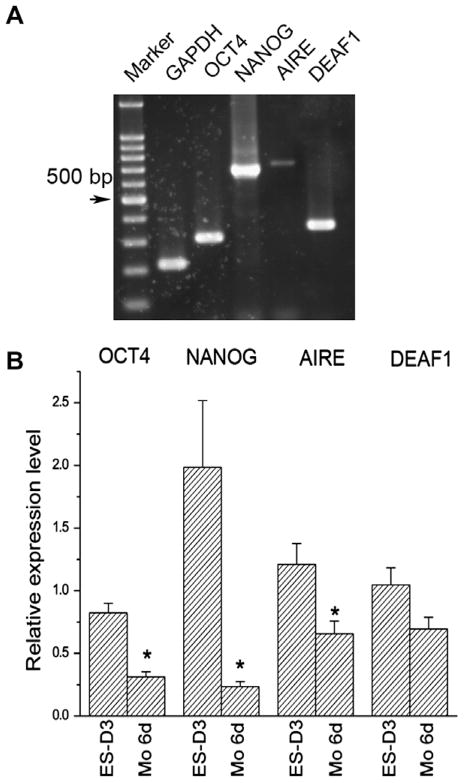
Since most genes in the hES genome are also in a transcriptional competent state [11], we assessed the expression of AIRE and DEAF1 in hES cells. As shown in Fig. 4A, undifferentiated hES cells expressed both AIRE and DEAF1 along with OCT4 and NANOG We also examined the effects of differentiation on the expression of AIRE and DEAF1 in hES cells by Real-time RT-PCR (Fig. 4B). The expression levels of the AIRE mRNA transcripts declined significantly with differentiation. DEAF1 expression also declined, although the decline was less prominent than the decrease in Aire expression (Fig. 4B). Taken together, our results support the idea that AIRE and DEAF1 may play significant roles in the regulation of global expression of differentiation-associated genes and self-renewal in hES cells.
Fig. 4.
Aire is expressed on hES cells (A) Expression of AIRE and DEAF1 on hES cells shown by RT-PCR. (B) The expression of AIRE and DEAF1 decreased with the differentiation of hES cells shown by Real-time RT-PCR (n = 4).
4. Discussion
The undifferentiated state of ES cells is thought to be partially maintained by sustaining the expression of pluripotency-associated genes and selective silencing of the lineage specific genes [4,5,32]. In contrast to that view, recent studies have shown that most of genes, including differentiation-associated genes in the genome were set in an expression competent state or expressed at low levels [11,12,17]. However, although mechanisms like high-level expression of epigenetic modifiers and chromatin remodelers that keep chromatin in an open state have been proposed to explain the global expression in ES cells [15,17], the underlying mechanisms are, for the most part, still poorly understood. In this study, we discovered that established mechanisms in mTECs and PLN stromal cells that control promiscuous gene expression may serve the same function in ES cells. We found that both mES and hES cells expressed Aire and Deaf1, and the expression decreased with differentiation of ES cells. Moreover, we demonstrated that Aire controlled the expression of some differentiation-associated genes in ES cells, like in mTECs. Aire had been shown to activate gene expression by several mechanisms, including facilitating transcriptional activation [33], promoting transcript elongation [34], stimulating transcription-activating epigenetic modifications [35], chromatin remodeling [36] and RNA maturation [36]. Thus, it is plausible that Aire promotes global gene transcription in ES cells by incorporating a variety of mechanisms at the epigenetical, transcriptional, and post-transcriptional levels.
The most exciting finding from the present study is the important role of Aire in the self-renewal and proliferation of ES cells. Although global expression of differentiation-associated genes has been considered an important mechanism for pluripotency maintenance in ES cells [15,17,18], such a model remains largely speculative. The theory is based on the substantial correlative observations, because it is so far unfeasible to perturb the global gene expression in ES cells. Since Aire is known to regulate promiscuous expression in medullary thymic epithelia cells, the defects in ES cells’ clonogenicity and proliferation observed in Aire knockdown cells are consistent with the notion that promiscuous gene expression is functionally important in pluripotency of ES cells.
5. Conclusions
In summary, we found that ES cells express Aire and Deaf1, two novel proteins mediating the promiscuous expression of tissue-restricted genes in lymphoid organs. We also found that Aire plays an important role both in the regulation of differentiation-associated gene expression and in self-renewal process in ES cells. Our studies suggest that mechanisms underlying the promiscuous expression of tissue-specific genes in the immune system may play a similar role in the very diverse gene expression in embryonic stem cells. Our results support the hypothesis that global expression of differentiation-associated genes is a mechanism for pluripotency maintenance in ES cells.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of the People’s Republic of China and the National Key Scientific Research Program of China (2007947804).
We thank Dr. David E. Root for his providing pLKO vectors, Dr. Didier Trono for his providing psPAX and pM2D.G vectors.
We thank Mr. Hong Yu and Miss Xiuli Song for technical support. We thank the Institute of Immunology, Zhejiang University for technical support in flow cytometry analyses. We thank Drs. Leif Nelin, Lyn Wancket, and Jing Yang for language editing and critical reading of the manuscript.
Abbreviations
- ES
embryonic stem
- mTECs
medullary thymic epithelial cells
- TRAs
tissue-restricted self-antigens
- HDP
hyperdiploid
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2010.03.042.
Footnotes
For interpretation of colour in Fig. 1, the reader is referred to the web version of this article.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 3.Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell. 1990;60:461–472. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- 4.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, Piao Y, Aiba K, Matoba R, Wang W, Ko MS. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 13.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 15.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 16.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, Gingeras TR, Misteli T, Meshorer E. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 20.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, Von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 21.Yip L, Su L, Sheng D, Chang P, Atkinson M, Czesak M, Albert PR, Collier AR, Turley SJ, Fathman CG, Creusot RJ. Deaf1 isoforms control the expression of genes encoding peripheral tissue antigens in the pancreatic lymph nodes during type 1 diabetes. Nat Immunol. 2009;10:1026–1033. doi: 10.1038/ni.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 23.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, Gu B, Wu R, Zhang J, Li Y, Zhang M. Development of a rabbit monoclonal antibody group against Smads and immunocytochemical study of human and mouse embryonic stem cells. Hybridoma (Larchmt) 2007;26:387–391. doi: 10.1089/hyb.2007.0517. [DOI] [PubMed] [Google Scholar]
- 25.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen LB, Calvin SA, Colvin KE, Wright M. FuGENE 6 Transfection Reagent: the gentle power. Methods. 2004;33:104–112. doi: 10.1016/j.ymeth.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 28.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 29.Kont V, Laan M, Kisand K, Merits A, Scott HS, Peterson P. Modulation of Aire regulates the expression of tissue-restricted antigens. Mol Immunol. 2008;45:25–33. doi: 10.1016/j.molimm.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giraud M, Taubert R, Vandiedonck C, Ke X, Levi-Strauss M, Pagani F, Baralle FE, Eymard B, Tranchant C, Gajdos P, Vincent A, Willcox N, Beeson D, Kyewski B, Garchon HJ. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 2007;448:934–937. doi: 10.1038/nature06066. [DOI] [PubMed] [Google Scholar]
- 31.Gavanescu I, Kessler B, Ploegh H, Benoist C, Mathis D. Loss of Aire-dependent thymic expression of a peripheral tissue antigen renders it a target of autoimmunity. Proc Natl Acad Sci USA. 2007;104:4583–4587. doi: 10.1073/pnas.0700259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitkanen J, Rebane A, Rowell J, Murumagi A, Strobel P, Moll K, Saare M, Heikkila J, Doucas V, Marx A, Peterson P. Cooperative activation of transcription by autoimmune regulator AIRE and CBP. Biochem Biophys Res Commun. 2005;333:944–953. doi: 10.1016/j.bbrc.2005.05.187. [DOI] [PubMed] [Google Scholar]
- 34.Oven I, Brdickova N, Kohoutek J, Vaupotic T, Narat M, Peterlin BM. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27:8815–8823. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Org T, Rebane A, Kisand K, Laan M, Haljasorg U, Andreson R, Peterson P. AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum Mol Genet. 2009;18:4699–4710. doi: 10.1093/hmg/ddp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abramson J, Giraud M, Benoist C, Mathis D. Aire’s partners in the molecular control of immunological tolerance. Cell. 2010;140:123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.