Abstract
Heroin addiction is a chronic complex disease with a substantial genetic contribution. This study was designed to identify genetic variants that are associated with susceptibility to develop heroin addiction, by analyzing 1350 variants in 130 candidate genes. All subjects had Caucasian ancestry. The sample consisted of 412 former severe heroin addicts in methadone treatment, and 184 healthy controls with no history of drug abuse. Nine variants, in six genes, showed the lowest nominal P values in the association tests (P < 0.01). These variants were in non-coding regions of the genes encoding the mu (OPRM1; rs510769, rs3778151), kappa (OPRK1; rs6473797), and delta opioid receptors, (OPRD1; rs2236861, rs2236857 and rs3766951), the neuropeptide galanin (GAL; rs694066), the serotonin receptor subtype 3B (HTR3B; rs3758987) and the casein kinase 1 isoform epsilon (CSNK1E; rs1534891). Several haplotypes and multi-locus genotype patterns showed nominally significant associations (e.g. OPRM1; P = 0.0006 and CSNK1E; P = 0.0007). Analysis of a combined effect of OPRM1 and OPRD1 showed that rs510769 and rs2236861 increase the risk of heroin addiction (P = 0.0005). None of these associations remained significant after adjustment for multiple testing. This study suggests the involvement of several genes and variants in heroin addiction that is worthy of future study.
Heroin addiction is a chronic relapsing disease characterized by compulsive drug seeking, drug abuse, tolerance and physical dependence. It is treated by methadone, buprenorphine and behavioral therapy. Heroin addiction is part of group of addictions (e.g. cocaine, alcohol and nicotine) that constitutes a worldwide public-health crisis. The genetic contribution to vulnerability to develop heroin addiction is 40–60%, suggesting a complex inheritance mode in which multiple genes exert a small effect, along with the environment (Kendler et al. 2003; Tsuang et al., 1996, 1998).
Several genetic variants have been shown to be associated with heroin addiction by family based linkage studies and association studies (for review see Kreek et al., 2005a, b, Kreek & LaForge, 2007 and also Cheng et al., 2005, Loh et al., 2007; Nielsen et al., 2008; Proudnikov et al., 2006; Szilagyi et al., 2005; Xu et al., 2004; Zou et al., 2007). These include variants in the genes encoding the mu and kappa opioid receptors, dopamine receptors D2 and D4, serotonin receptor 1B, GABA receptor subunit gamma 2, catechol-O-methyltransferase (COMT), period circadian protein (PER3), proenkephalin (PENK), proopiomelanocortin (POMC), tryptophan hydroxylase 2 (TPH2) and brain-derived neurotrophic factor (BDNF).
To identify genetic variants that underlie heroin addiction, we performed a candidate gene, case-control association study using a SNP array that was designed by the group of D. Goldman at the National Institute of Alcohol Abuse and Alcoholism (NIAAA). This approach is based on physiological hypotheses and the genes were selected based on their function (e.g. drug receptors, neurotransmitters, transporters and drug metabolism enzymes) and related pathways (e.g. reward modulation, behavioral control, cognitive function, signal transduction, and stress response). In order to maximize the power of the study, the cases were selected from the extreme margin of the specific phenotype range (e.g. severe heroin addicts in methadone maintenance treatment), and the controls were healthy volunteers that were selected by detailed personal interview and stringent criteria. To reduce the possible effect of population stratification, only subjects with Caucasian ancestry were included.
Materials and Methods
Subjects
Six hundred and twenty subjects participated in the study with the majority (90%) from USA (NYC and Las Vegas) and the minority from Israel with Jewish ancestry (cases only) (Table 1). Twenty-two DNA samples were too diluted or of low quality for genotyping and two samples were excluded from analysis based on low call rate or possible contamination. The patients (cases) were former severe heroin addicts treated at a methadone maintenance treatment program at the time of recruitment. Subjects were recruited at either the Rockefeller University Hospital or opiate substitution programs (e.g. Manhattan Campus of VA NY Harbor Health Care System, Weill Medical College of Cornell University, and the Dr. Miriam and Sheldon G. Adelson Clinics for Drug Abuse Treatment and Research, in Las Vegas and Israel). Ascertainment was made by personal interview, using several instruments: the Addiction Severity Index (ASI) (Mclellan et al., 1992), KMSK (Kellogg et al., 2003) and DSM-IV. All cases had a history of at least one year of daily multiple uses of heroin. The 184 healthy control subjects were recruited by posting of notices or referral by physicians. Each of the following was used as exclusion criteria from this category: a) At least one instance of drinking to intoxication, or any illicit drug use in the previous 30 days. b) A past history of alcohol drinking to intoxication, or illicit drug use, more than twice a week, for more than 6 consecutive months. c) Cannabis use for more than 12 days in the prior 30 days or past use for more than twice a week for more than 4 years. All subjects completed a family history questionnaire and were self-identified as Caucasians for three generations. Participants were excluded from the study if they had a relative in the study or if they had a mixed ancestry. The Institutional Review Boards of The Rockefeller University Hospital, the VA New York Harbor Healthcare System and the Tel-Aviv Sourasky Medical Center (Helsinki Committee), approved the study. All subjects signed informed consent for genetic studies.
Table 1.
Population demographic
| Cases |
Controls US | Total | |||
|---|---|---|---|---|---|
| US | Israel | Total | |||
| Males | 221 | 37 | 258 | 86 | 344 |
| Females | 129 | 25 | 154 | 98 | 252 |
| Total | 350 | 62 | 412 | 184 | 596 |
DNA and plates preparation
Blood samples were taken and DNA was extracted using the standard salting-out method (Miller et al., 1988). DNA was quantified using PicoGreen (Invitrogen, Carlsbad, CA).700 ng DNA (45 μL) was precipitated with ethanol by the following procedure: a 120 μl “ethanol mix” (4.5 μl of 3M sodium acetate, pH 4.6; 105 μl of ethanol, 100%; 10.5 μl of H2O and 0.044 μl of glycogen, 5 mg/ml) was dispensed into each well. The plate was sealed, vortexed and incubated at room temperature for 15 min. The plate was then spun at 3700 rpm (2400 g) for 30 min. The plate was inverted onto paper towels, followed by a short spin with the plate inverted, for 1 minute at 530 rpm (50 g). DNA pellets were washed with 150 μl 70% ethanol, followed by re-sealing and inverting the plate a few times. A spin at 3700 rpm for 10 min was followed by the inverting procedure (as described above), and the DNA was air dried for 15 min and re-suspended in 6–7 μl Tris-EDTA (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). It was then stored at 4°C for up to 2 days, or at −20°C, for a longer period.
Genotyping and quality assessment
Genotyping was performed on a 1,536-plex GoldenGate Custom Panel (GS0007064-OPA, Illumina, San Diego, CA). The micro-array was designed by Dr. D. Goldman’s group, NIAAA). 1350 SNPs were selected from 130 genes implicated in addiction (Supplement Table 1). In addition, the array includes 186 ancestry informative markers (AIMs) that were selected based on allele frequencies in the European, African and Chinese population of the HapMap project (Enoch et al., 2006, Supplement Table 1). Genotyping was performed at the Rockefeller University Genomics Resource Center according to the manufacture’s protocol (Illumina). Analysis was performed using BeadStudio genotyping software (Illumina) that provides automated genotype calling, cluster separation score (0–1) and quality check. Questionable SNPs were visually inspected and the genotype was determined. Genotype data was filtered based on SNP call rates (> 99.5%), MAF > 0.01, and deviation from Hardy-Weinberg Equilibrium (HWE) (P = 0.00005). Seventy-seven random samples were genotyped in duplicates and two identical samples were genotyped on each of the arrays for reproducibility control purposes.
Statistical analysis
Population stratification
Ancestry informative markers were employed to test for population stratification using the Structure software v2.2 (Pritchard et al., 2000). This software places populations in K clusters that have distinct marker frequencies. A no-admixture ancestry model with K set to 1 was used to infer the Dirichlet parameter, lambda, of the distribution of allele frequencies. The value of lambda thus obtained was later used under an admixture model with allele frequencies correlated using a burn-in length of 50,000 followed by 50,000 Markov Chain Monte Carlo iterations, which was sufficient to yield stable results. The genomic control method (Devlin et al., 2001) was also used to examine the potential impact of population structure in this study. In this method, markers are used to estimate a background association that can be used to infer the variance inflation factor, lambda, which is introduced when there is subdivision.
Association Analysis
Fisher’s exact test, as implemented in R v2.6.1 (The R Development Core Team 2007, http://www.R-project.org), was used to compare the distribution of marker alleles and genotypes in cases and controls as well as to estimate odds ratios (OR) with their corresponding 95% confidence intervals. Analyses were carried out using both US and Israel cases combined and US cases alone against the US controls. An exact test of difference in allelic proportions between the Israel and US cases was performed to account for any underlying allele frequency differences between these two samples. An uncorrected P value of 0.01 was considered significant for the association test.
Allelic correlations at the locus level were estimated using the exact test of deviation from the Hardy-Weinberg (DHW) proportions as implemented in the R package GeneticsBase v1.2.0 from the R genetics project (www.rgenetics.org). A P value of 0.05/1000 = 0.00005 was considered significant for the test of DHW.
Linkage Disequilibrium (LD) Blocks, Haplotype Analysis and Genotype Patterns
The pattern of pair-wise LD between SNPs was measured by the standardized disequilibrium value (D′), as implemented in Haploview v3.32 (Barrett et al., 2005). LD blocks were identified using the Gabriel’s method (Gabriel et al., 2002). The Score test was used to test for association between the LD blocks and disease state as implemented in the program haplo.score (using simulated p-values) from the R package haplo.stats v1.3.1 (Schaid et al., 2002). Differences in genotype pattern frequencies between cases and controls were evaluated by using Fisher’s exact test. Genotype patterns with frequencies less than 5% were pooled into a single class.
Correction for Multiple Testing
Results were adjusted for multiple testing by using the R package QVALUE v1.1 (Storey, 2003; Storey & Tibshirani, 2003, Storey et al., 2004). Instead of controlling the probability of one or more false positives in a family of tests (the family-wise error rate), QVALUE controls the expected proportion of false positives among all rejected hypotheses (the false discovery rate - FDR) (Benjamini & Hochberg, 1995). QVALUE takes a given set of P values and, for each test, estimates the minimum FDR that is incurred when calling a particular test significant (the q-value of the test). The q-value measures the significance of each of a family of tests performed simultaneously and holds under different forms of dependence.
The smallest nominal P value, of all tests performed (Pmin) was obtained from the multi-locus genotype pattern tests. The QVALUE program was run on the list of P values created by adding Pmin to the set of P values obtained from the single-locus tests. This estimated the experiment-wise significance of Pmin. An FDR of 0.05 was used as the significant level.
Results
One thousand three hundred fifty SNPs, from 130 candidate genes, were genotyped in 412 former severe heroin addicts, currently treated at a methadone maintenance treatment program (cases), and 184 healthy volunteers, with no history of drug abuse (controls, see methods). All individuals self reported Caucasian ancestry. The majority of cases (90%) were from the US (NYC and LV) and the minority from Israel (Table 1). Two hundred seventy five SNPs were excluded because of inadequate quality or low variability in our sample (MAF < 0.01) (Table 2). Genotyping reproducibility was 99.9%. The mean marker call rate for the analyzed markers was 99.8%. Nine SNPs showed significant deviation from HWE in controls (rs10809907, rs12473028, rs1426223, rs2014663, rs3779084, rs483021, rs7103679, rs6432224 and rs7785096) (P < 0.00005).
Table 2.
Summary of genotyped SNPs
| SNPs | Genotyped | MAF < 0.01 | Poor Quality | Analyzed |
|---|---|---|---|---|
| Auto-somal | 1261 | 197 | 61 | 1003 |
| X-Chr. | 89 | 3 | 6 | 80 |
| AIMs | 186 | 1 | 7 | 178 |
| Total | 1536 | 201 | 74 | 1261 |
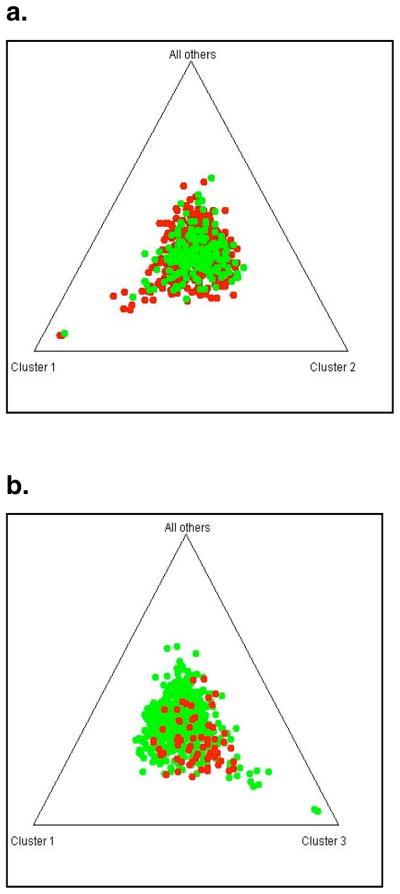
Using data from 178 ancestry-informative markers (AIMs), no evidence of population stratification was found within whole dataset (Fig. 1a), or within the Israeli and the US cases dataset (Fig. 1b), using the Structure program and the genomic control method (Lambda=1.00). To account for any subtle underlying difference that may not be detected by the AIMs used in this study, allele frequencies of all analyzed SNPs were compared between the US and the Israeli cases. The frequencies of three SNPs were significantly different using the Bonferroni correction (P < 0.00005), and of 18 SNPs (1.8%) using the q-value approach (q < 0.05).
Figure 1. Population stratification analysis.
a. Cases (US and Israel) (in red) and controls (in green).
b. Cases from the US (in green) and Israel (in red).
Genotypes and allele frequencies of 1083 SNPs were analyzed for association with heroin addiction. The analysis was performed in two steps: 1) 350 US cases and 184 US controls 2) 412 cases (350 US and 62 Israeli) and 184 US controls. The SNPs with significant different frequency between the US and the Israeli cases were excluded from this step. Listed in Table 3 are the SNPs that gave the lowest P values in the combined group (P < 0.01). The P values are given for the allele and genotype tests for the US only and the combined cases. Six SNPs gave nominal significant values in both analyses (Table 3, group 1). Three SNPs gave nominal significant values in the combined cohort and borderline values for the US cases only (group 2). Six SNPs gave nominal significant values in the US only group (Supplement Table 2). One of these SNPs (rs1229984) also showed the most significant difference in allelic proportions between the Israeli and the US cases (P = 7.2E-10). It is a nonsynonymous variant (Arg47His) in the alcohol dehydrogenase 1B gene (ADH1B) that is reported to be common in Asian and Jewish populations and rare in Caucasians (Shibuya & Yoshida, 1988, Thomasson et al., 1991, Neumark et al., 1998, Carr et al., 2002).
Table 3.
The most significant associations of single SNPs with heroin addiction
| a. | |||||||
|---|---|---|---|---|---|---|---|
| SNP | Gene | Allele test | Genotype testa | Effect of the minor allele | |||
| P - Value | OR (95% CI) | P- Value | |||||
| US only | Combined | Combined | US only | Combined | |||
| Group 1 | |||||||
| rs510769 | OPRM1 | 0.0003 | 0.0008 | 1.7 (1.2–2.2) | 0.001 | 0.0029 | risk |
| rs3778151 | OPRM1 | 0.0007 | 0.003 | 1.7 (1.2–2.4) | 0.003 | 0.015 | risk |
| rs6473797 | OPRK1 | 0.0009 | 0.004 | 0.7 (0.5–0.9) | 0.003 | 0.011 | protection |
| rs1534891 | CSNK1E | 0.0016 | 0.002 | 0.6 (0.4–0.8) | 0.002 | 0.001 | protection |
| rs694066 | GAL | 0.0019 | 0.001 | 2.1 (1.3–3.6) | 0.006 | 0.004 | risk |
| rs2236861 | OPRD1 | 0.0029 | 0.002 | 1.6 (1.7–2.2) | 0.003 | 0.003 | risk |
| Group 2 | |||||||
| rs3766951 | OPRD1 | 0.0125 | 0.009 | 1.4 (1.1–1.9) | 0.040 | 0.030 | risk |
| rs2236857 | OPRD1 | 0.0165 | 0.008 | 1.5 (1.1–1.9) | 0.019 | 0.005 | risk |
| rs3758987 | HTR3B | 0.0174 | 0.007 | 1.5 (1.1–2.0) | 0.049 | 0.019 | risk |
| b. SNP location | |||||
|---|---|---|---|---|---|
| SNP | Gene Symbol | Gene | Gene Location | Chr. | Position Build 36.2 |
| rs510769 | OPRM1 | mu opioid receptor | intron 1 | 6 | 154403712 |
| rs3778151 | OPRM1 | mu opioid receptor | intron 1 | 6 | 154435373 |
| rs6473797 | OPRK1 | kappa opioid receptor | intron 2 | 8 | 54315535 |
| rs1534891 | CSNK1E | casein kinase 1, epsilon | intron 7 | 22 | 37025045 |
| rs694066 | GAL | galanin | intron 2 | 11 | 68209561 |
| rs2236861 | OPRD1 | delta opioid receptor | intron 1 | 1 | 29012343 |
| rs2236857 | OPRD1 | delta opioid receptor | intron 1 | 1 | 29034196 |
| rs3766951 | OPRD1 | delta opioid receptor | intron 1 | 1 | 29042146 |
| rs3758987 | HTR3B | serotonin receptor 3B | −381 | 11 | 113280485 |
additive mode of inheritance.
OR were calculated for the minor allele. SNPs rs6473797 and rs1534891, showed a protective effect (Table 3a). Listed in Supplement Table 3 are the alleles and genotype counts and frequencies in cases and controls. No SNP on chromosome X was significantly associated with heroin addiction. Six association tests for haplotype and three tests for Multi locus genotype patterns (MLGPs) that include SNPs with the top signals, gave significant positive values (Table 4a,b).
Table 4.
Haplotype and genotype patterns association with heroin addiction
| a. Haplotypes | ||||
|---|---|---|---|---|
| SNPs | Gene | Haplotypea | P-value | Effect |
| rs510769-rs3778151 | OPRM1 | CT | 0.0006 | protection |
| rs1534891-rs6001093-rs135757 | CSNK1E | AGT | 0.0007 | protection |
| rs1893679-rs694066 | GAL | GA | 0.0012 | risk |
| rs510769-rs3778151 | OPRM1 | TC | 0.0033 | risk |
| rs3758987-rs11606194 | HTR3B | TT | 0.0064 | protection |
| rs204055-rs2236857-rs2298896 | OPRD1 | TCG | 0.0067 | risk |
| b. Multi locus genotype patterns | ||
|---|---|---|
| SNPs | Gene | P-value |
| rs510769-rs3778151 | OPRM1 | 0.005 |
| rs2236857-rs3766951 | OPRD1 | 0.013 |
| rs2236861-rs510769 | OPRD1/OPRM1 | 0.0005 |
Alleles are in the sense orientation of the relevant gene.
SNPs that gave significant P-values on single SNP analysis (Table 3) are in bold.
The top signals were from SNPs in the following genes: the opioid receptors mu, kappa and delta, galanin, the 3B subtype of the serotonin receptor, and the casein kinase epsilon. The information on the position of these SNPs is shown in Table 3b.
Mu opioid receptor
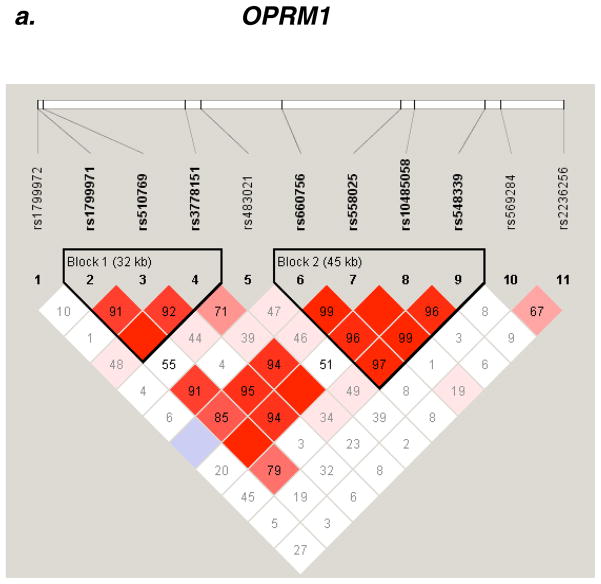
Two OPRM1 variants (rs510769, rs3778151) accounted for the two strongest signals in the association test (P = 0.0008, 0.003, respectively, Table 3). These SNPs are in strong LD (D′ = 0.92) and belong to a haplotype block of 32 kb (block 1, Fig. 2a). The two SNPs are located in intron 1, adjacent to rs1799971 (118A>G) on exon 1. SNP rs1799971 belongs to the same haplotype block (D′ = 0.91 with rs510769, D′ = 1 with rs3778151, Fig. 2a). No evidence for association was found with SNP rs1799971 (118A>G) in this cohort (P = 0.16, MAF = 0.12).
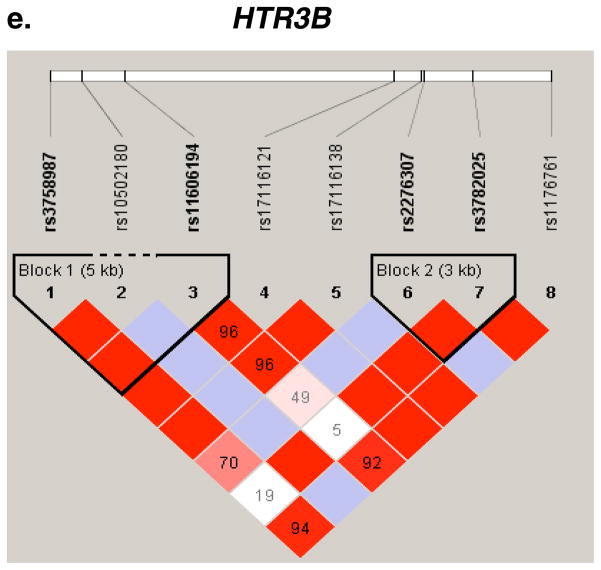
Figure 2.
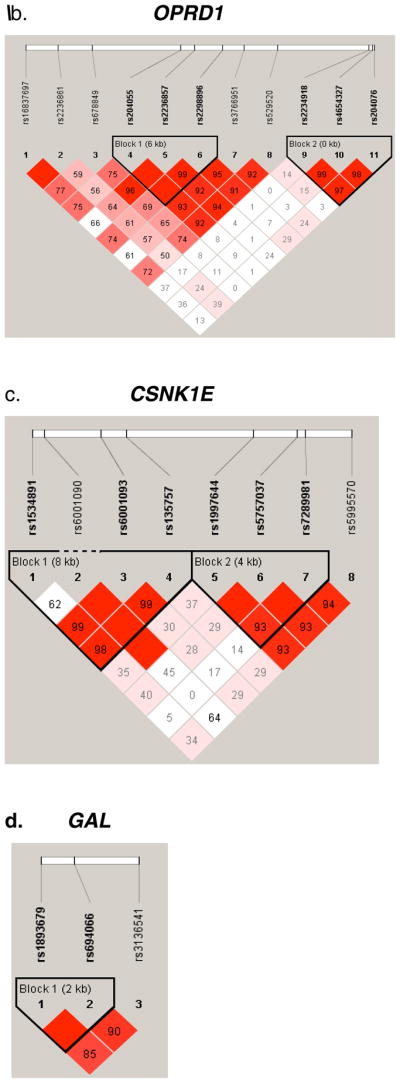
Pair-wise LD Analysis. LD between SNPs in seven genes was derived from genotypes of controls. The pair-wise correlation between SNPs was measured as D′ and is shown (x100) in each box. The color scheme indicates the magnitude of D′. Dark red indicated D′ >0.80 with D′ =1.0 when no number is given. Haplotypes were generated using the Gabriel rule and haplotype blocks are marked. a, OPRM1. b, OPRD1. c, CSNK1E. d, GAL. e, HTR3B.
Haplotype analysis, of OPRM1 SNPs rs510769 and rs3778151, suggested association of haplotypes CT (protection) and TC (risk) (P = 0.0006, 0.0033, respectively, Table 4a). Multi locus genotype patterns (MLGPs) analysis, of OPRM1 SNPs rs510769 and rs3778151, revealed significant differences in the genotypes distribution between cases and controls. Particularly, the CC-TT genotype pattern (rs510769-rs3778151, respectively) appears to be protective (P = 0.005, Table 4b).
Delta opioid receptor
Three OPRD1 SNPs (rs2236861, rs2236857 and rs3766951) showed suggestive associations with heroin addiction (P = 0.002, 0.008 and 0.009, respectively, Table 3). These SNPs are common and are located within a 30 kb region of intron 1. SNP rs2236857 is in strong LD with SNPs rs3766951 and rs678849 (Fig. 2b). An additional SNP in this region, rs2298896, that showed a trend for association (P = 0.015), is in strong LD with SNPs rs2236857 and rs3766951 (Fig. 2b). A 6 kb haplotype block, that includes SNPs rs204055, rs2236857 and rs2298896, showed suggestive association with heroin addiction (P = 0.0067, Table 4a). The genotype pattern distribution of the two loci rs2236857-rs3766951 was significantly different between cases and controls (P = 0.01, Table 4b).
Combined effect of the Mu and delta opioid receptors
In order to evaluate whether there is a combined effect of OPRM1 and OPRD1 variants, the genotypes pattern of rs510769 (OPRM1) and rs2236861 (OPRD1) was compared between cases and controls. The analysis revealed significant difference of the genotypes pattern between cases and controls (P = 0.0005, Table 4b).
Kappa opioid receptor
One common SNP (rs6473797) in OPRK1 showed suggestive association (P = 0.004, OR = 1.5 for the major C allele, Table 3a, Supplement Table 3). LD analysis with other OPRK1 SNPs, in our cohort, revealed that this SNP does not belong to any haplotype block.
Casein kinase 1 epsilon
A tentative association was also detected with SNP rs1534891 in CSNK1E with protection from heroin addiction (P = 0.001, Table 3a, Supplement Table 3). Two additional SNPs (rs6001093 and rs135757) showed a trend for association (P = 0.016, 0.019, respectively). Haplotype analysis revealed association with protection from heroin addiction of an 8 kb haplotype block (P = 0.0007, Table 4a, Fig. 2c).
Galanin
An association was detected for the SNP rs694066 in the galanin gene. (P = 0.001, Table 3). Haplotype analysis revealed suggestive association of a 2 kb haplotype block with heroin addiction (P = 0.0027, Table 3a, Fig. 2d).
Serotonin receptor 3B
A SNP (rs3758987) in the 5′ region (−381) near the serotonin receptor gene (HTR3B) gave a nominally significant signal for association (P = 0.007, Table 3). One additional SNP in this gene (rs11606194, in intron 2) showed a trend toward significance (P = 0.017). SNP rs3758987 is in complete LD with rs11606194 (Fig. 2e). A 5 kb haplotype block, spanning a region from 5′ upstream of the gene to intron 2, showed association with protection from heroin addiction (P = 0.0064, Table 4a).
Discussion
Our primary goal in this study was to identify variations, in candidate genes, contributing to vulnerability to heroin addiction. Our top signals are from genes encoding opioid receptors, a neuropeptide (galanin), a ligand-gated ion channel serotonin receptor, and a kinase. All the variants identified are from non-coding regions. As expected for a complex genetic disorder, each variant shows a small effect on the risk to (or protection from) develop heroin addiction.
The opioid system plays a central role in reward, drug craving and relapse, in part by altering stress physiology (for review see Koob and Kreek, 2007). Genes encoding opioid receptors are prime candidates for opioid addiction. The mu-opioid receptor is a G protein coupled receptor for beta-endorphin, and the major target for heroin and opioid analgesia. It may be a common component of several types of addictions by mediating drug reward. The OPRM1 gene variant 118A>G (Asn40Asp, rs1799971), was associated with increased affinity to beta-endorphin (Bond et al., 1998), increased basal levels of cortisol (Bart et al., 2006), exaggerated hypothalamic-pituitary-adrenal axis response to a mu-opioid receptor antagonist, naloxone, in healthy individuals (e.g. Wand et al., 2002) and enhanced opiate abuse vulnerability (Drakenberg et al., 2006), among other functions. 118A>G was associated with heroin addiction and alcohol dependence in certain populations, in some studies, but not in others (for review see Glatt et al., 2007; Kreek et al., 2005a, b). Association was also found between two OPRM1 haplotype blocks and drug or alcohol dependence in Caucasians, and between 3 tag SNPs and the response to heroin, in the first use, in Han Chinese (Zhang et al., 2006, 2007a). Hoehe et al., (2000) identified a haplotype pattern in the 5′ regulatory region that is associated with poly-substance dependence.
We did not detect association of 118A>G with heroin addiction in this diverse cohort of European origin. However, an association was suggested with two OPRM1 SNPs (rs510769, rs3778151) in intron 1, that are part of a haplotype block, that spans the 118A>G region. This block is similar to the block previously described (Zhang et al., 2007a). There is no evidence that these SNPs are causative, and they might be in LD with a causative variant.
The delta opioid receptor has been shown, in animal studies, to be involved in addictive processes, emotional response and antinociception (Quock et al., 1999; Filliol et al., 2000; Roberts et al., 2001; Nieto et al., 2005). The OPRD1 gene has only two coding sequence polymorphisms, the non-synonymous rs1042114 and the synonymous rs2234918. An association between rs2234918 and heroin dependence was reported in German Caucasians, but was not replicated by other studies (Franke et al., 1999; Mayer et al., 1997; Xu et al., 2002). This variant did not give a significant signal in our study (P > 0.5). In a recent study, significant association with opioid dependence was obtained for several OPRD1 markers (Zhang et al., 2007b). Our results imply association of three OPRD1 SNPs (rs2236861, rs2236857, and rs3766951) with heroin addiction. Further analysis is required to verify the role of these SNPs and potential additional SNPs on the risk haplotype.
The combined effect of OPRM1 and OPRD1 indicated in this study is intriguing considering the biological interaction of these receptors which are known to form heterodimers as well as homodimers (Gomes et al., 2000, Rozenfeld & Devi, 2007). It also fits the hypothesis of complex inheritance mode in which multiple genes exert a small effect.
The kappa opioid receptor (OPRK1) is activated by opioid neuropeptides to modulate dopaminergic tone. OPRK1 was also shown to be involved in pain sensitivity and response to stress, in mice (Simonin et al. 1998, Mclaughlin et al., 2003). The OPRK1 gene is comprised of four exons, with only two rare non-synonymous variants. In a report from our laboratory, Yuferov et al., found a point-wise significant association of the synonymous SNP rs1051660 (36G>T) with opioid addiction (Yuferov et al., 2004). Similar results were obtained in an Italian population (Gerra et al., 2007). Two studies of European-Americans found significant association of multiple SNPs (including rs6473797) with alcohol dependence (Xuei et al., 2006), but these results were not replicated in a Taiwanese population (Loh el et al., 2004). SNP rs1051660 (36G>T) was not included in our panel.
Galanin is a 30-amino acid peptide widely distributed in the peripheral and central nervous systems. Galanin and its receptors were shown to be involved in behavioral processes, morphine withdrawal, behavioral and neurochemical effects of opiates and high stress response, among others (Belfer et al., 2006, Hawes et al., 2007, Holmes & Picciotto, 2006, Picciotto et al., 2005, Unschuld et al., 2007, Zachariou et al., 2003). Galanin is considered a candidate for a protective factor against opiate addiction and galanin receptor agonists have been suggested as therapeutic targets. A haplotype association with alcoholism, in Finnish and Native American males, was reported, (Belfer et al., 2006). The variant rs694066 was shown to be associated with weight loss (Ruano et al., 2006). This is the first report indicating association between a GAL variant and opioid addiction. SNP rs694066 is located at intron 2, with relatively low MAF in our control population (0.06).
Serotonergic neurotransmission plays a major role in the regulation of affective states, pain perception, reward, anxiety and addiction (Soubrie et al., 1986; Lucki et al., 1998). Serotonin receptors affect the release of other neurotransmitters such as glutamate, dopamine and GABA. The serotonin receptor 3, subunit B (HTR3B) belongs to the super-family of ligand-gated ion channels and is co-expressed with subunit HTR3A in the amygdala, caudate and the hippocampus. A recent study showed association between a HTR3B haplotype block and major depression in Japanese women (Yamada et al., 2006). The variant rs3758987, that gave a nominally significant signal for association in our analysis, is located at the regulatory region (−381C>T) and may impact gene expression.
Casein kinase 1 epsilon (CSNK1E) is the human homolog of the Drosophila circadian-associated protein DOUBLETIME (Kloss et al., 1998). Observations in animal models suggest interactions between the brain circadian and reward systems (see commentary by Yuferov et al., 2005). For example, it was shown in mice that another circadian-associated gene (clock) regulates the brain rewarding response to cocaine (McClung et al., 2005). CSNK1E participates in important signaling pathways: a) Phosphorylation of DARPP-32 (the dopamine-and cyclic AMP regulated phosphoprotein) that mediates the behavioral effects of serotonin (Svenningsson et al., 2002); b) Modulation of the activation of glycogen synthase kinase-3 (GSK-3), a downstream target of DARPP-32 that is important in stimulant drug response (Beaulieu et al., 2004); c) Regulation of the circadian clock gene PER1, whose expression has been linked to drug dependence and reward (Yuferov et al., 2003, Liu et al., 2005, 2007). A recent study found a different expression level of Csnk1e in mice with low- and high- sensitivity to locomotor stimulation that is a characteristic response to drugs of abuse (Palmer et al., 2005). A follow up study showed association between CSNK1E SNP rs135745 and sensitivity to the euphoric effects of amphetamine, in healthy human volunteers, suggesting that this variant might facilitate the development of drug abuse (Veenstra-Vanderweele et al., 2006). Interestingly, SNP rs1534891 that shows association with protection from heroin addiction, in our study, is in close proximity (8 kb) to rs135757 and in almost complete LD. The two SNPs are part of the same haplotype block, suggesting an interesting similarity between the findings of the two studies.
Evaluation of previous studies in relation to this study is complex for a few reasons; some variants from previous reports were not genotyped in this study, the phenotype was often different (e.g. multiple dependencies, less stringent criteria for defining specific addiction or different criteria for controls definition), or the population was different. On the other hand, alcohol, cocaine and heroin dependence may share genetic influences, and some populations may share similar risk variants, that justify comparison between studies. We cannot determine if the negative findings in this study are true negatives because the SNP coverage of some of the genes may be insufficient to exclude real associations, and due to the relatively small number of subjects.
The candidate gene approach has advantages and disadvantages. It is hypothesis-driven, based on genes, with a known or inferred biological function. As such, it is limited by the current knowledge. In contrast, genome-wide association study is an exploratory approach in which the whole genome is scanned in an unbiased fashion and as such, it has the potential to identify novel susceptibility factors. The candidate gene approach allows a scan of a limited number of SNPs thus it is more economical and requires less testing.
This study suggests an extension to the list of susceptibility genes and variants underlying heroin addiction. These results warrant further exploration to confirm the tentative associations and to elucidate the involved mechanisms. Although the variants identified in this study are suggested to produce low relative risk for the vulnerability to develop heroin addiction, they may uncover novel mechanisms of addictive behavior.
Supplementary Material
Acknowledgments
We thank all the clinical staff that enrolled and assessed subjects for this study, including Elizabeth Ducat, N.P., Brenda Ray, N.P., Dorothy Melia, R.N., Lisa Borg, M.D., and Scott Kellogg, M.D. We are grateful to David Goldman M.D. and his group from the NIH/NIAAA, for the design of the micro-array and their support; Connie Zhao, Ph.D. and Bin Zhang from the Rockefeller Genomic Resource Center, for their excellent assistance in genotyping. We thank Ann Ho, Ph.D. and Vadim Yuferov, Ph.D. from our laboratory for discussion and comments on the manuscript. We would like to express our profound gratitude to the late K. Steven LaForge, Ph.D. for his invaluable role in setting the foundation for this study. This work was supported in part by NIDA-P60-05130 (MJK), NIDA-K05-00049 (MJK), NIH/NCRR-CTSA UL1-RR024143 (The Rockefeller University Center for Clinical and Translational Science), NIMH-R01-44292 (JO) and NSFC grant 30730057 from the Chinese Government (JO).
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bart G, LaForge KS, Borg L, Lilly C, Ho A, Kreek MJ. Altered levels of basal cortisol in healthy subjects with a 118G allele in exon 1 of the Mu opioid receptor gene. Neuropsychopharmacol. 2006;31:2313–2317. doi: 10.1038/sj.npp.1301128. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Sotnikova TD, Yao WD, Kockeritz L, Woodgett JR, Gainetdinov RR, Caron MG. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase 3 signaling cascade. Proc Natl Acad Sci U S A. 2004;101:5099–5104. doi: 10.1073/pnas.0307921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfer I, Hipp H, McKnight C, Evans C, Buzas B, Bollettino A, Albaugh B, Virkkunen M, Yuan Q, Max MB, Goldman D, Enoch MA. Association of galanin haplotypes with alcoholism and anxiety in two ethnically distinct populations. Mol Psychiatry. 2006;11:301–311. doi: 10.1038/sj.mp.4001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, Tischfield JA, Kreek MJ, Yu L. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J of the Roy Statist Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- Carr LG, Foroud T, Stewart T, Castelluccio P, Edenberg HJ, Li TK. Influence of ADH1B polymorphism on alcohol use and its subjective effects in a Jewish population. Am J Med Genet. 2002;112:138–143. doi: 10.1002/ajmg.10674. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- Drakenberg K, Nikoshkov A, Horvath MC, Fagergren P, Gharibyan A, Saarelainen K, Rahman S, Nylander I, Bakalkin G, Rajs J, Keller E, Hurd YL. Mu opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. Proc Natl Acad Sci U S A. 2006;103:7883–7888. doi: 10.1073/pnas.0600871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20:19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- Filliol D, Ghozland S, Chluba J, Martin M, Matthes HW, Simonin F, Befort K, Gaveriaux-Ruff C, Dierich A, LeMeur M, et al. Mice deficient for delta- and mu-opioid receptors exhibit opposing alterations of emotional responses. Nat Genet. 2000;25:195–200. doi: 10.1038/76061. [DOI] [PubMed] [Google Scholar]
- Franke P, Nothen MM, Wang T, Neidt H, Knapp M, Lichtermann D, Weiffenbach O, Mayer P, Hollt V, Propping P, Maier W. Human delta-opioid receptor gene and susceptibility to heroin and alcohol dependence. Am J Med Genet. 1999;88:462–464. doi: 10.1002/(sici)1096-8628(19991015)88:5<462::aid-ajmg4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gerra G, Leonardi C, Cortese E, D′Amore A, Lucchini A, Strepparola G, Serio G, Farina G, Magnelli F, Zaimovic A, Mancini A, Turci M, Manfredini M, Donnini C. Human kappa opioid receptor gene (OPRK1) polymorphism is associated with opiate addiction. Am J Med Genet B Neuropsychiatr Genet. 2007;144:771–775. doi: 10.1002/ajmg.b.30510. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Bousman C, Wang RS, Murthy KK, Rana BK, Lasky-Su JA, Zhu SC, Zhang R, Li J, Zhang B, Li J, Lyons MJ, Faraone SV, Tsuang MT. Evaluation of OPRM1 variants in heroin dependence by family-based association testing and meta-analysis. Drug Alcohol Depend. 2007;90:159–165. doi: 10.1016/j.drugalcdep.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes I, Jordan BA, Gupta A, Trapaidze N, Nagy V, Devi LA. Heterodimerization of mu and delta opioid receptors: A role in opiate synergy. J Neurosci. 2000;20:RC110. doi: 10.1523/JNEUROSCI.20-22-j0007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes JJ, Brunzell DH, Narasimhaiah R, Langel U, Wynick D, Picciotto MR. Galanin Protects Against Behavioral and Neurochemical Correlates of Opiate Reward. Neuropsychopharmacol. 2007 doi: 10.1038/sj.npp.1301579. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehe MR, Kopke K, Wendel B, Rohde K, Flachmeier C, Kidd KK, Berrettini WH, Church GM. Sequence variability and candidate gene analysis in complex disease: association of mu opioid receptor gene variation with substance dependence. Hum Mol Genet. 2000;9:2895–2908. doi: 10.1093/hmg/9.19.2895. [DOI] [PubMed] [Google Scholar]
- Holmes A, Picciotto MR. Galanin: a novel therapeutic target for depression, anxiety disorders and drug addiction? CNS Neurol Disord Drug Targets. 2006;5:225–232. doi: 10.2174/187152706776359600. [DOI] [PubMed] [Google Scholar]
- Kellogg SH, McHugh PF, Bell K, Schluger JH, Schluger RP, LaForge KS, Ho A, Kreek MJ. The Kreek-McHugh-Schluger-Kellogg scale: a new, rapid method for quantifying substance abuse and its possible applications. Drug Alcohol Depend. 2003;69:137–150. doi: 10.1016/s0376-8716(02)00308-3. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. Am J Psychiatry. 2007;164:1149–59. doi: 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev. 2005a;57:1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci. 2005b;8:1450–1457. doi: 10.1038/nn1583. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, LaForge KS. Stress responsivity, addiction, and a functional variant of the human mu-opioid receptor gene. Mol Interv. 2007;7:74–78. doi: 10.1124/mi.7.2.7. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Wan C, Zhou W, Peng T, Liu Y, Wang Z, Li G, Cornelisson G, Halberg F. The role of mPer1 in morphine dependence in mice. Neurosci. 2005;130:383–388. doi: 10.1016/j.neuroscience.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Jiang Z, Wan C, Zhou W, Wang Z. The extracellular signal-regulated kinase signaling pathway is involved in the modulation of morphine-induced reward by mPer1. Neurosci. 2007;146:265–271. doi: 10.1016/j.neuroscience.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Loh el W, Fann CS, Chang YT, Chang CJ, Cheng AT. Endogenous opioid receptor genes and alcohol dependence among Taiwanese Han. Alcohol Clin Exp Res. 2004;28:15–19. doi: 10.1097/01.ALC.0000106303.41755.B8. [DOI] [PubMed] [Google Scholar]
- Loh EW, Tang NL, Lee DT, Liu SI, Stadlin A. Association analysis of GABA receptor subunit genes on 5q33 with heroin dependence in a Chinese male population. Am J Med Genet B Neuropsychiatr Genet. 2007;144:439–443. doi: 10.1002/ajmg.b.30429. [DOI] [PubMed] [Google Scholar]
- Lucki I. The spectrum of behaviors influenced by serotonin. Biol Psychiatry. 1998;44:151–162. doi: 10.1016/s0006-3223(98)00139-5. [DOI] [PubMed] [Google Scholar]
- Mayer P, Rochlitz H, Rauch E, Rommelspacher H, Hasse HE, Schmidt S, Hollt V. Association between a delta opioid receptor gene polymorphism and heroin dependence in man. Neuroreport. 1997;8:2547–2550. doi: 10.1097/00001756-199707280-00025. [DOI] [PubMed] [Google Scholar]
- McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, Nestler EJ. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc Natl Acad Sci U S A. 2005;102:9377–81. doi: 10.1073/pnas.0503584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin JP, Marton-Popovici M, Chavkin C. κ Opioid Receptor Antagonism and Prodynorphin Gene Disruption Block Stress-Induced Behavioral Responses. J Neurosci. 2003;23:5674–5683. doi: 10.1523/JNEUROSCI.23-13-05674.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan AT, Kushner H, Metzger D, Peters R, Smith I, Grissom G, Pettinati H, Argeriou M. The Fifth Edition of the Addiction Severity Index. J Subst Abuse Treat. 1992;9:199–213. doi: 10.1016/0740-5472(92)90062-s. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumark YD, Friedlander Y, Thomasson HR, Li TK. Association of the ADH2*2 allele with reduced ethanol consumption in Jewish men in Israel: a pilot study. J Stud Alcohol. 1998;59:133–139. doi: 10.15288/jsa.1998.59.133. [DOI] [PubMed] [Google Scholar]
- Nielsen DA, Barral S, Proudnikov D, Kellogg S, Ho A, Ott J, Kreek MJ. TPH2 and TPH1: Association of Variants and Interactions with Heroin Addiction. Behav Genet. 2008;38:133–50. doi: 10.1007/s10519-007-9187-7. [DOI] [PubMed] [Google Scholar]
- Nieto MM, Guen SL, Kieffer BL, Roques BP, Noble F. Physiological control of emotion-related behaviors by endogenous enkephalins involves essentially the delta opioid receptors. Neuroscience. 2005;135:305–13. doi: 10.1016/j.neuroscience.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Verbitsky M, Suresh R, Kamens HM, Reed CL, Li N, Burkhart-Kasch S, McKinnon CS, Belknap JK, Gilliam TC, et al. Gene expression differences in mice divergently selected for methamphetamine sensitivity. Mamm Genome. 2005;16:291–305. doi: 10.1007/s00335-004-2451-8. [DOI] [PubMed] [Google Scholar]
- Picciotto MR, Hawes JJ, Brunzell DH, Zachariou V. Galanin can attenuate opiate reinforcement and withdrawal. Neuropeptides. 2005;39:313–315. doi: 10.1016/j.npep.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quock RM, Burkey TH, Varga E, Hosohata Y, Hosohata K, Cowell SM, Slate CA, Ehlert FJ, Roeske WR, Yamamura HI. The delta-opioid receptor: molecular pharmacology, signal transduction, and the determination of drug efficacy. Pharmacol Rev. 1999;51:503–32. [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: 2007. [Google Scholar]
- Roberts AJ, Gold LH, Polis I, McDonald JS, Filliol D, Kieffer BL, Koob GF. Increased ethanol self-administration in delta-opioid receptor knockout mice. Alcohol Clin Exp Res. 2001;25:1249–1256. [PubMed] [Google Scholar]
- Rozenfeld R, Devi LA. Receptor heterodimerization leads to a switch in signaling: beta-arrestin2-mediated ERK activation by mu-delta opioid receptor heterodimers. Faseb J. 2007;21:2455–2465. doi: 10.1096/fj.06-7793com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano G, Windemuth A, Kocherla M, Holford T, Fernandez ML, Forsythe CE, Wood RJ, Kraemer WJ, Volek JS. Physiogenomic analysis of weight loss induced by dietary carbohydrate restriction. Nutr Metab (Lond) 2006;3:20. doi: 10.1186/1743-7075-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya A, Yoshida A. Genotypes of alcohol-metabolizing enzymes in Japanese with alcohol liver diseases: a strong association of the usual Caucasian-type aldehyde dehydrogenase gene (ALDH1(2)) with the disease. Am J Hum Genet. 1988;43:744–748. [PMC free article] [PubMed] [Google Scholar]
- Simonin F, Valverde O, Smadja C, Slowe S, Kitchen I, Dierich A, Le Meur M, Roques BP, Maldonado R, Kieffer BL. Disruption of the kappa-opioid receptor gene in mice enhances sensitivity to chemical visceral pain, impairs pharmacological actions of the selective kappa-agonist U-50,488H and attenuates morphine withdrawal. Embo J. 1998;17:886–97. doi: 10.1093/emboj/17.4.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Statist. 2003;31:2013–2035. [Google Scholar]
- Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J of the Royal Statist Society: Ser B (Statist Method) 2004;66:187–205. [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubrie P. Reconciling the role of central serotonin neurons in human and animal behavior. Behav Brain Sci. 1986;9:319–364. [Google Scholar]
- Svenningsson P, Tzavara ET, Liu F, Fienberg AA, Nomikos GG, Greengard P. DARPP-32 mediates serotonergic neurotransmission in the forebrain. Proc Natl Acad Sci U S A. 2002;99:3188–93. doi: 10.1073/pnas.052712699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, Meyer JM, Toomey R, Faraone SV, Eaves L. Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet. 1996;67:473–477. doi: 10.1002/(SICI)1096-8628(19960920)67:5<473::AID-AJMG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Unschuld PG, Ising M, Erhardt A, Lucae S, Kohli M, Kloiber S, Salyakina D, Thoeringer CK, Kern N, Lieb R, Uhr M, Binder EB, Muller-Myhsok B, Holsboer F, Keck ME. Polymorphisms in the galanin gene are associated with symptom-severity in female patients suffering from panic disorder. J Affect Disord. 2007;105:177–84. doi: 10.1016/j.jad.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Qaadir A, Palmer AA, Cook EH, Jr, de Wit H. Association between the casein kinase 1 epsilon gene region and subjective response to D-amphetamine. Neuropsychopharmacol. 2006;31:1056–1063. doi: 10.1038/sj.npp.1300936. [DOI] [PubMed] [Google Scholar]
- Wand GS, McCaul M, Yang X, Reynolds J, Gotjen D, Lee S, Ali A. The mu-opioid receptor gene polymorphism (A118G) alters HPA axis activation induced by opioid receptor blockade. Neuropsychopharmacol. 2002;26:106–114. doi: 10.1016/S0893-133X(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Xu K, Liu XH, Nagarajan S, Gu XY, Goldman D. Relationship of the delta-opioid receptor gene to heroin abuse in a large Chinese case/control sample. Am J Med Genet. 2002;110:45–50. doi: 10.1002/ajmg.10374. [DOI] [PubMed] [Google Scholar]
- Xu K, Lichtermann D, Lipsky RH, Franke P, Liu X, Hu Y, Cao L, Schwab SG, Wildenauer DB, Bau CH, Ferro E, Astor W, Finch T, Terry J, Taubman J, Maier W, Goldman D. Association of specific haplotypes of D2 dopamine receptor gene with vulnerability to heroin dependence in 2 distinct populations. Arch Gen Psychiatry. 2004;61:597–606. doi: 10.1001/archpsyc.61.6.597. [DOI] [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L, Goate A, Bucholz K, Schuckit M, Nurnberger J, Jr, Tischfield J, Kuperman S, Porjesz B, Begleiter H, Foroud T, Edenberg HJ. Association of the kappa-opioid system with alcohol dependence. Mol Psychiatry. 2006;11:1016–1024. doi: 10.1038/sj.mp.4001882. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hattori E, Iwayama Y, Ohnishi T, Ohba H, Toyota T, Takao H, Minabe Y, Nakatani N, Higuchi T, Detera-Wadleigh SD, Yoshikawa T. Distinguishable haplotype blocks in the HTR3A and HTR3B region in the Japanese reveal evidence of association of HTR3B with female major depression. Biol Psychiatry. 2006;60:192–201. doi: 10.1016/j.biopsych.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Fussell D, LaForge KS, Nielsen DA, Gordon D, Ho A, Leal SM, Ott J, Kreek MJ. Redefinition of the human kappa opioid receptor gene (OPRK1) structure and association of haplotypes with opiate addiction. Pharmacogenet. 2004;14:793–804. doi: 10.1097/00008571-200412000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuferov V, Kroslak T, LaForge KS, Zhou Y, Ho A, Kreek MJ. Differential gene expression in the rat caudate putamen after “binge” cocaine administration: advantage of triplicate micro array analysis. Synapse. 2003;48:157–169. doi: 10.1002/syn.10198. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Butelman ER, Kreek MJ. Biological clock: biological clocks may modulate drug addiction. Eur J Hum Genet. 2005;13:1101–3. doi: 10.1038/sj.ejhg.5201483. [DOI] [PubMed] [Google Scholar]
- Zachariou V, Brunzell DH, Hawes J, Stedman DR, Bartfai T, Steiner RA, Wynick D, Langel U, Picciotto MR. The neuropeptide galanin modulates behavioral and neurochemical signs of opiate withdrawal. Proc Natl Acad Sci U S A. 2003;100:9028–9033. doi: 10.1073/pnas.1533224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, Zvartau E, Gelernter J. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15:807–819. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shao C, Shao M, Yan P, Wang Y, Liu Y, Liu W, Lin T, Xie Y, Zhao Y, Lu D, Li Y, Jin L. Effect of mu-opioid receptor gene polymorphisms on heroin-induced subjective responses in a Chinese population. Biol Psychiatry. 2007a;61:1244–1251. doi: 10.1016/j.biopsych.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2007b doi: 10.1038/sj.mp.4002035. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Liao G, Liu Y, Wang Y, Yang Z, Lin Y, Shen Y, Li S, Xiao J, Guo H, Wan C, Wang Z. Association of the 54-nucleotide repeat polymorphism of hPer3 with heroin dependence in Han Chinese population. Genes Brain Behav. 2007;7:26–30. doi: 10.1111/j.1601-183X.2007.00314.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.