Abstract
Autophagy initiation is strictly dependent on phosphatidylinositol 3-phosphate (PI3P) synthesis. PI3P production is under tight control of PI3Kinase, hVps34, in complex with Beclin-1. Mammalian cells express several PI3P phosphatases that belong to the myotubularin family. Even though some of them have been linked to serious human diseases, their cellular function is largely unknown. Two recent studies indicate that PI3P metabolism involved in autophagy initiation is further regulated by the PI3P phosphatases Jumpy and MTMR3. Additional pools of PI3P, upstream of mTOR and on the endocytic pathway, may modulate autophagy indirectly, suggesting that other PI3P phosphatases might be involved in this process. This review sums up our knowledge on PI3P phosphatases and discusses the recent progress on their role in autophagy.
Keywords: autophagy, myotubularin, PI3P, phosphatase, Jumpy, MTMR14
1.Introduction
PI3P synthesis has long been recognized as one of the key events in the initiation of the autophagic process (1). The phosphorylation of phosphatidylinositol is catalyzed by the well known, type III phosphatidylinositol 3-kinase, Vps34, in complex with the autophagy proteins, Beclin-1 (Atg6) and Atg14 (2–4) (Fig. 1). This highly regulated complex (see chapter in this issue), conserved from yeast to humans, allows spatio-temporal control of autophagy initiation (5). Besides, and in contrast, to this proximal action at the early stage of autophagy, PI3P may also regulate autophagy upstream of mTOR, a negative regulator autophagy. Amino-acids have been shown to stimulate mTOR activity via Vps34 activation (6, 7), therefore, this pool of PI3P could potentially inhibit autophagy, even though at the moment, evidence is lacking to support this model. Finally, PI3P plays a major role in the endocytic pathway where it promotes fusion of endosomes and receptor degradation (8). Since autophagic and endocytic pathways merge, it is likely that this third pool of PI3P modulates either directly or indirectly the last stages of autophagy (9, 10).
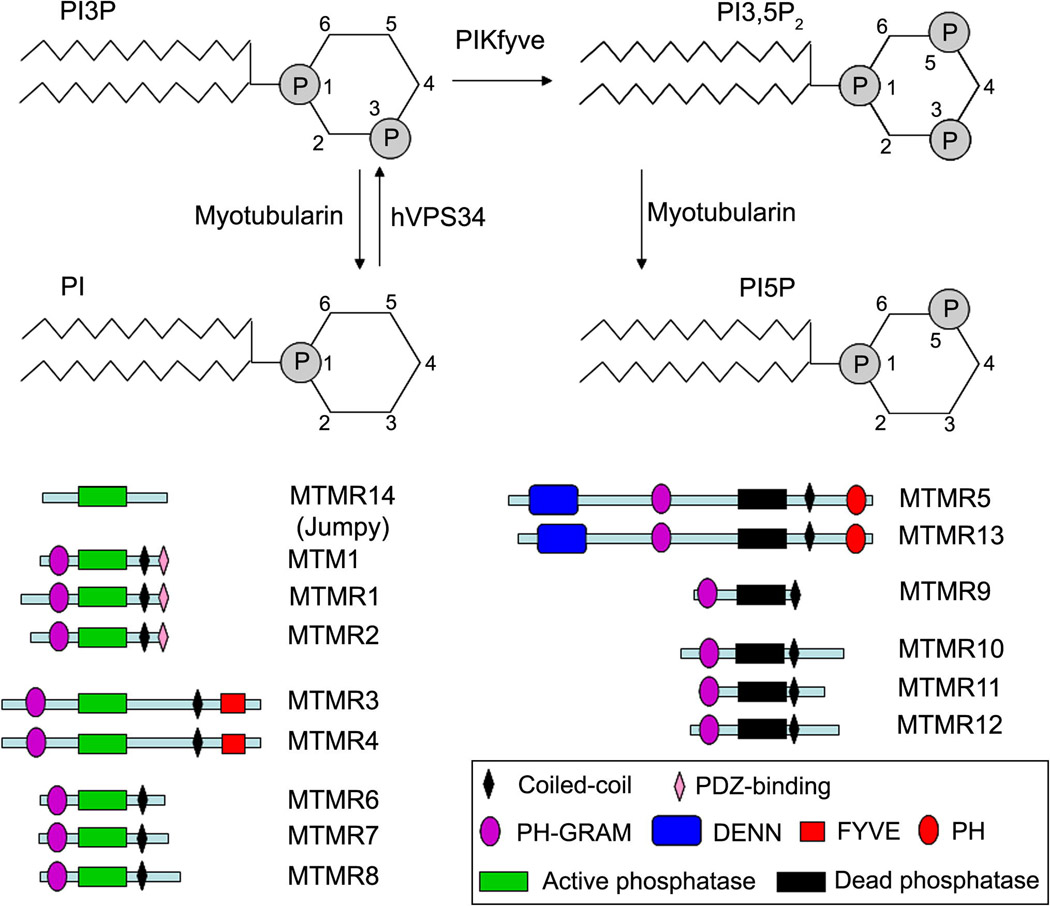
Figure 1. Myotubularin family: activity and different members with their domains.
hVps34 catalyzes phosphatidylinositol (PI) phosphorylation at position 3 on inositol ring to form phosphatidylinositol 3-phosphate (PI3P). PI3P can be further phosphorylated by PYKfyve to generate PI3,5,P2. Myotubularins remove 3-phosphate on PI3P and PI3,5P2 resulting in generation of PI and PI5P, respectively. Nine active and six inactive phosphatases are present in humans. See text for domain description.
Although, the synthesis of PI3P and its regulation have been well studied, how PI3P is turned-over and how autophagy is turned-off once PI3P is produced, remained to be elucidated. The PI3P phosphatases were one of the likely candidates as it is known that the phosphoinositide, PI3,4,5P3 and its signaling can be down-regulated by PI3,4,5P3 phosphatase, PTEN. In this review we will summarize briefly the structure and functions of PI3P phosphatases (for reviews see, (11–13)) culminating in a discussion of the recently developing role of PI3P phosphatases in autophagy.
2. Structure and expression of PI3P phosphatases
In mammalian cells PI3P phosphatases belong to the myotubularin family (11–13). This subgroup of phosphatases contains 15 members in humans of which all have a characteristic phosphatase domain with a C×5R motif. Only 9 of them, MTM1 (also called myotubularin), MTMR1, MTMR2, MTMR3, MTMR4, MTMR6, MTMR7, MTMR8 and Jumpy (also known as MTMR14), possess an active phosphatase domain, that specifically dephosphorylates PI3P and phosphatidylinositol (3,5)-bisphosphate (PI3,5P2) at position 3 on inositol ring. However, MTMR7 seems to have preference for inositiol (1,3)-bisphosphate as a soluble substrate (14). The six remaining myotubularins are inactive pseudophosphatases due to a substitution of Cys and Arg residues in their catalytic site. Besides the catalytic domain, all myotubularins, with the exception of MTMR14, contain two other protein domains, PH-GRAM (Pleckstrin Homology-Glucosyl transferases, Rab-like GTPase activators and myotubularins) and coiled-coil, that are believed to be important for lipid-protein or protein-protein interactions. Subgroups of myotubularins present additional domains such as PH and FYVE which are predicted to bind to phosphoinositides, PDZ-binding site likely to mediate protein-protein interaction and DENN in inactive myotubularins MTMR5 and MTMR13 (Fig. 1) (11, 12). Myotubularins are ubiquitously expressed, however, they can be enriched in certain tissues. For instance, MTM1 and Jumpy are highly expressed in skeletal muscles whereas MTMR2 is abundant in neurons and Schwann cells (15–19).
S. cerevisiae genome encodes only one myotubularin, Ymr1p. However, yeast expresses two additional unrelated phosphatases, members of the synaptojanin-like family, Sjl2p and Sjl3p that account also for PI3P dephosphorylation (20). Only ymr1Δ sjl3Δ and ymr1Δ sjl3Δ double mutants show a significant increase in PI3P level, suggesting that these phosphatases may have redundant function in yeast (20).
3. PI3P phosphatases and diseases
Four of the fifteen PI3P phosphatases present in mammals have been linked to serious human diseases. MTM1 defects cause X-linked myotubular myopathy (XLMTM) also known as centronuclear myopathy, a severe congenital disorder affecting the physiology of skeletal muscle fibers and characterized by centrally localized nuclei and hypotonia (12, 13). More than two hundred different mutations, truncations and missenses, in MTM1 have been reported in XLMTM patients that result in loss or decrease of MTM1 level (21). A severe case of centronuclear myopathy was observed in one patient with an inactive MTM1 suggesting that XLMTM is likely due to defect in PI3P metabolism (21). MTM1 knockout mice studies indicate that myotubularin is essential for muscle growth and maintenance but not for myogenesis (22). Histopathological analyses of MTM1-deficient mice reveal skeletal hypotrophic myofibers with numerous vacuoles (22). However, ultrastructural analyses show that these vacuoles do not seem to contain cellular debris (22).
Recently, another gene encoding for a PI3P phosphatase, jumpy, has been identified as mutated in some patients with centronuclear myopathy (15). The mutations lead to a reduced or total loss of phosphatase activity and they are likely the basis for the disease as observed for MTM1 (15). Indeed, a recent study reports that MTMR14−/− mice display muscle weakness and fatigue similar to patients with centronuclear myopathy (16).
MTMR2 and MTMR13 have been found mutated in two forms of Charcot-Marie-Tooth (CMT) Type 4B disease, an autosomal recessive disorder of the peripheral nervous system characterized by nerve demyelination and myelin outfoldings (13, 21). CMT type 4B1 is caused by missense or deletion mutations in MTMR2 gene that result in MTMR2 loss of function. In addition to CMT4B1, one patient manifests azoospermia suggesting that MTMR2 also plays a role in testis. MTMR2-deficient mice develop CMT4B1-like neuropathy and azoospermia (21). Interesting, inactive myotubularin MTMR5, which interacts with MTMR2, has been shown to be essential for spermatogenesis in mice (13). Cell type-specific gene knockout indicates that loss of MTMR2 in Schwann cells but not in neurons lead to CMT4B1-like neuropathy. CMT type 4B2 is due to mutations in MTMR13gene which encodes a pseudophosphatase (12, 13). MTMR13-deficient mice manifest myelin outfoldings in peripheral nerves very similar to ones found in CMT4B2 (13). It has been suggested that MTMR2 might form two complexes, one with MTMR5, in Sertoli or germ cells, involved in spermatogenesis and a second one with MTMR13, in Schwann cells, important in myelin homeostasis (13).
4. Regulation and cellular functions of PI3P phosphatases
One important feature of myotubularins is their ability to form homo- and heterodimers (23) (Table 1). These interactions often lead to an enhancement of phosphatase activity or to regulation of myotubularin intracellular localization. For instance, MTMR9, an inactive phosphatase, binds to both MTMR6 and MTMR7 resulting in a significant increase of their activities (14, 24) whereas MTMR13 binds and activates MTMR2 (25). Myotubularin coiled-coil domains play a major role in oligomer formation (14, 26). Generally, these interactions take place between active and inactive myotubularins (23), however, it is interesting to note that MTMR3 and MTMR4, two active phosphatases also form a heterodimer, which has been postulated to play a role in their redistribution within cells (23). Importantly, phosphatase activity does not seem to be required for myotubularin interactions (23, 27). Several other myotubularin interacting proteins have been identified and they are summarized in Table 1.
Table 1.
Protein Interacting partners of active PI3P phosphatases
| Myotubularins | Interacting partners | Effects or Functions | References |
|---|---|---|---|
| MTMR1 | Myotubularins: 1, R10, R12 |
- MTM1 activity increase with MTM1 oligomerization - MTM1 redistribution with MTMR12 |
(23,27,53) |
| p150 (hVps34) | Unknown | (35) | |
| MTMR1 | Unknown | - | - |
| MTMR2 | Myotubularins: R2,R5, R10, R12, R13 |
- MTMR2 activity increase with MTMR13 - MTMR2 redistribution with MTMR5 |
(23, 25, 26, 53, 54) |
| p150 (hVps34) | - MTMR2 activity decrease - Indirect PI3P inhibition |
(33) | |
| Neurofilament light chain, Disk large 1 Dlg1/SAP97 |
Myelin outfoldings regulation with Dlg1 |
(38, 55, 56) | |
| Nonreceptor protein tyrosine kinase c-Src |
Possibly cell adhesion regulation |
(57) | |
| MTMR3 | Myotubularins: R3, R4 | MTMR3 redistribution with MTMR4 |
(23) |
| MTMR4 | Myotubularins: R3, R4 | MTMR4 redistribution with MTMR3 |
(23) |
| Ubiquitin ligase Need4 | Unknown | (58) | |
| MTNR6 | Myotubularins: R9 | MTMR6 activity increase MTMR6 stability increase |
(23, 24) |
| Ca2+-activated K+ channel (K(Ca)3.1) |
Indirect K(Ca)3.1 inhibition | (42) | |
| MTMR7 | Myotubularins: R9 | MTMR7 activity increase | (14, 23) |
| MTMR8 | Myotubularins: R8, R9 | Unknown | (23) |
| MTMR14 | Unknown | - | - |
In addition to being regulated by other myotubularins, MTM1, MTMR3 and MTMR6 have been shown to be activated by phosphatidylinositol 5-phosphate (PI5P), the product of PI3,5P2 dephosphorylation (27). PI5P binds to PH-GRAM, a domain important for MTM1 and MTMR3 phosphatase activities, and for MTMR2, MTMR3 and MTMR6 association to intracellular membranes (27–30). PI5P is a specific allosteric activator of MTM1 that promotes oligomerization of MTM1 into a heptamer (27).
For the most part, the biological functions of myotubularins are poorly understood. When overexpressed most myotubularins promote a decrease in PI3P levels on endosomal compartments (23). Exogenously expressed myotubularins are found in cytosol and on intracellular structures that display different partners depending on the myotubularin investigated, suggesting that each myotubularin is targeted to a specific intracellular compartment where it dephosphorylates a definite pool of PI3P (23). A recent siRNA screen identified several myotubularins, MTMR1, MTMR6, MTMR7, MTMR8 and MTMR12, as being important for cellular survival, yet, the molecular mechanisms behind this effect remain to be clarified (31). Since both are associated with debilitating human illnesses, MTM1 and MTMR2 have been the most studied. MTM1 is present on ruffles at the plasma membrane, on early endosomes and partially on late endosomes where they regulate intracellular trafficking of Glut-4 and EGF receptor, respectively (32–35). In muscle fibers lacking MTM1, the structure of T-tubules, invaginations originating from the plasma membrane, is impaired, supporting the idea of MTM1 being involved in membrane traffic (36, 37). MTMR2 localizes on Rab7 positive compartment and similar to MTM1 its depletion results in reduction of EGFR degradation (33). In Schwann cells MTMR2 is believed to play an important role in membrane homeostasis at the plasma membrane during the process of myelination (38). Interestingly, MTMR2 binds to type III PI3K regulatory subunit, hVps15 (p150), which results in inhibition of phosphatase activity and in diminution of hVps34-Rab7 interaction important for PI3P synthesis (33) (Table 1). A similar binding was also observed between MTM1 and hVps15 (35). Finally, a recent study reports that in muscle fibers, MTMR14 regulates Ca2+ homeostasis by dephosphorylating PI3,5P2, an activator of Ca2+ release channel ryanodine receptor 1 (16).
5. PI3P phosphatases and autophagy
The majority of PI3P phosphatase functions described so far take place along the endocytic pathway. Given that PI3P plays a central role in autophagy, we wondered whether PI3P phosphatase(s) were also involved in regulating this pathway. A recent siRNA-based screen identified three PI3P phosphatases, Jumpy, MTMR6 and MTMR7, that affect level of autophagic marker LC3-II (39).
Jumpy-depleted mammalian cells show an increase in number of autophagic vacuoles in nutrient-rich and starvation conditions as well as an upregulation of long-lived protein proteolysis (39). In contrast cells overexpressing Jumpy but not MTM1 or MTMR2, fail to degrade autophagic substrate p62 leading to accumulation of protein aggregates (39). Jumpy colocalizes with LC3 positive compartments in particular with isolation membranes marked with Atg12 and Atg16 indicating that this phosphatase acts directly, and at an early stage of autophagic process (39). Importantly, Jumpy regulates Atg9 intracellular distribution, a PI3P-dependent event important for initiating autophagy (39–41). In yeast, Atg9 retrieval from autophagosomes requires the PI3P-binding protein Atg18 (41). Interestingly, Jumpy depletion in mammalian cells increases WIPI-1 (Atg18 ortholog) on membranes (39) suggesting that Jumpy dephosphorylates a pool of PI3P involved in WIPI-1 binding and Atg9 retrieval. In the future it would be important to elucidate the molecular mechanisms involved in Jumpy targeting to autophagic vacuoles and its regulation. In addition to increasing autophagy initiation, Jumpy depletion also promotes maturation of autophagosomes and LC3-II degradation (39). At the moment is unclear, whether this later effect is coupled to the initiation event or whether Jumpy acts at two different stages of the pathway. Inactive Jumpy variant, R336Q, found in one patient with centronuclear myopathy, is unable to inhibit autophagy suggesting that deregulation of autophagic pathway might be an important contributing factor to the disease (39).
In macrophages, MTMR6 and MTMR7 knockdown increase LC3-II level suggesting that additional phosphatases besides Jumpy may be important in regulating autophagy, however their mechanism of action remains to be investigated (39). MTMR6 is known to prevent activation of Ca2+-activated K+ channel, KCa3.1, which results in decreased Ca2+ influx during T-cell activation (42). Even though the role of Ca2+ in autophagy is still controversial (43–45), it is possible that MTMR6 modulates indirectly the autophagic response through change in Ca2+ homeostasis (Fig. 2).
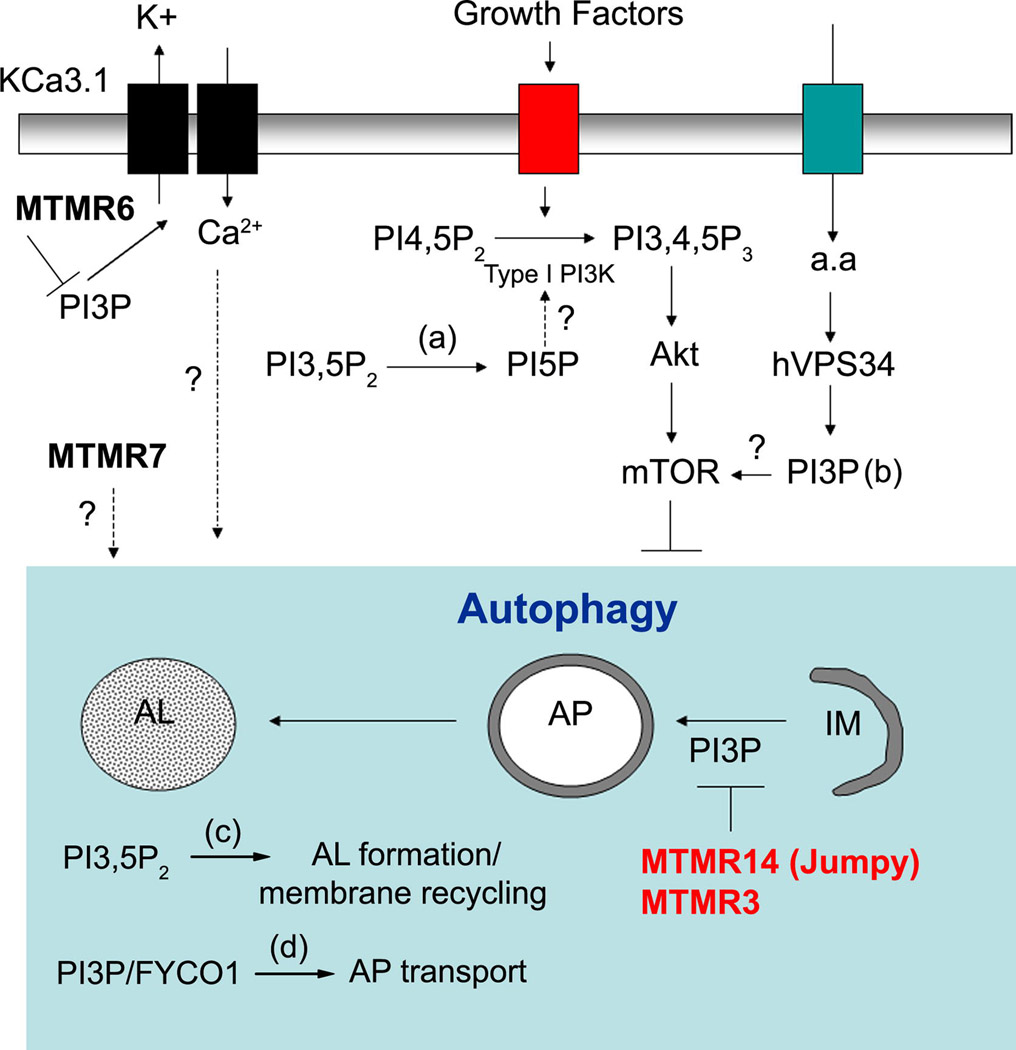
Figure 2. Intracellular PI3P pools and PI3P phosphatases in autophagy regulation.
MTMR14 and MTMR3 inhibit autophagy by dephosphorylating PI3P involved in autophagosome formation. MTMR6 and MTMR7 depletions increase LC3-II via unknown mechanisms. Since MTMR6 regulates KCa3.1, we propose that MTMR6 might modulate autophagy by acting on Ca2+ influx. We speculate that some PI3P phosphatases might regulate autophagy by dephosphorylating pools of PI3P and/or PI3,5P2 involved in Akt and mTOR activation (a and b), autolysosome formation/membrane recycling (c) and autophagosome transport (d) (see text and (52)). AP: autophagosome; AL: autolysosome. a.a.:amino-acids. In red: PI3P phosphatases inhibiting autophagy. IM: isolation membrane; In bold: PI3P phosphatases whose knockdown affect LC3-II level. Question marks indicate that the role of these pathways in regulating autophagy is speculative and remains to be investigated.
Walker and al., 2001 reported that an inactive form of MTMR3, mutant C413S, is present inside compartment resembling autophagosomes in Hela cells (46). More recently, MTMR3 was reported to regulate constitutive autophagy initiation and autophagosome size in epithelial cells (47). However, MTMR3 knockdown does not affect steady-state level of LC3-II in macrophages (39). One possible explanation for this apparent discrepancy is that MTMR3 and Jumpy might play a redundant role during constitutive autophagy depending on cell types and levels of expression. Yet, given that Jumpy lacks most of MTMR3 domains, theirs modes of regulation and recruitment are likely to be different.
PI3P is known to be a substrate for PIP5-kinase, Fab1 (also named PIKfyve), an enzyme that generates phosphoinositide PI3,5P2 (Fig. 1). Although PI3,5P2 does not appear to be important for autophagy in yeast, several studies indicate that PI3,5P2 plays a key role in maturation of autophagosomes in mammals and flies (9, 10, 48). Mice deficient for Fig4 and Vac14, two regulatory proteins required for PI3,5P2 synthesis, accumulate p62 in Lamp-2 positive compartments in neurons and astrocytes (48). Chemical inhibition of PYKfyve activity was shown to block LC3-II turn-over, and in drosophila, with impaired Fab1 function, an accumulation of autolysosomes was observed (9, 10). Since myotubularins are also efficient PI3,5P2 phosphatases, one can speculate that they might also be involved in late stages of autophagy.
Dephosphorylation of PI3,5P2 by myotubularins gives rise to PI5P (Fig. 1). In particular, cells overexpressing MTM1 show an increase in the amount of PI5P which was not observed in cells expressing an inactive mutant D278A (49). This rare phospholipid, PI5P, has been found to activate the Akt pathway, though its mechanism of action remains elusive (50). It is important to note that MTMR2 overexpression also leads to sustained Akt activation (51). Thus, myotubularins may potentially activate mTOR via generation of PI5P and consequently block autophagy (Fig. 2). On the other hand, as we mentioned in the introduction, amino-acid-mediated PI3P synthesis induces mTOR activation (6, 7). Therefore, if any myotubularin acts on this PI3P pool, it might affect mTOR activity and therefore autophagic response (Fig. 2). Anyhow, it would be of interest to investigate whether myotubularins play a role in regulating Akt/mTOR pathway and hence autophagy.
6. Conclusions and perspectives
The discovery of the PI3P phosphatases, Jumpy and more recently, MTMR3, as regulators of autophagy, shed some light on an unappreciated regulatory mechanism of the autophagic process. The initiation of autophagy is not only controlled by hVps34/Beclin-1-mediated synthesis of PI3P but also by its turn-over via action of specific phosphatases. These findings add Jumpy and MTMR3 to the list of potential therapeutic targets that could be used to modulate autophagy in the context of human diseases. The recent work on PI3P phosphatases and autophagy also indicate that other myotubularins could be implicated in this process. Elucidating their biological functions will undoubtedly improve our understanding of autophagy pathway and its regulation.
Acknowledgements
This work was supported by grants AI45148 from the National Institutes of Health to VD. This project was supported in part by the Dedicated Health Research Funds from the University of New Mexico School of Medicine to IV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 2.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Tassa A, Roux MP, Attaix D, Bechet DM. Class III phosphoinositide 3-kinase--Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J. 2003;376:577–586. doi: 10.1042/BJ20030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 8.Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 9.de Lartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbe S, Clague MJ. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 11.Clague MJ, Lorenzo O. The myotubularin family of lipid phosphatases. Traffic. 2005;6:1036–1069. doi: 10.1111/j.1600-0854.2005.00338.x. [DOI] [PubMed] [Google Scholar]
- 12.Laporte J, Bedez F, Bolino A, Mandel JL. Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum Mol Genet. 2003;12:R285–R292. doi: 10.1093/hmg/ddg273. Spec No 2. [DOI] [PubMed] [Google Scholar]
- 13.Robinson FL, Dixon JE. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 2006;16:403–412. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Mochizuki Y, Majerus PW. Characterization of myotubularin-related protein 7 and its binding partner, myotubularin-related protein 9. Proc Natl Acad Sci U S A. 2003;100:9768–9773. doi: 10.1073/pnas.1333958100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tosch V, Rohde HM, Tronchere H, Zanoteli E, Monroy N, Kretz C, Dondaine N, Payrastre B, Mandel JL, Laporte J. A novel PtdIns3P and PtdIns(3,5)P2 phosphatase with an inactivating variant in centronuclear myopathy. Hum Mol Genet. 2006;15:3098–3106. doi: 10.1093/hmg/ddl250. [DOI] [PubMed] [Google Scholar]
- 16.Shen J, Yu WM, Brotto M, Scherman JA, Guo C, Stoddard C, Nosek TM, Valdivia HH, Qu CK. Deficiency of MIP/MTMR14 phosphatase induces a muscle disorder by disrupting Ca(2+) homeostasis. Nat Cell Biol. 2009;776:769–776. doi: 10.1038/ncb1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laporte J, Blondeau F, Buj-Bello A, Tentler D, Kretz C, Dahl N, Mandel JL. Characterization of the myotubularin dual specificity phosphatase gene family from yeast to human. Hum Mol Genet. 1998;7:1703–1712. doi: 10.1093/hmg/7.11.1703. [DOI] [PubMed] [Google Scholar]
- 18.Berger P, Bonneick S, Willi S, Wymann M, Suter U. Loss of phosphatase activity in myotubularin-related protein 2 is associated with Charcot-Marie-Tooth disease type 4B1. Hum Mol Genet. 2002;11:1569–1579. doi: 10.1093/hmg/11.13.1569. [DOI] [PubMed] [Google Scholar]
- 19.Bolino A, Marigo V, Ferrera F, Loader J, Romio L, Leoni A, Di Duca M, Cinti R, Cecchi C, Feltri ML, et al. Molecular characterization and expression analysis of Mtmr2, mouse homologue of MTMR2, the Myotubularin-related 2 gene, mutated in CMT4B. Gene. 2002;283:17–26. doi: 10.1016/s0378-1119(01)00876-9. [DOI] [PubMed] [Google Scholar]
- 20.Parrish WR, Stefan CJ, Emr SD. Essential role for the myotubularin-related phosphatase Ymr1p and the synaptojanin-like phosphatases Sjl2p and Sjl3p in regulation of phosphatidylinositol 3-phosphate in yeast. Mol Biol Cell. 2004;15:3567–3579. doi: 10.1091/mbc.E04-03-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicot AS, Laporte J. Endosomal phosphoinositides and human diseases. Traffic. 2008;9:1240–1249. doi: 10.1111/j.1600-0854.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buj-Bello A, Laugel V, Messaddeq N, Zahreddine H, Laporte J, Pellissier JF, Mandel JL. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc Natl Acad Sci U S A. 2002;99:15060–15065. doi: 10.1073/pnas.212498399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzo O, Urbe S, Clague MJ. Systematic analysis of myotubularins: heteromeric interactions, subcellular localisation and endosome related functions. J Cell Sci. 2006;119:2953–2959. doi: 10.1242/jcs.03040. [DOI] [PubMed] [Google Scholar]
- 24.Zou J, Chang SC, Marjanovic J, Majerus PW. MTMR9 increases MTMR6 enzyme activity, stability, and role in apoptosis. J Biol Chem. 2009;284:2064–2071. doi: 10.1074/jbc.M804292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger P, Berger I, Schaffitzel C, Tersar K, Volkmer B, Suter U. Multi-level regulation of myotubularin-related protein-2 phosphatase activity by myotubularin-related protein-13/set-binding factor-2. Hum Mol Genet. 2006;15:569–579. doi: 10.1093/hmg/ddi473. [DOI] [PubMed] [Google Scholar]
- 26.Kim SA, Vacratsis PO, Firestein R, Cleary ML, Dixon JE. Regulation of myotubularin-related (MTMR)2 phosphatidylinositol phosphatase by MTMR5, a catalytically inactive phosphatase. Proc Natl Acad Sci U S A. 2003;100:4492–4497. doi: 10.1073/pnas.0431052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaletzky J, Dove SK, Short B, Lorenzo O, Clague MJ, Barr FA. Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr Biol. 2003;13:504–509. doi: 10.1016/s0960-9822(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 28.Berger P, Schaffitzel C, Berger I, Ban N, Suter U. Membrane association of myotubularin-related protein 2 is mediated by a pleckstrin homology-GRAM domain and a coiled-coil dimerization module. Proc Natl Acad Sci U S A. 2003;100:12177–12182. doi: 10.1073/pnas.2132732100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenzo O, Urbe S, Clague MJ. Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J Cell Sci. 2005;118:2005–2012. doi: 10.1242/jcs.02325. [DOI] [PubMed] [Google Scholar]
- 30.Choudhury P, Srivastava S, Li Z, Ko K, Albaqumi M, Narayan K, Coetzee WA, Lemmon MA, Skolnik EY. Specificity of the myotubularin family of phosphatidylinositol-3-phosphatase is determined by the PH/GRAM domain. J Biol Chem. 2006;281:31762–31769. doi: 10.1074/jbc.M606344200. [DOI] [PubMed] [Google Scholar]
- 31.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 32.Laporte J, Blondeau F, Gansmuller A, Lutz Y, Vonesch JL, Mandel JL. The PtdIns3P phosphatase myotubularin is a cytoplasmic protein that also localizes to Rac1-inducible plasma membrane ruffles. J Cell Sci. 2002;115:3105–3117. doi: 10.1242/jcs.115.15.3105. [DOI] [PubMed] [Google Scholar]
- 33.Cao C, Backer JM, Laporte J, Bedrick EJ, Wandinger-Ness A. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol Biol Cell. 2008;19:3334–3346. doi: 10.1091/mbc.E08-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaussade C, Pirola L, Bonnafous S, Blondeau F, Brenz-Verca S, Tronchere H, Portis F, Rusconi S, Payrastre B, Laporte J, et al. Expression of myotubularin by an adenoviral vector demonstrates its function as a phosphatidylinositol 3-phosphate [PtdIns(3)P] phosphatase in muscle cell lines: involvement of PtdIns(3)P in insulin-stimulated glucose transport. Mol Endocrinol. 2003;17:2448–2460. doi: 10.1210/me.2003-0261. [DOI] [PubMed] [Google Scholar]
- 35.Cao C, Laporte J, Backer JM, Wandinger-Ness A, Stein MP. Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic. 2007;8:1052–1067. doi: 10.1111/j.1600-0854.2007.00586.x. [DOI] [PubMed] [Google Scholar]
- 36.Dowling JJ, Vreede AP, Low SE, Gibbs EM, Kuwada JY, Bonnemann CG, Feldman EL. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5:e1000372. doi: 10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Qusairi L, Weiss N, Toussaint A, Berbey C, Messaddeq N, Kretz C, Sanoudou D, Beggs AH, Allard B, Mandel JL, et al. T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc Natl Acad Sci U S A. 2009;106:18763–18768. doi: 10.1073/pnas.0900705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolis A, Coviello S, Visigalli I, Taveggia C, Bachi A, Chishti AH, Hanada T, Quattrini A, Previtali SC, Biffi A, et al. Dlg1, Sec8, and Mtmr2 regulate membrane homeostasis in Schwann cell myelination. J Neurosci. 2009;29:8858–8870. doi: 10.1523/JNEUROSCI.1423-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase jumpy. EMBO J. 2009;28:2244–2258. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 41.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 42.Srivastava S, Li Z, Lin L, Liu G, Ko K, Coetzee WA, Skolnik EY. The phosphatidylinositol 3-phosphate phosphatase myotubularin- related protein 6 (MTMR6) is a negative regulator of the Ca2+-activated K+ channel KCa3.1. Mol Cell Biol. 2005;25:3630–3638. doi: 10.1128/MCB.25.9.3630-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HJ, Soyombo AA, Tjon-Kon-Sang S, So I, Muallem S. The Ca(2+) channel TRPML3 regulates membrane trafficking and autophagy. Traffic. 2009;10:1157–1167. doi: 10.1111/j.1600-0854.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 45.Gordon PB, Holen I, Fosse M, Rotnes JS, Seglen PO. Dependence of hepatocytic autophagy on intracellularly sequestered calcium. J Biol Chem. 1993;268:26107–26112. [PubMed] [Google Scholar]
- 46.Walker DM, Urbe S, Dove SK, Tenza D, Raposo G, Clague MJ. Characterization of MTMR3. an inositol lipid 3-phosphatase with novel substrate specificity. Curr Biol. 2001;11:1600–1605. doi: 10.1016/s0960-9822(01)00501-2. [DOI] [PubMed] [Google Scholar]
- 47.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of Local PtdIns3P Levels by the PI Phosphatase MTMR3 Regulates Constitutive Autophagy. Traffic. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 48.Ferguson CJ, Lenk GM, Meisler MH. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum Mol Genet. 2009;18:4868–4878. doi: 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tronchere H, Laporte J, Pendaries C, Chaussade C, Liaubet L, Pirola L, Mandel JL, Payrastre B. Production of phosphatidylinositol 5-phosphate by the phosphoinositide 3-phosphatase myotubularin in mammalian cells. J Biol Chem. 2004;279:7304–7312. doi: 10.1074/jbc.M311071200. [DOI] [PubMed] [Google Scholar]
- 50.Pendaries C, Tronchere H, Arbibe L, Mounier J, Gozani O, Cantley L, Fry MJ, Gaits-Iacovoni F, Sansonetti PJ, Payrastre B. PtdIns5P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. EMBO J. 2006;25:1024–1034. doi: 10.1038/sj.emboj.7601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger P, Tersar K, Ballmer-Hofer K, Suter U. The CMT4B disease-causing proteins MTMR2 and MTMR13/SBF2 regulate AKT signaling. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nandurkar HH, Layton M, Laporte J, Selan C, Corcoran L, Caldwell KK, Mochizuki Y, Majerus PW, Mitchell CA. Identification of myotubularin as the lipid phosphatase catalytic subunit associated with the 3-phosphatase adapter protein, 3-PAP. Proc Natl Acad Sci U S A. 2003;100:8660–8665. doi: 10.1073/pnas.1033097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson FL, Dixon JE. The phosphoinositide-3-phosphatase MTMR2 associates with MTMR13, a membrane-associated pseudophosphatase also mutated in type 4B Charcot-Marie-Tooth disease. J Biol Chem. 2005;280:31699–31707. doi: 10.1074/jbc.M505159200. [DOI] [PubMed] [Google Scholar]
- 55.Bolino A, Bolis A, Previtali SC, Dina G, Bussini S, Dati G, Amadio S, Del Carro U, Mruk DD, Feltri ML, et al. Disruption of Mtmr2 produces CMT4B1-like neuropathy with myelin outfolding and impaired spermatogenesis. J Cell Biol. 2004;167:711–721. doi: 10.1083/jcb.200407010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Previtali SC, Zerega B, Sherman DL, Brophy PJ, Dina G, King RH, Salih MM, Feltri L, Quattrini A, Ravazzolo R, et al. Myotubularin-related 2 protein phosphatase and neurofilament light chain protein, both mutated in CMT neuropathies, interact in peripheral nerve. Hum Mol Genet. 2003;12:1713–1723. doi: 10.1093/hmg/ddg179. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J, Wong CH, Xia W, Mruk DD, Lee NP, Lee WM, Cheng CY. Regulation of Sertoli-germ cell adherens junction dynamics via changes in protein-protein interactions of the N-cadherin--beta-catenin protein complex which are possibly mediated by c-Src and myotubularin-related protein 2: an in vivo study using an androgen suppression model. Endocrinology. 2005;146:1268–1284. doi: 10.1210/en.2004-1194. [DOI] [PubMed] [Google Scholar]
- 58.Plant PJ, Correa J, Goldenberg N, Bain J, Batt J. The inositol phosphatase MTMR4 is a novel target of the ubiquitin ligase Nedd4. Biochem J. 2009;419:57–63. doi: 10.1042/BJ20081866. [DOI] [PMC free article] [PubMed] [Google Scholar]