Abstract
Autophagy, a critical process for bulk degradation of proteins and organelles, requires conjugation of Atg8 proteins to phosphatidylethanolamine on the autophagic membrane. At least eight different Atg8 orthologs belonging to two subfamilies (LC3 and GATE-16/GABARAP) occur in mammalian cells, but their individual roles and modes of action are largely unknown. In this study, we dissect the activity of each subfamily and show that both are indispensable for the autophagic process in mammalian cells. We further show that both subfamilies act differently at early stages of autophagosome biogenesis. Accordingly, our results indicate that LC3s are involved in elongation of the phagophore membrane whereas the GABARAP/GATE-16 subfamily is essential for a later stage in autophagosome maturation.
Keywords: Atg8, autophagy, GABARAP, GATE-16, LC3
Introduction
Autophagy is an intra-cellular pathway for bulk degradation of proteins and organelles within the lysosomes/vacuole. This pathway takes part in multiple physiological functions, such as cell development, programmed cell death, cancer, pathogen infection, and degradation of ubiquitinated protein aggregates, which are formed in many pathological conditions (Cecconi and Levine, 2008; Mizushima et al, 2008). Autophagy is initiated by the formation of a cup-shaped membrane that has been recently suggested to originate from the endoplasmic reticulum (Hayashi-Nishino et al, 2009; Yla-Anttila et al, 2009). This membrane is termed phagophore or isolation membrane. Next, the phagophore enwraps parts of the cytoplasm to form a double-membrane vesicle, called autophagosome, which eventually fuses with the lysosomes/vacuole.
The formation of the phagophore requires two ubiquitin-like conjugation systems: the conjugation of Atg12–Atg5, which is localized together with Atg16 to the phagophore and, downstream to it, the Atg8 conjugation to phosphatidylethanolamine (PE), which decorates both the phagophore and the autophagosomal membrane. In both systems Atg7 functions as an E1-like enzyme, whereas Atg10 and Atg3 act as E2-like enzymes for Atg12 and Atg8, respectively (Geng and Klionsky, 2008). Moreover, the Atg12–Atg5 conjugate may act as an E3-like enzyme for the conjugation of Atg8s to PE and, together with Atg16, is responsible for their recruitment to the phagophore (Hanada et al, 2007; Fujita et al, 2008b). Atg8-PE undergoes deconjugation by the Atg4 protease, a step regulated by reactive oxygen species, which allows the recycling of this protein. Atg4 is also responsible for the priming of Atg8 by cleaving its C-terminus to expose a glycine residue (Kirisako et al, 2000; Scherz-Shouval et al, 2007).
In yeast, Atg8 has a crucial role in the autophagic process possibly in the elongation of the phagophore membrane by mediating hemi-fusion events (Abeliovich et al, 2000; Nakatogawa et al, 2007; Xie et al, 2008). At least eight Atg8 orthologs were identified in mammals; however, to date only LC3B has been extensively studied. This autophagic factor is known to decorate autophagosomes and recruit factors such as p62 and NBR1 (Bjorkoy et al, 2005; Kirkin et al, 2009). Other Atg8 orthologs such as GATE-16 and GABARAP were initially characterized as intra-cellular trafficking factors (Sagiv et al, 2000; Kittler et al, 2001; Nakamura et al, 2008) and later were shown to be localized to starvation-induced autophagosomes (Kabeya et al, 2004). Mammalian Atg8s can be divided into two subgroups based on their amino acid sequence homology, where LC3A-C (LC3A has two variants originating from alternative splicing event) constitute the LC3 subfamily and GABARAP, GABARAPL1, GATE-16 (also known as GABARAPL2), and GABARAP-L3 constitute the GABARAP/GATE-16 subfamily (Xin et al, 2001; He et al, 2003). The occurrence of several Atg8 orthologs in the mammalian system raises the question whether each of these has a distinct and crucial role in autophagy. In this study, we used different experimental approaches to examine the role of the two Atg8 subfamilies in mammalian cells. We show that both LC3 and GABARAP/GATE-16 subfamilies are indispensable for the autophagic process, acting differentially at early stages of autophagosome biogenesis.
Results
Both LC3 and GABARAP/GATE-16 subfamilies are crucial for the autophagic process
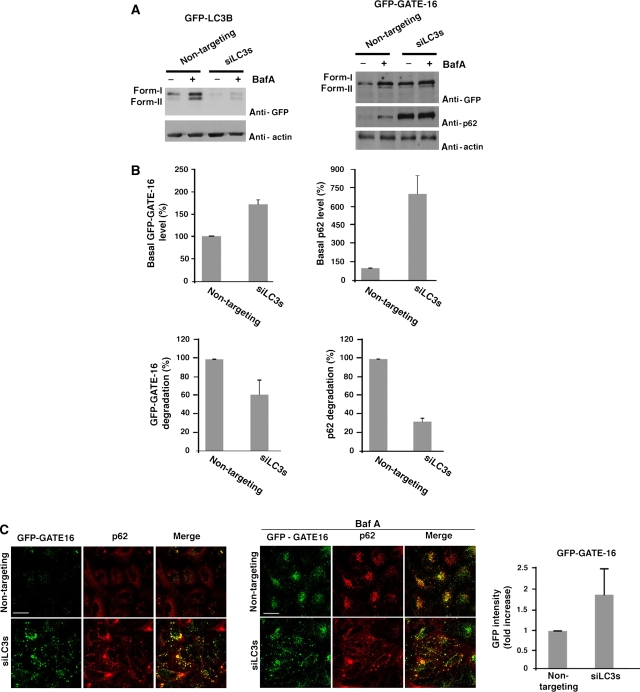
The yeast Atg8 has eight orthologs in mammals that can be divided into LC3 and GABARAP/GATE-16 subfamilies according to their sequence homology (Xin et al, 2001; He et al, 2003). Thus far, LC3B is the only Atg8 mammalian ortholog that has been mainly studied and identified as a factor associated with autophagic membranes; however, the essentiality of either the LC3 or GABARAP/GATE-16 in the autophagic process is yet unknown. To assure that both LC3B and GATE-16, representing the two subfamilies, are present on the same autophagosomes, cells stably expressing GFP-LC3B and dsRed-GATE-16 were examined by live microscopy. As presented in Supplementary Movie 1 and consistent with a previous report by Kabeya et al (2004), these proteins co-localized on dynamic puncta representing autophagosomes. To determine whether both subfamilies are essential for autophagy we used siRNA approach by which we knocked down all three isoforms of the LC3 subfamily in HeLa cells stably expressing GFP-GATE-16 (Figure 1A; Supplementary Figure S1-A). Cells transfected with the LC3 s siRNA pools were induced for autophagy by incubation in a starvation medium (EBSS) in the presence or absence of the lysosomal inhibitor Bafilomycin A1 (Baf A); autophagic flux was then detected by western blot and confocal microscopy (Figure 1). We first showed that the level of p62, an indicator of autophagic activity (Pankiv et al, 2007; Shvets et al, 2008b), is elevated on knock down of the LC3 subfamily (Figure 1A and B). Moreover, knock down of the LC3 isoforms led to the accumulation of GFP-GATE-16 both in its lipidated and unlipidated forms (Figure 1A and B). Notably, treatment with Baf A had no effect on the level of either GFP-GATE-16 or p62 in cells transfected with the siRNA pools, indicating that the LC3 subfamily is essential for the autophagic process (Figure 1A and B). The effect of silencing the LC3 subfamily was confirmed by confocal microscopy. As shown in Figure 1C, both p62 and GFP-GATE-16 were accumulated in the LC3s knockdown cells. Moreover, though addition of Baf A led to an increased signal of both GFP-GATE-16 and p62 in the control cells, the level of these proteins in the knockdown cells remained unchanged (Figure 1C).
Figure 1.
The LC3 protein subfamily is essential for autophagy. (A) HeLa cells stably expressing GFP-LC3B (left panel) or GFP-GATE-16 (right panel) were transfected with either non-targeting siRNA (control siRNA) or a pool of LC3 siRNAs (A, B, C), using DharmaFect reagent. After 72 h interval, the cells were incubated for 2 h in EBSS medium in the absence or presence of 0.1 μM Baf A and subjected to western blot analysis after lysis with RIPA extraction buffer. (B) Quantification of relative p62 and GFP-GATE-16 level (upper panel) and degradation (lower panel) was performed as described in ‘Materials and methods'. Mean±s.d. of three independent experiments is presented. (C) HeLa cells stably expressing GFP-GATE-16 were transfected with non-targeting siRNA or a pool of LC3 siRNAs and treated as in (A). The cells were subjected to immunostaining with anti-p62 after fixation, and analysed by confocal microscopy. The GFP intensity of three independent experiments was measured as described in ‘Materials and methods' (right panel). Scale bar: 20 μm.
We next tested whether the GABARAPGATE-16 subfamily members, which were previously characterized mostly as intra-cellular transport factors (Sagiv et al, 2000; Kittler et al, 2001; Nakamura et al, 2008), are crucial for autophagy contributing in a specific manner to this process. For this purpose HeLa cells stably expressing GFP-LC3B were knocked down with specific siRNA pools directed against the isoforms of the GABARAPGATE-16 subfamily (excluding GABARAP-L3) (Figure 2A; Supplementary Figure S1-A). Cells transfected with the siRNA pools were incubated in EBSS in the presence or absence of Baf A and the autophagic flux was tested by monitoring the autophagic markers p62 and GFP-LC3B. As depicted in Figure 2A and B, both p62 and GFP-LC3B accumulated after GABARAPs knockdown and was not effected by the Baf A treatment. We confirmed the above results using immunofluorescence analysis of these cells (Figure 2C). After the silencing of GABARAPs the cells exhibited elevated levels of p62, as well as elevated GFP-LC3B signal intensity, which was about two-folds higher in the knockdown cells compared with control cells. In addition, these proteins were not further accumulated after Baf A treatment (Figure 2C), indicating an inhibition in the autophagic flux. Thus, our data suggest that the GABARAPGATE-16 subfamily has an essential role in the autophagic process. Similar effect of GABARAPs knockdown was observed in cells that do not express GFP-LC3B (Supplementary Figures S1-B and S1-C), hence excluding the possibility that overexpression of this protein affected our system. Notably, our attempt to eliminate individual members of each subfamily using siRNA against only one protein led to a partial effect on autophagy measured by p62 levels in the presence or absence of Baf A (Supplementary Figure S2). This result supports our initial strategy to knock down all members of each Atg8 subfamily.
Figure 2.
The GABARAP/GATE-16 protein subfamily is essential for autophagy. (A) HeLa cells stably expressing GFP-GATE-16 (left panel) or GFP-LC3B (right panel) were transfected with either non-targeting siRNA (control siRNA) or a pool of GABARAP siRNAs (GABARAP, GABARAPL1, and GATE-16) using DharmaFect reagent. After 72 h interval, the cells were incubated for 2 h in EBSS medium in the absence or presence of 0.1 μM Baf A and subjected to western blot analysis after lysis with RIPA extraction buffer. (B) Quantification of relative p62 and GFP-LC3B level (upper panel) and degradation (lower panel) were performed as described in ‘Materials and methods'. Mean±s.d. of three independent experiments is presented. (C) HeLa cells stably expressing GFP-LC3B were transfected with non-targeting siRNA or a pool of GABARAP siRNAs and treated as in (A). The cells were subjected to immunostaining with anti-p62 after fixation, and analysed by confocal microscopy. The GFP intensity of three independent experiments was measured as described in ‘Materials and methods' (right panel). Scale bar: 20 μm.
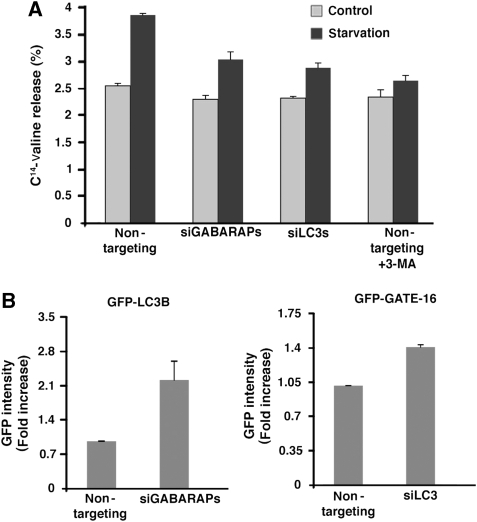
To verify the inhibition of autophagy in both LC3 and GABARAPGATE-16 subfamilies knockdown cells, overall bulk protein degradation was examined. As shown in Figure 3A, LC3s and GABARAPs silencing led, respectively, to about 40 and 60% decrease in the starvation-induced bulk protein degradation compared with the non-targeting transfected control cells. The effect of LC3s or GABARAPs knockdown on autophagy was also tested by FACS analysis (Shvets et al, 2008a). Consistently, both LC3 and GATE-16 accumulated in the knockdown cells, as demonstrated by the elevated levels of GFP intensity measured by FACS (Figure 3B). These experiments further indicated that knock down of either of the Atg8 subfamilies blocks the autophagic flux.
Figure 3.
Both GABARAP/GATE-16 and LC3 protein subfamilies are essential for autophagic flux. (A) Bulk protein degradation was tested in HeLa cells that were transfected with non-targeting siRNA (control siRNA), a pool of GABARAP/GATE-16 siRNAs, or a pool of LC3 siRNAs. The rate of long-lived protein degradation was measured in cells incubated in either αMEM (control) or EBSS (starvation) medium, in the presence or absence of 10 mM of 3-MA, 72 h after siRNA transfection. Values expressing the protein degradation percentage are represented as the mean±s.d. of three determinations. (B) HeLa cells stably expressing GFP-LC3B (left panel) or GFP-GATE-16 (right panel) were transfected with a pool of GABARAP/GATE-16 siRNAs or a pool of LC3, respectively, and with non-targeting siRNA. Seventy-two hours after transfection, the cells were collected after 5 h of starvation and the relative level of GFP-LC3B or GFP-GATE-16 was measured using flow cytometry. Values represent the mean±s.d. of three experiments.
Notably, knock down of each subfamily did not impair lysosomal structure nor the ability to acidify, as tested with anti-LAMP1 antibodies and Lysotracker, respectively (Supplementary Figures S3-A and S3-B). Furthermore, to rule out that the GABARAPs knockdown cells phenotype resulted from an effect on the secretory pathway (Sagiv et al, 2000; Kittler et al, 2001), cells were treated with brefeldin A (BFA), an agent known to retain the Golgi apparatus in the ER (Orci et al, 1991). As shown in Supplementary Figure S3-C, the autophagy flux was not affected by this treatment, thus even in the absence of intact Golgi autophagy was not impaired. Taken together our data indicate that both LC3 and GABARAP/GATE-16 protein subfamilies are crucial for the autophagic process.
Both LC3 and GABARAP/GATE-16 subfamilies promote autophagosome biogenesis
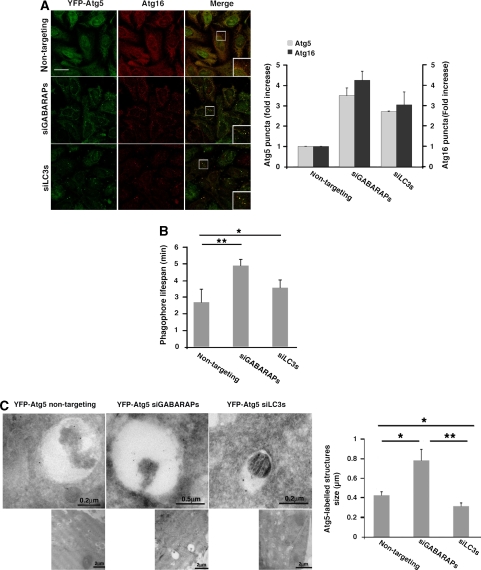
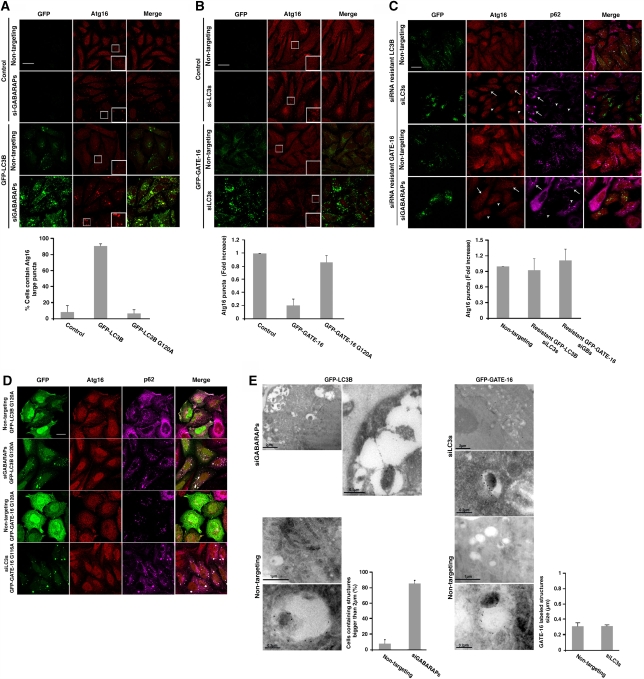
Formation of phagophores is the initial step in autophagosome biogenesis. Atg12–Atg5 conjugate and Atg16, forming together a multisubunit complex, act before the conjugation of Atg8s and recruit them to the membrane, thus serving as bona fide marker for phagophores (Geng and Klionsky, 2008). To test whether knock down of LC3 or GABARAP/GATE-16 subfamilies affected autophagy downstream to phagophore formation, cells stably expressing YFP-Atg5 were transfected with siRNA against LC3s or GABARAPs and stained with anti-Atg16 antibodies after 2 h amino acid starvation. Knock down of either LC3s or GABARAPs led to a three- to four-fold increase in the number of punctate structures labelled with Atg5 or Atg16 (Figure 4A). The elevation in the number of Atg5 positive puncta observed in response to LC3s or GABARAPs knockdown resulted, in part, from increase in their lifespan (Figure 4B; Supplementary Movie 2). Clearly, knock down of any of the Atg8 subfamilies does not affect the recruitment of the complex but rather inhibits autophagy downstream to this step. As the Atg12–Atg5–Atg16 complex associates with phagophores but not with mature autophagosomes, this phenotype suggests an abnormal maturation of autophagosomes. Notably, the effect of Atg8s silencing was not dependent on p62, as additional knock down of this protein did not alter the phenotype described above (Supplementary Figure S4).
Figure 4.
Both Atg8 subfamilies are required for autophagosome maturation. (A) HeLa cells stably expressing YFP-Atg5 were transfected with non-targeting siRNA (control siRNA), a pool of LC3 siRNAs, or a pool of GABARAP/GATE-16 siRNAs using DharmaFect reagent. Seventy-two hours after transfection, the cells were incubated for 2 h in EBSS medium, fixed, and immunostained with anti-Atg16 antibodies. Quantification of Atg5 and Atg16-labelled puncta structures from three independent experiments is presented on the right panel. Scale bar: 20 μm. (B) HeLa cells stably expressing YFP-Atg5 were treated as in (A) and monitored by live microscopy. Quantification of the lifespan of Atg5-labelled puncta structures is presented. Mean±s.d. of three independent experiments is presented. *P<0.05, **P<0.001. (C) YFP-Atg5 cells were transfected with siRNA pool and starved as in (A). Cryo sections of fixed cells were immunolabelled using anti-GFP antibodies and analysed by TEM as described in ‘Materials and methods'. Quantification of the Atg5-labelled structures size is presented at the right panel. *P<0.05, **P<0.001.
Immunoelectron microscopic analysis was used to gain higher resolution of the YFP-Atg5-labelled structures on knock down of GABARAP/GATE-16 or LC3 subfamilies. Apparently, knock down of either subfamily exhibited a different effect on the phagophore appearance; when GABARAPs were knocked down the Atg5-labelled structures appeared significantly larger than in the control cells. However, silencing of the LC3 proteins led to the accumulation of smaller Atg5-labelled structures in comparison to control cells (Figure 4C).
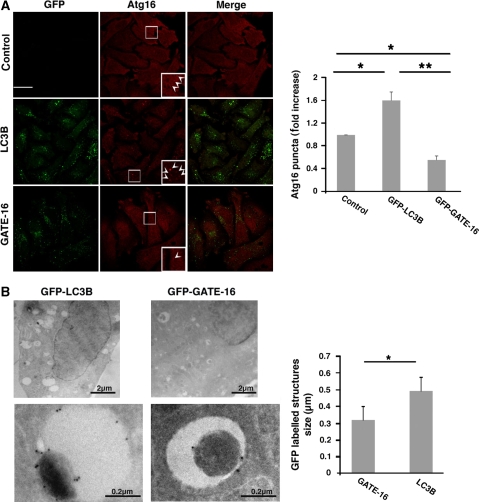
We next tested whether overexpression of LC3B or GATE-16 alters the appearance of phagophores under starvation conditions. Apparently, overexpression of LC3B led to ∼60% increase in the number of visible Atg16-labelled structures whereas GATE-16 overexpression resulted in 40% reduction in the number of these structures (Figure 5A). The effect of overexpressed GFP-LC3B or GFP-GATE-16 was also analysed by immunoelectron microscopy (Figure 5B). Consistently, the membranal structures labelled by GFP-LC3B were larger than those labelled by GFP-GATE-16. The fact that both knock down and overexpression of each Atg8 subfamily led to an opposite effect on the size and number of phagophores, respectively, raises the possibility that LC3s and GABARAPs act in different steps during autophagosomes maturation.
Figure 5.
LC3B and GATE-16 overexpression differently affects phagophore appearance. (A) Control HeLa cells and HeLa cells stably expressing GFP-LC3B or GFP-GATE-16 were starved for 2 h in EBSS medium fixed and immunostained with anti-Atg16 antibodies. Quantification of Atg16-labelled puncta structures from three independent experiments is presented at the right panel. Arrowheads represent Atg-16-labelled puncta. Scale bar: 20 μm. (B) HeLa cells stably expressing GFP-LC3B (left panel) or GFP-GATE-16 (middle panel) were starved for 2 h in EBSS medium. The cells were fixed and their cryo sections were immunolabelled as in (Figure 4B). Quantification of the size of GFP-labelled structures (right panel) from three independent experiments is presented. *P<0.05, **P<0.001.
LC3 and GABARAP/GATE-16 subfamilies act at different steps of autophagosome biogenesis
To further test the hypothesis that the LC3 and GABARAP/GATE-16 subfamilies act differently in the autophagic process we investigated whether overexpression of one Atg8 subfamily member during silencing of the reciprocal subfamily extends the phenotype. As depicted in Figure 6A, the expression of GFP-LC3B enhanced the effect obtained by the GABARAP/GATE-16 subfamily silencing, leading to an accumulation of large structures labelled by Atg16 in about 90% of the cells. GFP-GATE-16 expression, however, reversed the effect of LC3 subfamily knockdown, reducing the Atg16-labelled puncta to 20% in comparison to control cells (Figure 6B). Notably, as shown in Figures 1 and 2, overexpression of either GFP-LC3B or GFP-GATE-16 in this system did not overcome the inhibition of autophagy as detected by the accumulation of LC3B- and GATE-16-labelled puncta, further supporting the specificity of the different siRNA.
Figure 6.
LC3B and GATE-16 act distinctively in autophagosomal biogenesis. (A, B) Control HeLa cells and HeLa cells stably expressing GFP-LC3B (A) or GFP-GATE-16 (B) were transfected with the reciprocal siRNA pools using DharmaFect reagent. After 72 h interval, the cells were incubated for 2 h in EBSS, subjected to immunostaining with anti-Atg16 after fixation, and analysed by confocal microscopy. Quantification of cells containing Atg16 structures larger than 2 μm (A) or Atg16-labelled puncta structures (B) from three independent experiments in comparison to control and to G to A mutants (D) is presented at the lower panel. Scale bar: 20 μm. (C) HeLa cells stably expressing silent GFP-LC3B or GFP-GATE-16 were transfected with their siRNA pools using DharmaFect reagent. After 72-h interval, the cells were incubated for 2 h in EBSS medium and subjected to immunostaining with anti-Atg16 and anti-p62 antibodies followed by fixation. The cells were analysed by confocal microscopy. Arrows represent cells, which do not express GFP proteins whereas arrowheads represent cells, which express GFP-proteins. Quantification of Atg16-labelled puncta structures from three independent experiments is presented at the right panel. Scale bar: 20 μm. (D) HeLa cells stably expressing GFP-LC3BG120A or GFP-GATE-16G116A were transfected with either pool of GABARAP/GATE-16 siRNAs or LC3 siRNAs, respectively, and a non-targeting siRNA. The cells were starved for 2 h, fixed, and immunostained with anti-Atg16 and anti-p62 antibodies. Quantification is presented in (A, B) at the lower panels. Scale bar: 20 μm. (E) HeLa cells stably expressing GFP-LC3B (left panel) or GFP-GATE-16 (right panel) were transfected with the reciprocal siRNA pool or with non-targeting siRNA and starved for 2 h in EBSS medium. The cells were fixed and their cryo sections were immuno-labelled using anti-GFP antibodies and analysed by TEM as described in ‘Materials and methods'. Quantification of cells containing Atg16 structures larger than 2 μm (left panel) or the size of Atg16-labelled structures (right panel) is presented.
To rule out the possibility that the different effects of the two subfamilies on Atg16 accumulation were due to differences in their mRNA levels after siRNA transfection, the effect of overexpression of GFP-LC3B and GFP-GATE-16 in cells knocked down of the same subfamily was examined. Cells stably expressing siRNA resistant GFP-LC3B or GFP-GATE-16 were transfected with the LC3s and GABARAPs siRNA pool, respectively, and the accumulation of phagophores was tested by labelling the cells with Atg16 (Figure 6C). Unlike the effect of these proteins overexpression in cells knocked down to the reciprocal subfamily, and overexpression of GFP-LC3B or GFP-GATE-16 in cells knocked down to the same subfamily led to a recovery of the inhibited autophagic process (Figure 6C). This result further supports the idea that the two subfamilies have different roles in autophagosome formation.
To test whether GFP-LC3B and GFP-GATE-16 activity is dependent on their conjugation to PE, cells stably expressing GFP-LC3BG120A or GFP-GATE-16G116A mutants were transfected with control siRNA or siRNA directed against the reciprocal subfamily. As depicted in Figure 6D, neither GFP-LC3B nor GFP-GATE-16 mutants affected Atg16 localization. Apparently, the effect of LC3B and GATE-16 on Atg16 depends on their lipidation.
The consequence of Atg8s overexpression in the knockdown cells was further tested by immunoelectron microscopy, in which LC3B and GATE-16 were visualized by immunogold labelling using anti-GFP antibodies. As shown in Figure 6E, overexpression of LC3B in cells knocked down of GABARAPs led to an accumulation of large membranal structures (>2 μm), whereas overexpression of GATE-16 in cells knocked down of LC3s did not increased the size of the labelled structures. Taken together, our results are consistent with the idea that the two Atg8 subfamilies are essential for the autophagic process and, more significantly, indicate that LC3B mediate the elongation of the phagophore membrane whereas GATE-16 may be involved in a downstream step along the maturation of autophagosome.
The role of the two Atg8 subfamilies in autophagosome biogenesis was further determined by using an alternative approach bypassing the need for siRNA. It has been recently reported that overexpression of a dominant negative mutant of Atg4B (Atg4BC74A) leads to inhibition of autophagy accompanied by accumulation of phagophores labelled with Atg16 (Fujita et al, 2008a). As Atg4B is a promiscuous protease that recognizes all mammalian Atg8s (Hemelaar et al, 2003), this system cannot dissect the function of the different Atg8 family members. To selectively address the role of the GABARAPs/GATE-16 family members in this process, cells were transfected with the dominant negative mutant of Atg4A (Atg4AC77A), a protease specific to GABARAPs (Scherz-Shouval et al, 2003; Kabeya et al, 2004). As depicted in Figure 7A, overexpression of Atg4AC77A resulted in accumulation of membranal structures labelled with Atg16, consistent with the notion that GABARAPs are essential for autophagosome biogenesis. Overexpression of GFP-LC3B in cells ectopically expressing Atg4AC77A led to a significant increase in the size of the Atg16-labelled structures (Figure 7A). Notably, overexpression of GFP-GATE-16 in cells expressing Atg4AC77A had no such effect, supporting the selectivity of this protease. Importantly, overexpression of Atg4AC77A led to an increase in the number of open autophagic membranes as detected by morphological electron microscopic analysis (Figure 7B). Moreover, the size of both open and closed autophagic membranes formed in cells expressing this mutant was significantly larger (Figure 7B). In sum, the results are consistent with the data obtained after the siRNA approach and further indicate that GABARAPs act during late steps of autophagosome formation and downstream to the elongation of the membrane, possibly in sealing of autophagosomes.
Figure 7.
LC3B mediates phagophore elongation whereas GATE-16 acts downstream. (A) Control HeLa cells and HeLa cells stably expressing GFP-LC3B or GFP-GATE-16 were transfected with HsAtg4AC77A-Myc-His6. After 48 h interval, the cells were incubated for 2 h in EBSS, and subjected to immunostaining with anti-Atg16 and anti-myc followed by confocal microscopy analysis. Quantification of cells containing Atg16 structures larger than 2 μm from three independent experiments is presented at the right panel. Scale bar: 20 μm. (B) HeLa cells were transfected with HsAtg4AC77A-Myc-His6 or empty vector (mock). After a 48 h interval, the cells were incubated for 2 h in EBSS, fixed, and their ultrathin sections were analysed by TEM as described in ‘Materials and methods'. *P<0.05.
Discussion
The occurrence of multiple autophagy-related Atg8 proteins in higher eukaryotes raises the question of whether family members have different roles in the autophagic process. After dissecting the mammalian Atg8s into two subfamilies based on their amino acid similarity, we found that LC3s and GABARAPs have distinct roles in this process. We show that both subfamilies are essential for the overall autophagic process. Moreover, we show that both LC3s and GABARAPs participate in autophagosome biogenesis. By combining different experimental approaches to selectively delete each of the subfamilies we show that LC3s are involved in elongation of the phagophore membrane whereas GABARAPs, represented by GATE-16, act at a later stage possibly in sealing of autophagosomes.
Although in yeast Atg8 is represented by a single protein, almost all other eukaryote ubiquitously express at least two homologs. Previous attempts to determine the role of the mammalian Atg8s in autophagy involved the elimination of the entire family by knocking out of the conjugation enzyme Atg3 (Sou et al, 2008) or overexpressing the protease dominant negative mutant Atg4BC74A (Fujita et al, 2008a), which led to the accumulation of small (Sou et al, 2008), unsealed phagophores (Sou et al, 2008; Fujita et al, 2008a). By using a siRNA approach, we selectively knocked down each of the Atg8 subfamilies and showed that both are required to complete the maturation of autophagosomes. The GATE-16/GABARAP subfamily, previously implicated in intra-cellular protein trafficking processes (Sagiv et al, 2000; Kittler et al, 2001; Nakamura et al, 2008), was recently linked to autophagy (Betin and Lane, 2009; Novak et al, 2010). In this study, we show that although similar in structure, the two subfamilies did not complement each other indicating that these proteins are acting differentially in the autophagic process.
The molecular mechanisms of autophagosome biogenesis is largely unknown, however, evidence showed that this process involves the recruitment of the Atg5/Atg12/Atg16 complex to the phagophore membrane (Fujita et al, 2008b). Consistently, it has been recently suggested that this extremely conserved complex acts as an E3-like factor facilitating the conjugation of Atg8 to the autophagic membrane (Hanada et al, 2007). The Atg5/Atg12/Atg16 complex is restricted to the phagophore and is excluded from mature autophagosomes (Mizushima et al, 2003). In this study, we show that both LC3B and GATE-16 undergo lipidation in the absence of the reciprocal subfamily whereas the overall autophagy is inhibited. Moreover, we show in our system that Atg8s act only on conjugation to PE, implying that both Atg8 subfamilies act downstream to the Atg5/Atg12/Atg16 complex. We also show that overexpression of LC3B in cells deleted of functional GABARAPs leads to an accumulation of the Atg5/Atg12/Atg16 complex on the phagophore membrane, a process accompanied by a significant enlargement of these membranes. In contrast, overexpression of GATE-16 (in its lipidation form) in cells knocked down of LC3s leads to the dissociation of the Atg5/Atg12/Atg16 complex from the phagophore membrane. This is consistent with the notion that GATE-16 (a representative of the GABARAP subfamily) acts at a late stage of the autophagosome biogenesis process.
A better insight into the role of the GABARAP subfamily in autophagosome formation was gained from the experiments using the dominant negative mutant of Atg4A. Notably, this protease specifically recognizes GABARAPs (Scherz-Shouval et al, 2003; Kabeya et al, 2004; Figure 7A), thus serving as an excellent tool to study their specific function. In agreement with Yoshimori and co-workers (Fujita et al, 2008a; Hayashi-Nishino et al, 2009), who sequestered all ATG8s using the dominant negative form of Atg4B, we show here that selective removal of GABARAPs by the Atg4A mutant leads to the accumulation of open autophagic membranes. On the basis of both studies we conclude that the GABARAPs subfamily is involved in the regulation of the sealing process needed for the maturation of autophagosomes. As the Atg4A mutant does not affect the LC3s, our system also reflects on the function of this subfamily. The accumulation of larger phagophores under these conditions further emphasizes the role of LC3s in the elongation process. In agreement with our model, removal of both subfamilies by Atg4B led to the accumulation of small autophagic membranes (Fujita et al, 2008a; Hayashi-Nishino et al, 2009).
The exact role of the Atg8s in the autophagic process is yet unclear. The yeast Atg8 was shown to act in autophagosome biogenesis and according to a recent report to mediate membrane fusion. In mammals Atg8s have a dual role: LC3 was shown by several groups to act as a selective recruiter of p62 and NBR1, which bind to polyubiquitinated proteins (Bjorkoy et al, 2005; Kirkin et al, 2009). Importantly, this function is not essential for the formation of autophagosomes, as knock down of both p62 and NBR1 does not affect autophagy (Shvets et al, 2008b; Kirkin et al, 2009). As indicated above, removal of all Atg8s in mammalian systems blocks autophagosome biogenesis (Sou et al, 2008; Fujita et al, 2008a). It is possible that some of the mammalian Atg8s are involved in cargo recruiting whereas others may function specifically in autophagosome biogenesis. Our results, however, indicate that both LC3s and GABARAPs are essential for autophagosome formation and therefore suggest a dual role for individual members of this protein family.
Materials and methods
Cell culture and transfection
HeLa cells were grown on αMEM medium supplemented with 10% foetal calf serum (FCS) and 1% penicillin–streptomycin (Sigma) at 37°C in 5% CO2. All stable clones of transfected HeLa cells were selected in 1 mg/ml geneticin (G418). Transfection was conducted using LT1 transfection reagent (Mirus) and the transfected rates were over 85%. For siRNA silencing subconfluent HeLa cells were transfected using DharmaFect 1 (Dharmacon) with different pools of siRNA (50 nM from each siRNA SMARTpool). For example, to knock down an entire subfamily a total concentration of 150 nM siRNA was used. siRNA SMARTpools, consisting of four RNA duplexes, each targeting LC3A (M-013579-00), LC3B (M-012846-01), LC3C (M-32399-01), GABARAP (M-012368-01), GABARAPL1 (M-014715-01) or GABARAPL2 (M-006853-02) (GABARAPL3 was not used because it was not commercially available), p62 (SQSTM1) (M-010230-00), and non-targeting siRNAs control (D-001206-14) were purchased from Dharmacon (Lafayette, CO, USA).
siRNA resistant GFP-LC3B and GFP-GATE-16 were constructed by the introduction of silent mutations in all regions of these cDNAs that are complementary to the RNA duplexes from each pool.
Cells were grown to densities of 5 × 104 per 4-cm dish in 2 ml αMEM without antibiotics, and transfected with DharmaFect according to the manufacturer's instructions. Experiments were performed 72 h after transfection. To obtain starvation conditions, cells were washed three times with PBS and incubated in EBSS medium at 37°C for 2 h. To inhibit lysosomal degradation cells were starved in the presence of 100 nM Baf A. BFA was used at a final concentration of 2 μg/mg. Total cell extracts were made using RIPA extraction buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 1.5 mM MgCl2, 1% DOC, 1% Triton X-100) with a protease inhibitors mixture (Sigma-Aldrich).
Antibodies
The following antibodies were used: rabbit polyclonal anti-Atg16 (Cosmo Bio Co.); mouse monoclonal anti-LAMP1 (Developmental Studies Hybridoma Bank, University of Iowa); mouse monoclonal anti-GFP (Babco); rabbit polyclonal anti-GFP (ImmunoEM) (Abcam); mouse monoclonal anti-myc mouse and monoclonal anti-p62 antibodies (Santa-Cruz Biotechnology); rhodamine-conjugated goat anti-rabbit IgG and Cy™5-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch). LysoTracker Red DND-99 was obtained from Molecular Probes. Anti-LC3 antibody was produced by immunization of a rabbit with a peptide corresponding to the 14 amino acids of the N-terminus of LC3 with an additional cysteine (PSEKTFKQRRTFEQC). The above antibodies were used in western blot analyses in a dilution of 1:1000 and in immunofluorescence analyses in a dilution of 1:200.
Real-time PCR analysis
The levels of each Atg8 homolog were tested on cDNA from cells transfected with the siRNA pool against its subfamily and control non-targeting siRNA. Real-time PCR, reverse transcription, and cell lysis were carried out with Power SYBR Green Cells-to-CT kit (Applied Biosystems) using human TBP, GABARAP, GABARAP-L1, GABARAP-L2, and LC3B-specific primers. The levels of LC3A and LC3C were tested on the above cDNAs using Taq-man master mix (Applied Biosystems).
Bulk protein degradation
Cells were transfected with different siRNA pools as described earlier and grown to 70–80% confluence in 12-well plates (35 mm). Cells were then labelled for 16 h in medium containing [14C]valine (0.5 mCi/ml) and 5% FCS in valine-free αMEM. After three rinses with PBS, cells were incubated in valine-free αMEM 10 mM cold valine. After 3-h incubation, the medium was replaced with either αMEM or EBSS medium, also containing 0.1% BSA and 10 mM cold valine, and cells were incubated for additional 4 h. When indicated, 10 mM of 3-MA was added to the medium. The medium was precipitated in 10% TCA, and TCA-soluble radioactivity was measured. Total cell radioactivity was measured after lysis with 0.1 M NaOH. [14C]-valine release was calculated as the percentage of the radioactivity in the TCA-soluble supernatant relative to total cell radioactivity.
Statistical analysis
Results from separate western blots experiments (three at least) were analysed using ImageQuant image program and quantified as follows:
The percentages of p62, LC3B, or GATE-16 degradation were calculated as the difference in total (lipidated and unlipidated formes) p62/LC3B/GATE-16 amount (normalized to actin as loading control) in the presence or absence of Baf A. The amount obtained from the control siRNA cells was set to 100%. The relative levels of p62, LC3B, or GATE-16 proteins were calculated as the difference in total p62/LC3B/GATE-16 amount (normalized to actin as loading control) with the control siRNA set to 100%.
For analysis of immunofluorescence experiments, images of ∼50–60 cells were taken in each experiment and 3–5 experiments were analysed, bringing the total number of cells to 150–300 per one determination.
GFP intensity quantification was performed using ImageJ program (NIH Image).
Atg5 and Atg16 were quantified by counting the labelled puncta structures and in GFP-LC3B expressing cells by counting only structures that are larger than 2 μm.
GFP intensity was also measured by FACS analysis (described in Shvets et al, 2008a) of three experiments, collecting in each experiment ∼30 000 cells. The level of GFP fluorescence intensity in LC3s or GABARAPs knockdown cells was normalized to the level in control cells, which was set to 1.
Fluorescence microscopy
HeLa cells were plated on sterile coverslips (13 mm diameter) and cultured under the conditions indicated. Cells were fixed with 3% paraformaldehyde in PBS for 10 min and permeabilized with 0.1% Triton-X100 for 8 min. When anti-LC3 antibodies were used, cells were fixed with cold methanol for 5 min at −20°C and permeabilized by quick washing with cold acetone. Cells were blocked by incubation with 10% FCS in PBS for 30 min at room temperature, followed by 1 h incubation with the primary antibody. Cells were then incubated with the secondary antibody for 30 min. LysoTracker Red DND-99 (Molecular Probes) was added to live cells. Confocal images were taken by a FV500 laser-scanning confocal microscope equipped with a PLAPO 60 × 1.4 NA oil immersion lens and analysed by Fluoview software (Olympus).
Immunoelectron microscopy
HeLa cells were fixed in 4% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M cacodylate buffer, pH=7.4, washed in the same buffer, embedded in 10% gelatin and sliced into thin pieces. The samples were then infused with 2.3 M sucrose in 0.1 M cacodylate buffer overnight on a rotary stirrer and rapidly frozen in liquid nitrogen. Frozen ultrathin (60–80 nm) sections were cut with a diamond knife at −120°C on Leica EM FC6 ultra microtome with a cryo chamber. The sections were collected on 200-mesh formvar-coated nickel grids and floated on PBS, after blocking with 1% BSA, 1% gelatin, 0.1% glycine, and 0.1% Tween-20. Immunolabelling was performed using rabbit polyclonal anti-GFP (dilution of 1:50) and goat anti-rabbit IgG conjugated to 10 nm gold particles (dilution of 1:20). Contrasting and embedding was performed by Tokyashu technique (Tokuyasu, 1986) in methylcellulose and 0.2% uranyl acetate. The embedded sections were scanned and analysed under 120 kV by Tecnai 12 Transmission Electron Microscope with EAGLE CCD camera using TIA software (FEI).
Morphology electron microscopy analysis
HeLa cells were fixed with 3% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) washed in the same buffer and post fixed with 1% osmium tetroxide. After en bloc staining with 2% uranylacetate in water for 1 h at RT, the slices were dehydrated in graded ethanol solutions and embedded in Epon 812. Ultrathin sections (70–90 nm thickness) were prepared using Ultramicrotome Leica UCT and analysed under 120 kV at Spirit Transmission Electron Microscope (FEI).
Supplementary Material
Acknowledgments
We thank Noboru Mizushima for the anti- Atg16 antibodies. We also thank Y Avivi for discussions and critical evaluation of the manuscript, and V Kiss for technical assistance. ZE is an incumbent of the Harold Korda Chair of Biology. This work was supported in part by the Israel Science Foundation, the Israeli Cancer Research Foundation, and the Weizmann Institute Minerva Center. The electron microscopy studies were conducted at the Irving and Cherna Moskowitz Center for Nano and Bio-Nano Imaging at The Weizmann Institute of Science.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abeliovich H, Dunn WA Jr, Kim J, Klionsky DJ (2000) Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol 151: 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betin VM, Lane JD (2009) Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci 122: 2554–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Levine B (2008) The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 15: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T (2008a) An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 19: 4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T (2008b) The Atg16 L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ (2008) The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ′Protein modifications: beyond the usual suspects′ review series. EMBO Rep 9: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282: 37298–37302 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, Zhao S, Liu JO, Yu L (2003) Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem 278: 29278–29287 [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL (2003) A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8 L. J Biol Chem 278: 51841–51850 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117: 2805–2812 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Kittler JT, Rostaing P, Schiavo G, Fritschy JM, Olsen R, Triller A, Moss SJ (2001) The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABA(A) receptors. Mol Cell Neurosci 18: 13–25 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T (2003) Mouse Apg16 L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 116: 1679–1688 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Hayashi T, Nasu-Nishimura Y, Sakaue F, Morishita Y, Okabe T, Ohwada S, Matsuura K, Akiyama T (2008) PX-RICS mediates ER-to-Golgi transport of the N-cadherin/beta-catenin complex. Genes Dev 22: 1244–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130: 165–178 [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 11: 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Tagaya M, Amherdt M, Perrelet A, Donaldson JG, Lippincott-Schwartz J, Klausner RD, Rothman JE (1991) Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell 64: 1183–1195 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Sagiv Y, Legesse-Miller A, Porat A, Elazar Z (2000) GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J 19: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Sagiv Y, Shorer H, Elazar Z (2003) The COOH terminus of GATE-16, an intra-Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J Biol Chem 278: 14053–14058 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvets E, Fass E, Elazar Z (2008a) Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy 4: 621–628 [DOI] [PubMed] [Google Scholar]
- Shvets E, Fass E, Scherz-Shouval R, Elazar Z (2008b) The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci 121: 2685–2695 [DOI] [PubMed] [Google Scholar]
- Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, Kominami E, Tanaka K, Komatsu M (2008) The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 19: 4762–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT (1986) Application of cryoultramicrotomy to immunocytochemistry. J Microsc 143: 139–149 [DOI] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19: 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y, Yu L, Chen Z, Zheng L, Fu Q, Jiang J, Zhang P, Gong R, Zhao S (2001) Cloning, expression patterns, and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics 74: 408–413 [DOI] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL (2009) 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5: 1180–1185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.