Abstract
Epithelial–mesenchymal transition (EMT) is a transdifferentiation programme. The mechanism underlying the epigenetic regulation of EMT remains unclear. In this study, we identified that Snail1 interacted with histone lysine-specific demethylase 1 (LSD1). We demonstrated that the SNAG domain of Snail1 and the amine oxidase domain of LSD1 were required for their mutual interaction. Interestingly, the sequence of the SNAG domain is similar to that of the histone H3 tail, and the interaction of Snail1 with LSD1 can be blocked by LSD1 enzymatic inhibitors and a histone H3 peptide. We found that the formation of a Snail1–LSD1–CoREST ternary complex was critical for the stability and function of these proteins. The co-expression of these molecules was found in cancer cell lines and breast tumour specimens. Furthermore, we showed that the SNAG domain of Snail1 was critical for recruiting LSD1 to its target gene promoters and resulted in suppression of cell migration and invasion. Our study suggests that the SNAG domain of Snail1 resembles a histone H3-like structure and functions as a molecular hook for recruiting LSD1 to repress gene expression in metastasis.
Keywords: chromatin, demethylation, EMT, LSD1, Snail1
Introduction
The increased motility and invasiveness of tumour cells are reminiscent of the processes at the epithelial–mesenchymal transition (EMT), which is essential for morphogenetic events in embryonic development, tissue remodelling, wound healing and metastasis (Wu and Zhou, 2008; Yang and Weinberg, 2008; Thiery et al, 2009). In these EMT processes, epithelial cells acquire fibroblast-like properties and show reduced intercellular adhesion and increased motility. These acquired traits confer cancer cells with the distinct advantages of invasion, metastatic dissemination and acquisition of drug resistance (Wu and Zhou, 2008; Yang and Weinberg, 2008; Thiery et al, 2009). Interestingly, EMT is a reversible process (Thiery, 2002; Weinberg, 2008). When cancer cells disseminate to distant sites in the body, they no longer encounter the signals that they experienced in the primary tumour and they can revert to an epithelial state by a mesenchymal–epithelial transition (MET). The phenotypic plasticity of cancer cells at EMT/MET suggests that an epigenetically regulated differentiation programme is involved (Feinberg, 2007).
In the eukaryotic nucleus, DNA is packaged with core histones and other chromosomal proteins to form chromatin (Bernstein et al, 2007; Downs et al, 2007; Fraser and Bickmore, 2007). The basic repeating unit of chromatin, nucleosome, is composed of two copies of each of the four core histones, H2A, H2B, H3 and H4, wrapped by 146 base pairs of DNA (Bernstein et al, 2007; Downs et al, 2007; Fraser and Bickmore, 2007). Chromatin harbours not only genetic information encoded in the DNA but also epigenetic information carried by the reversible covalent modifications at the N-terminal tails of histones (Esteller, 2007). Histone modifications, such as acetylation, phosphorylation and methylation, control gene expression by altering chromatin structure (Berger, 2007; Bhaumik et al, 2007; Ruthenburg et al, 2007). Methylation at specific residues of histone H3, in particular, has a major role in the maintenance of the active or silent state of gene expression (Martin and Zhang, 2005; Bhaumik et al, 2007). For example, methylated lysine 9 or lysine 27 on histone H3 (denoted as H3K9 and H3K27) is generally associated with genes the transcription of which is repressed, whereas methylated H3K4, H3K36 and H3K79 are found in active chromatin. Although histone methylation was long considered irreversible, the recent identification of several site-specific histone demethylases provides compelling evidence that this modification is dynamically regulated (Klose and Zhang, 2007; Shi and Whetstine, 2007; Cloos et al, 2008). The reversible methylation of histones allows gene expression to be rapidly switched on at differentiation, while retaining cellular plasticity in response to developmental and microenvironmental signals (Reik, 2007). The mechanism underlying the epigenetic regulation of EMT at metastasis remains unclear, and thus, an understanding of this process will provide important insight into the early steps of cancer metastasis.
A hallmark of EMT is the loss of E-cadherin expression (Nieto, 2002; Thiery, 2002), which is often inversely correlated with tumour grade and stage (Cowin et al, 2005). Several transcription factors, such as Snail1, Twist and ZEB1, have been implicated in the transcriptional repression of E-cadherin and in the induction of EMT (Nieto, 2002; Thiery and Sleeman, 2006; Peinado et al, 2007; Yang and Weinberg, 2008). Snail1, a DNA-binding factor, which was identified in Drosophila as a suppressor of the transcription of shotgun (an E-cadherin homologue), controls large-scale cell movement during formation of the mesoderm and neural crest (Nieto, 2002). Expression of Snail1 suppresses E-cadherin expression and induces EMT in MDCK and breast cancer cells, indicating that Snail1 has a fundamental role in EMT and breast cancer metastasis (Batlle et al, 2000; Cano et al, 2000; Zhou et al, 2004). Our findings, and those of others, clearly show that expression of Snail1 correlates with high tumour grade and nodal metastasis and is predictive of a poor outcome in patients with breast cancer (Cheng et al, 2001; Blanco et al, 2002; Zhou et al, 2004; Martin et al, 2005). Besides being a critical regulator of EMT, Snail1, when overexpressed, induces resistance to apoptosis and tumour recurrence in breast cancer (Kajita et al, 2004; Vega et al, 2004; Moody et al, 2005). To better understand the mechanism underlying the epigenetic regulation of EMT at metastasis, we undertook the unbiased approach of tandem array purification (TAP) coupled with mass spectrometry analysis to identify the chromatin-modifying enzymes that interact with Snail1. We found that Snail1 interacted with lysine-specific demethylase 1 (LSD1), a key component of several co-repressor complexes including CoREST, CtBP and HDAC1/2, which remove the methylation of H3K4 (Shi, 2007). In this paper, we characterize the functional interaction of Snail1 with LSD1 and investigate their roles in mediating chromatin modification during metastasis.
Results
Snail1 interacts with LSD1 through the SNAG domain
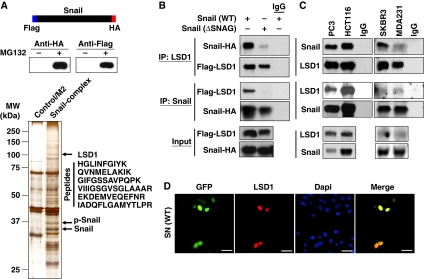
To identify the potential proteins that interact with Snail1, we generated a stable HEK293 cell line expressing dual-tagged Snail1 (Figure 1A). After enriching the nuclear extracts, we carried out a two-step sequential protein purification process with Flag and hemagglutinin (HA) affinity columns (Shi et al, 2003). The final immunocomplexes were separated on SDS–PAGE and subjected to silver staining (Figure 1A). Bound proteins were excised from the gel and subjected to mass spectrometry analysis. Three known proteins that interact with Snail1, GSK-3β, β-Trcp and PRMT5 (Zhou et al, 2004; Hou et al, 2008), were found in the complexes, which validated the specificity of this system. Lysine-specific demethylase 1 (Shi, 2007), an H3K4 demethylase, was also identified as a protein that associated with Snail1. Six trypsin-digested peptides from a protein with a molecular weight of about 110 kDa were perfectly matched to the protein sequence of LSD1 (Figure 1A).
Figure 1.
Snail1 interacts with LSD1 through the SNAG domain. (A) The schematic diagram shows the stable expression of dual-tagged Snail1 in HEK293 cells (top and middle panels). The Snail1 complex was isolated by two-step immunopurification, separated on SDS–PAGE and visualized by silver staining. A protein with molecular weight close to 110 kDa was excised and identified as LSD1 by mass spectrometry (bottom panel). (B) Flag-tagged LSD1 and HA-tagged wild-type or SNAG-deleted Snail1 were co-expressed in HEK293 cells. After immunoprecipitation, bound Snail1 or LSD1 was examined by western blotting. (C) Endogenous Snail1 and LSD1 were immunoprecipitated from PC3, HCT116, SKBR3 and MDA-MB231 cells and bound endogenous LSD1 and Snail1 were examined by western blotting. (D) GFP-tagged Snail1 was expressed in HEK293 cells. After fixation, the cellular localization of Snail1 (green) and LSD1 (red) was examined by immunofluorescent staining. Scale bar=50 μm.
To validate the physical interaction of Snail1 with LSD1, we co-expressed Snail1–HA and Flag–LSD1 in HEK293 cells and conducted a co-immunoprecipitation experiment. After immunoprecipitating LSD1, we detected the associated Snail1, and vice versa (Figure 1B), indicating that these two molecules are associated. Interestingly, deletion of the SNAG (Snail1/GFI) domain (Batlle et al, 2000; Nieto, 2002; Peinado et al, 2004; Barrallo-Gimeno and Nieto, 2009), a highly conserved repressive domain present at the N-terminus of several transcription factors (such as Snail1 and GFI1), significantly reduced the interaction of Snail1 with LSD1, indicating that the SNAG domain is required for this interaction (Figure 1B). We also immunoprecipitated endogenous Snail1 and LSD1 from PC3, HCT116, MDA-MB 231 and SKBR3 cells and detected the presence of endogenous LSD1 and Snail1, respectively (Figure 1C). Consistent with these data, when GFP–Snail1 was expressed in HEK293 cells, we found that Snail1 was co-localized with endogenous LSD1 in the nucleus (Figure 1D). Taken together, our results indicate that Snail1 interacts with LSD1 and that the SNAG domain of Snail1 is required for mediating their interaction.
The SNAG domain is essential for the stability of Snail1
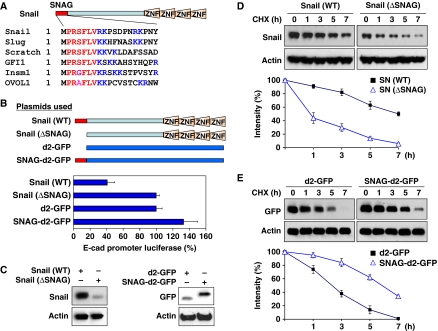
In addition to Snail1 and GFI1, several other transcription factors, such as Slug, Scratch, insulinoma-associated protein IA-1 (Insm1) and Ovo-like 1 (OVOL1), also contain a similar SNAG domain at their N-terminus (Figure 2A). This motif is highly conserved among species and is important for the repressive activity of these transcription factors in mammalian cells (Batlle et al, 2000; Nieto, 2002; Peinado et al, 2004; Barrallo-Gimeno and Nieto, 2009). We, along with others, previously showed that Snail1 is a highly unstable protein and is regulated by protein stability and subcellular localization (Dominguez et al, 2003; Zhou et al, 2004; Yook et al, 2005). To test whether the SNAG domain regulates the protein stability, subcellular localization and repressive activity of Snail1, we generated a Snail1 mutant with the SNAG domain deleted (ΔSNAG–Snail1) and a SNAG-fused destabilized d2-GFP (SNAG–d2-GFP; Figure 2B). Consistent with previous findings, deletion of the SNAG domain significantly reduced the repressive activity of Snail1 on the E-cadherin promoter, whereas adding the SNAG domain to d2-GFP had no effect (Figure 2B), suggesting that the SNAG domain is required but is not sufficient for the transcriptional repressive activity of Snail1. Interestingly, ΔSNAG-Snail1 became less stable than wild-type Snail1, whereas SNAG-d2-GFP became stabilized in comparison with d2-GFP (Figure 2C). The failure of ΔSNAG-Snail1 to suppress E-cadherin promoter activity (Figure 2B) was not due to the instability of this molecule, as treatment with MG132 did not enhance the suppressive function of this mutant (Supplementary Figure S1). To further extend this finding, we investigated the degradation of these proteins with treatment of the protein synthesis inhibitor, cycloheximide, for various time intervals. We found that the protein level of ΔSNAG–Snail1 significantly decreased after 1 h in comparison with that of wild-type Snail1 (Figure 2D). In contrast, SNAG–d2-GFP became more stable than d2-GFP (Figure 2E), indicating that the SNAG domain is critical for maintaining the protein stability of Snail1, in addition to its transcriptional repressive function.
Figure 2.
The SNAG domain is essential for the stability of Snail1. (A) Sequence alignment of the SNAG domain from several transcriptional repressors. The consensus sequence is shown in red and the lysine and arginine residues are highlighted in blue. (B) Scheme showing the SNAG constructs used in this study (top panel). WT or SNAG-deleted Snail1, d2-GFP or SNAG-d2-GFP was co-expressed with the E-cadherin promoter luciferase construct in MCF7 cells. After 48 h, luciferase activity was measured by using a Dual-Luciferase Reporter Assay (Promega) (mean±s.d. of three separate experiments; bottom panel). (C) WT and SNAG-deleted Snail1, d2-GFP and SNAG–d2-GFP were expressed in HEK293 cells and analysed by western blotting. (D) WT or SNAG-deleted Snail1 was expressed in HEK293 cells then treated with cycloheximide (10 μg/ml) for different time intervals. The level of Snail1 was analysed by western blotting. Densitometry results from three independent experiments were statistically analysed and plotted (bottom panel). A representative western blotting experiment is shown in the top panel. (E) d2-GFP or SNAG-fused d2-GFP was expressed in HEK293 cells and treated with cycloheximide as described above. The level of d2-GFP was analysed by western blotting. Densitometry results from three independent experiments were statistically analysed and plotted (bottom panel) and a representative blot is shown on the top panel.
We next examined whether the SNAG domain affects the subcellular location of these proteins. Although the intensity of SNAG–d2-GFP was enhanced because of its increased stability, d2-GFP and SNAG–d2-GFP were distributed equally in the nucleus and cytoplasm (Supplementary Figure S2A). We also expressed wild-type Snail1– and ΔSNAG–Snail1–GFP in HEK293 cells and found that both were localized predominantly in the nucleus (Supplementary Figure S2B). Although ΔSNAG–Snail1 has significantly decreased protein stability, the loss of the SNAG domain did not affect its nuclear localization. We also performed subcellular fractionation analysis and found that the addition or deletion of the SNAG domain did not change the subcellular localization of d2-GFP or Snail1, respectively (Supplementary Figure S3). Taken together, our results indicate that the SNAG domain is critical in controlling the protein stability but not the subcellular location of Snail1.
The SNAG domain of Snail1 interacts with LSD1 by mimicking the structure of the histone H3 tail
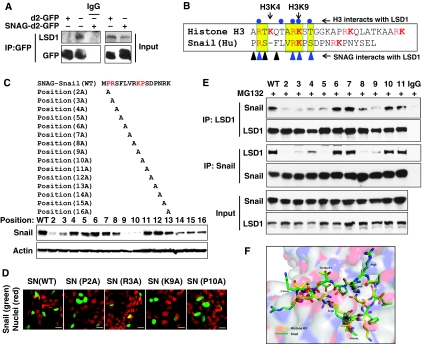
We also expressed d2-GFP and SNAG–d2-GFP in HEK293 cells. Immunoprecipitation of SNAG–d2-GFP, but not of d2-GFP, revealed the association of endogenous LSD1, indicating that the SNAG domain is sufficient for Snail1 to interact with LSD1 (Figure 3A). Lysine-specific demethylase 1 is a demethylase for H3K4me2 (Shi, 2007). Although it can bind to the histone H3 tail for demethylation, it does not contain a DNA-binding motif. The manner in which LSD1 binds to a specific promoter chromatin remains unclear. The X-ray structure of the LSD1–CoREST–Histone H3 peptide complex has been determined (Forneris et al, 2007, 2008). Forneris et al (2008) demonstrated that residues of Arg2, Thr6, Arg8, Lys9 and Thr11 of histone H3 (highlighted with blue dots at the top of Figure 3B) are critical for establishing the contact interactions of histone H3 within the catalytic cavity of LSD1. We noticed that the sequence of the SNAG domain is highly similar to that of the N-terminus of histone H3 and contains arginine- and lysine-rich residues (Figure 3B). Interestingly, the SNAG domain of Snail1 contains almost identical residues at four of these five positions (Arg3, Arg8, Lys9 and Ser11). To identify the critical residues on the SNAG domain required for interaction with LSD1, we performed alanine scan mutagenesis on the SNAG domain of Snail1 (Figure 3C). Among the 15 Snail1 mutants screened, we found that mutations at Pro2, Arg3, Lys9 and Pro10 decreased the protein stability of Snail1 (Figure 3C; Supplementary Figure S4). However, treatment with proteasome inhibitor, MG132, restored the protein stability of these mutants (Supplementary Figure S4), indicating that these four residues are critical for controlling the protein stability of Snail1. This is consistent with the finding that the SNAG domain is important for the protein stability of Snail1 (Figure 2). Similar to the SNAG deletion mutant of Snail1, mutation of these four residues did not alter the nuclear localization of Snail1 (Figure 3D).
Figure 3.
The SNAG domain of Snail1 interacts with LSD1 by mimicking the structure of the tail of histone H3. (A) d2-GFP or SNAG–d2-GFP was expressed in HEK293 cells. After immunoprecipitation of d2-GFP, bound endogenous LSD1 was examined by western blotting. (B) The schematic diagram shows the sequence alignment of the SNAG domain with the histone H3 tail. The conserved sequence is highlighted in yellow and arginine and lysine resides are shown in red. Residues of Arg2, Thr6, Arg8, Lys9 and Thr11 of histone H3 (highlighted in blue dots on the top) are important for establishing the critical interaction of histone H3 within the catalytic cavity of LSD1 (Forneris et al, 2008). Triangles at the bottom represent the residues on the SNAG domain that are important for interacting with LSD1. (C) Schematic diagram shows the position of the alanine mutations in the SNAG domain (top panel). The Snail1 mutants were expressed in HEK293 cells and the level of Snail1 was examined by western blotting (bottom panel). (D) EGFP-tagged WT or mutant Snail1 was expressed in HEK293 cells and the subcellular localization of Snail1 (green) was visualized by immunofluorescence microscopy (DAPI for nuclei, red). Scale bar=20 μm. (E) WT or mutant Snail1 was expressed in HEK293 cells treated with MG132 (10 μM) for 6 h. Endogenous LSD1, WT or Snail1 mutants were immunoprecipitated, and the bound Snail1 or endogenous LSD1 was examined by western blotting, respectively. Input lysates were shown in the bottom panel. (F) A ball-and-stick model structure of the LSD1–SNAG–Snail1 complex (green) superposed to the LSD1–histone H3 peptide complex (yellow; PDB access code 2V1d) showing the position of key interacting residues and the substrate lysine residue (Lys4). The LSD1 molecule is shown as a partially transparent surface representation.
We next examined the interaction of these 15 mutants with LSD1 by immunoprecipitating endogenous LSD1. We found that Pro2, Arg3, Ser4, Phe5, Arg8 and Lys9 mutants completely lost their ability to interact with LSD1 (Figure 3E; data not shown). The loss of interaction of these mutants with LSD1 was not due to the instability of these mutants, as cells were treated with the proteasome inhibitor, MG132, to prevent Snail1 from degrading (input lysates on Figure 3E). Consistent with these data, when Snail1 was immunoprecipitated, the association of these mutants with LSD1 was also abolished (Figure 3E). Interestingly, mutants that cannot interact with LSD1 also lost their ability to inhibit E-cadherin promoter luciferase activity, suggesting that the interaction with LSD1 is critical for the suppressive function of Snail1 (Supplementary Figure S5).
We also performed protein modelling analysis on the basis of the structure of the LSD1–CoREST–Histone H3 complex. We found that the SNAG domain of Snail1 adopted a conformation that was superimposed by the histone H3 tail at the catalytic cavity of LSD1 (Figure 3F). Noticeably, Arg3, Arg8 and Lys9 of the SNAG domain of Snail1 participate in similar critical contacts within the catalytic cavity of LSD1, compared with those of the histone H3 tail. This is consistent with the finding that these mutants lose their interaction with LSD1 and their suppressive function on the E-cadherin promoter. As methylation of arginine and lysine residues has been reported on other non-histone proteins (Huang and Berger, 2008), and because Arg3, Arg8 and Lys9 in the SNAG domain of Snail1 are critical for the interaction with LSD1, we speculate that methylation of these three residues may regulate their interaction with LSD1. To test this idea, we immunoprecipitated Snail1 and subjected it to western blot analysis using antibodies against H3K4, H3K9 and H3K27 methylation, as well as against pan-lysine and pan-arginine methylation (Supplementary Figure S6; data not shown). Unfortunately, we were unable to detect the methylation of Snail1 because of the lack of a specific methylation antibody on these three residues. Together, our results indicate that the SNAG domain of Snail1 interacts with LSD1 by adopting a conformation similar to that of the N-terminal tail of histone H3.
The amine oxidase domain of LSD1 is responsible for its interaction with Snail1
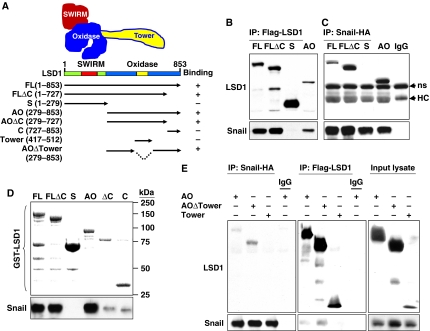
The N-terminal one-third of LSD1 contains a SWIRM (Swi3p, Rsc8p and Moira) domain that is commonly found in chromatin-remodelling complexes with unknown functions (Figure 4A). The C-terminal two-thirds of LSD1 comprise an amine oxidase (AO) domain that shares extensive sequence homology with FAD-dependent AO. To identify the region that is responsible for the LSD1 interaction with Snail1, we generated LSD1 domain-deletion mutants (Figure 4A) and co-expressed them with Snail1 in HEK293 cells. As expected, immunoprecipitation of full-length LSD1 revealed the association with Snail1. A small C-terminal deletion mutant of LSD1 and the AO domain retained the ability to interact with Snail1 (lanes 2 and 4; Figure 4B; Supplementary Figure S7). The N-terminal region of the SWIRM domain, however, was completely incapable of interacting with Snail1 (lane 3; Figure 4B). In the reciprocal immunoprecipitation experiment, immunoprecipitation of Snail1 revealed the associations between full-length and small C-terminal-deleted LSD1 and the AO domain, but not with the SWIRM domain (Figure 4C). These results indicate that the AO domain is required for the interaction of LSD1 with Snail1. Consistent with this result, when wild-type and deletion mutants of glutathione-S-transferase (GST)–LSD1 were pulled down from cell lysates, the SWIRM domain of LSD1 failed to interact with Snail1 (lane 3; Figure 4D), confirming that the AO domain but not the SWIRM domain is required for mediating the interaction of LSD1 with Snail1.
Figure 4.
AO domain of LSD1 is responsible for its interaction with Snail1. (A) The cartoon and schematic diagram shows the structure of LSD1 and the different deletion constructs used in this study. (B, C) HA-tagged Snail1 and Flag-tagged full-length or deletion mutants of LSD1 were co-expressed in HEK293 cells treated with MG132 for 6 h. After immunoprecipitation, bound Snail1 (B) and LSD1 (C) were examined by western blotting. (D) GST-tagged full-length or deletion mutants of LSD1 were incubated with lysate from HEK293 cells expressing Snail1. The GST pull-down complex was eluted with SDS–PAGE buffer and about one-tenth of these elutents were analysed for the association of Snail1 by western blotting (bottom panel). The rest of the elutents were examined for the presence of purified GST–LSD1 by Coomassie staining (top panel). (E) Flag-tagged AO domain, AO domain without Tower (AOΔTower) or Tower domain of LSD1 was co-expressed with Snail1 in HEK293 cells treated with MG132 for 6 h. After immunoprecipitation, the bound deletion mutants of LSD1 and Snail1 were examined by western blotting.
Although the AO domain of LSD1 shares high sequence homology with other FAD-containing AOs, it contains a unique 100-amino-acid insertion that forms a tower-like structure (tower domain) protruding away from the AO domain (yellow insertion; Figure 4A; Chen et al, 2006; Stavropoulos et al, 2006; Yang et al, 2006; Forneris et al, 2007). The tower domain divides the AO domain of LSD1 into two functional lobes, the FAD-binding lobe and the substrate-binding and -recognition lobe. CoREST binds to the tower domain and allosterically modulates the interaction of the FAD-binding lobe with the substrate-binding lobe of LSD1 and thus controls the demethylase activity of LSD1 (Chen et al, 2006; Stavropoulos et al, 2006; Yang et al, 2006; Forneris et al, 2007). To further narrow down the region that is required for the association of the AO domain with Snail1, we generated a tower domain-only LSD1 and a tower-deleted AO domain (AOΔTower) by linking FAD- and substrate-binding lobes with a polyglycine linker as described previously (Figure 4A; Chen et al, 2006). When these deletion mutants were co-expressed with Snail1 in HEK293 cells, the AO and AOΔTower mutants were still able to interact with Snail1 (Figure 4E), whereas the tower domain failed to interact with Snail1. Conversely, immunoprecipitation of AO and AOΔTower, but not of the Tower domain, revealed the association of Snail1. These results indicate that the AO domain, but not the tower region of LSD1, is responsible for the interaction of LSD1 with Snail1.
On the basis of the structural analysis of LSD1 (Chen et al, 2006; Stavropoulos et al, 2006; Yang et al, 2006; Forneris et al, 2007), aspartic acid 375 and glutamic acid 379 form a charge interaction with arginine 8 of histone H3, whereas glutamic acids at positions 553, 555 and 556 interact with arginine 2 of histone H3 (Supplementary Figure S8A). Mutation of these critical residues within the catalytic domain of LSD1 abolished its catalytic activity on H3K4 demethylation (Chen et al, 2006; Stavropoulos et al, 2006). Interestingly, mutation of these residues completely compromised the interaction of LSD1 with Snail1 (Supplementary Figure S8B). Taken together, our data indicate that the SNAG domain of Snail1 adopts a structural conformation similar to that of the histone H3 tail and interacts with the catalytic domain of LSD1 in a manner similar to that of histone H3.
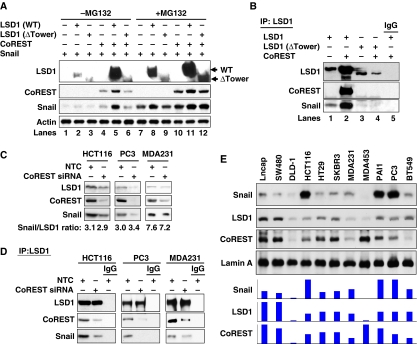
CoREST enhances the interaction of LSD1 with Snail1 and the stability of the ternary complex
The tower region of LSD1 is required for its interaction with CoREST (Chen et al, 2006; Stavropoulos et al, 2006; Yang et al, 2006; Forneris et al, 2007), a SANT domain-containing co-repressor. Binding of CoREST not only protects LSD1 from proteasomal degradation in vivo but also modulates the structure of the AO domain of LSD1 to control the interaction of LSD1 with its substrate, histone H3 (Lee et al, 2005; Shi et al, 2005). As the sequence of the SNAG domain is similar to that of the N-terminus of histone H3 (Figure 3B), and because the SNAG domain of Snail1 interacts with the AO domain of LSD1 (Figures 1B, 3A, 4; Supplementary Figure S8), we reasoned that the association of CoREST with LSD1 may affect the binding and stability of Snail1. To test this idea, we expressed Snail1, LSD1 or CoREST in HEK293 cells. Consistent with previous findings (Zhou et al, 2004; Shi et al, 2005), treatment with the proteasome inhibitor MG132 significantly stabilized Snail1 and LSD1 (lanes 1 and 2 versus lanes 7 and 8, Figure 5A). Co-expression of Snail1 with wild-type LSD1, ΔTower LSD1 or CoREST did not change the stability of Snail1 (lanes 2, 3 and 4, Figure 5A). As expected, CoREST enhanced the stability of wild-type LSD1 but not of ΔTower LSD1 (lane 5 versus lane 6, Figure 5A). Noticeably, expression of Snail1, LSD1 and CoREST together significantly enhanced the protein stability of each in the complex, as much as treatment with proteasome inhibitor MG132 (lane 5 versus lane 11, Figure 5A). These results indicate that the formation of the Snail1–LSD1–CoREST ternary complex is critical for their protein stabilization and function in vivo. Consistent with this finding, when LSD1 was immunoprecipitated, co-expression of CoREST significantly enhanced the association of LSD1 with Snail1 (lane 2 versus lane 1, Figure 5B; Supplementary Figure S9). However, ΔTower LSD1, which does not interact with CoREST, could not enhance the interaction with Snail1 in the presence of CoREST (lane 3 versus lane 4, Figure 5B). These results suggest that the binding of CoREST with the tower domain of LSD1 modulated the AO structure of LSD1, and thus enhanced the interaction of LSD1 with Snail1. To further confirm our finding, we knocked down the expression of endogenous CoREST in HCT116, PC3 and MDA-MB231 cells, which slightly decreased the levels of endogenous LSD1 and Snail1 in these cells (Figure 5C). Again, when equal amounts of LSD1 were immunoprecipitated (top panel, Figure 5D), knockdown of CoREST expression significantly decreased the interaction of LSD1 with Snail1 in these cell lines (Figure 5D). As LSD1 resides mainly in the nucleus and Snail1 is localized in the nucleus when it is stabilized (Zhou et al, 2004), we measured the expression of Snail1, LSD1 and CoREST in nuclear extracts from 11 different cancer cell lines. We found that the level of Snail1 was highly correlated with the levels of LSD1 and CoREST in these cells (Figure 5E). Together, our data indicate that the association of Snail1, LSD1 and CoREST is important for their stability as a functional complex; the binding of CoREST with LSD1 is not only critical for the stability of LSD1 but also modulates the association of LSD1 with Snail1.
Figure 5.
CoREST enhances the interaction of LSD1 with Snail1 and the stability of the ternary complex. (A) Snail1, WT or Tower-deleted LSD1, and CoREST were co-expressed in HEK293 cells. Cells were treated with or without MG132 for 6 h before being collected. The levels of LSD1, Snail1 and CoREST were analysed by western blotting. (B) WT or Tower-deleted LSD1 and CoREST were co-expressed in Snail1/HEK293 cells. After immunoprecipitation of LSD1, the bound Snail1 and CoREST were examined by western blotting. (C) CoREST siRNA or non-target control (NTC) was expressed in HCT116, PC3 and MDA-MB231 cells. Endogenous LSD1, CoREST and Snail1 were examined by western blotting. (D) Cells were prepared as described above in (C). After endogenous LSD1 was immunoprecipitated, the bound CoREST and Snail1 were examined by western blotting. (E) Nuclear extracts from various cancer cell lines were analysed for the levels of Snail1, LSD1 and CoREST by western blotting. Lamin A served as the nuclear marker. The expression intensity of these proteins was plotted and shown (bottom panel).
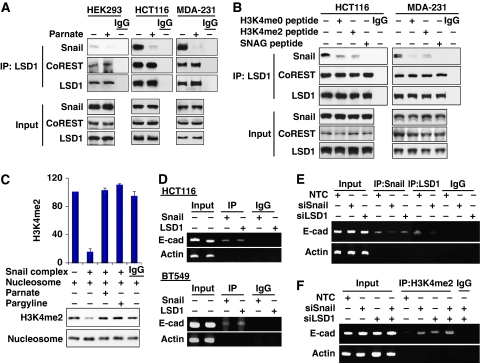
Interaction with Snail1 is required for LSD1 recruitment to the E-cadherin promoter in vivo
We found that the SNAG domain of Snail1 and the AO domain of LSD1 were required for their association, and this interaction was regulated by CoREST. Given a high sequence similarity between the SNAG domain and the N-terminus of histone H3 (Figure 3B), we reasoned that Snail1 used its SNAG domain as a pseudo-substrate for LSD1 by mimicking the structure of the histone H3 tail, which allowed for the recruitment of LSD1 to its targeted gene promoter. To test this idea, we immunoprecipitated endogenous LSD1 from HEK293 (with exogenously expressed Snail1), HCT116 and MDA-MB231 cells in the presence or absence of Parnate (tranylcypromine), a mechanism-based inhibitor that binds to the catalytic cavity of LSD1 (Schmidt and McCafferty, 2007; Yang et al, 2007b). We found that Parnate significantly disrupted the interaction of LSD1 with Snail1 (Figure 6A). This inhibitory effect is specific, as Parnate did not alter the association of LSD1 with CoREST. A similar finding was also obtained using another LSD1 inhibitor, Pargyline (data not shown; Yang et al, 2007a). To further extend our observation, we added H3K4me0 (1–21 amino acids), H3K4me2 (1–21 amino acids) and SNAG (1–17 amino acids) peptides for competition binding during the immunoprecipitation of endogenous LSD1 in HCT116 and MDA-MB231 cells (Figure 6B). We found that H3 peptides significantly disrupted the interaction of LSD1 with Snail1. Noticeably, the SNAG peptide completely abolished the interaction of LSD1 with Snail1, suggesting that the SNAG peptide has higher affinity for binding to LSD1 than does the histone H3 peptide. Together, these results indicate that agents (inhibitors, histone H3 and SNAG peptides) competing for the binding of the catalytic cavity of LSD1 can disrupt the association of LSD1 with Snail1.
Figure 6.
Interaction with Snail1 is required for LSD1 recruitment to the E-cadherin promoter in vivo. (A) The LSD1 enzymatic inhibitor Parnate (100 μM) was added to the immunoprecipitation buffer during immunoprecipitation. After endogenous LSD1 was immunoprecipitated from HEK293 (expressing exogenous Snail1), MDA-MB231 and HCT 116 cells, bound Snail1 and CoREST were detected by western blotting. (B) H3K4me0 (1–21 amino acids), H3K4me2 (1–21 amino acids) and SNAG peptides (60 μg/ml) were added to the immunoprecipitation buffer during immunoprecipitation. After endogenous LSD1 was immunoprecipitated, bound Snail1 and CoREST were detected by western blotting as described above. (C) In vitro histone demethylation assay was carried out as described in the Materials and methods section using mononucleosomes as substrate. After the reaction, demethylation of histone H3K4 was examined by western blotting. The statistical analysis for the demethylation from three independent experiments is shown on the bar graph (top panel). A representative demethylation assay is shown in the bottom panel. (D) The association of endogenous Snail1 and LSD1 with the E-cadherin promoter was analysed by chromatin immunoprecipitation (ChIP) assay in HCT116 and BT549 cells. (E) Snail1, LSD1 or non-target control (NTC) siRNA was expressed in HCT116 cells, the association of endogenous Snail1 and LSD1 with the E-cadherin promoter was analysed by ChIP assay. (F) Snail1, LSD1 or non-target control siRNA was expressed in HCT116 cells; methylation of H3K4 on the E-cadherin promoter was analysed by CHIP assay using anti-H3K4me2 antibody.
To test whether the Snail1 complex contains LSD1 demethylase activity (Shi et al, 2005), we immunoprecipitated Snail1 and incubated the complex with purified mononucleosome. We found that the Snail1 complex was able to demethylate H3K4me2 in vitro and that this enzymatic activity was suppressed by LSD1 inhibitors Parnate and Pargyline (Figure 6C). To further examine whether the Snail1–LSD1 complex is associated with the E-cadherin promoter and mediates transcriptional repression during EMT in vivo, we performed chromatin immunoprecipitation (ChIP) assays on two cancer cell lines. As expected, Snail1 and LSD1 can interact with the E-cadherin promoter in these two cell lines (Figure 6D). Interestingly, the interaction of LSD1 with the E-cadherin promoter was significantly decreased when Snail1 was specifically knocked down in HCT116 cells (lane 7 versus lane 8, Figure 6E), indicating that the interaction of LSD1 with the E-cadherin promoter is mediated through the association of Snail1 that binds to the E-box on the E-cadherin promoter through zinc-finger motifs. Consistent with this idea, no H3K4 methylation on the E-cadherin promoter could be detected in HCT116 cells (lane 5, Figure 6E). However, knockdown of Snail1 or LSD1 expression enhanced the methylation of H3K4 (lanes 6, 7 and 8, Figure 6F). Taken together, our results indicate that the interaction with LSD1 is critical for the suppressive activity of Snail1 and that Snail1 is required for LSD1 recruitment to the E-cadherin promoter in vivo.
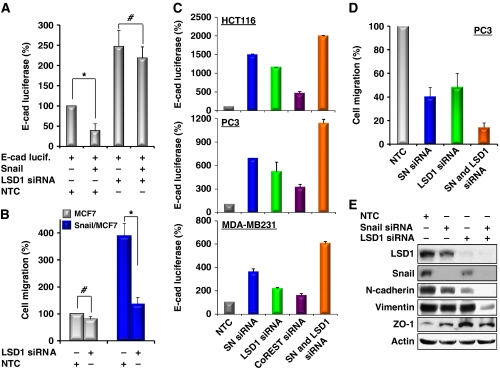
Knockdown of Snail1 and LSD1 expression suppresses cell migration
To examine the functional relationship of Snail1 and LSD1 in vivo, we expressed an E-cadherin promoter luciferase construct in MCF7 cells that expresses no detectable endogenous Snail1. As expected, co-expression of Snail1 suppressed E-cadherin promoter luciferase activity (Figure 7A; Supplementary Figure S10A). However, knockdown of LSD1 expression blocked the suppressive effect of Snail1 on the E-cadherin promoter in these cells. We also measured the cell migration of isogenic MCF7 and Snail1/MCF cells (Figure 7B; Supplementary Figure S10B). As we have shown previously, stable expression of Snail1 in MCF7 cells enhances the migratory ability of these cells (Wu et al, 2009a). However, knockdown of LSD1 expression abolished the enhancement of Snail1-mediated cell migration in Snail1/MCF7 cells (Figure 7B), whereas knockdown of LSD1 expression in MCF7 cells has no significant effect on cell migration. These results suggest that the repressive function of Snail1 depends on LSD1. Similarly, we knocked down the expression of Snail1 or LSD1 or CoREST in HCT116, PC3 and MDA-MB231 cells, which express different levels of endogenous Snail1, LSD1 and CoREST, and measured E-cadherin promoter luciferase activity. Knockdown of CoREST expression partially derepressed the suppression of the E-cadherin promoter. Knockdown of either Snail1 or LSD1 expression significantly derepressed the suppression of the E-cadherin promoter (Figure 7C; Supplementary Figure S11). Knockdown of both Snail1 and LSD1 expression further enhanced the activity of the E-cadherin promoter luciferase in all three cell lines tested, suggesting that the functional interaction of Snail1 and LSD1 is important for E-cadherin promoter suppression. Consistent with this finding, knockdown expression of Snail1 or LSD1 suppressed the migration of HCT116, PC3 and MDA-MB231 cells, and knockdown of both molecules further enhanced this suppressive effect on migration (Figure 7D; Supplementary Figure S12). Furthermore, knockdown of Snail1, LSD1, CoREST or both Snail1 and LSD1 in PC3 and MDA-MB231 cells impaired the invasiveness of these cells (Supplementary Figure S13). In line with these functional changes, knockdown of either Snail1 or LSD1 expression decreased the expression of the mesenchymal markers N-cadherin and vimentin and enhanced the expression of the tight-junction molecule ZO-1 (Figure 7E). Knockdown of both molecules can significantly enhance this effect. We speculate that this additive effect was due to the higher knockdown efficiency on the functional Snail1–LSD1 repressor complex when both molecules were targeted at the same time. Together, our results indicate that the functional interaction of Snail1 with LSD1 is critical in mediating E-cadherin promoter suppression and enhancing cell migration.
Figure 7.
Knockdown of Snail1 and LSD1 expression suppresses cell migration. (A) An E-cadherin promoter luciferase construct was co-expressed with or without Snail1, non-target control (NTC) or LSD1 siRNA in MCF7 cells. After 48 h, luciferase activities were normalized and determined (mean±s.d. of three separate experiments). Luciferase activities for each sample were statistically analysed by Student's t-test (P<0.01). *P<0.001; #, no significance. (B) LSD1 or NTC siRNA was expressed in isogenic MCF7 and Snail1/MCF7 cells. After 48 h, a scratch (‘wound') was induced in a cell monolayer and cell culture was continued for an additional 48 h. Images were obtained at the beginning and at the 48 h time point to monitor the cell migration for the closure of the wound. The percentage of cell migration was calculated on the basis of the migration of MCF7 cells (experiments were conducted at least twice in duplicate). Statistical analysis was performed by Student's t test (P<0.01). *P<0.001; #, no significance. (C) Snail1 (SN), LSD1, CoREST and NTC siRNA were co-expressed with the E-cadherin promoter luciferase construct in HCT116, PC3 and MDA-MB231 cells. After 48 h, luciferase activities were normalized and determined (mean±s.d. of three separate experiments). (D) Snail1 (SN), LSD1 and NTC siRNA were expressed in HCT116, PC3 and MDA-MB231 cells as described in (C). After 48 h, a scratch (‘wound') was induced in a cell monolayer and cell culture was continued for 48 h to measure cell migration as described in (B). The statistical analysis for the migration of PC3 cells is shown on the bar graph, whereas statistical results for HCT116 and MDA-MB231 cells are shown in Supplementary Figure S12A. Representative images from PC3 cells are shown in Supplementary Figure S12B. (E) PC3 cells were treated as described above and expressions of LSD1, Snail1, N-cadherin, vimentin, ZO-1 and Actin were examined by western blotting.
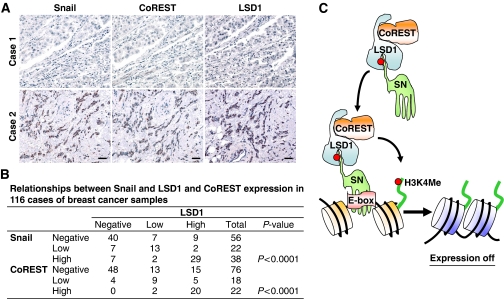
Expression of Snail1 correlates with the level of LSD1 and CoREST in tumour tissues
Having established that the interaction of Snail1 and LSD1 is critical for their stability and for the suppressive function of Snail1, we further extended our findings in vivo by examining the correlation between the expression of Snail1 and LSD1 in 116 resected human breast tumour specimens. In agreement with previous findings (Shi, 2007), expression of LSD1 was significantly correlated with the expression of CoREST. Again, consistent with our results obtained from cell lines (Figure 5E), the expression of Snail1 was highly correlated with the expression of LSD1 and CoREST in tumour samples (Figure 8A and B). These results in human breast cancer tissues confirm our observations in cell culture, lending further support to our hypothesis that the interaction of Snail1 with LSD1 is required for their stability and for the suppressive function of Snail1 during cell invasion and metastasis.
Figure 8.
Expression of Snail1 correlates with the level of LSD1 and CoREST in tumour tissues. (A) The 116 surgical specimens of breast cancer were immunostained using antibodies against Snail1, LSD1, CoREST, and the control serum (data not shown). Representative stainings from the same tumour samples are shown. Scale bar=100 μm. (B) The expression patterns of Snail1, LSD1 and CoREST in the 116 breast tumour samples were determined and summarized. Correlation of Snail1 with LSD1 and CoREST was analysed using Fisher's exact test (P<0.001). P<0.05 was set as the criterion for statistical significance. (C) A proposed model to illustrate how Snail1 recruits the LSD1–CoREST complex to the E-cadherin promoter. The SNAG domain of Snail1 assembles a histone H3-like structure and serves as a molecular ‘hook' (or pseudo-substrate, a red dot indicates the critical residues in Snail1 and histone H3 that interact with LSD1) to interact with the LSD1–CoREST complex. The formation of this complex stabilizes the individual components from potential proteasomal degradation. Snail1 brings this complex to its targeted gene promoters through the binding of the E-box through the zinc-finger motifs. An overabundant amount of histone H3 at the chromatin region outcompetes the binding of the SNAG domain with the catalytic core of LSD1 and results in the demethylation of histone H3K4.
Discussion
In this study, we showed that the interaction of Snail1 with LSD1 is critical for their stability and for the transcriptional repressive function of Snail1. Our study provides several new insights into the regulation of EMT and metastasis. First, Snail1 may represent the first unique example of a transcription factor that uses a histone-mimicking motif, SNAG domain in this case, for recruiting chromatin-modifying enzymes. Although transcription factors contain DNA-binding motifs, they do not modify chromatin. On the contrary, chromatin-modifying enzymes contain many histone-binding modules (such as the Bromo, Chromo and PhD domains) and are capable of changing the chromatin structure through acetylation or methylation, although they lack specific DNA-binding motifs. The manner in which transcription factors recruit specific chromatin-modifying enzymes to their target gene promoters remains a subject of major interest and an area of intensive investigation. We found that the SNAG domain is required and sufficient for Snail1 to interact with LSD1 (Figures 1B and 3A). Interestingly, the sequence of the SNAG domain is highly similar to that of the N-terminus of histone H3 and adopts a conformation similar to that of histone H3 at the catalytic cavity of LSD1 (Figure 3B and F). Mutation of the critical contact residues (Arg3, Arg8 and Lys9) of the SNAG domain of Snail1 completely disrupts the interaction of Snail1 with LSD1 and blocks its suppressive activity on the E-cadherin promoter (Figure 3E; Supplementary Figure S4). We also found that the SNAG domain of Snail1 interacts with the AO domain of LSD1 (Figure 4), which consists of the catalytic core of LSD1 and is required to interact with the tail of histone H3 for H3K4 demethylation (Chen et al, 2006; Stavropoulos et al, 2006; Yang et al, 2006; Forneris et al, 2007). The interaction of Snail1 with LSD1 is disrupted by LSD1 enzymatic inhibitors and histone H3 peptides (Figures 6A and B). In addition, the Snail1–LSD1 interaction is regulated by CoREST (Figure 5), which is known to bind to the tower region and modulates the structure of the AO domain of LSD1 for controlling its interaction with the histone H3 tail (Chen et al, 2006; Stavropoulos et al, 2006; Yang et al, 2006; Forneris et al, 2007). We showed that expression of CoREST enhanced the formation of the Snail1–LSD1–CoREST ternary complex and significantly stabilized individual components of the complex, whereas knockdown of CoREST expression decreased the interaction of LSD1 with Snail1 (Figure 5). This result is further supported by the finding that expressions of Snail1, LSD1 and CoREST are highly correlated in multiple cancer cell lines and in the 116 primary breast cancer samples (Figures 5E and 8B). The region connecting the SNAG domain and the zinc-fingers in Snail1 contains a serine-rich motif. We, along with others, previously showed that this serine-rich motif is highly phosphorylated and controls the subcellular localization and stability of Snail1 (Dominguez et al, 2003; Zhou et al, 2004; Yook et al, 2005). As Snail1 is a labile protein, we speculate that deletion or mutation of the SNAG domain disrupts the interaction of Snail1 with the LSD1–CoREST complex, and thus leads to the accessibility of phosphorylation and consequent proteasome degradation by GSK-3β and β-Trcp (Dominguez et al, 2003; Zhou et al, 2004; Yook et al, 2005). This is supported by the finding that the SNAG domain does not alter subcellular localization but controls the stability of Snail1 (Figure 2; Supplementary Figures S2 and S3). Our results also indicate that the association of LSD1 with the E-cadherin promoter requires the binding of Snail1. Knockdown of Snail1 expression inhibits the association of LSD1 with the E-cadherin promoter (Figure 6). Our results, together with previous findings (Shi, 2007), lead us to propose a plausible model for how Snail1 recruits the LSD1–CoREST complex to its targeted gene promoter for transcriptional repression (Figure 8C). The SNAG domain of Snail1 resembles a histone H3-like structure and functions as a molecular ‘hook' (or pseudo-substrate) to interact with LSD1–CoREST. The formation of this ternary complex stabilizes the individual components from potential proteasome degradation. Snail1 brings this complex to its targeted gene chromatin through the binding of the E-box through zinc-finger motifs. An overabundant amount of histone H3 at the chromatin region outcompetes the binding of the SNAG domain with the catalytic core of LSD1 and results in the demethylation of histone H3K4. Our study is supported by the finding that the SNAG domain of GFI1 interacts with LSD1 and CoREST and that this interaction is required for hematopoietic cell differentiation (Saleque et al, 2007). Although our model provides a new mechanism for the manner in which Snail1 recruits chromatin-modifying enzymes to target gene promoters, there are many questions remaining for further investigation. For example, future studies should address whether Arg3, Arg8 and Lys9 on the SNAG domain are methylated in vivo and how the SNAG domain is released from the LSD1-binding pocket after Snail1 interacts with promoter DNA. Elucidation of these questions will provide new insights into the regulation of EMT and metastasis.
Second, our study indicates that Snail1 uses either the SNAG domain or the CtBP-binding motif for recruiting the LSD1–CoREST complex. All vertebrate Snail1 genes contain the SNAG domain, but they do not have a CtBP-binding motif. However, Drosophila Snail1 does not contain the SNAG domain, but it has a consensus PxDLSx motif that is required to interact with the co-repressor CtBP (Chinnadurai, 2002; Nieto, 2002; Barrallo-Gimeno and Nieto, 2009). Both the SNAG domain and the CtBP motif are critical for the function of vertebrate and Drosophila Snail1, respectively. Deletion or mutation of the SNAG domain abolishes the repressive function of SNAG domain-containing transcriptional repressors (Okubo et al, 2001; Zhu et al, 2002; Fiolka et al, 2006). Similarly, mutation or deletion of the CtBP-binding motif in Drosophila Snail1 eliminates its repressive activity (Hemavathy et al, 2004). Interestingly, LSD1 was initially identified as a protein associated with CtBP1 during protein purification and it has been confirmed that the interaction of LSD1 and CtBP1 is critical for many repressor complexes (Shi et al, 2003). Thus, our results suggest that vertebrate and Drosophila Snail1 recruit the same LSD1–CoREST co-repressor complex for their repressive function through either the SNAG domain or the CtBP-binding motif.
Third, our study indicates that Snail1 uses different co-repressor complexes to achieve its repressor function during the EMT process. The reciprocal relationship of H3K4 and H3K27 methylation in gene regulation is exemplified by the antagonistic action of TrxG and PcG proteins (Sparmann and van Lohuizen, 2006; Schuettengruber et al, 2007). The polycomb group (PcG) and trithorax group (trxG) genes were first discovered in Drosophila as a repressor and activator of Hox gene expression in controlling cell identity during the establishment of body segmentation. The trxG complex has H3K4 trimethylase activity for activating gene expression. However, PcG proteins form three different classes of complexes. The PRC2 protein contains the four core components of E(z) (enhancer of zeste), Esc (extra sex combs), Su(z)12 (suppressor of zeste 12) and Nurf-55 (in humans, EZH2, EED, SUZ12 and RbAp46/48). The SET domain-containing EZH2 of PRC2 trimethylates H3K27 (H3K27me3) and results in gene silencing (Sparmann and van Lohuizen, 2006; Schuettengruber et al, 2007). In our study, Snail1 interacted with the LSD1–CoREST complex, resulting in demethylation of H3K4me2 at the E-cadherin promoter to suppress its expression. Interestingly, Snail1 has been shown to interact with EZH2 and Suz12 of PRC2 and induce the trimethylation of H3K27 at the E-cadherin promoter (Herranz et al, 2008). Knockdown of PRC2 proteins compromised the ability of Snail1 to repress E-cadherin expression. The manner in which Snail1 corporately induces the demethylation of H3K4 and the methylation of H3K27 requires further investigation. The finding that Snail1 also interacts with PRMT5 adds another layer of intricacy to the regulation of E-cadherin expression during EMT (Hou et al, 2008). Hou et al. showed that PRMT5 interacts with the SNAG domain of Snail1 through Ajuba and is translocated into the nucleus in a Snail1- and Ajuba-dependent manner. The Snail1–Ajuba–PRMT5 ternary complex is found at the proximal promoter region of the E-cadherin gene, where it increases arginine methylation of H4R3. Furthermore, Snail1 interacts with the Sin3A–HDAC1/2 complex through the SNAG domain to deacetylate histone H3 and H4, leading to the suppression of E-cadherin expression (Peinado et al, 2004). In Drosophila, transcriptional silencing is initiated through the removal of H3K4 methylation by the LSD1 homologue SU(VAR)3-3 (Rudolph et al, 2007), indicating that the demethylation of H3K4 by LSD1 is the critical initiating step for gene silencing. However, E-cadherin is frequently silenced through DNA hypermethylation on its promoter in many cancer cell lines and tumour samples (Lombaerts et al, 2006). The manner in which the demethylation of H3K4 by the LSD1–CoREST complex cooperates with methylation (H3K27 and H4R3) and deacetylation (H3 and H4), and subsequently leads to E-cadherin promoter methylation remains an important area for further investigation.
In summary, we demonstrated that the interaction with the LSD1–CoREST complex is critical for the stability and function of Snail1. Our results provide a new model for how a transcription factor recruits chromatin-modifying enzymes to target gene promoter during transcription. This finding has important clinical ramifications in that chemical compounds that mimic the structure of the SNAG domain of Snail1 may hold great promise for inhibiting the function of Snail1 in mediating EMT induction and cancer metastasis.
Materials and methods
Antibodies and plasmids
Human cDNA for LSD1 and CoREST was amplified from HeLa cDNA and cloned into pCMV-Tag2B and pcDNA3 with the N-terminal Flag or HA tag, respectively. Deletion mutants for LSD1 were generated by PCR and subcloned into the GST expression vector pGEX-6P-3-TEV that contains two protease cleavage sites of tobacco etch virus. Snail1 mutants were generated using the QuikChange Mutagenesis kit (Stratagene, La Jolla, CA) as described previously (Wu et al, 2009b). All sequences were verified by DNA sequencing. Antibodies against Snail1, LSD1 and CoREST were purchased from Cell Signaling Technology (Danvers, MA). Anti-Flag and anti-HA antibodies were obtained from Sigma-Aldrich (St Louis, MO) and Roche Molecular Biochemicals (Indianapolis, IN), respectively. An antibody against H3K4me2 was purchased from Millipore (Bedford, MA).
Cell cultures, transfections and reporter assays
The human embryonic kidney HEK293, breast cancer MCF7, MDA-MB231 and SKBR3, prostate cancer PC3 and colon cancer HCT116 cell lines were purchased from the American Type Culture Collection (Manassas, VA) and grown in Dulbecco's modified Eagle's/F12 medium plus 10% foetal bovine serum as described previously (Wu et al, 2009a). Lysine-specific demethylase 1, CoREST and Snail1 were transiently transfected into cells using FuGENE 6 (Roche Molecular Biochemicals). For luciferase assay, cells were plated in six-well plates at a density of 2 × 105 cells per well. Cells were transfected with 0.3 μg of the pGL3–E-cadherin promoter–luciferase plasmid along with 0.1 nM of Snail1 and/or LSD1 siRNA in each well. To normalize transfection efficiency, cells were also co-transfected with 0.1 μg of pRL-CMV (Renilla luciferase). At 48 h after transfection, luciferase activity was measured using the Dual-Luciferase Assay kit (Promega, Madison, WI). Three independent experiments were performed, and the calculated means and s.d. values are presented.
GST pull-down assay
Glutathione-S-transferase proteins were expressed as described previously (Wu et al, 2009b). Cells were subjected to lysis in GST pull-down buffer (20 mM Tris, 150 mM NaCl and 1% Nonidet P-40 with protease cocktail) and rotated with glutathione–Sepharose-bound wild-type or deletion mutants of GST–LSD1. The binding complexes were eluted with SDS–PAGE sample buffer. About one-tenth of these elutents were analysed for the association of Snail1 by western blotting and the rest were examined for the presence of purified GST–LSD1 by Coomassie staining.
Histone demethylation assay
Mononucleosomes were purified from HeLa S3 nuclear extract (Utley et al, 1996). The histone demethylation assay was performed as described previously using mononucleosomes as a substrate (Lee et al, 2005; Shi et al, 2005; Yang et al, 2006). Briefly, HEK293 cells were co-transfected with constructs encoding LSD1, CoREST and Snail1. After immunoprecipitation of Snail1, complexes were incubated with mononucleosomes in demethylation buffer (50 mM Tris–HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2, 2 mM DTT with proteosome inhibitors) with or without Parnate or Pargyline at 37°C for 12 h. Reactions were stopped with SDS sample buffer. Western blot analysis was performed with an antibody against H3K4me2 (Upstate). The same membrane was then stripped and re-probed with α-histone H3 (Abcam, Cambridge, MA, ab1791) as a substrate-loading control.
Western blot analysis and immunoprecipitation
For protein extraction, 5 × 105 cells per well were plated onto six-well plates and transiently transfected with 0.5 μg of pcDNA3-Snail1 and 0.5 μg of pCMV-Tag2B-LSD1 or vector. At 48 h after transfection, cells were incubated with or without the proteasome inhibitor MG132 (10 μmol/l) for an additional 6 h before protein extraction and western blot analysis. Primary antibodies against Flag (M2, 1:1000) and HA (3F10, 1:4000) were used for protein detection. For immunoprecipitation, HEK293 cells transfected with the indicated expression plasmids were lysed in buffer (50 mmol/l Tris (pH 7.5), 150 mmol/l NaCl, 5 μg/ml aprotinin, 1 μg/ml pepstatin, 1% Nonidet P-40, 1 mmol/l EDTA and 0.25% deoxycholate). Total cell lysates (1000 μl) were incubated overnight with 1 μg of anti-HA or anti-Flag antibody conjugated to agarose beads (Roche Molecular Biochemicals) at 4°C. The beads were then washed with lysis buffer, and the immunoprecipitated protein complexes were resolved by 10% SDS–PAGE.
Invasion assay
Invasion assays were performed as previous described (Wu et al, 2009a). Briefly, Boyden chambers were coated with Matrigel according to the manufacturer's protocol (BD Biosciences, San Jose, CA). Cancer cells were seeded on top of the Matrigel in the upper chamber, and the bottom chamber was filled with culture medium containing LPA (10 μM) as the chemoattractant. Cells that invade through the Matrigel-coated membrane after 4 or 24 h were fixed with paraformaldehyde, followed by staining with crystal violet. All experiments were conducted at least twice in triplicate. Statistical analysis was performed with Student's t-test; a P-value of <0.05 was considered significant.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed according to the protocol described by Nowak et al (2005) with some modifications. The cells were crosslinked with disuccimidyl glutarate (Pierce, Rockford, IL) and formaldehyde at room temperature. Cells were subjected to lysis with L1 buffer (50 mM Tris, 2 mM EDTA, 0.1% IGEPAL, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor mixture (pH 8.0)) on ice. After centrifugation, the nuclear pellet was resuspended in ChIP lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris and protease inhibitor mixture (pH 8.0)). Cell lysates were subjected to sonication and then incubated with 4 μg of Snail1 (Abcam) or LSD1 (Sigma-Aldrich) antibody overnight, followed by incubation with a 50% slurry of protein A–agarose/Salmon sperm DNA (Upstate Biotechnology, Lake placid, NY) for 3 h at 4°C. Bound DNA–protein complexes were eluted and crosslinks were reversed after a series of washes. Purified DNA was resuspended in TE buffer (10 mM Tris–HCl and 1 mM EDTA (pH 8.0)) for PCR. The primers for the E-cadherin promoter were 5′-ACTCCAGGCTAGAGGGTCACC-3′ and 5′-CCGCAAGCTCACAGGTGCTTTGCAGTTCC-3′.
Immunofluorescence imaging
For immunofluorescence microscopy, cells were grown on coverslips, fixed with 4% paraformaldehyde and incubated overnight with anti-LSD1 monoclonal antibody. Proteins were visualized by incubation with goat anti-mouse conjugated with Alexa fluor 568 (Invitrogen, Carlsbad, CA). Finally, coverslips were incubated with 4′,6′-diamidino-2-phenylindole (Sigma-Aldrich) for 20 min and visualized under a fluorescent microscope.
Immunohistochemical staining
All the breast tumour samples used in this study are invasive ductal carcinoma. The average age for patients in this group is 56.8 (±13.4) years with either negative (53.3%) or positive (N1=34.6; N2=12.1%) lymph node metastasis. These samples are of stage I (12.5%), stage II (38.2%), stage III (41.1%) or stage IV (8.2%). Tissue samples were stained with anti-Snail1 (1:200 dilution, Abcam), anti-LSD1 (1:150, Abcam) and anti-CoREST (1:150, BD Biosciences) antibodies, and each sample was scored by an H-score method that combines the values of immunoreaction intensity and the percentage of tumour cell staining as described previously (Wu et al, 2009a). Chi-square analysis was used to analyse the relationship between Snail1 and LSD1 and CoREST expression; statistical significance was defined as P<0.05.
Subcellular fractionation
Cytoplasmic and nuclear fractions were prepared as described previously (Wu et al, 2009b). Briefly, cultured cells were suspended in buffer A (10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM EDTA and protein inhibitor cocktail) on ice and lysed using a Dounce homogenizer. The nuclear pellet was washed and isolated. Nuclei were subjected to lysis in RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris (pH 8.0), 25 mM NaF, 2 mM Na3VO4, 5 mM PMSF and 2 μg/ml of aprotinin). Poly (ADP-ribose) polymerase and β-tubulin were used as markers of the nuclear and cytoplasmic fractions, respectively.
Protein model structure
The three-dimensional (3D) model of the LSD1–SNAG–Snail1 complex was built by comparative protein structure modelling using the program MODELLER 9v3 (Eswar et al, 2006). The input consisted of the template structure and the alignment of the target sequence with this structure. The output was a 3D model of the target including all non-hydrogen atoms. This model was derived by minimizing violations of distance and dihedral angle restraints extracted from the template structure while maintaining favourable interactions. The template structure was the LSD1–CoREST–Histone H3 peptide complex structure (Protein Data Bank, 2V1D). The figure was prepared using PyMol (http://www.pymol.org).
Supplementary Material
Acknowledgments
We thank Dr Nathan L Vanderford for critical reading and editing of this paper. This study was supported by grants from NIH (RO1CA125454), from the Susan G Komen Foundation (KG081310), the Mary Kay Ash Foundation (to BPZ) and from the American Cancer Society (IRG-85-001-22; to YW).
Footnotes
The authors declare that they have no conflict of interest.
References
- Barrallo-Gimeno A, Nieto MA (2009) Evolutionary history of the Snail/Scratch superfamily. Trends Genet 25: 248–252 [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2: 84–89 [DOI] [PubMed] [Google Scholar]
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES (2007) The mammalian epigenome. Cell 128: 669–681 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A (2007) Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 14: 1008–1016 [DOI] [PubMed] [Google Scholar]
- Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J, Nieto MA (2002) Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene 21: 3241–3246 [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA (2000) The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2: 76–83 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M (2006) Crystal structure of human histone lysine-specific demethylase 1 (LSD1). Proc Natl Acad Sci USA 103: 13956–13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CW, Wu PE, Yu JC, Huang CS, Yue CT, Wu CW, Shen CY (2001) Mechanisms of inactivation of E-cadherin in breast carcinoma: modification of the two-hit hypothesis of tumor suppressor gene. Oncogene 20: 3814–3823 [DOI] [PubMed] [Google Scholar]
- Chinnadurai G (2002) CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell 9: 213–224 [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22: 1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowin P, Rowlands TM, Hatsell SJ (2005) Cadherins and catenins in breast cancer. Curr Opin Cell Biol 17: 499–508 [DOI] [PubMed] [Google Scholar]
- Dominguez D, Montserrat-Sentis B, Virgos-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Franci C, Garcia de Herreros A (2003) Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol Cell Biol 23: 5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A (2007) Chromatin dynamics and the preservation of genetic information. Nature 447: 951–958 [DOI] [PubMed] [Google Scholar]
- Esteller M (2007) Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8: 286–298 [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A (2006) Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics Chapter 5: Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP (2007) Phenotypic plasticity and the epigenetics of human disease. Nature 447: 433–440 [DOI] [PubMed] [Google Scholar]
- Fiolka K, Hertzano R, Vassen L, Zeng H, Hermesh O, Avraham KB, Duhrsen U, Moroy T (2006) Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep 7: 326–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A (2007) Structural basis of LSD1–CoREST selectivity in histone H3 recognition. J Biol Chem 282: 20070–20074 [DOI] [PubMed] [Google Scholar]
- Forneris F, Binda C, Battaglioli E, Mattevi A (2008) LSD1: oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem Sci 33: 181–189 [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W (2007) Nuclear organization of the genome and the potential for gene regulation. Nature 447: 413–417 [DOI] [PubMed] [Google Scholar]
- Hemavathy K, Hu X, Ashraf SI, Small SJ, Ip YT (2004) The repressor function of snail is required for Drosophila gastrulation and is not replaceable by Escargot or Worniu. Dev Biol 269: 411–420 [DOI] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, Garcia de Herreros A, Peiro S (2008) Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 28: 4772–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ III (2008) The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol 28: 3198–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Berger SL (2008) The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev 18: 152–158 [DOI] [PubMed] [Google Scholar]
- Kajita M, McClinic KN, Wade PA (2004) Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol Cell Biol 24: 7559–7566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8: 307–318 [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R (2005) An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437: 432–435 [DOI] [PubMed] [Google Scholar]
- Lombaerts M, van Wezel T, Philippo K, Dierssen JW, Zimmerman RM, Oosting J, van Eijk R, Eilers PH, van de Water B, Cornelisse CJ, Cleton-Jansen AM (2006) E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer 94: 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- Martin TA, Goyal A, Watkins G, Jiang WG (2005) Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol 12: 488–496 [DOI] [PubMed] [Google Scholar]
- Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA (2005) The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8: 197–209 [DOI] [PubMed] [Google Scholar]
- Nieto MA (2002) The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol 3: 155–166 [DOI] [PubMed] [Google Scholar]
- Nowak DE, Tian B, Brasier AR (2005) Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques 39: 715–725 [DOI] [PubMed] [Google Scholar]
- Okubo T, Truong TK, Yu B, Itoh T, Zhao J, Grube B, Zhou D, Chen S (2001) Down-regulation of promoter 1.3 activity of the human aromatase gene in breast tissue by zinc-finger protein, snail (SnaH). Cancer Res 61: 1338–1346 [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A (2004) Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 24: 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7: 415–428 [DOI] [PubMed] [Google Scholar]
- Reik W (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, Reuter G (2007) Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell 26: 103–115 [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S, Kim J, Rooke HM, Orkin SH (2007) Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell 27: 562–572 [DOI] [PubMed] [Google Scholar]
- Schmidt DM, McCafferty DG (2007) trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry 46: 4408–4416 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G (2007) Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Shi Y (2007) Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet 8: 829–833 [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR (2007) Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 25: 1–14 [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19: 857–864 [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M (2006) Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6: 846–856 [DOI] [PubMed] [Google Scholar]
- Stavropoulos P, Blobel G, Hoelz A (2006) Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol 13: 626–632 [DOI] [PubMed] [Google Scholar]
- Thiery JP (2002) Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer 2: 442–454 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial–mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP (2006) Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol 7: 131–142 [DOI] [PubMed] [Google Scholar]
- Utley RT, Owen-Hughes TA, Juan LJ, Cote J, Adams CC, Workman JL (1996) In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol 274: 276–291 [DOI] [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA (2004) Snail blocks the cell cycle and confers resistance to cell death. Genes Dev 18: 1131–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA (2008) Mechanisms of malignant progression. Carcinogenesis 29: 1092–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP (2009a) Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell 15: 416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Evers BM, Zhou BP (2009b) Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J Biol Chem 284: 640–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou BP (2008) New insights of epithelial–mesenchymal transition in cancer metastasis. Acta Biochim Biophys Sin (Shanghai) 40: 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weinberg RA (2008) Epithelial–mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14: 818–829 [DOI] [PubMed] [Google Scholar]
- Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H (2007a) Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol 14: 535–539 [DOI] [PubMed] [Google Scholar]
- Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H (2007b) Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry 46: 8058–8065 [DOI] [PubMed] [Google Scholar]
- Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H (2006) Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell 23: 377–387 [DOI] [PubMed] [Google Scholar]
- Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ (2005) Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem 280: 11740–11748 [DOI] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC (2004) Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial–mesenchymal transition. Nat Cell Biol 6: 931–940 [DOI] [PubMed] [Google Scholar]
- Zhu J, Guo L, Min B, Watson CJ, Hu-Li J, Young HA, Tsichlis PN, Paul WE (2002) Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity 16: 733–744 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.