Abstract
The plant hormone gibberellin (GA) is crucial for multiple aspects of plant growth and development. To study the relevant regulatory mechanisms, we isolated a rice mutant earlier flowering1, el1, which is deficient in a casein kinase I that has critical roles in both plants and animals. el1 had an enhanced GA response, consistent with the suppression of EL1 expression by exogenous GA3. Biochemical characterization showed that EL1 specifically phosphorylates the rice DELLA protein SLR1, proving a direct evidence for SLR1 phosphorylation. Overexpression of SLR1 in wild-type plants caused a severe dwarf phenotype, which was significantly suppressed by EL1 deficiency, indicating the negative effect of SLR1 on GA signalling requires the EL1 function. Further studies showed that the phosphorylation of SLR1 is important for maintaining its activity and stability, and mutation of the candidate phosphorylation site of SLR1 results in the altered GA signalling. This study shows EL1 a novel and key regulator of the GA response and provided important clues on casein kinase I activities in GA signalling and plant development.
Keywords: casein kinase I, EL1, flowering time, GA response, rice
Introduction
The switch from vegetative to reproductive growth is a critical event in the life cycle of flowering plants and is essential for their maximum reproductive success (Bernier, 1988). In Arabidopsis, a model plant for eudicots, four major pathways are involved in the control of flowering time: the photoperiod, autonomous, vernalization, and gibberellin (GA) pathways. In contrast to Arabidopsis, rice is a short-day plant. Several rice genes in the photoperiod pathway controlling heading date (Hd) (flowering time) have been genetically characterized, including Se1 (photoperiod sensitivity 1; Yokoo et al, 1980; Yamagata et al, 1986), Se3–Se7 (Yamagata et al, 1986; Poonyarit et al, 1989; Sano, 1992; Yokoo and Okuno, 1993), and E1–E3 (Hd 1–3, Tsai, 1995; Kinoshita, 1998). Some rice genes encode proteins similar to those in the Arabidopsis long-day pathway. Specifically, the products of Hd 1, 3a, and 6 are homologous to Arabidopsis CO (Yano et al, 2000), FT (FLOWERING LOCUS T, Kojima et al, 2002), and a subunit of kinase CK2 (a regulator of circadian rhythm and flowering time, Takahashi et al, 2001), respectively. In addition, rice phytochromes are also involved in the regulation of flowering time in response to day length (Izawa et al, 2000). Similar to Arabidopsis, circadian-regulated OsGI expression in transgenic rice has striking effects on flowering time (Hayama et al, 2003).
Arabidopsis mutants defective in GA biosynthesis or signalling show severe delayed flowering (Moon et al, 2003; Yu et al, 2004); however, the role of GA in rice flowering is less clear. Biochemical and genetic studies have characterized key components, especially DELLA proteins, in the GA signalling cascade in both Arabidopsis (Peng et al, 1997; Dill and Sun, 2001; Lee et al, 2002; Hussain et al, 2005) and rice (Ikeda et al, 2001). The Arabidopsis DELLA proteins GAI, RGA, and RGL1 negatively regulate flowering time in the absence of GA (Mouradov et al, 2002). Recently, the GA receptor GID1 was identified in Arabidopsis (Griffiths et al, 2006; Nakajima et al, 2006) and rice (Ueguchi-Tanaka et al, 2005), and found to interact with DELLA proteins (GAI, RGA, or SLR1) in a GA-dependent manner both in vitro and in vivo (Willige et al, 2007; Ueguchi-Tanaka et al, 2007a, 2007b). The GA–GID1–SLR1 complex is targeted for ubiquitination by SCFGID2, an F-box protein, and degraded by the ubiquitin-dependent proteasome pathway (Sasaki et al, 2003; Gomi et al, 2004; Ueguchi-Tanaka et al, 2005, 2007a, 2007b), which in turn results in the activated GA response.
Casein kinase I, a serine/threonine protein kinase, is a multifunctional protein kinase detected in most eukaryotic cells (Gross and Anderson, 1998). In mammalian cells, there are five isoforms of casein kinase I: α, β, γ, δ, and ɛ (Fish et al, 1995). They are involved in multiple signalling pathways, including vesicular trafficking (Panek et al, 1997; Murakami et al, 1999), growth and morphogenesis (Robinson et al, 1993), circadian rhythm (Kloss et al, 1998; Peters et al, 1999), DNA-repair (Dhillon and Hoekstra, 1994), and cell cycle progression, and cytokinesis (Behrend et al, 2000). Previous studies showed that CKI regulates BR signalling in rice (Liu et al, 2003) and cell-to-cell communication (Lee et al, 2005), as well as modifying cell shape by phosphorylating tubulins in Arabidopsis (Ben-Nissan et al, 2008, 2010; Lee, 2009). Although a previous report showed that Hd6 (CK2) participates in rice Hd control, there has been no further study on how casein kinase is involved in the regulation of rice flowering time.
From a rice mutant population that we have generated (Fu et al, 2009), an early flowering mutant, earlier flowering1 (el1), was identified. Functional characterization revealed that EL1, which encodes a casein kinase I, could phosphorylate the rice DELLA protein SLR1 in vitro and in vivo, stabilize SLR1, and sustain its activity in vivo, to negatively regulate the GA signalling.
Results
Identification of the rice mutant early flowering1 (el1)
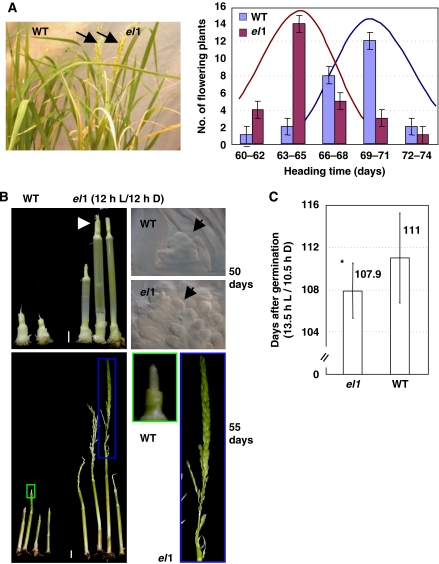
To understand the molecular mechanisms underlying heading, the rice mutant early flowering1 (el1) was identified from the Shanghai rice T-DNA insertion population (http://ship.plantsignal.cn, Fu et al, 2009). Under normal growth conditions (12 h light/12 h dark cycle), el1 flowered 5–6 days earlier than the wild type (WT), with a much slower leaf emergence rate (Figure 1A; Supplementary Figure 1A). At various time points, the el1 plants showed stimulated development of young panicles and the formation of inflorescence meristems (Figure 1B), which is consistent with the analysis on the transcription of floral organ identity genes that revealed advanced or enhanced expression of Hd6, Hd1, and OsMADS1 at 40, 45, 50, and 55 days of cultivation in el1 (Supplementary Figure 1B). This further confirmed the earlier floral initiation of el1 plants. Interestingly, although most rice early flowering mutants are regulated by photoperiod, measurement of flowering time under long-day treatment (13.5 h light/10.5 h dark) showed similar responses of el1 and WT plants (Figure 1C), indicating that the function of EL1 is independent of the photoperiod.
Figure 1.
Phenotypic analysis of rice early flowering 1 (el1). (A) Phenotypic observation (left) and analysis of the frequency distribution of heading date (right) indicated the earlier flowering of el1 plants. The plants were grown in a greenhouse with a 12 h light (L)/12 h dark (D) period. (B) Observations of the inflorescence development of WT and el1 plants at 50 or 55 days confirmed the earlier flowering of el1 (Bar=5 mm). The internodes of el1 were more elongated and the young panicles in the main culms were more obvious at 50 days of cultivation compared with that of WT. After 55 days of cultivation, the young panicles of el1 plants were almost fully developed—much earlier than WT. The floral meristems were observed through DIC or highlighted (squared). (C) Analysis of the heading date of el1 and WT plants under long-day treatment (13.5 h L/10.5 h D) showed that flowering of el1 is not modulated by photoperiod. The heading time was calculated and statistically analysed using a heteroscedastic t-test (*P<0.05, n=10).
EL1 encodes a casein kinase I
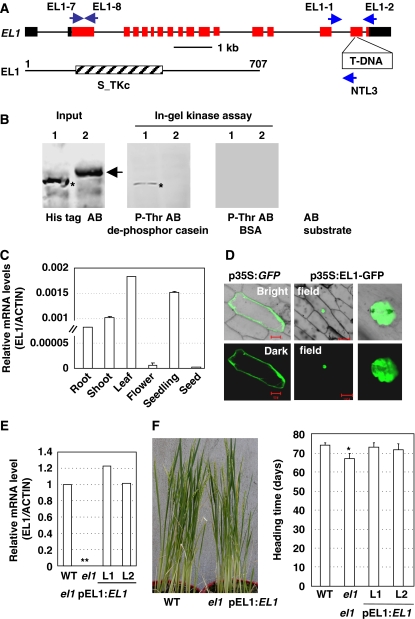
Segregation analysis (χ2[3:1]=3.44, 0.1<P<0.5) indicated that el1 was a recessive mutation. Further analysis of the DNA sequence flanking the T-DNA insertion, obtained by thermal asymmetric interlaced (TAIL)–PCR analysis, revealed a T-DNA insertion in the 15th exon of rice gene Os03g57940, which is designated EL1 and encodes a putative casein kinase I (a 707-amino acid polypeptide, Mw 79.8 kDa, Figure 2A).
Figure 2.
EL1 encodes a casein kinase I and is regulated by GA. (A) Scheme of the EL1 gene. Exons (red boxes), introns (lines), and UTR (untranslated region, black boxes) are indicated. The T-DNA is inserted at the 15th exon of EL1. Positions of primers used for confirming the T-DNA insertion (EL1-1 and NTL3), for identifying the homozygous mutant lines (EL1-1 and EL1-2), and examining the transcripts of EL1 by qRT–PCR analysis (EL1-7 and EL1-8) are indicated. The Thr_Ser protein kinase domain is located in the middle region (lower panel). (B) In-gel kinase assay revealed that recombinant expressed EL1 shows the casein kinase I activity, which could specifically phosphorylate the dephosphorylated casein (middle, asterisk) in vitro. Protein of EL1 (lane 1, asterisk) or AtPIP5K9 (lane 2, arrowhead, a kinase control) was used for the assay. Dephosphorylated casein or BSA was used as substrate. The input of EL1 and AtPIP5K9 was detected by His tag antibody (left) and the kinase assay was detected by Thr-P antibody (AB). (C) qRT–PCR analysis revealed the expression of EL1 in roots, shoots, leaves, and seedlings and relatively lower expression in flowers and seeds. The rice ACTIN gene was amplified and used as an internal positive control. (D) Transient expression of EL1–GFP fusion protein in onion epidermis cells through particle bombardment revealed that EL1 is localized in the nuclei. Bar=20 μm (left) or 50 μm (middle). (E) qRT–PCR analysis on the EL1 expression in homozygous el1 plants and el1 plants transformed with EL1 under its own promoter (pEL1:EL1) revealed the deficient or complemented expression of EL1 in el1 plants or el1 plants transformed with pEL1:EL1. The rice ACTIN gene was amplified and used as an internal positive control. (F) Phenotypic observation (left) and calculation (right) revealed that complementary expression of EL1 recovered the earlier flowering time of el1 plants (compared with WT), confirming the role of EL1 in controlling the heading time. The heading time was calculated and statistically analysed using a heteroscedastic t-test (*P<0.05, n=10).
Homologous analysis showed that EL1 shows high similarity to other casein kinase I members. Analysis of the phylogenetic relationship between EL1 and other CKIs indicated that EL1 is closest to yeast YCK2 and CKI (Supplementary Figure 2A, upper panel). Structural organization analysis of EL1 revealed the presence of a nuclear localization signal (NLS), a highly conserved S/T kinase domain, and conserved motifs in CKI (Figure 2A, lower panel; Supplementary Figure 2B) that is identical to that previously identified rice CKI1 (Liu et al, 2003). Biochemical studies of enzymatic activity showed that recombinant EL1 from Escherichia coli was able to phosphorylate the partially dephosphorylated casein but not for BSA in vitro (Figure 2B), indicating that EL1 is likely an active CKI.
Expression pattern analysis by quantitative real-time RT–PCR (qRT–PCR) showed that EL1 was transcribed in various tissues, including root, stem, leaf, seedling, and ripened seeds (Figure 2C). Although EL1 was expressed at relatively low levels in flowers, it was specifically transcribed at stage 2 during floral development (Supplementary Figure 3).
Transient expression of an EL1–GFP fusion protein in onion epidermal cells revealed that EL1 protein is accumulated in the nucleus (Figure 2D), consistent with the presence of an NLS sequence in EL1.
T-DNA insertion resulted in the defective expression of EL1 (Figure 2E) and expression of the full-length EL1 cDNA driven by its own native promoter in el1 (Figure 2E) rescued the early flowering phenotype (Figure 2F) showing the negative effects of EL1 in controlling rice flowering time.
EL1 is regulated by GA and el1 shows an increased response to GA
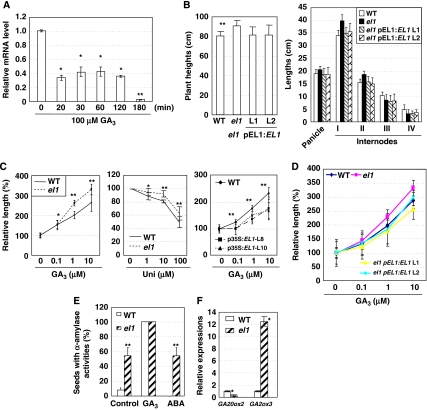
As GA is involved in controlling flowering time in Arabidopsis, we tested whether EL1 expression was regulated by hormones. The results showed that EL1 expression was evidently and rapidly suppressed by GA treatment (100 μM GA3) and declined to a minimal level after 3 h treatment (Figure 3A), suggesting that enzymatic activity of EL1 is regulated in a GA-dependent manner.
Figure 3.
el1 shows increased response to GA. (A) qRT–PCR analysis revealed that the expression of EL1 is rapidly suppressed by exogenous GA3 (100 μM) treatment for 20, 30, 60 120, or 180 min. The EL1 expression without GA3 treatment is set as 1.0. The rice ACTIN gene was amplified and used as an internal positive control. Seven-day-old seedlings were used for treatment and analysis. (B) Mature el1 plants are higher than WT (whereas complemented expression of El1 rescued the phenotype, left panel) and have more elongated uppermost internodes (right panel). The lengths of internodes and panicles are calculated and statistically analysed using the heteroscedastic t-test (compared with WT, *P<0.05; **P<0.01, n=6–10). Data are presented as mean±s.e. (C) Relative length of the second leaf sheaths of 7-day-old el1 seedlings in the presence of exogenous GA3 (0, 0.1, 1, or 10 μM, left) or uniconazole (0, 1, 10, 100 μM, middle), or that of transgenic plants overexpressing EL1 (p35S:EL1) in the presence of exogenous GA3 (right). The lengths of the second leaf sheaths were measured and the relative length was calculated and statistically analysed using a heteroscedastic t-test (*P<0.05; **P<0.01, n=15–20). (D) Calculation of the relative length of the second leaf sheaths of 7-day-old WT, el1 plants, and el1 with complementary expression of EL1 (pEL1:EL1, lines L1 and L2) in the presence of exogenous GA3 (0, 0.1, 1, or 10 μM). Error bars represent s.d. (n=15–20). (E) Analysis of α-amylase activities using a starch-containing plate in the presence of GA3 (1 mM) or ABA (10 μM) for 2 days. The number of seeds secreting α-amylase was calculated and statistically analysed using a heteroscedastic t-test (**P<0.01, n=20–30). (F) qRT–PCR analysis revealed the suppressed expression of GA20ox2 and enhanced expression of GA2ox3 in el1.
GA stimulates plant height and indeed el1 plants were consistently taller than WT, especially the uppermost internode (UI) (Figure 3B). Further measurement of cell lengths and widths under uniconazole treatment showed that, compared with WT, the elongated longitudinal lengths of the epidermal cells (at the middle section of the second leaf sheath) were less suppressed (Table 1). Leaf sheath growth exhibited an enhanced response to GA3 (Figure 3C, left panel; Supplementary Figure 5A) and a suppressed response to uniconazole (Figure 3C, middle panel; Supplementary Figure 5B), whereas the transgenic WT plants overexpressing EL1 consistently resulted in suppressed responses to GA3 (Figure 3C, right panel; Supplementary Figures 4 and 5C), further supporting the negative effects of EL1 on GA response. Being consistent, calculation of the relative length of the second leaf sheaths of 7-day-old seedlings showed that complementary expression of EL1 in el1 rescued the enhanced GA response of el1 (Figure 3D; Supplementary Figure 5D).
Table 1. Length of the epidermal cells in the second leaf sheath of 1-week-old el1 and WT plants under uniconazole treatment.
| Length (μm) | Uniconazol (μM) | |||
|---|---|---|---|---|
| 0 | 1 | 10 | 100 | |
| WT | 111.23±27.99 | 93.84±26.58 | 86.69±28.73 | 39.32±31.81 |
| el1 | 105.45±21.69 | 109.54±30.59* | 101.21±37.26** | 99.46±27.47** |
| Width (μm) | ||||
| WT | 12.54±2.42 | 8.63±1.52 | 11.09±1.89 | 10.80±1.88 |
| el1 | 11.57±2.16 | 9.60±2.00 | 10.59±1.83 | 12.87±1.93 |
| Length/width | ||||
| WT | 8.87±2.79 | 8.16±3.54 | 7.82±2.91 | 3.64±2.09 |
| el1 | 9.11±3.86 | 9.99±3.09* | 9.56±2.54** | 7.73±2.48** |
| Data are presented as mean±s.e. (n=10). Statistical analysis indicated significant differences (*P<0.05; **P<0.01). | ||||
This is further demonstrated by the observation that el1 seeds had much higher α-amylase activity in the absence of GA3 and that the inhibitory effects of ABA were significantly suppressed (Figure 3E; Supplementary Figure 6), consistent with the altered transcription of rice GA-biosynthesis-related genes GA20ox2 (suppressed expression) and GA2ox3 (enhanced expression, Figure 3F). These results confirm the enhanced responses of el1 to GA and hence the negative role of EL1 in GA signalling.
EL1 phosphorylates the rice DELLA protein SLR1
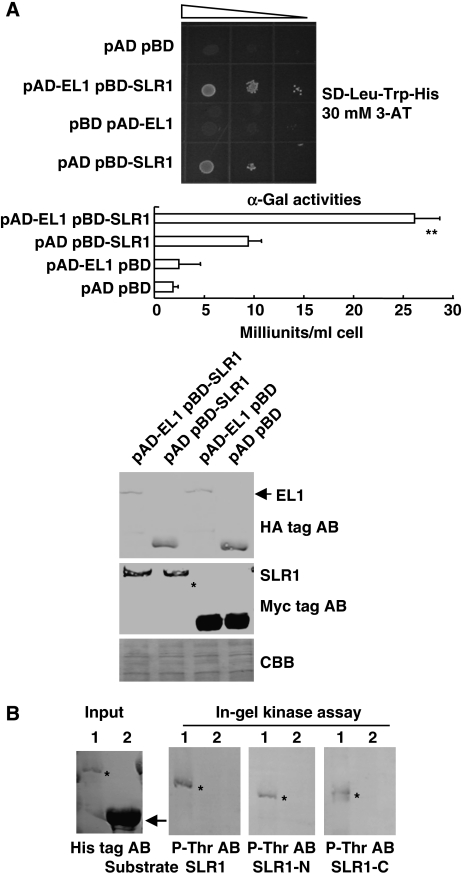
Previous studies have shown that CKI could regulate the relevant signalling pathway by phosphorylating the downstream proteins; we thus searched for candidate substrates of EL1 in GA signalling with the help of a computational prediction program (http://scansite.mit.edu/motifscan_seq.phtml). The rice DELLA protein SLR1, which has been shown as a key regulator in GA signalling, was identified. Computational analysis indicated the predicted phosphorylation sites at Ser196 and Ser510 (Supplementary Figure 7). Indeed, yeast two-hybrid analysis revealed that EL1 and SLR1 could interact with each other (Figure 4A), and an in-gel kinase assay indicated that SLR1 can be phosphorylated by EL1 in vitro in an EL1-specific manner (Figure 4B). Interestingly, either the N- or the C-termini of SLR1 can be phosphorylated by EL1 (Figure 4B), which is consistent with the presence of the predicted phosphorylation sites at Ser196 and Ser510 and suggests that EL1 may differentially regulate SLR1 by phosphorylating its N- or C-termini.
Figure 4.
EL1 phosphorylates rice DELLA protein SLR1. (A) Interaction of the EL1 protein with SLR1 (upper panel) and quantitative assay of interactions (bottom panel). Yeast transformants were diluted 100, 102, and 104 times and grown on SD-Leu-Trp-His plates (supplemented with 30 mM 3-AT). For the α-galactosidase assay, yeast cells were cultured in liquid medium and values are the mean of two independent assays (10 randomly selected clones were measured each time) and bars indicate the standard deviation (s.d.). Statistical analysis was performed using a heteroscedastic t-test (**P<0.01, n=10). Western blot analysis using HA tag or Myc tag antibodies indicated that equal amounts of proteins were used for the assay. (B) In-gel kinase assay revealed that EL1 specifically phosphorylated SLR1 in vitro, as well as the N- or the C-terminus of SLR1 (asterisks, recombinantly expressed N- or C-terminus of SLR1 were used as substrate.). The protein extracts of EL1 (lane 1) and OsCKI1 (another member of the rice casein kinase I family, lane 2) were used for the assay. Input of EL1 and OsCKI1 was detected by His tag antibody and the kinase assay was detected by using Thr-P antibody (AB).
EL1 phosphorylation of SLR1 is critical for sustaining its activity and stability
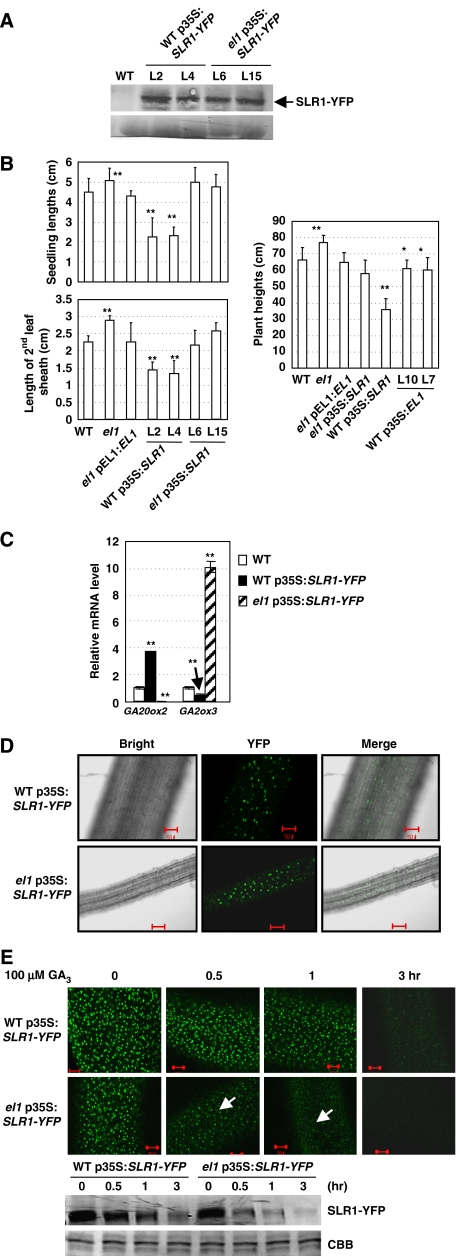
Further in vivo analysis using transgenic WT or el1 plants overexpressing SLR1 (Supplementary Figure 8A) indicates that although the protein level of SLR1–YFP fusion protein was almost same (Figure 5A), the effects of SLR1 were significantly suppressed under EL1 deficiency. Analysis of the lengths of seedlings, mature plants, and second leaf sheaths showed that SLR1 overexpression resulted in a severe dwarf phenotype and shortened second leaf sheaths in WT plants; these effects were suppressed in el1 plants (Figure 5B; Supplementary Figure 8B). This trend was more significant in mature plants (Figure 5B, right panel; Supplementary Figure 8B, bottom panel), indicating that phosphorylation of SLR1 by EL1 is critical for its function. In addition, analyses of the expression levels of GA20ox2 and GA2ox3 showed that GA20ox2 expression was increased and GA2ox3 expression was decreased when SLR1 was overexpressed in WT plants, whereas the opposite expressions were detected in the el1 mutant (Figure 5C) showing the effects of EL1 phosphorylation on SLR1 activity. In addition, the el1 plants overexpressing SLR1 (el1 p35S:SLR1–YFP) were much taller than WT plants overexpressing SLR1 (WT p35S:SLR1–YFP), correlating with the reduced GA20ox2 and increased GA2ox3 expressions in these lines, which suggests that EL1 may take part in the feedback regulation of these genes.
Figure 5.
EL1 phosphorylates SLR1 to sustain the activity and stability of SLR1. (A) Western blot analysis confirmed the similar levels of SLR1–YFP protein in WT and el1 plants. Equal amounts of proteins (∼10 μg) were used for the blotting and staining by Coomassie brilliant blue (CBB) showed the similar loading of proteins (lower panel). (B) Statistical analysis on the lengths of 7-day-old seedlings (left-upper panel), second leaf sheaths (left-bottom panel), and mature plants (right panel), which indicated the negative effects of SLR1 on plant growth were evidently suppressed under EL1 deficiency. Error bars represent s.d. (n=15). Heteroscedastic t-test analysis indicated a significant difference (compared with WT, *P<0.05, **P<0.01). (C) qRT–PCR analysis revealed that the increased expression of rice GA20ox2 and suppressed expression of rice GA2ox3 in WT plants overexpressing SLR1 were oppositely regulated in el1 plants overexpressing SLR1, indicating the crucial role of EL1 in SLR1 functions. Error bars represent s.d. (n=15). Heteroscedastic t-test analysis indicated a significant difference (**P<0.01). (D) Phosphorylation of EL1 on SLR1 does not affect the sub-cellular localization of SLR1. Compared with the nuclear localization, deficiency of EL1 does not alter SLR1 sub-cellular localization. Bar=50 μm. (E) GA-dependent SLR1 degradation was evidently enhanced under EL1 deficiency under GA3 treatment (100 μM, for 0.5, 1, or 3 h), revealing the crucial role of EL1 in SLR1 stability. SLR1–YFP fusion protein of 7-day-old WT or el1 plants (p35S:SLR1–YFP) was observed under a confocal microscope (up panel) and analysed by western blot analysis using GFP antibody (bottom panel, staining CBB showed the similar amounts of loading proteins). Bar=50 μm.
Aside from the effects of phosphorylation by EL1 on SLR1 activity, possible effects of EL1 on SLR1 protein localization and stability were studied. Compared with the nuclear localization in WT, reduced phosphorylation of SLR1 in el1 did not alter its sub-cellular localization (Figure 5D). However, detailed studies showed that GA-mediated SLR1 degradation was greatly enhanced in el1. Fluorescence observation and western blot analysis revealed that in comparison with WT the degradation of SLR1–YFP fusion protein after GA3 treatment was significantly enhanced in el1 (Figure 5E). This indicates that phosphorylation of SLR1 by EL1 is crucial for its stability.
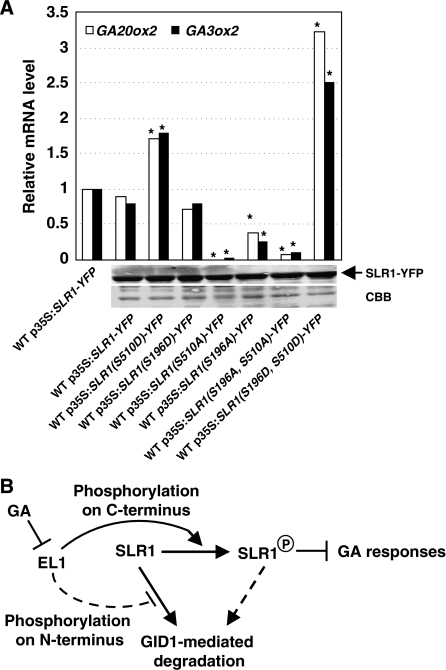
Pervious studies showed that the SLR1 N-terminal is the regulatory domain and C-terminal is responsible for the repression activity (Itoh et al, 2002). To further confirm the effects of phosphorylation on SLR1, point mutation of Ser196 and Ser510 into either A (short for Ala, which results in the defective phosphorylation on this site) or D (short for Asp, which results in the constitutive phosphorylation on this site) was performed and GA signalling of plants containing the corresponding mutated SLR1 was examined by detecting the transcripts of GA synthesis genes GA20ox2 and GA3ox2. The results showed that after GA3 treatment, suppressed phosphorylation of SLR1 at either Ser196, Ser510, or both Ser196/Ser510 indeed resulted in the significantly enhanced GA signalling (evidenced suppressed expression of GA20ox2 and GA3ox2), whereas the constitutive phosphorylation of SLR1 at Ser510 or both Ser196/Ser510 would result in the suppressed GA signalling (enhanced expression of GA20ox2 and GA3ox2, Figure 6A). These further confirmed the importance of phosphorylation of SLR1, especially at sites Ser196 or Ser510, on the SLR1 effects in GA signalling. The constitutive phosphorylation of SLR1 at Ser196 did not affect the GA signalling, which might be due to the N-terminal of SLR1, which is mainly for the regulatory domain (Itoh et al, 2002).
Figure 6.
Phosphorylation of EL1 on SLR1 is crucial for GA signalling. (A) After GA3 treatment (100 μM, 1 h), expressions of GA20ox2 and OsGA3ox2 were decreased in WT plants overexpressing mutated SLR1m–YFP (S196A, S510A, and S196A/S510A, which results in the suppressed phosphorylation of these sites), revealing the enhanced GA responses. On the opposite, the expressions of GA20ox2 and OsGA3ox2 were increased in WT plants overexpressing mutated SLR1m–YFP (S510D, S196D/S510D, which results in the constitutive phosphorylation of these sites), revealing the suppressed GA responses. qRT–PCR analysis was performed and heteroscedastic t-test analysis indicated a significant difference (**P<0.01). Expression of the corresponding genes in WT expressing SLR1–GFP without GA3 treatment was set as 1.0. Western blot analysis confirmed the similar levels of proteins (SLR1–YFP, and SLR1m–YFP) in all of the transgenic lines before GA treatment (bottom panel). (B) Hypothetical model of EL1 function in GA signalling. EL1 is repressed by GA and phosphorylation of EL1 on the SLR1 C-terminus sustains its active form (in red). Phosphorylation on the SLR1 N-terminus suppresses the GID1–GA-mediated degradation of SLR1, resulting in the suppressed GA responses.
Discussion
EL1, a casein kinase, is a novel regulator of GA signalling and has important functions in controlling rice flowering time by regulating GA responses
As an important process in plant reproduction, flowering time is finely controlled by a complex network. In Arabidopsis, the key regulators, FT and SOC1, regulate the expression of floral identity genes including AP1, LFY, and AG (Mouradov et al, 2002); most of these genes are MADS box genes and determine the identity of floral organs (Yanofsky et al, 1990; Weigel et al, 1992; Jofuku et al, 1994). In rice, the major floral initiation pathway is the photoperiod pathway; five QTLs (Hd1, Hd2, Hd3, Hd5, and Hd6) were found to confer photoperiod sensitivity (Lin et al, 2000; Takahashi et al, 2001). Genetic analysis of the relationship between EL1 and Hd genes showed that hd1, hd3a, or hd6 plants with suppressed EL1 expressions still flower early, similar to el1 (Supplementary Figure 9). This finding indicates that EL1 is epistatic to the Hd genes, consistent with the phenotypic observation that el1 has similar photoperiod responses to WT.
GA is crucial for rice flower development and deficiency of GA results in severe dwarfism and failed grain setting in rice (Sakamoto et al, 2003). Although GA is critical in flowering control in the Arabidopsis, there is no report of its role in rice flowering. Deletion of a DELLA domain (such as the gai allele) will cause delayed flowering even in the presence of GA in Arabidopsis (Peng et al, 1997); however, no difference was detected in the flowering time of rice. In the GAMYB-deficient mutant, despite shortened internodes and defects in floral organ development, a notable defect in pollen development was observed (Kaneko et al, 2004). There was no change in the heading date in the slr1 mutant (Itoh et al, 2002) or Slr1-d3 (a GA-insensitive mutant, Chhun et al, 2007). Recent studies revealed that GA-deficient plants exhibit shortened UI and panicles enclosed by flag leaves (Yin et al, 2007), suggesting that GA may influence rice heading time by modulating UI elongation. This study provides direct evidence that GA signalling regulates rice flowering and identifies EL1 as a novel regulator of flowering time by affecting GA responses.
DELLA proteins are important in GA signal transduction (Peng et al, 1997). Truncating the DELLA domain of rice SLR1 suppresses SLR1 degradation under GA treatment (Itoh et al, 2002). Recent studies showed that suppression of SLR1 activity could be accomplished by GA and GID1 alone, without the F-box protein GID2 (Ueguchi-Tanaka et al, 2008). These results suggest that other unknown factors might interact with SLR1 to induce its suppressive activity (Ueguchi-Tanaka et al, 2008). This study is the first direct report showing that SLR1 can be phosphorylated by casein kinase I and that EL1 modulates GA signalling by phosphorylating the rice DELLA protein SLR1.
Phosphorylation of SLR1 is important for the regulation of its activity and stability
Although DELLA proteins have inhibitory roles in the GA response (Itoh et al, 2005), it remains unclear how phosphorylation of DELLA proteins affects their function and/or stability, although this question has been addressed by several research groups (Itoh et al, 2002, 2005; Ueguchi-Tanaka et al, 2008).
It was reported that in the GA signalling pathway, phosphorylation may inactivate DELLA function (Itoh et al, 2002). Furthermore, some kinases can phosphorylate SLR1 within the polyS/T/V, DELLA, and TVHYNP domains (Itoh et al, 2005). However, phosphorylation of SLR1 is independent of its degradation and is not necessary for the interaction of SLR1 with the GID2/F-box protein (Itoh et al, 2005). The relationship between SLR1 degradation and phosphorylation remains to be elucidated, as do the identities of the protein kinases involved. We showed that both the N- and the C-termini of SLR1 can be phosphorylated by EL1 and confirmed that in el1, the effects of SLR1 are suppressed (the GA signalling is enhanced) and the GA-mediated degradation of SLR1 is greatly enhanced (Figure 5E). This is consistent with the previous reports that the SLR1 N-terminus and C-terminus are responsible for the degradation and activity of SLR1, respectively (Itoh et al, 2002). Constitutive or suppressed phosphorylation of SLR1 at site Ser196 or Ser510 resulted in the suppressed or enhanced GA signalling, which further confirmed the critical role of phosphorylation of SLR1 on its negative effects in GA signalling. As a sum, these findings indicate that phosphorylation by EL1 is crucial for SLR1 stability at the N-terminus and SLR1 activity at the C-terminus, and hence the regulation of GA responses (Figure 6B). However, phosphorylation of SLR1 by other kinases cannot be excluded, which need further studies and will be helpful to clarify the regulatory mechanism of SLR1 and GA signalling.
In addition, it is worth to notice that the el1 plants expressing SLR1 are much taller than WT plants expressing SLR1, which nicely correlates with the reduced GA20ox2 and increased GA2ox3 expression in the corresponding lines. However, observation of plant height (Figure 5B) showed that el1 plants expressing SLR1 is intermediate between WT and WT plants expressing SLR1, and expression levels of GA20ox2 and GA2ox3 are much changed compared to the WT, which may suggest that EL1 has a role in feedback regulation of the GA biosynthetic genes.
Although CKIs modulate the activity of a broad variety of substrates, and multiple isoforms exist in both animals and plants, they have specific roles at certain developmental stages. Concerning the presence of multiple isoforms of CKI in Arabidopsis and rice, specific phosphorylation of rice SLR1 by EL1 to modulate the GA response suggests the presence of a specific regulatory mechanism. In addition, distinct domain structures in EL1 compared with those of broccoli (Klimczak and Cashmore, 1993) or yeast (Robinson et al, 1992) further suggest the existence of another regulatory mechanism (aside from the conserved kinase domain and characteristic peptides, EL1 has a more expanded region at the N-terminus).
In summary, the studies presented not only shed light on the mechanisms of GA signalling control, but also provide important clues regarding the mechanism and effects mediated by casein kinase I. CKI is a multifunctional protein kinase involved in hierarchical protein phosphorylation, modulating signal transduction through second-messenger-responsive protein kinases throughout the plant and animal kingdoms. This study will surely expand our knowledge and facilitate a comprehensive understanding of the underlying mechanisms in plants, as well as in animals.
Materials and methods
Isolation of rice early flowing 1 (el1) mutant
A recessive mutant, el1, was identified in rice (Oryza sativa, Zhonghua11) T-DNA insertion mutant population (SHIP, http://ship.plantsignal.cn, Fu et al, 2009). Surface-sterilized seeds of el1 mutant and WT plants were soaked in water for 7 days and then placed in soil and grown in a greenhouse. The flanking sequence was obtained by TAIL-PCR(Liu et al, 1995). Primers EL1-1 (5′-CTTCTGGTGTCCTTCCATTAG-3′) and EL1-2 (5′-TGGGAGAGCTGAAGATGTATG-3′) were used for PCR amplification to confirm the T-DNA insertion using genomic DNA as template.
Measurement of shoot elongation
Shoot elongation was quantified by a modification of the method described by Matsukura et al (1998). Seeds from WT and el1 plants (n=15–25) were surface sterilized for 30 min with a 3% NaClO solution, washed four times with sterile distilled water, soaked in the distilled water for 24 h in the presence or absence of different concentration of uniconazole, and then placed in sterile distilled water for another 24 h. The seeds were then placed on a 1% agar plate and grown under fluorescent lamps at 28°C (12 h light/12 h dark). To investigate the role of GA on the elongation of the second leaf sheath, seeds of WT and el1 mutant plants (n=15–25) were germinated on 1% agar plates supplemented with various concentrations of GA3. All plants were grown at 28°C (12 h light/12 h dark) for 7 days. For analysis of surface anatomy, the second leaf sheath was dissected from third-leaf-stage plants and observed using differential interference contrast (DIC) microscopy. The middles of the second leaf sheath sections were incubated in 9:1 (v:v) ethanol:acetic acid overnight, rinsed with water, and placed in chloral hydrate (glycerol:chloral hydrate:water, 1:8:1, v:w:v). Samples were visualized using a Leica DMR microscope (Germany). The plant heights were measured and calculated when the rice plants had ripened.
Agar plate assay of α-amylase
The agar plate assay was performed essentially as described by Yamachuchi (1998). Seeds were cut transversely and endosperm half-seeds were surface sterilized with 1% NaClO for 15 min, washed with sterile water six times, and placed on 2% agar plates containing soluble starch (0.2%) (Linhu Food Products Factory, China). Plates with 16 or more endosperms were incubated at 30°C in the dark for 48 h, and the endosperms were then removed to examine the α-amylase activity by staining agar with a solution containing 0.1% I2 and 1% KI.
Protein expression and in-gel kinase assay
The coding region of EL1 was amplified with primers EL1-5 (5′-CATGCCATGGCTATGCCAGAGTTGCGGGGT-3′, added NcoI site underlined) and EL1-6 (5′-GGACTAGTGCATACGGTCCGGCCGTAGC-3′, added SpeI site underlined), and the resultant DNA fragment was sub-cloned into vector pET32c (Novagen, USA). The inducement of recombinant protein expression was carried out by supplement with 1 mM IPTG (28°C, 3 h). The full-length cDNA of SLR1 was amplified with primers SLR1-1 (5′-GGGGTACCATGAAGCGCGAGTACCAAGAAG-3′, added KpnI site underlined) and SRL1-2 (5′-CGGAATTCGACGCGCCATGCCGAGGTGG-3′, added EcoRI site underlined), and then sub-cloned into vector pET32c. The cDNA fragments encoding the N-terminal or C-terminal of SLR1 were amplified with primers SLR-N-1 (5′-CGGAATTCATGAAGCGCGAGTACCAAGAAG-3′, added EcoRI site underlined) and SLR-N-2 (5′-CCCAAGCTTGAAGTGGGCGAACTTGAGGTAG-3′, added HindIII site underlined), and SLR-3 (5′-CATGCCATGGAAACCGCAAATCAAGCCATCCTCG-3′ added NcoI site underlined) and SLR-4 (5′-CGGAATTCGGCGACGCGCCATGCCGAGGTG-3′, added EcoRI site underlined), respectively. The vectors pET32a-AtPIP5K9 (Lou et al, 2007) and pET32a-OsCKI1 (Liu et al, 2003) were used for recombination expression of AtPIP5K9 and OsCKI1, respectively.
The in-gel kinase assay was performed mainly as described by Murray (http://www.biocompare.com/protocols/protocol/163/In-Gel-Kinase-Assay.html) with few modifications. A 10% SDS–PAGE gel containing purified SLR1 protein (0.5 mg/ml) was prepared, and the crude EL1 protein was diluted in 2 × sample buffer [2 mM DTT, 20% glycerol, 100 mM Tris–Cl (pH6.8), 0.2% bromophenol blue (w/v), 4% SDS (w/v)] and loaded into the gel after boiling for 2 min. After electrophoresis, the gel was washed in wash buffer 1 [50 mM Tris–HCl, pH 8.0, 20% (v/v) 2-propanol] and wash buffer 2 (50 mM Tris–Cl, pH 8.0; 6 M urea; 5 mM 2-mercaptoethanol) each for 1 h at room temperature, then incubated in the wash buffer 3 [50 mM Tris–Cl, pH 8.0; 5 mM 2-mercaptoethanol; 0.04% (v/v) Tween20] at 4°C overnight with gentle rocking. Furthermore, the gel was incubated in the kinase buffer (40 mM Tris–Cl, pH 7.5; 2 mM DTT; 0.1 mM EGTA; 10 mM magnesium acetate) for 30 min at room temperature and then incubated in the buffer containing 20 μM ATP (with rocking) for another 60–90 min. The gel was then transferred to PVDF membrane (PerkinElmer Life Science, USA) by semi-dry blotting and the blot was incubated with rabbit anti-phosphorylation Thr-antiserum (Cell Signaling, USA), then the bovin anti-rabbit IgG alkaline phosphatase (AP)-conjugated secondary antibody (Santa Cruz, Germany). The AP activity was detected by BCIP/NBT solution (BBI Company, USA).
Semi-quantitative RT–PCR analysis
Total RNAs were extracted and used to synthesize the first-strand cDNAs by reverse transcription. Equal amounts of first-strand cDNAs were used as templates for PCR amplification. Primers EL1-7 (5′-ATGCCAGAGTTGCGGGGTGGTG-3′) and EL1-8 (5′-AGGTTGATCCTTACACAAATC-3′) were used to study the expression pattern of EL1 in various tissues of WT plants. Primes Hd1-1 (5′-TTCTCCTCTCCAAAGATTCCG-3′) and Hd1-2 (5′-AGCAGGTGTCAGGATTCTGG-3′); Hd6-1 (5′-TTGTCAGGAAAGTTGGAAGAGG-3′) and Hd6-2 (5′-TCCCTGGATGATAGAACTCAGC-3′); OsMADS1-1 (5′-TGCTCAAGAAGGCCTACGAG-3′) and OsMADS1-2 (5′-TGATGATACCCAATCTGCAGG-3′) were used to examine the expression of corresponding genes at different floral stages. The ACTIN gene was amplified using primers actin-1 (5′-GAACTGGTATGGTCAAGGCTG-3′) and actin-2 (5′-ACACGGAGCTCGTTGTAGAAG-3′) and served as an internal positive control.
qRT–PCR analysis
qRT–PCR analyses were performed to study the transcription of EL1 in WT (various tissues or under GA3 treatment), el1, and transgenic el1 plants with complementary expression of EL1, or the expressions of genes involved in GA biosynthesis in WT, el1, and transgenic WT or el1 lines overexpressing SLR1 or mutated SLR1. Total RNAs were extracted and used to synthesize cDNAs by reverse transcription. Primers EL1-7 and EL1-8 were used to examine the expression of EL1. Primers used to analyse the transcripts of GA20ox2 (Sd1), GA3ox2, and GA2ox3 were as previous description (Sakamoto et al, 2004). The ACTIN gene was amplified using primers actin-1 and actin-2, and served as an internal positive control.
EL1–GFP fusion studies
The whole coding region of EL1 was PCR amplified using primers EL1-5 and EL1-6, and the resultant products were sub-cloned into pCambia1302 vector to generate p35S:EL1–GFP. The construct was sequenced to confirm the in-frame fusion of EL1 to GFP and positive clone was used for transient transformation in onion epidermal cells through particle bombardment (Bio-Rad, USA). Transformed onion cells were culture for 24 h and observed under a confocal microscope (Zeiss LSM 510 META; argon laser excitation wavelength 488 nm for GFP observation).
Constructs and rice transformation
The whole coding region of YFP was amplified with primer YFP-1 (5′-GGACTAGTATGGTGAGCAAGGGCGAGGA, added SpeI site underlined) and YFP-2 (5′-GCGTCGACAAGTTGGGTAACGCCAGGGT-3′, added SalI site underlined), and sub-cloned into pBluescript SK(+) (Stratagene, USA). The coding region of EL1 was amplified with primers EL1-5 and EL1-6, and then sub-cloned into vector pBSK-YFP. The DNA fragment containing EL1–YFP was then sub-cloned into pCambia2301 (Cambia, Australia), resulting in construct p35S:EL1–YFP.
The EL1 promoter was amplified with primer EL1-9 (5′-ACGCGT CGACTGGCATTATCGCCCCATGC-3′, added SalI site underlined) and EL1-10 (5′-AAAACTGCAGTCACGATCTAGAGAAATTAC-3′, added PstlI site underlined), and sub-cloned into pBluescript SK(+). The coding region of EL1 was amplified with primers EL1-5 and EL1-6, and then sub-cloned into vector pBSK-pEL1. The DNA fragment containing pEL1-EL1 was then sub-cloned into pCambia2301 (Cambia, Australia), resulting in construct pEL1:EL1.
The full-length cDNA of SLR1 was PCR amplified using primers SLR1-1 and SLR1-4 (5′-GGACTAGTGACGCGCCATGCCGAGGTGG-3′, added SpeI site underlined), and then sub-cloned into pBSK-YFP vector. The fused SLR1–YFP fragment was then sub-cloned into pCambia2301, resulting in the construct p35S:SLR1–YFP.
The partial cDNA of Hd genes Hd1, Hd3a, and Hd6 were amplified by primers Hd1-3 (5′-ACGCGTCGACCGACCAGGAGGTTGGAGTT-3′, added SalI site underlined) and Hd1-4 (5′-CGGGATCCGGAGCTGAAGTGAAGGGACA-3′, added BamHI site underlined); Hd3a-3 (5′-GCTCTAGAGGTTGGTAGGGTTGTGGGT-3′, added XbaI site underlined) and Hd3a-4 (5′-GGGGTACCCATGCTGGATGATGATAGTGAG-3′, added KpnI site underlined); Hd6-3 (5′-ACGCGTCGACGGTGAGCAGGATGACTATGA-3′, added SalI site underlined) and Hd6-4 (5′-CGGGATCCTGGAGGAAGTACGGATGTG-3′, added BamHI site underlined), respectively. The amplified fragments were sub-cloned into pCambia2301, resulting in constructs p35S:A-Hd1, p35S:A-Hd3a, and p35S:A-Hd6.
The full-length cDNA of SLR1 were amplified by primers SLR1-10 (5′-TCGTCGTCCTCATCGTCGGACGACGAC-3′) and SLR1-11 (5′-GCGAGGCCCCACCGCCCAGGTCGTCGTC-3′) for the replacement of Ser196 to Asp; SLR1-9 (5′-AGAGAGCTCGGCCTGGCCGGAGTCGCC-3′) and SLR1-17 (5′-CCCTCGAGGGCGGCGACTCCGGCCAG-3′) were used for replacement of Ser510 to Asp; SLR1-15 (5′-ACGTCGGCAGCAGCAGCAGCAGCAGCA-3′) and SLR1-16 (5′-AGGCCCCACCGCCCAGTGCTGCTGCTGC-3′) were used for replacement of Ser196 to Ala; SLR1-18 (5′-AGAGAGCTCGGCCTGGCCTGCTGCTGCTGCCTC-3′) and SLR1-19 (5′-GTTCGATTCCCTCGAGGCAGCAGCAGCAGGC-3′) were used for replacement of Ser510 to Ala. The amplified cDNA fragments were sub-cloned into pBSK-YFP vector, and then the fused SLR1m–YFP fragment was sub-cloned into pCambia2301, resulting in construct p35S:SLR1m–YFP (including S196A, S196D, S510A, S510D, S196A/S510A, and S196D/S510D).
All the positive constructs were introduced into rice WT, el1 plants, respectively, by Agrobacterium tumefaciens-mediated transformation using immature embryos as materials.
Preparation of protein extracts and immunoblot analysis
Total proteins were extracted by grinding rice leaves with liquid nitrogen; the ground tissues were then resuspended in the extraction buffer [20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5% Tween-20, 1 mM EDTA, 1 mM dithiothreitol (DTT)] containing the protease inhibitor cocktail (Roche, Mannheim, Germany). After the addition of an equal volume of 2 × sample buffer [1 × sample buffer: 67.5 mM Tris–HCl (pH 6.8), 2% (w/v) SDS, 10% (w/v) glycerol, 0.01% (w/v) bromophenol blue, and 0.1 M DTT], the samples were boiled for 5 min, separated by 8% SDS–PAGE, and transferred to a PVDF membrane by semi-dry blotting. The blots were incubated with mouse anti-GFP antiserum (Neo-Marker, UK) and then with goat anti-mouse IgG AP-conjugated secondary antibody (Santa Cruz, Germany). AP activity was detected by BCIP/NBT solution (BBI Company, USA).
Yeast two-hybrid analysis and α-galactosidase assay
The yeast Gal4 system was used for two-hybrid analysis of EL1 and SLR1 protein interactions. Primers EL1-11 (5′-GATGCCAGAGTTGCGGGGTGGT-3′) and EL1-12 (5′-CCGCTCGAGGCATACGGTCCGGCCGTAGCAG-3′, added XhoI site underlined) were used to amplify the EL1 coding region, which was sub-cloned into vector pGADT7 (Clontech, USA). Primers SLR1-11 (5′-GGAATTCCATATGAAGCGCGAGTACCAAGAAG-3′, added NdeI site underlined) and SLR1-12 (5′-CGGAATTCCGCCGCGGCGACGCGCCATGCC-3′, added EcoRI site underlined) were used to amplify the SLR1 coding region, which was sub-cloned into vector pGBKT7 (Clontech, USA). The yeast strain AH109 was used as the host strain and transformed using a modified lithium acetate method.
The α-galactosidase assay was performed essentially according to the user manual provided by the manufacturers. Ten clones were randomly selected for measurement in each experiment. To confirm that equal amounts of proteins were used for the assay, western blot was performed using extracts prepared from yeast cells as described by Nam and Li (2002). The yeast cells were collected and ground to a fine powder in liquid nitrogen, and further ground in cold grinding buffer [50 mM HEPES (pH 7.4), 10 mM EDTA, 0.1% Triton X-100, 1 mM PMSF]. After addition of an equal volume of 2 × sample buffer, the samples were boiled for 5 min, separated by 10% SDS–PAGE and transferred to a PVDF membrane by semi-dry blotting. The blots were incubated with mouse anti-Myc antiserum (Neo-Marker, UK), or with rabbit anti-HA antiserum (Santa Cruz, Germany), and then with goat anti-mouse IgG or bovine anti-rabbit IgG AP-conjugated secondary antibody (Santa Cruz, Germany). AP activity was detected by BCIP/NBT solution (BBI Company, USA).
Phylogenetic analysis
Homolog sequences in A. thaliana, O. sativa, Saccharomyces cerevisiae, and Homo sapiens were obtained at the NCBI Web site (http://www.ncbi.nlm.nih.gov/). The conserved domains of proteins were analysed at the website: http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. As all casein kinase I homologs had S/T protein kinase domains, we used the conserved domains for phylogenetic analyses. Sequence alignments were generated with CLUSTALX 1.83, and the alignments between Arabidopsis, O. sativa, S. cereviiae, and H. sapiens were adjusted before the tree was constructed. Neighbour-joining analyses were performed using MEGA3 (Kumar et al, 2004) with the pair-wise deletion option, with Poisson correction set for distance model, and 1000 bootstrap replicates selected.
The sequence accession number was obtained from NCBI, as follows: CKIepsilon: AAQ02559 (H. sapiens); CKIdelta isoform 2: BAB23405 (H. sapiens); CKIbeta isoform: AAS46020 (Toxoplasma gondii); CKIalpha (DmCK1): NP_511140; EL1: NP_001051531; CKI: AAA19019; YCK2: NP_014245; Hhp1: NP_595760; OsCKI: CAD32377; AtCKI1: NP_193170; AtCKI2: NP_188976; CKIgamma 2: AAP36921 (H. sapiens); Os01g51200: BAB92346; Os01g13060: NP_001042496; Os01g38950: NP_001043372; Os02g40860: NP_001047465; Os02g56560: EAZ25030; Os02g17910: NP_001046556; Os04g43490: NP_001053309; Os05g51560: NP_001056503; Os10g33650: NP_001064847; CKL3: NP_194617; CKL4: NP_194615; CKL11: AAY24540; CKL9a: AAY24537; CKL2: AAY24533; CKL6: NP_567812; CKL7: NP_199223; CKL1: NP_194340.
Supplementary Material
Acknowledgments
This study was supported by the state key project of basic research (2005CB120803), and National Science Foundation of China (90717001, 30721061). We thank Shu-Ping Xu for rice transformation and Professor Hong Ma (Fudan University, China) and Prof Xiang-Dong Fu (Institute of Genetics and Developmental Biology, CAS) for helpful discussions.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bernier G (1988) The control of floral evocation and morphogenesis. Annu Rev Plant Physiol Plant Mol Biol 39: 175–219 [Google Scholar]
- Behrend L, Stoter M, Kurth M, Rutter G, Heukeshoven J, Deppert W, Knippschild U (2000) Interaction of casein kinase 1 delta (CK1delta) with post-Golgi structures, microtubules and the spindle apparatus. Eur J Cell Biol 79: 240–251 [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G, Cui W, Kim DJ, Yang Y, Yoo BC, Lee JY (2008) Arabidopsis casein kinase 1-like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiol 148: 1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Nissan G, Yang Y, Lee JY (2010) Partitioning of casein kinase 1-like 6 to late endosome-like vesicles. Protoplasma 240: 45–56 [DOI] [PubMed] [Google Scholar]
- Chhun T, Aya K, Asano K, Yamamoto E, Morinaka Y, Watanabe M, Kitano H, Ashikari M, Matsuoka M, Ueguchi-Tanaka M (2007) Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19: 3876–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N, Hoekstra MF (1994) Characterization of two protein kinases from Schizosaccharomyces pombe involved in the regulation of DNA repair. EMBO J 13: 2777–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun TP (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish KJ, Cegielska A, Getman ME, Landes GM, Virshup DM (1995) Isolation and characterization of human casein kinase I epsilon (CKI), a novel member of the CKI gene family. J Biol Chem 270: 14875–14883 [DOI] [PubMed] [Google Scholar]
- Fu FF, Ye R, Xu SP, Xue HW (2009) Studies on rice seed quality through analysis of a large-scale T-DNA insertion population. Cell Res 19: 380–391 [DOI] [PubMed] [Google Scholar]
- Gomi K, Sasaki A, Itoh H, Ueguchi-Tanaka M, Ashikari M, Kitano H, Matsuoka M (2004) GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J 37: 626–634 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun T-P, Thomas SG (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SD, Anderson RA (1998) Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal 10: 699–711 [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Hussain A, Cao D, Cheng H, Wen Z, Peng JR (2005) Identification of the conserved serine/ threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J 44: 88–99 [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Sasaki A, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Hasegawa Y, Minami E, Ashikari M, Matsuoka M (2005) Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol 46: 1392–1399 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22: 391–399 [DOI] [PubMed] [Google Scholar]
- Jofuku KD, Boer BG, Montagu MV, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, Matsuoka M (2004) Loss-of-function mutations of the rice GAMYB gene impair a-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T (1998) Report of the committee on gene symbolization, nomenclature and linkage groups. II. Linkage mapping using mutant genes in rice. Rice Genet Newsl 15: 13–74 [Google Scholar]
- Klimczak LJ, Cashmore AR (1993) Purification and characterization of casein kinase I from broccoli. Biochem J 293: 283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94: 97–107 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA 3, integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lee JY (2009) Versatile casein kinase 1: multiple locations and functions. Plant Signal Behav 4: 652–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Taoka KI, Yoo BC, Ben-Nissan G, Kim DJ, Lucas WJ (2005) Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng JR (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following inhibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HX, Yamamoto T, Sasaki T, Yano M (2000) Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines. Theor Appl Genet 101: 1021–1028 [Google Scholar]
- Liu W, Xu ZH, Luo D, Xue HW (2003) Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J 36: 189–202 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lou Y, Gou JY, Xue HW (2007) PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19: 163–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura C, Itoh S, Nemoto K, Tanimoto E, Yamaguchi J (1998) Promotion of leaf sheath growth by gibberellic acid in a dwarf mutant of rice. Planta 205: 145–152 [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami A, Thurlow J, Dickson C (1999) Retinoic acid-regulated expression of fibroblast growth factor 3 requires the interaction between a novel transcription factor and GATA-4. J Biol Chem 274: 17242–17248 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuk H, Katoh E, Iuchi S, Kobayashi M, Maeda T, Matsuoka M, Yamaguch I (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li JM (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Panek HR, Stepp JD, Engle HM, Marks KM, Tan PK, Lemmon SK, Robinson LC (1997) Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J 16: 4194–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JR, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, McKay RM, McKay JP, Graff JM (1999) Casein kinase I transduces Wnt signals. Nature 401: 345–350 [DOI] [PubMed] [Google Scholar]
- Poonyarit M, Mackill DJ, Vergara BS (1989) Genetics of photoperiod sensitivity and critical daylength in rice. Crop Sci 29: 647–652 [Google Scholar]
- Robinson LC, Hubbard EJ, Graves PR, DePaoli-Roach AA, Roach PJ, Kung C, Haas DW, Hagedorn CH, Goebl M, Culbertson MR (1992) Yeast casein kinase I homologues: an essential gene pair. Proc Natl Acad Sci USA 89: 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LC, Menold MM, Garrett S, Culbertson MR (1993) Casein kinase I-like protein kinases encoded by YCK1 and YCK2 are required for yeast morphogenesis. Mol Cell Biol 13: 2870–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, Miyao A, Hirochika H, Kitano H, Ashikari M, Matsuoka M (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ishiyama K, Kobayashi M, Itoh H, Kayano T, Iwahori S, Matsuoka M, Tanaka H (2003) Genetic manipulation of gibberellin metabolism in transgenic rice. Nat Biotechnol 21: 909–913 [DOI] [PubMed] [Google Scholar]
- Sano Y (1992) Genetic comparisons of chromosome 6 between wild and cultivated rice. Jpn J Breed 42: 561–572 [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, Matsuoka M (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Shomura A, Sasaki T, Yano M (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc Natl Acad Sci USA 98: 7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KH (1995) Genetic analysis for heading time in wild rice strains. Jpn J Genet 70: 555–562 [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, Matsuoka M (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Xiang H, Ashikari M, Kitano H, Yamaguchi I, Matsuoka M (2007a) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M (2007b) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58: 183–198 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M (2008) Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamachuchi J (1998) Analysis of embryo-specific a-amylase using isolated mature rice embryos. Breed Sci 48: 365–370 [Google Scholar]
- Yamagata H, Okumoto Y, Tanisaka T (1986) Analysis of Genes Controlling Heading Time in Japanese Rice. Los Baos, Philippines: International Rice Research Institute, pp 351–359 [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Yin C, Gan L, Ng D, Zhou X, Xia K (2007) Decreased panicle-derived indole-3-acetic acid reduces gibberellin A1 level in the uppermost internode, causing panicle enclosure in male sterile rice Zhenshan 97A. J Exp Bot 58: 2441–2449 [DOI] [PubMed] [Google Scholar]
- Yokoo M, Kikuchi F, Nakane A, Fujimaki H (1980) Genetical analysis of heading time by aid of close linkage with blast, Pyricularia oryzae, resistance in rice. Bull Natl Inst Agric Sci Ser D 31: 95–126 [Google Scholar]
- Yokoo M, Okuno K (1993) Genetic analysis of earliness mutations induced in the rice cultivar Norin 8. Jpn J Breed 43: 1–11 [Google Scholar]
- Yu H, Ito T, Zhao Y, Peng J, Kumar P, Meyerowitz EM (2004) Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci USA 101: 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.