Abstract
In mammalian zygotes, the 5-methyl-cytosine (5mC) content of paternal chromosomes is rapidly changed by a yet unknown but presumably active enzymatic mechanism. Here, we describe the developmental dynamics and parental asymmetries of DNA methylation in relation to the presence of DNA strand breaks, DNA repair markers and a precise timing of zygotic DNA replication. The analysis shows that distinct pre-replicative (active) and replicative (active and passive) phases of DNA demethylation can be observed. These phases of DNA demethylation are concomitant with the appearance of DNA strand breaks and DNA repair markers such as γH2A.X and PARP-1, respectively. The same correlations are found in cloned embryos obtained after somatic cell nuclear transfer. Together, the data suggest that (1) DNA-methylation reprogramming is more complex and extended as anticipated earlier and (2) the DNA demethylation, particularly the rapid loss of 5mC in paternal DNA, is likely to be linked to DNA repair mechanisms.
Keywords: γH2A.X, DNA demethylation, DNA repair, epigenetic reprogramming, mouse zygote
Introduction
The mammalian development is characterized by major phases of DNA methylation reprogramming. Particularly striking are two phases of development in which an extensive loss of 5-methyl-cytosines (5mC) is found which at least partially is regarded as independent of DNA replication and hence referred to as ‘active DNA demethylation' (Mayer et al, 2000; Ooi and Bestor, 2008). The first phase is found during early germ cell development and the second in the zygote shortly after fertilization (Reik et al, 2001). The genome-wide DNA demethylation in primordial germ cells affects both parental genomes simultaneously and includes reprogramming of parental imprints, the X chromosome and some repetitive elements (Surani et al, 2008). This initial phase of DNA methylation erasure is followed by extensive de novo methylation during spermio- and oogenesis in which sex-specific imprints and repeat methylation are re-established in a sex-specific manner (Bourc'his and Proudhon, 2008; Sasaki and Matsui, 2008).
A second wave of demethylation after fertilization predominantly affects the paternal chromosomes (Rougier et al, 1998; Mayer et al, 2000; Oswald et al, 2000). Immunofluorescence (IF) analysis using antibodies (Ab's) against 5mC suggests a rapid, replication independent, active demethylation process, which initiates shortly after protamine-histone exchange and is supposed to be completed before replication commences in late G1 (Santos et al, 2002; Santos and Dean, 2004). The precise staging, timing and extent of this zygotic DNA demethylation before and after replication has not been determined by direct methods such as bisulphite sequencing, that is the relative contribution of active and passive mechanisms remains elusive. At the end of the first cell cycle, few regions of the paternal genome such as long terminal repeats of intracisternal A-particle transposons and the differentially methylated regions of paternally imprinted genes remain methylated, whereas others such as long interspersed nuclear element 1 (Line1) and early retrotransposons (ETn) retrotransposable elements are largely demethylated (Lane et al, 2003; Kim et al, 2004). The differential reprogramming of DNA methylation during early embryogenesis might be necessary to allow the activation of certain early developmental genes of the paternal genomes such as the pluripotency factors Oct4 and Nanog (Aoki et al, 1997; Farthing et al, 2008) while retaining epigenetic silencing at active transposable elements and maintaining paternal imprints (Morgan et al, 2005).
The factors and mechanisms responsible for both processes, that is the factors and mechanisms promoting active DNA demethylation and those protecting against DNA demethylation marks, remain largely unknown in mammals. In mouse, all data support the notion that both are provided by the oocyte cytoplasm but that induction of active DNA demethylation of the paternal genome is triggered by specific chromatin modifications and its composition (Lepikhov and Walter, 2004; Nakamura et al, 2007). To date, the experimental evidence for the existence of a bona fide demethylase catalysing the direct removal of methyl groups directly from 5mC within DNA has failed to be reproducible (Ooi and Bestor, 2008). Indirect demethylation mechanisms involving base removal by DNA repair are currently discussed as the most likely strategy for active, replication-independent DNA demethylation processes (Gehring et al, 2009). Repair-coupled DNA demethylation events have been found to occur in other organisms such as plants and zebrafish, strongly suggesting that analogous mechanisms might be conserved in mammals (Gehring et al, 2009).
A very suggestive mechanism for repair-coupled DNA demethylation includes the deamination of 5mC to thymine by cytidine deaminases such as AID or Apobec's followed by base excision repair (BER) of the T:G mismatch performed by TDG like glycosylases. A very recent study by Popp et al (2010) shows the influence of AID knockout on genome-wide methylation levels of mouse primordial germ cells. AID knockdown of interspecies heterokaryons interferes with active DNA demethylation of promoter regions of OCT4 and NANOG (Bhutani et al, 2010). Furthermore, experiments in zebrafish zygotes (Rai et al, 2008) indicate that DNA demethylation can indeed be enhanced by co-transfecting AID or Apobec's together with DNA glycosylases and/or Gadd45. However, although Gadd45 was previously shown to enhance DNA demethylation in cell transfection experiments (Barreto et al, 2007; Schmitz et al, 2009), AID and Gadd45 knockout mice show no effects on DNA methylation levels (Engel et al, 2009) and other results do not support the influence of Gadd45 on DNA demethylation (Jin et al, 2008; Okada et al, 2010).
Also glycosylases responsible for a direct removal of 5mC from the DNA creating abasic sites and/or nicks in the DNA, such as ROS1 and DEMETER in plants (Ponferrada-Marin et al, 2009), are apparently unique to plants and no homologues have been found in animals. Possible mammalian candidates such as the thymine DNA glycosylase TDG and the methyl-binding domain protein 4 (MBD4), a uridine glycosylase, showed only weak catalytic activity on the demethylation of 5mC:G dinucleotides (Hardeland et al, 2003) and oocytes derived from MBD4 knockout mice are still able to demethylate the paternal genome (Santos and Dean, 2004). It seems more likely that indeed active removal of 5mC in mammalian DNA is induced by a secondary modification that induces a yet unknown glycosylase activity. The very recent detection of 5-hydroxy-methyl-cytosine (5hmC) (Kriaucionis and Heintz, 2009; Tahiliani et al, 2009) in mammalian DNA fuels such an idea. This modification could be a direct target for novel 5hmC-specific (direct or deamination induced) glycosylase. A very elegant paper by Liutkeviciute et al (2009) suggests the possibility that DNA methyltransferases themselves could trigger the reverse reaction, that is the direct removal of the hydroxy-methyl group from DNA. However, so far it remains unclear whether this modification has an important function in zygotic demethylation—as 5hmC has only been found in Purkinje brain cells and in mouse ES cells (Kriaucionis and Heintz, 2009; Tahiliani et al, 2009).
In conclusion, most data point towards indirect repair-coupled pathways as possible active DNA demethylation mechanisms. All such mechanisms involve the induction of DNA single-strand breaks (SSBs) as an intermediate step. A well-known marker for DNA breaks is the phosphorylation of Serine139 in the histone H2A.X, termed γH2A.X (Fernandez-Capetillo et al, 2004). γH2A.X was initially described as a histone modification marking DNA double-strand breaks (DSBs) (Rogakou et al, 1999) to recruit DSB repair factors. The phosphorylation of H2A.X is detected by specific Ab's as nuclear localized foci. Such foci appear minutes after exposure of DNA to damaging events like ionizing radiation (Rogakou et al, 1998). At γ irradiation induced DSBs γH2A.X co-localizes with DNA repair and checkpoint proteins such as Rad50, Rad51 and BRCA1 (Paull et al, 2000). Later, it was shown that γH2A.X is not only a sensor for DNA DSBs but also participates in nucleotide excision repair (NER) by marking DNA single-strand gaps (Matsumoto et al, 2007). However, the full spectrum of γH2A.X-associated effects is certainly not completely understood.
In this study, we identify a very dynamic pattern of γH2A.X in the developing mouse zygote and relate this to the presence of strand breaks and possible BER repair processes. A comparison of the developmental dynamics of such repair processes to the dynamics of paternal DNA methylation, monitored by Ab staining and bisulphite sequencing, indeed suggests a link between active DNA demethylation and DNA repair in the zygote.
Results
Developmental timing and dynamics of DNA demethylation in zygotes
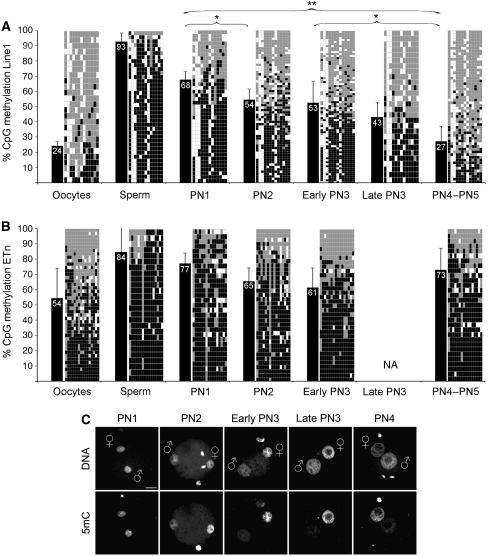
IF analyses using Ab's against 5mC suggested that the major phase of DNA demethylation in the paternal pronucleus is already completed at 7–8 h post-fertilization (hpf) (Figure 1C), that is around the mid-to-late stage of pronuclear development (pronuclear stage 3, PN3) (Santos et al, 2002). Complementary bisulphite analysis was performed on late stage zygotes derived by natural matings around 23 h post-hCG, that is at later stages when DNA replication was most likely completed (Lane et al, 2003; Kim et al, 2004). To precisely correlate the dynamics of DNA demethylation to the pronuclear development, we collected in vitro fertilized zygotes at defined pronuclear stages (PN1–PN5) and subjected their DNA to bisulphite conversion, amplification, cloning and sequencing of two types of retrotransposable elements Line1 (about 2200 elements/genome) and ETn (about 160 elements/genome). For comparison, we also included DNA of sperm and oocytes. The results obtained on sperm, oocyte and late zygotic stages (>PN4) confirm earlier data showing that Line1 methylation is indeed significantly reduced at the end of the first cell cycle (Figure 1A; Lane et al, 2003). A detailed stage-specific analysis revealed remarkable novel aspects of the dynamics of DNA demethylation. The total CpG methylation of Line1 elements at pronuclear stage 1 (PN1) (i.e. shortly after histone-protamine exchange) is on average 68%, corresponding to the mean of methylation levels observed between sperm and oocytes. Hence, at PN1 (approximately up to 4 hpf) no DNA demethylation has occurred. During the next stages (PN2 to early PN3), the DNA methylation decreases by 15% to an average of 53%. A final reduction to a mean of 27% is found when replication is completed at late PN4/PN5 (Figure 1A). In summary, we detect distinct effects of Line1 CpG methylation reduction in pre-replicative (up to PN3) and post-replicative developmental windows (PN4/5) (see below). ETn elements show a similar tendency in the first pre-replicative window, with a decrease from 77% at PN1 to 61% at early PN3. However, in contrast to Line1 elements, the CpG methylation of ETn's is even increasing again to 73% at PN4–PN5 (Figure 1B) in the second post-replicative developmental window almost to the level found at PN1.
Figure 1.
Line1/ETn CpG methylation and loss of 5mC antibody signal during zygotic development. (A) Line1 CpG methylation of gametes and zygotes at distinct pronuclear stages reveals a stepwise demethylation process. Black bars indicate average methylation status. Diagrams specify analysed CpG positions where each row depicts an individual chromosomal Line1 pattern (black=methylated, grey=unmethylated, white=not analysable/mutated). Note the increasing mosaicism of methylated CpG positions over time. Asterisks indicate significant methylation changes between different pronuclear stages. (B) ETn CpG methylation of gametes and zygotes at distinct pronuclear stages. The methylation level also decreases from PN1 up to early PN3 likewise Line1. After completion of S-phase, the average methylation level increases again. (C) Representative IF analysis with a monoclonal Ab against 5mC during pronuclear stages (see also Santos et al, 2002). The major loss of Ab signal in the paternal pronucleus occurs between PN2 and PN3. Scale bar=20 μm.
Timing of DNA replication in the mouse zygote
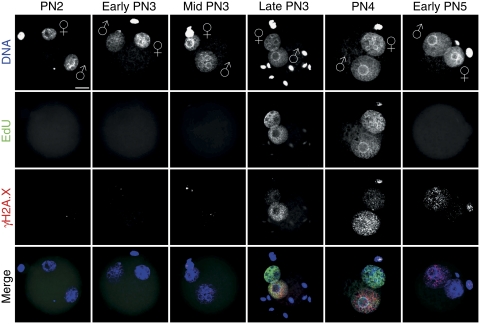
To precisely determine the onset and duration of DNA replication relative to pronuclear maturation and the methylation dynamics, we performed staged pulse labelling experiments using 5-bromo-2-deoxyuridine (BrdU) or 5-ethynyl-2′-deoxyuridine (EdU) and visualized incorporation by indirect IF (BrdU) or click-iT chemistry (EdU). BrdU or EdU incorporation starts around 9 hpf in embryos at late PN3 or early PN4 stages (Figure 2; Supplementary Figure S1) simultaneously in both pronuclei. We never found incorporation of BrdU or EdU earlier than late PN3 stage (n>60, at least three independent in vitro fertilization (IVF) experiments per PN stage). This finding is in contrast to earlier reports that suggested a slightly delayed onset of replication in the maternal pronucleus (Bouniol-Baly et al, 1997; Ferreira and Carmo-Fonseca, 1997; Aoki and Schultz, 1999). The EdU signals remain equally intense in both maternal and paternal pronuclei throughout late PN3 up to early PN5 stages (i.e. about 9–12 hpf) (Figure 2). We conclude that the S-phase in mouse zygotes (1) starts and ends rather synchronously in both paternal and maternal pronuclei and (2) covers only a short period of the first cycle as it lasts just about 4 h.
Figure 2.
Timing of DNA replication in mouse zygotes. Representative images of click-iT-labelled EdU incorporation during pronuclear stages (classified according to Adenot et al, 1997) (n>60, at least three independent IVFs per PN stage). Pulse labelling of IVF zygotes with EdU was performed for 30 min before fixation. EdU incorporation occurs synchronously in male and female pronuclei at late PN3–PN4. The lower two panels show the co-staining with anti-γH2A.X and the merge with EdU signals, which depict the replication-associated γH2A.X foci formation predominantly in paternal pronuclei. Note the few pre-replicative γH2A.X foci in the paternal pronucleus and the stronger staining at early PN5. Scale bar=20 μm.
Dynamics of γH2A.X in the mouse zygote
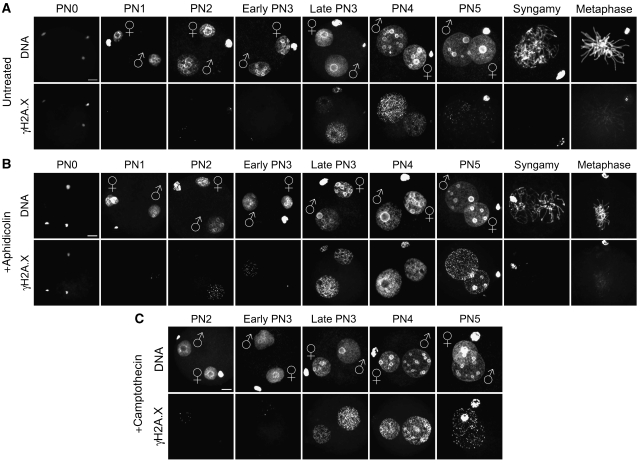
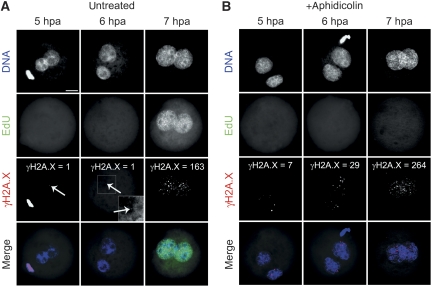
Having precisely determined the stages and extent of DNA demethylation and their relation to replicative phases, we next asked whether DNA repair processes accompany the dynamics of DNA demethylation, particularly in the early pre-replicative events. In a series of experiments, we analysed the presence of DNA strand break marker γH2A.X at various developmental stages of mouse zygotes. IF analysis using specific Ab's directed against γH2A.X, the Serine139 phosphorylated form of H2AX, revealed dynamic focal patterns at different pronuclear stages (Figures 2 and 3A; Supplementary Figure S2). In contrast to the dynamic γH2A.X patterns, Ab's against the histone variant H2A.X itself stained both pronuclei uniformly (Supplementary Figure S3) through all (PN1–PN5) pronuclear stages (see also Ziegler-Birling et al, 2009). Hence, the presence of this histone variant does not change in both female and male pronuclei during zygotic development. Intense γH2A.X staining appears immediately after fertilization at PN0 with both decondensing parental genomes showing an equally strong reactivity with the γH2A.X antibody. Around mid-PN1 (at about 4 hpf), the IF signal then disappears entirely from both pronuclei. At PN2 and early PN3 (5–7 hpf), few γH2A.X foci can be counted in paternal pronuclei only (Figure 4). At 8–9 hpf, that is around late PN3 to early PN4, the γH2A.X foci massively accumulate in early replicating nuclei. γH2A.X foci now also become strongly visible in the maternal pronucleus. However, the paternal pronucleus always contains more γH2A.X foci as compared with the maternal (note that both pronuclei show an equal distribution and intense immunostaining against H2A.X (Supplementary Figure S3). In further development, that is from mid-PN4 to early PN5, the intensity of γH2A.X signal declines. Again the paternal pronucleus seems to retain more γH2A.X foci (see Figure 3A). γH2A.X foci disappear from both pronuclei at late PN5, with no apparent foci at syngamy and a few signals on metaphase plate chromosomes. The parental identity of the pronuclei was evident by morphology but was also verified by H3K9me2 staining, which is exclusively found in maternal pronuclei (see Lepikhov and Walter, 2004; Supplementary Figure S4). To examine the possible influence of IVF on timing and developmental dynamics of γH2A.X foci, we compared the γH2A.X dynamics of naturally mated and IVF-derived zygotes at respective pronuclear stages. At all stages, we did not detect any differences between both groups (data not shown). We would like to note that a recent study also shows an asymmetric appearance of γH2A.X foci in mouse zygotes at PN2 (corresponding to late PN3 in our classification) and PN4 (Ziegler-Birling et al, 2009).
Figure 3.
Dynamics of γH2A.X signal during zygotic development. Representative images of indirect immunostainings using antibodies against γH2A.X in IVF zygotes at distinct pronuclear stages (PN0–PN5, syngamy and metaphase). (A) γH2A.X dynamics in untreated controls. (B) IVF zygotes incubated with aphidicolin for 2 h before fixation. (C) IVF zygotes incubated with camptothecin for 2 h prior the fixation. Scale bar=20 μm. A colour version of this figure is available at The EMBO Journal Online (Supplementary Figure S2).
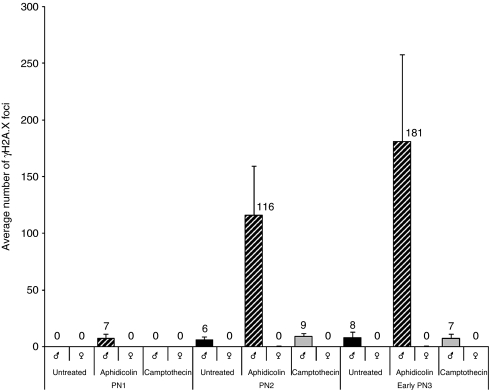
Figure 4.
Number of γH2A.X foci in paternal and maternal pronuclei during G1-phase. The graph shows the average number of γH2A.X foci (n>20 zygotes/analysis) in both parental pronuclei in untreated, aphidicolin-treated and camptothecin-treated IVF zygotes. In aphidicolin-treated zygotes at G1-phase, a significant increase of γH2A.X foci can be detected only in paternal pronuclei.
In summary, we find a very dynamic pattern of γH2A.X appearance during zygotic pronuclear development. Particularly, the occurrence of γH2A.X marked strand breaks at pre-replicative stages in the paternal pronucleus is remarkable with respect to the timing of DNA demethylation.
Inhibition of DNA polymerases enhances pre-replicative and replicative γH2A.X foci
The dynamic pattern of γH2A.X before and during DNA replication tempted us to examine the effect DNA polymerase inhibition at various pronuclear stages. We used aphidicolin (Aph), a nucleotide analogue competing with dCTP incorporation, which is known to inhibit all replicative and most repair-associated DNA polymerases (Berger et al, 1979; Krokan et al, 1981). IVF-derived mouse zygotes were exposed to Aph in the medium for 2 hours before fixation at defined pronuclear stages (Figure 3B). Strikingly, Aph strongly enhances the number, appearance and intensity of γH2A.X foci in paternal pronuclei at PN2 and early PN3 stage (Figure 4), that is before replication. γH2A.X foci reach their maximum density at late PN3 and at PN4 (Figure 3B). At early stages, counting of γH2A.X foci reveals an about 20-fold higher number in the paternal pronuclei compared with maternal and such asymmetry remains pronounced throughout S-phase. Incubation with Aph at late replicative/post-replicative syngamy to metaphase stages (15–18 hpf) has no effect on γH2A.X signals. Aph treatment during the phase of active DNA demethylation from late PN1 to early PN3 does not influence 5mC antibody signal (Supplementary Figure S5). In summary, the blocking of DNA polymerases by Aph results in distinct pre-replicative (G1-phase) (paternal only) and replication (S-phase, both pronuclei)-associated enhancement of γH2A.X foci. The strong pre-replicative effect in G1-phase seems to be unique for zygotes, as experiments in two-cell embryos do not show Aph-enhanced γH2A.X foci outside of S-phase (Supplementary Figure S6).
The origins of pre-replicative and replicative γH2A.X foci can be separated
The strong enhancement of γH2A.X in G1- and S-phase suggests distinct causes for the accumulating strand breaks in Aph-treated zygotes. To further dissect the connection between γH2A.X foci and DNA replication, we inhibited topoisomerases by camptothecin (Cpt) in defined time windows during pronuclear maturation. Cpt inhibits topoisomerase I resulting in stalled replication forks and the accumulation of DSBs (Furuta et al, 2003). IVF-derived mouse zygotes were exposed to Cpt for 2 hours in successive time windows before fixation at defined pronuclear stages (Figure 3C). When comparing the effects of Cpt to Aph-treated zygotes and controls, we observed some enhancement of replicative γH2A.X foci but no change in the developmental dynamics and patterning of γH2A.X foci at early pronuclear stages. Hence, the pre-replicative enhancement of γH2A.X foci by Aph (through polymerase blocking) is distinct from a replication fork-associated effect induced by Cpt (Figure 4).
In summary, we observe an intriguing overlap of γH2A.X foci with phases of DNA demethylation in early-to-mid zygotic stages, suggesting that these focal DNA strands breaks are marking repair particularly in paternal pronuclei at time points when DNA methylation is lost.
Dynamics of DNA demethylation and γH2A.X are comparable between IVF and cloned embryos
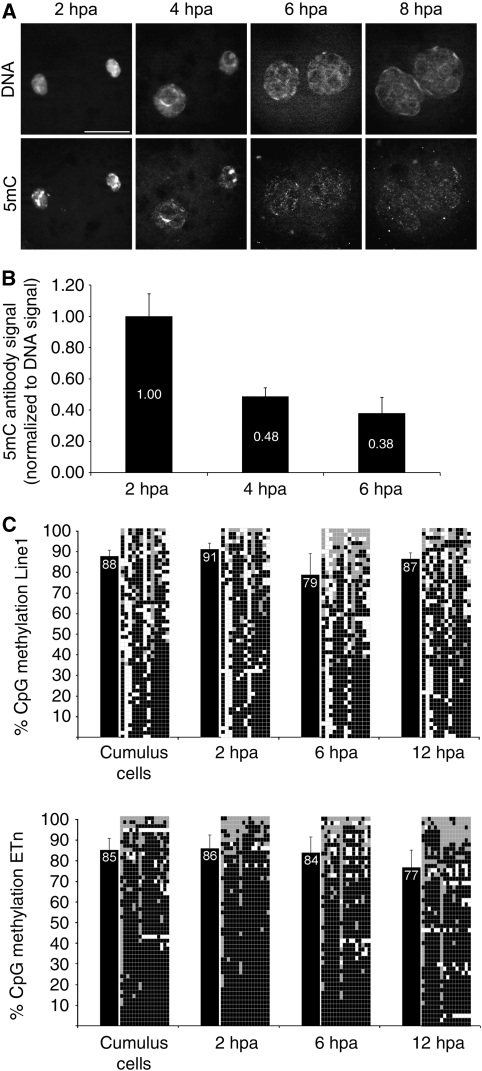
To investigate the correlation between γH2A.X strand breaks and DNA demethylation further, we analysed the dynamics of both at the relevant developmental stages in cloned embryos. Cloned one-cell stage embryos were derived after somatic cell nuclear transfer (SCNT) of cumulus cells into enucleated oocytes, followed by zygotic activation and in vitro culturing. First, we had to determine the extent and dynamics of DNA demethylation in cloned embryos at 2–8 h post-activation (hpa). Ab's against 5mC show a strong IF signal in activated SCNT embryos fixed at 2 hpa (Figure 5A). Note that the somatic chromosomes form new pseudo-pronuclei a process induced by the spindles left in the enucleated eggs (Sun and Schatten, 2007). The signal intensities in the pronuclei decreases at 4 hpa remaining detectable at pericentric heterochromatin. At 6 hpa, the IF signal is most strongly reduced and remains so up to 8 hpa. The overall intensity of the IF signal indicates a 50% decrease of 5mC signal intensity between 2 and 4 hpa and an additional minor decrease at 6 hpa (Figure 5B).
Figure 5.
Loss of 5mC-antibody signal and Line1/ETn methylation in cloned embryos. (A) Indirect immunostainings with antibodies against 5mC in SCNT embryos at 2, 4, 6 and 8 hpa. Note the dynamic loss of antibody signal in both pronuclei at 6 hpa. (B) Quantification of 5mC antibody signal normalized to DNA signal (n=4 per stage). At 4 hpa, the 5mC antibody signal decreases to 48% compared with 2 hpa. (C) Line1 bisulphite sequencing of cloned one-cell embryos at distinct pronuclear stages reveals only a minor drop of DNA methylation levels at 6 hpa, which then slightly increases at 12 hpa. ETn methylation levels remain rather constant with the tendency to decrease after replication. Black bars indicate average methylation status. Diagrams specify analysed CpG positions where each row depicts an individual chromosomal Line1 pattern (black=methylated, grey=unmethylated, white=not analysable/mutated).
Pulse-labelled EdU incorporation shows that replication in cloned embryos starts around 7 hpa (i.e. about 2 hours earlier compared with IVF zygotes) (Figure 6A). Hence, a considerable amount of DNA demethylation apparently occurs before the onset of DNA replication.
Figure 6.
Dynamics of γH2A.X signal in cloned embryos. (A) SCNT embryos incubated for 1 hour with EdU before fixation at 5, 6 and 7 hpa. S-phase starts at 7 hpa coinciding with the appearance of increased γH2A.X signals. (B) Aphidicolin blocks EdU incorporation and enhances the γH2A.X signal in both G1- (up to 6 hpa) and S-phase (7 hpa). Scale bar=20 μm; arrows indicate single γH2A.X foci.
Bisulphite sequencing of Line1 and ETn elements of cloned one-cell embryos reveals only minor effects of CpG DNA demethylation compared with zygotes. (Figure 5C). Whereas the overall methylation of Line1 elements seems to decrease by 12% at 6 hpa, the ETn methylation level does not show such pre-replicative reduction. Moreover, after completion of S-phase, we observe an increased CpG methylation for Line1 but not for ETn.
In SCNT embryos, we also find very few distinct γH2A.X foci in both ‘pronuclei' at pre-replicative stages (i.e. 5/6 hpa) but to a 5–10-fold lesser extent compared with corresponding early PN3 stages of paternal pronuclei in zygotes (Figure 6A). As in IVF zygotes, these foci are amplified when treating cloned embryos with Aph (Figure 6B), and the γH2A.X signal is highest during early replicative phases. In summary, DNA demethylation (5mC signal decrease), replication timing and γH2A.X dynamics follow similar patterns in cloned one-cell embryos and IVF zygotes. The bisulphite sequencing shows that in cloned embryos, demethylation of these elements is less extensive (Line1) or nearly not existing (ETn).
PARP-1 co-localizes with γH2A.X in G1 paternal pronuclei
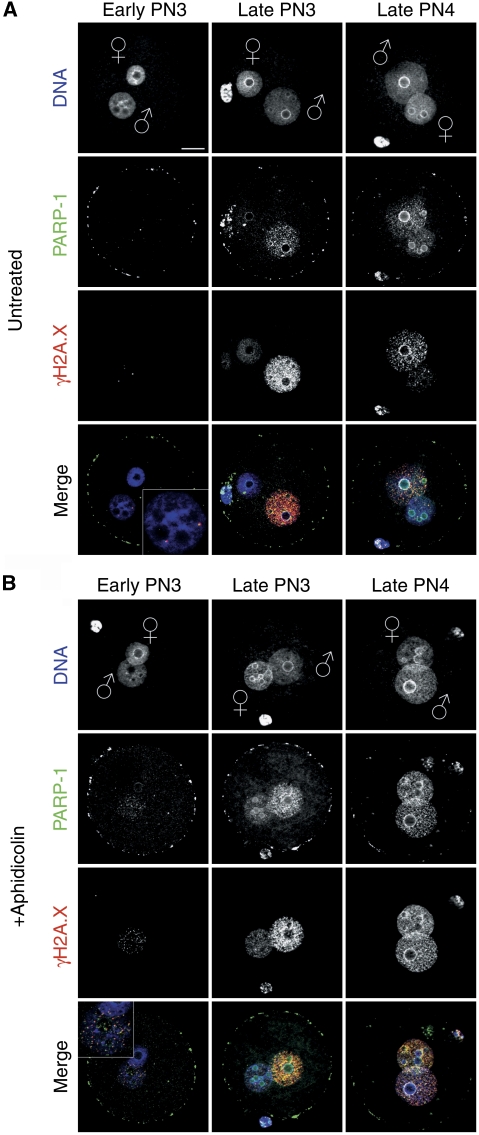
To understand the link of DNA strand breaks at pre- and replicative pronulear stages linked to specific repair pathways, we analysed several putative BER and NER indicators by IF. In IVF zygotes, we found a particular pattern for PARP-1, a sensor of SSBs in DNA and a component of the BER signalling cascade (Durkacz et al, 1980; Malanga and Althaus, 2005; Godon et al, 2008). Using Ab's against PARP-1, we find the protein in pre-replicative early PN3 paternal pronuclei co-localizing with γH2A.X in a focal pattern (Figure 7A). Treatment with Aph simultaneously enhances both the PARP-1 and γH2A.X signals (Figure 7B). Moreover, γH2A.X and PARP-1 signals co-localize and are most strongly enhanced in pre-replicative stages when early zygotes are treated with methylmethanesulfonate (MMS) (Supplementary Figure S7), an alkylating agent inducing BER (Lundin et al, 2005), which generates SSBs as intermediate step.
Figure 7.
PARP-1 co-localization with γH2A.X. Representative indirect immunostainings using antibodies against PARP-1 and γH2A.X in pre-replicative and replicative stages. The picture shows computed z-stacks to reduce unspecific background staining of PARP-1 immunostainings on the surface of the zygotes, which is still visible as ‘ring' on the oolemma. (A) PARP-1 co-localization with γH2A.X at pre-replicative and replicative pronuclear stages. (B) Aphidicolin treatment enhances the PARP-1 signal co-localized with also enhanced γH2A.X foci. Scale bar=20 μm.
DNA SSBs in G1-phase zygotes
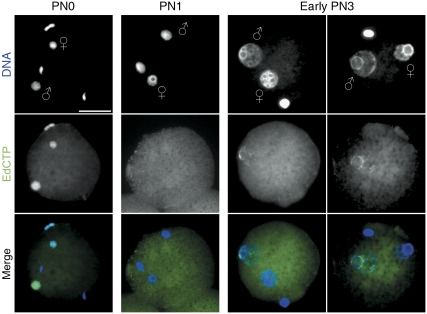
The co-localization of PARP-1 and γH2A.X foci in pre-replicative phases suggests the existence of transient SSBs in paternal DNA before replication. To verify the presence of such SSBs in early G1 mouse zygotes, we performed a modified nick translation assay. In brief, we introduced a mild chromatin decondensation step (see Materials and methods) prior the fixation of early zygotes to gain access to the nicked DNA in earlier PN stages (Note: we do not observe DNA polymerase I-mediated nucleotide incorporation on undecondensed fixed chromatin, data not shown).
At PN0, that is shortly after the fertilization (1–2 h), we find a strong incorporation of nucleotides by PolI in both parental genomes (Figure 8). This correlates with the increased γH2A.X signals (Figure 3A) at the phase of chromatin decondensation and protamine-histone exchange (note that this does not coincide with DNA demethylation). At PN1, this nucleotide incorporation completely disappears in both pronuclei and reappears at early PN3 again only in paternal pronuclei. Again the induction of DNA damage by MMS causes a massive enhancement of such labelling in both pronuclei (Supplementary Figure S8A and B).
Figure 8.
DNA single-strand breaks in pre-replicative zygotes. Modified nick translation assay using the 5-ethynyl-2′-deoxycytidine-triphosphate (EdCTP) for click-iT fluorescence labelling. PolI-mediated incorporation of EdCTP in mildly decondensed G1-phase zygotes. At PN0, both parental genomes show the presence of SSBs, whereas at PN1 stage, no nicks can be detected. In early PN3 zygotes, the signal exclusively appears in paternal pronuclei (two examples). Scale bar=20 μm.
We conclude that two distinct phases of single-stranded DNA break appearance are found in the early mouse G1 zygotes. During the first phase, nicks are introduced in the course of chromatin decondensation in both pronuclei. The accompanying repair is apparently not affecting the DNA methylation status in both pronuclei. In the second phase, nicks are only found in the paternal pronucleus, that is a phase and location where extensive DNA methylation changes are observed.
Discussion
Our study is the first to comprehensively describe and correlate the dynamics of two striking molecular processes in the DNA of mouse zygote: a stepwise DNA demethylation and a concomitant accumulation of DNA strand breaks/DNA repair events at corresponding developmental stages. This developmental correlation is not only attributed to fertilized zygotes but is also found in cloned embryos. Together, our data strongly suggest that DNA demethylation may be mediated to a large extent by DNA repair-induced mechanisms. The co-localization of PARP-1 with the γH2A.X marked strand breaks indicates that BER may have a function in this process.
One key observation of our staged bisulphite analysis is that the DNA demethylation kinetics in the zygote is more complex than anticipated earlier (Reik et al, 2001; Morgan et al, 2005). Whereas the loss of Ab staining against 5mC in the paternal pronucleus suggested a rapid and almost complete loss of the DNA methylation epitope at PN3 (Figure 1C; Santos et al, 2002), the staged bisulphite sequencing of Line1 and ETn elements reveals that DNA demethylation in the pre-replicative phase is clearly detectable but not very pronounced. For LINE elements, the pre-replicative phase of apparently active demethylation is followed by a second wave of demethylation during S-phase. This demethylation could be explained by simple absence of maintenance methylation. However, the methylation patterns of Line1 elements at late PN4/early PN5 (i.e. in early post-replicative phases) show that the overall reduction does not coincide with an increase in the number of completely demethylated clones (Figure 1A). Instead, we observe an increase in the number of mosaic patterns. Finally, we even detect a mild increase in the overall methylation of ETn elements, suggesting that dependent on the sequence context, the DNA can even be de novo methylated during S-phase. In summary, the bisulphite sequencing of repetitive elements in zygotes shows a complex scenario of dynamic DNA methylation changes for the two types of repetitive elements. Moreover, in cloned embryos, the DNA demethylation is much weaker (at Line1) or even barely detectable (at ETns). Although there is clear evidence for an active phase of demethylation at PN2/3 stages as shown by 5mC immunostaining, the bisulphite data do not allow to draw direct conclusions on the genome-wide scale of demethylation, as the observed effects at repetitive elements may be rather specific.
In line with a number of earlier observations by other groups (Reik, 2007; Popp et al, 2010), we favour the idea that the observed active DNA demethylation is linked to an indirect repair-coupled process. The signal for such repair could indeed be either a deamination-induced base change or a yet unidentified secondary modification of the methylated cytosine such as hydroxylation. Such modifications could trigger BER or NER repair-coupled demethylation caused by a DNA glycosylase recognizing the specific base changes. A modification that does not alter the base pairing could even induce such processes throughout pre- and post-replicative phases of the cell cycle. It is very suggestive to assume that such a modification could be the recently described hydroxylation of 5mC (Kriaucionis and Heintz, 2009; Tahiliani et al, 2009). The proposed secondary modification of 5mC would also explain the apparent discrepancy between the rapid loss of 5mC staining at PN2 and PN3, which contrasts the moderate reduction seen at Line1 elements in the respective phases (note that both 5hmC and 5mC cannot be distinguished in bisulphite sequencing (Hayatsu and Shiragami, 1979; Huang et al, 2010)). Still while the discussed scenarios remain rather hypothetical they would all imply the involvement of DNA repair events.
The second major finding of our study shows that the dynamic appearance of γH2A.X foci coincides with phases and locations (paternal pronucleus) of DNA demethylation in both zygotes and cloned embryos. The dynamic changes of γH2A.X foci at early, middle and late zygotic stages reveal that the zygotic development is marked by several waves of appearing and disappearing DNA strand breaks. Our comprehensive analysis includes all stages and extents earlier reports, which discuss the presence of γH2A.X foci at only few pronuclear stages (Derijck et al, 2006; Ziegler-Birling et al, 2009). At earliest pronuclear stage PN0, we observe a strong γH2A.X marking of both parental chromosomes. The strand breaks at this stage (also shown by nick translation assay; Figure 8) are linked to chromatin decondensation (PN0). DNA methylation remains unaffected at this stage (Figure 1; see also Santos et al, 2002). Both nicks and the γH2A.X foci completely disappear at PN1 when the decondensation and protamine-histone exchange is finished. Few but reproducible and distinct γH2A.X foci are again detected at the pre-replicative PN2 and early PN3 stages exclusively in the paternal pronuclei. At the beginning of the replication (late PN3), the signal intensity strongly increases and remains predominant in the paternal pronucleus, reaching its maximum at PN4. It is conceivable that the γH2A.X foci in IVF zygotes and at 7–8 hpa in cloned embryos mark replication-dependent strand breaks in the short S-phase (late PN3 to late PN4). However, the increased paternal γH2A.X signal throughout DNA replication (late PN3 to early PN5) points towards more extensive DNA repair events in the paternal genome. This may either be due to a higher number of replication forks (and errors) or continued replication-independent repair in the paternal pronuclei. It is remarkable in this context that the paternal pronuclei show stronger γH2A.X signals before and after completion of the replicative phase (early PN5) (Figures 2 and 3A).
In conclusion, the dynamics of γH2A.X foci suggest the existence of distinct phases and types of accumulated strand breaks: those marking decondensing chromatin (PN0), stalled replication forks (PN4) and others that are associated with non-replicative SSB-induced repair (PN2–4). The possible distinction of replication-independent and replication-dependent γH2A.X foci is supported by effects induced by Aph and Cpt, respectively. Although Aph treatment results in enhanced accumulation of unrepaired DNA strand breaks at pre-replicative and replicative phases, Cpt exerts an enhancing effect only during DNA replication. Aph inhibits replication and repair-associated polymerases causing the accumulation of temporarily unrepaired DNA breaks. It remains to be clarified which of the repair-associated DNA polymerases are indeed present in the zygote and hence blocked by Aph. Aph is known to have no inhibitory effect on DNA polymerase β, the major BER DNA polymerase at least in somatic cells. IF with Ab's against Pol β suggests that the enzyme is present in the zygote but surprisingly excluded from both pronuclei during early zygotic development (Supplementary Figure S9).
A major question of our analysis concerns the nature of the strand breaks we observe. γH2A.X is known as a marker for DSBs. Several lines of evidence indicate that the γH2A.X marked strand breaks are also SSBs and not only DSBs (see also Matsumoto et al, 2007). First, our modified nick translation assay detects SSBs at early PN3 (Figure 8) only in the paternal pronucleus, where we also find γH2A.X foci. Second, in the zygote, we can artificially induce SSBs by MMS and this induction strongly enhances the γH2A.X and PARP-1 signal (Supplementary Figure S7). The induction of SSBs but not DSBs by MMS is shown by positive nick translation signal (Supplementary Figure S8A) and by the absence of positive terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL assay) (data not shown).
In conclusion, the data support the notion that phosphorylation of H2A.X at PN2 and early PN3 stages marks SSB DNA lesions and this may help to recruit DNA repair enzymes such as PARP-1, an important factor in the BER pathway (Malanga and Althaus, 2005; Godon et al, 2008). As PARP-1/γH2A.X co-localization continues until the end of S-phase of late PN3 up to PN4/early PN5 stages in both pronuclei (Figure 7) such repair processes may continue throughout pre- and post-replicative stages. Indeed, PARP-1 was shown to serve as a general DNA damage survey factor (nick sensor) in the context of replication fork progression (Dantzer et al, 1998).
The overall spatiotemporal correlation of DNA demethylation and DNA repair processes in fertilized and cloned embryos suggest a very fast and efficient process of DNA demethylation accompanied by waves of DNA repair. Our staged bisulphite analysis defines a particular time window (PN2 to mid-PN3) in which active demethylation clearly occurs replication independently and is linked to the presence of DNA nicks, γH2A.X and PARP-1 in the paternal pronucleus. Particularly, this more than suggestive developmental overlap of both events makes it highly likely that active DNA demethylation processes are indeed directly linked to repair. Although the inducing mechanisms and enzymes for DNA demethylation are still to be identified, our findings already point to a strong involvement of repair (BER) processes in zygotic DNA demethylation in mammals.
Materials and methods
All animal experiments were carried out according to the German Animal Welfare law in agreement with the authorizing committee.
IVF of mouse oocytes and manipulation of zygotic development
Spermatozoa collection and IVF procedures were carried out as described in Nagy (2003). Sperm was isolated from the cauda epididymis of adult (C57BL/6 X CBA)F1 male mice and capacitated by pre-incubation for 1.5 h in pre-gassed modified KSOM medium supplemented with 30 mg BSA/ml. Mature oocytes were collected 14 h post-hCG injection of adult (C57BL/6 X CBA)F1 female mice according to the standard procedures (Nagy, 2003). Cumulus–oocyte complexes were placed into a 400 μl drop of KSOM medium with capacitated sperm and incubated at 37°C in a humidified atmosphere of 5% CO2 and 95% air. For the inhibition of DNA polymerases or topoisomerase I, the embryos were treated 2 h prior the fixation with 3 μg/ml Aph or 10 μM Cpt. For the induction of DNA damage with MMS, the zygotes were treated with 5 mM MMS for 1 h before fixation.
Bisulphite sequencing of mouse zygotes and SCNT-derived one-cell embryos
To isolate sperm DNA, somatic contamination of the sperm sample was first diminished by a soft Proteinase K digestion (200 μl 17.5 mM EDTA, 37.5 mM NaCl, 5 mM Tris–HCl pH 8.0, 0.5% SDS, 10 μl Proteinase K (20 mg/ml) for 3 h at 55°C). The undigested part was treated with 0.01 M Tris–HCl pH 8.0, 0.01 M EDTA pH 8.0, 0.1 M NaCl, 3% SDS, 44 mM DTT and 0.4 mg/ml Proteinase K (over night at 55°C), followed by standard ethanol precipitation. IVF-derived zygotes and SCNT-derived one-cell embryos were collected at different pronuclear stages. The correct PN staging and contamination with sperm or cumulus cells were controlled by Hoechst staining and embryo-by-embryo selection under a fluorescent microscope. For bisulphite analysis, we used a procedure described by Tierling et al with some modifications. In brief, 4–40 PN staged zona-free zygotes and cloned one-cell embryos were boiled for 30 s with 7 μl of molten low melting point agarose (2% in TE). The followed steps were performed according to the protocol (Tierling et al). PCR amplification of Line1 5′-UTR region, described in Lane et al (2003), was performed using the following conditions: primers used were 5′ TAGGAAATTAGTTTGAATAGGTGAGAGGT 3′ and 5′ TCAAACACTATATTACTTTAACAATTCCCA 3′. PCR conditions were: 97°C 15 min, 45 cycles [95°C 1 min, 56°C 1 min 72°C 1:30 min], 72°C 10 min. PCR amplification of ETn elements was done according to Kim et al (2004), with slightly modified reverse primer: 5′ GTTAGYGTTAGTATGTGTATTTGTT 3′. The PCR products were cloned into pGEM-T vector (Promega) and sequenced. On average, 13 single sequences per bisulphite treatment were obtained. The results shown are combinations of 2–6 individual bisulphite treatments for each stage.
IF staining
Zygotes were harvested at several time points corresponding to pronuclear stages from PN0 up to metaphase stage. The classification of PN stages was done according to Adenot et al (1997) and [50]Santos et al (2002), where the pronuclear morphology and hours post-fertilization are taken into consideration. After brief washing in M2 medium, zona pellucida was removed by treatment with Acidic Tyrodes solution (Nagy, 2003). Subsequently, the zygotes were fixed for 20 min in 3.7% paraformaldehyde in PBS at room temperature (RT) and permeabilized with 0.2% Triton X-100 in PBS for 10 min at RT. The fixed zygotes were blocked overnight at 4°C in 1% BSA, 0.1% Triton X-100 in PBS, and the embryos were incubated in the same blocking solution for 4 h at RT with either anti-γH2AX (phosphorylated Ser 139) (rabbit polyclonal, US Biological; mouse monoclonal, Abcam), anti-H3K9me2 (rabbit polyclonal, gift from Thomas Jenuwein), anti-BrdU (fluorescein conjugated, rat monoclonal, Abcam), anti-PARP-1 (mouse monoclonal, Biolegend) or anti-5mC (mouse monoclonal, Eurogentec). For 5mC immunostaining, fixed zygotes were incubated in 4 M HCl solution at RT for 10 min. Following neutralization in Tris–Cl pH 8.0 for 10 min and second fixation with 3.7% paraformaldehyde, the embryos were blocked overnight at 4°C. After several washes, embryos were incubated at RT for 2 h with mouse secondary Ab's coupled with Alexa Fluor 488 (Molecular Probes) and anti-rabbit secondary Ab's coupled with Rhodamine Red-X (Jackson ImmunoResearch Laboratories Inc.). Zygotes were then washed and mounted on slides with a small drop of Vectashield (VectorLab) mounting medium containing 0.5 μg/ml 4,6-diamino-2-phenylindole or 2 μg/ml propidium iodide. For each experimental group, we analysed at least three IVFs containing at least 20 zygotes.
EdU and BrdU incorporation experiments
For the incorporation of EdU or BrdU during S-phase of zygotic development, 50 μM EdU or 25 μg/ml BrdU were added to the culture medium 30 min prior the fixation. The detection of incorporated EdU was done using click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen) according to the manufacturer's protocol. For BrdU detection, fixed zygotes were incubated in 4 M HCl solution at RT for 10 min. Following neutralization for 10 min in Tris–Cl, pH 8.0 and a second fixation with 3.7% paraformaldehyde embryos were blocked overnight at 4°C. Adjacent IF stainings were performed as described.
Modified nick translation assay
IVF-derived embryos were washed in M2 medium and were permeabilized with 0.2% Triton X-100 in PBS for 1 min at RT and then incubated in M2 containing 0.3 mM spermine, 2.5 mM spermidine and 1 mg/ml heparine for 20 min at 37°C and 5% CO2. Subsequent fixation was performed as described. Next, the fixed zygotes were washed twice in NEB2 buffer (New England Biolabs), with BSA for 30 min at RT. For positive control, some of the zygotes were treated with DNAseI (Promega, 0.5 μ/100 μl) for 10 min at RT. Afterwards, the zygotes were incubated in nick translation mix containing 5-ethynyl-2′-deoxycytidine-triphosphate (EdCTP), TTP, GTP, ATP, NEB2 buffer, BSA and DNA polymerase I (New England Biolabs) for 2 h at RT. Adjacent click-iT labelling and IF was done as described. EdCTP was synthesized in the group of Professor Michal Hocek, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Czech Republic.
Somatic cell nuclear transfer
MII oocytes were collected from superovulated B6C3F1 mice, 14 h post-hCG administration. Oviducts were dissected in Hepes-CZB (H-CZB) containing 50 I.U./ml hyaluronidase for 20′; denuded MII oocytes were washed in α-MEM, BSA 0.2% and cultured for 30′ in α-MEM at 37°C in 5% CO2. Enucleation was performed in H-CZB containing 5 μg/ml cytochalasin B and PVP 0.1%. Groups of 20 oocytes were enucleated in a time window of 10′ and then extensively washed and returned to the incubator in a drop of α-MEM, BSA 0.2%. Nuclear transfer was done in 90% H-CZB, PVP 1%. Reconstructed embryos were left to recover for at least 1 h in α-MEM in the incubator before being chemically activated in Ca2+free α-MEM, SrCl2 10 mM in the presence of 5 μg/ml cytochalasin B and DMSO 0.05% (Boiani et al, 2002). At different time points, embryos were collected and transferred into either activation media or α-MEM containing EdU in the presence or absence of Aph (3 μg/ml).
IF microscopy
The mounted embryos were analysed on Zeiss Axiovert 200 M inverted microscope equipped with the fluorescence module, ApoTome and B/W digital camera for imaging. The images were captured, pseudocoloured and merged using AxioVision software (Zeiss).
Supplementary Material
Acknowledgments
We thank Professor Michal Hocek for synthesizing 5-ethynyl-2′-deoxycytidine-triphosphate. Furthermore, we thank Professor Thomas Jenuwein for H3K9me2 antibodies. This work was supported by the grant from Deutsche Forschungsgemeinschaft (DFG) WA 1029/3-2 and /4-1 and by the EPIGENOME Network of Excellence LSHG-CT-2004-503433.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adenot PG, Mercier Y, Renard JP, Thompson EM (1997) Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 124: 4615–4625 [DOI] [PubMed] [Google Scholar]
- Aoki E, Schultz RM (1999) DNA replication in the 1-cell mouse embryo: stimulatory effect of histone acetylation. Zygote 7: 165–172 [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM (1997) Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 181: 296–307 [DOI] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C (2007) Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445: 671–675 [DOI] [PubMed] [Google Scholar]
- Berger NA, Kurohara KK, Petzold SJ, Sikorski GW (1979) Aphidicolin inhibits eukaryotic DNA replication and repair—implications for involvement of DNA polymerase alpha in both processes. Biochem Biophys Res Commun 89: 218–225 [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM (2010) Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M, Eckardt S, Scholer HR, McLaughlin KJ (2002) Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev 16: 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouniol-Baly C, Nguyen E, Besombes D, Debey P (1997) Dynamic organization of DNA replication in one-cell mouse embryos: relationship to transcriptional activation. Exp Cell Res 236: 201–211 [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Proudhon C (2008) Sexual dimorphism in parental imprint ontogeny and contribution to embryonic development. Mol Cell Endocrinol 282: 87–94 [DOI] [PubMed] [Google Scholar]
- Dantzer F, Nasheuer HP, Vonesch JL, de Murcia G, Menissier-de Murcia J (1998) Functional association of poly(ADP-ribose) polymerase with DNA polymerase alpha-primase complex: a link between DNA strand break detection and DNA replication. Nucleic Acids Res 26: 1891–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijck AA, van der Heijden GW, Giele M, Philippens ME, van Bavel CC, de Boer P (2006) gammaH2AX signalling during sperm chromatin remodelling in the mouse zygote. DNA Repair (Amst) 5: 959–971 [DOI] [PubMed] [Google Scholar]
- Durkacz BW, Omidiji O, Gray DA, Shall S (1980) (ADP-ribose)n participates in DNA excision repair. Nature 283: 593–596 [DOI] [PubMed] [Google Scholar]
- Engel N, Tront JS, Erinle T, Nguyen N, Latham KE, Sapienza C, Hoffman B, Liebermann DA (2009) Conserved DNA methylation in Gadd45a(−/−) mice. Epigenetics 4: 98–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W (2008) Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet 4: e1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A (2004) H2AX: the histone guardian of the genome. DNA Repair (Amst) 3: 959–967 [DOI] [PubMed] [Google Scholar]
- Ferreira J, Carmo-Fonseca M (1997) Genome replication in early mouse embryos follows a defined temporal and spatial order. J Cell Sci 110 (Pt 7): 889–897 [DOI] [PubMed] [Google Scholar]
- Furuta T, Takemura H, Liao ZY, Aune GJ, Redon C, Sedelnikova OA, Pilch DR, Rogakou EP, Celeste A, Chen HT, Nussenzweig A, Aladjem MI, Bonner WM, Pommier Y (2003) Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J Biol Chem 278: 20303–20312 [DOI] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S (2009) DNA demethylation by DNA repair. Trends Genet 25: 82–90 [DOI] [PubMed] [Google Scholar]
- Godon C, Cordelieres FP, Biard D, Giocanti N, Megnin-Chanet F, Hall J, Favaudon V (2008) PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res 36: 4454–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland U, Bentele M, Jiricny J, Schar P (2003) The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res 31: 2261–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu H, Shiragami M (1979) Reaction of bisulfite with the 5-hydroxymethyl group in pyrimidines and in phage DNAs. Biochemistry 18: 632–637 [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A (2010) The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5: e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Guo C, Pfeifer GP (2008) GADD45A does not promote DNA demethylation. PLoS Genet 4: e1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kang YK, Koo DB, Kang MJ, Moon SJ, Lee KK, Han YM (2004) Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem Biophys Res Commun 324: 58–63 [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan H, Wist E, Krokan RH (1981) Aphidicolin inhibits DNA synthesis by DNA polymerase alpha and isolated nuclei by a similar mechanism. Nucleic Acids Res 9: 4709–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W (2003) Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88–93 [DOI] [PubMed] [Google Scholar]
- Lepikhov K, Walter J (2004) Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S (2009) Cytosine-5-methyltransferases add aldehydes to DNA. Nat Chem Biol 5: 400–402 [DOI] [PubMed] [Google Scholar]
- Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T (2005) Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res 33: 3799–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanga M, Althaus FR (2005) The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol 83: 354–364 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, Inobe M, Matsunaga T (2007) Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J Cell Sci 120: 1104–1112 [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T (2000) Demethylation of the zygotic paternal genome. Nature 403: 501–502 [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W (2005) Epigenetic reprogramming in mammals. Hum Mol Genet 14 Spec No 1: R47–R58 [DOI] [PubMed] [Google Scholar]
- Nagy A (2003) Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T (2007) PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64–71 [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y (2010) A role for the elongator complex in zygotic paternal genome demethylation. Nature 463: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH (2008) The colorful history of active DNA demethylation. Cell 133: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J (2000) Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478 [DOI] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10: 886–895 [DOI] [PubMed] [Google Scholar]
- Ponferrada-Marin MI, Roldan-Arjona T, Ariza RR (2009) ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res 37: 4264–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR (2008) DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135: 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J (2001) Epigenetic reprogramming in mammalian development. Science 293: 1089–1093 [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Rougier N, Bourc'his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E (1998) Chromosome methylation patterns during mammalian preimplantation development. Genes Dev 12: 2108–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Dean W (2004) Epigenetic reprogramming during early development in mammals. Reproduction 127: 643–651 [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W (2002) Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 241: 172–182 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y (2008) Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 9: 129–140 [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C (2009) TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell 33: 344–353 [DOI] [PubMed] [Google Scholar]
- Sun QY, Schatten H (2007) Centrosome inheritance after fertilization and nuclear transfer in mammals. Adv Exp Med Biol 591: 58–71 [DOI] [PubMed] [Google Scholar]
- Surani MA, Durcova-Hills G, Hajkova P, Hayashi K, Tee WW (2008) Germ line, stem cells, and epigenetic reprogramming. Cold Spring Harb Symp Quant Biol 73: 9–15 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by the MLL fusion partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierling S, Reither S, Walter J. Bisulfite sequencing of small DNA/cell samples http://www.epigenome-noe.net/researchtools/protocol.php?protid=35, 12 Nov 2007
- Ziegler-Birling C, Helmrich A, Tora L, Torres-Padilla ME (2009) Distribution of p53 binding protein 1 (53BP1) and phosphorylated H2A.X during mouse preimplantation development in the absence of DNA damage. Int J Dev Biol 53: 1003–1011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.