Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) expresses numerous intronless mRNAs that are unable to access splicing-dependent cellular mRNA nuclear export pathways. To circumvent this problem, KSHV encodes the open reading frame 57 (ORF57) protein, which orchestrates the formation of an export-competent virus ribonucleoprotein particle comprising the nuclear export complex hTREX, but not the exon-junction complex (EJC). Interestingly, EJCs stimulate mRNA translation, which raises the intriguing question of how intronless KSHV transcripts are efficiently translated. Herein, we show that ORF57 associates with components of the 48S pre-initiation complex and co-sediments with the 40S ribosomal subunits. Strikingly, we observed a direct interaction between ORF57 and PYM, a cellular protein that enhances translation by recruiting the 48S pre-initiation complex to newly exported mRNAs, through an interaction with the EJC. Moreover, detailed biochemical analysis suggests that ORF57 recruits PYM to intronless KSHV mRNA and PYM then facilitates the association of ORF57 and the cellular translation machinery. We, therefore, propose a model whereby ORF57 interacts directly with PYM to enhance translation of intronless KSHV transcripts.
Keywords: KSHV, mRNA export, translation
Introduction
The cellular machineries involved in processing a nascent mRNA are both numerous and complex (Kohler and Hurt, 2007). Of these, the spliceosome is the largest and perhaps the most intricate, functioning to remove non-coding intronic sequence from multi-exon genes, which constitute the majority of genes expressed in higher eukaryotes (Jurica and Moore, 2003). Intriguingly, the act of splicing is itself coupled to the deposition of two distinct multi-protein complexes on a burgeoning mRNA, namely the hTREX complex and the exon-junction complex (EJC) (Reed and Cheng, 2005). The hTREX complex is deposited at the 5′ end of the first exon, where it facilitates the polarized nuclear export of the spliced mRNA (Cheng et al, 2006). In contrast, the EJC is deposited 20–24 nucleotides upstream of each exon–exon boundary on a spliced mRNA (Le Hir et al, 2000) and has key functional roles in mRNA surveillance (Chang et al, 2007) and translational enhancement (Nott et al, 2004; Gudikote et al, 2005).
The observation that a spliced mRNA exhibits enhanced translation was initially made over two decades ago (Callis et al, 1987) and has subsequently been supported by numerous studies (Braddock et al, 1994; Matsumoto et al, 1998; Lu and Cullen, 2003). Furthermore, it is clear that deposition of the EJC is both necessary and sufficient for the observed splicing-dependent increase in translation (Nott et al, 2004). Several mechanisms have been proposed providing insight into how the EJC may influence translational efficiency. One example is through the adapter protein, SKAR, recently reported to associate with the EJC and recruit the mTOR-target kinase, S6K1 (Ma et al, 2008). S6K1 has been shown to associate with and phosphorylate components of the pre-initiation complex, leading to an increase in translation (Peterson and Sabatini, 2005). A second study investigating the EJC-associated protein, PYM, provides a mutually exclusive mechanism for EJC-mediated translational enhancement. PYM functions to recruit the 40S ribosomal subunit to spliced mRNAs through a cytoplasmic interaction with the EJC components Y14 and Magoh (Bono et al, 2004; Diem et al, 2007). In each case, siRNA-mediated depletion of SKAR or PYM leads to a significant decrease in the translation of a spliced reporter mRNA (Diem et al, 2007; Ma et al, 2008).
Kaposi's sarcoma-associated herpesvirus (KSHV) is a γ-2 herpesvirus associated with a number of AIDS-related malignancies (Chang and Moore, 1996). In contrast to higher eukaryotes, all herpesviruses express numerous lytic intronless mRNAs. As herpesviruses rely on host-cell machinery for mRNA export and translation, a caveat exists concerning the mechanisms by which these viral intronless mRNAs are exported from the nucleus and efficiently translated in the cytoplasm, given that splicing is an essential pre-requisite of both these events. In KSHV, the nuclear export of intronless viral mRNA is facilitated by the virus-encoded, intron-containing, open reading frame 57 (ORF57) protein (Swaminathan, 2005). KSHV ORF57 interacts directly with the export adapter, Aly, and shuttles between the nucleus and the cytoplasm promoting the nuclear export of intronless viral mRNAs through a TAP-mediated pathway (Malik et al, 2004). These properties are also conserved in ORF57 homologues throughout herpesviruses such as ICP27 from Herpes simplex virus type-1 (HSV-1), SM protein from Epstein–Barr virus (EBV) (Koffa et al, 2001; Ruvolo et al, 2001; Chen et al, 2005; Sergeant et al, 2008) and Herpesvirus saimiri (HVS) ORF57 protein (Goodwin and Whitehouse, 2001; Williams et al, 2005; Boyne et al, 2008a; Colgan et al, 2009).
We recently investigated the composition of an export-competent intronless KSHV ribonucleoprotein particle (vRNP) and showed that ORF57 functions to recruit the hTREX complex to intronless viral mRNA, an event that is essential for viral intronless mRNA export and KSHV replication (Boyne et al, 2008a). In contrast, ORF57 failed to recruit an EJC to intronless KSHV mRNA. Given the function of the EJC in translation enhancement, its absence from intronless viral mRNA is intriguing and we hypothesized that the lack of an EJC may negatively impact on the translational efficiency of these intronless viral mRNAs. Therefore, we set out to test whether, in the absence of an EJC, the ORF57 protein was able to enhance translation of intronless KSHV mRNAs.
We show that ORF57 is able to enhance the translation of an intronless KSHV mRNA in vitro. Sucrose gradient analysis revealed that ORF57 sediments predominantly with the 40S ribosomal subunit and interacts with the components of the translation machinery. Intriguingly, we report a novel interaction between ORF57 and the recently identified translation enhancement factor, PYM. Biochemical analysis revealed that PYM only binds to intronless KSHV mRNA in the presence of ORF57 and that siRNA-mediated depletion of PYM resulted in a marked reduction in the association of ORF57 with components of the translational machinery. To determine whether PYM is essential for the observed enhancement of intronless KSHV mRNA translation, functional assays were carried out in PYM depleted or cells expressing an ORF57-specific dominant negative mutant form of PYM. In both cases, a decrease in functional PYM resulted in a specific decrease in the efficiency of intronless KSHV mRNA translation. Together, these data suggest that ORF57 functions to enhance the translation of intronless KSHV mRNAs by recruiting the translational enhancement protein, PYM and promoting the recruitment of the pre-initiation complex to intronless KSHV mRNA. Importantly, this work provides the first mechanistic model for how KSHV ensures the efficient translation of viral intronless mRNAs.
Results
ORF57 enhances the translation of an intronless KSHV mRNA
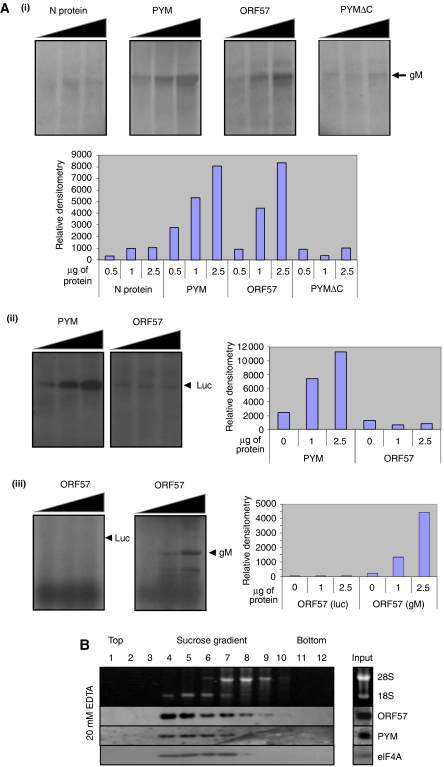
To determine whether ORF57 is capable of enhancing the translation of an intronless KSHV mRNA, an in vitro translation assay was used. The KSHV ORF39 (gM) gene was in vitro transcribed from a plasmid template and equal amounts of RNA used in radio-labelled in vitro translation reactions. A rate-limiting amount of gM mRNA (2.5 ng) template was purposefully added to each translation assay to ensure that any enhancement effect was detectable. Each assay was then spiked with increasing amounts of either a negative control (bunyamwera virus nucleocapsid protein; Rodgers et al, 2006), a known translational enhancer (glutathione S-transferase (GST)-PYM; Diem et al, 2007), a deletion mutant of PYM unable to enhance translation (GSTPYMΔC), or GST-ORF57 protein (Supplementary Figure S1). Results show that the level of gM translation observed in samples spiked with the negative controls, bunyamwera virus nucleocapsid protein or GSTPYMΔC remained consistent, showing no increase in translation. In contrast, samples spiked with PYM displayed a dose-dependent increase in translation of gM (up to eight-fold) in relation to the amount of GST-PYM added (Figure 1Ai). Strikingly, addition of GST-ORF57 led to a similar dose-dependent increase in the translation of gM mRNA (Figure 1Ai).
Figure 1.
ORF57 enhances translation of an intronless viral mRNA. (Ai) Intronless KSHV gM RNA was transcribed in vitro and equal amounts carried over into 35S-methionine-labelled in vitro rabbit reticulocyte translation reactions, spiked with increasing amounts (0.5, 1 or 2.5 μg) of either purified bunyamwera virus nucleocapsid protein, GST-PYM, GST-ORF57 or GST-PYMΔC. (Aii) Luciferase RNA was transcribed in vitro and equal amounts carried over into 35S-methionine-labelled in vitro rabbit reticulocyte translation reactions, spiked with increasing amounts (0, 1 or 2.5 μg) of GST-PYM or GST-ORF57. (Aiii) gM or luciferase RNA was transcribed in vitro and equal amounts were carried over into 35S-methionine-labelled in vitro human cell extract translation reactions, spiked with increasing amounts (0, 1 or 2.5 μg) of GST-ORF57. Translation reactions were separated by SDS–PAGE and proteins detected by autoradiography and quantified by densitometry. (B) Cytoplasmic lysate from reactivated BCBL-1 cells treated with cyclohexamide was separated by sucrose gradient fractionation in the presence of 20 mM EDTA. Fractions were analysed for rRNA and protein content by agarose gel electrophoresis or western blot analysis, respectively.
To determine the specificity of ORF57's translation enhancement, the in vitro translation assay was repeated using a rate-limiting amount of a control mRNA. Results show that PYM displayed a dose-dependent translation enhancement of luciferase; however, in contrast, no increase in translation was observed in samples spiked with increasing amounts of ORF57 protein (Figure 1Aii). This suggests that ORF57-mediated translation enhancement is not merely a non-specific enhancement due to increasing RNA stability but is specific to viral intronless mRNAs. Moreover, to ensure that ORF57 enhances cap-dependent translation, rate-limiting amounts of control luciferase and gM mRNAs were in vitro translated using human cell line extracts. Results show that ORF57 displays a dose-dependent translation increase of gM; however, no increase was observed with the control luciferase (Figure 1Aiii). Therefore, these data suggest that ORF57 is able to enhance the translation of the intronless gM mRNA.
ORF57 sediments with the 40S ribosomal subunit
Given that ORF57 is able to enhance the translation of a KSHV intronless mRNA in vitro, we next sought to determine whether ORF57 associates with ribosomal subunits during lytic replication. BCBL-1 cells are latently infected with KSHV, and treatment of these cells with the phorbel ester, TPA, triggers lytic reactivation. To determine whether ORF57 associates with the components of the cellular translation machinery, BCBL-1 cells were treated with TPA for 16 h to induce KSHV lytic replication and then cyclohexamide was added for 2 h to stabilize polysomes. Cytoplasmic cell lysate was isolated from the reactivated BCBL-1 cells and separated across a 5–20% linear sucrose gradient, to determine the sedimentation profile of ORF57 in relation to the large and small ribosomal subunits. Sedimentation was performed in the presence of 20 mM EDTA, which elicits the dissociation of ribosomes into the large (60S) and small (40S) subunits. Following centrifugation, fractions were collected and analysed for ORF57 protein and RNA content. The presence of 28S and 18S rRNA served as a marker for the large and small ribosomal subunits respectively, as described earlier (Diem et al, 2007). The majority of ORF57 fractionated with the 18S rRNA, indicating that a significant fraction of ORF57 co-sediments with the 40S ribosomal subunit (Figure 1B), suggesting that ORF57 may be associated with the pre-initiation complex, although we cannot conclusively determine whether ORF57 sediments with 40S subunits or binds RNA associated with 40S subunits. However, to confirm that components of the pre-initiation complex were also present in the 18S rRNA-rich fractions, western blot analysis was also performed against the translational enhancement factor, PYM and eIF4A. As shown in Figure 1B, PYM and eIF4A, like ORF57, are present predominantly in the 18S-rich fractions, confirming that these fractions contain components of the cellular translation machinery.
ORF57 interacts with the components of the translation machinery
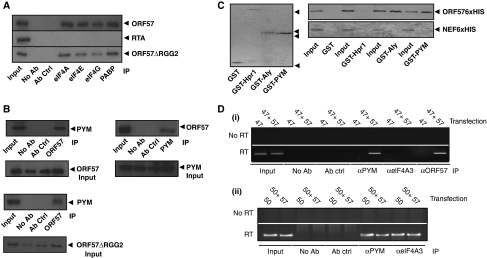
Having established that ORF57 enhances the translation of a KSHV intronless mRNA and co-sediments with the 40S ribosomal subunit, we next sought to determine whether ORF57 interacts with components of the translation machinery. BCBL-1 cells were treated with TPA for 16 h to activate lytic gene expression, and co-immunoprecipitations (Co-IPs) were performed using a panel of antibodies specific for the translation factors eIF4A-, eIF4E-, eIF4G- and PABP, in addition to a negative control antibody (α-SC35). ORF57 was readily co-precipitated by each of the translation factor-specific antibodies in contrast to negative controls (Figure 2A). As a further control, we assessed whether these translation factors precipitated another KSHV lytic protein, the viral transcription factor RTA. No interaction was observed with RTA (Figure 2A), suggesting that the interaction between ORF57 and these translation factors was not due to a non-specific interaction. Moreover, the observed interaction between ORF57 and the above translation factors was still detected when Co-IPs were performed in the presence of RNase, suggesting that these interactions may not be a result of RNA bridging (Supplementary Figure S2A). To ensure appropriate RNase activity, a control Co-IP experiment was performed assessing the interaction between two RNA-binding proteins, previously shown to be RNA dependent, namely Aly and UIF (Hautbergue et al, 2009). Supplementary Figure S3 shows that the RNase treatment was efficient, as in the presence of RNase, the interaction between Aly and UIF was abolished. However, although this analysis suggests that the interactions between ORF57 and translation factors are RNA independent, this analysis only shows that the interactions are RNase resistant. Therefore, the Co-IP experiments were repeated in transfected 293T cells using an ORF57 mutant, ORF57ΔRGG2, which has been shown earlier to be unable to bind RNA (Nekorchuk et al, 2007) and Supplementary Figure S2B. Results show that this ORF57 RNA-binding mutant can still interact with translation factors (Figure 2A), suggesting an RNA-independent interaction.
Figure 2.
ORF57 associates with components of the 48S pre-initiation complex and the EJC adapter protein, PYM. (A) Immunoprecipitations using eIF4A-, eIF4E-, eIF4G- or PABP-specific antibodies were performed on reactivated BCBL-1 cell or ORF57ΔRGG2-transfected cell lysates, in addition to no antibody and negative control antibody (α-SC35) controls. Precipitated ORF57 or RTA protein was detected by western blot analysis using ORF57- or RTA-specific antibodies, total reactivated or transfected cell lysate served as a positive control (input=10%). (B) Immunoprecipitations were performed on reactivated BCBL-1 or ORF57ΔRGG2-transfected cell lysates using either an ORF57- or PYM-specific antibody, in addition to no antibody and negative control antibody (α-SC35) controls. Western blot analysis was performed using either a PYM- or ORF57-specific antibody, total reactivated or transfected cell lysate served as a positive control (input=10%). Loading controls for each co-immunoprecipitation are also shown. (C) Recombinant GST-, GST-Hpr1-, GST-Aly- and GST-PYM-bound glutathione-agarose beads (indicated by the solid triangles) were incubated with purified baculovirus-expressed recombinant ORF57-6xHis or HIV-Nef-6xHis. After washes, bound proteins were analysed by western blot using a His-specific antibody. Purified ORF57-6xHis or HIV-Nef-6xHis protein served as a positive control (input). (D) Cells were transfected with (i) pORF47 or (ii) pORF50 in the absence or presence of pGFP or pORF57GFP. After UV crosslinking, RNA-IPs were performed using no antibody, negative antibody control (α-p53), PYM-, eIF4A3- and ORF57-specific antibodies. Total RNA served as a positive control for the PCR (input).
ORF57 interacts with PYM
Given recent data showing that EJCs are coupled to the translational machinery through PYM (Diem et al, 2007), we hypothesized that ORF57 may link intronless KSHV mRNAs to the translation machinery by interacting with PYM. To test this hypothesis, BCBL-1 cells were reactivated and RNase-treated cell lysates used in Co-IP experiments with either ORF57- or PYM-specific antibodies. Moreover, Co-IP assays were performed in 293T cells transfected with ORF57ΔRGG2. We were able to detect an interaction between ORF57 and PYM in reactivated BCBL-1 cells and in ORF57ΔRGG2-transfected cells (Figure 2B). Furthermore, to establish whether the observed interaction between ORF57 and PYM was direct, GST-pull-down experiments were performed using purified baculovirus-expressed recombinant ORF57-6xHis protein and a negative control baculovirus-expressed protein, HIV-Nef-6xHis (Harris and Coates, 1993). ORF57-6xHis protein was precipitated by GST-Aly, but failed to interact with GST-Hpr1 or GST alone (Figure 2C). Critically, ORF57-6xHis was also clearly precipitated by GST-PYM, suggesting that ORF57-6xHis and PYM interact directly in vitro (Figure 2C). No interaction was observed with the negative control HIV-Nef-6xHis. Although these experiments do not completely rule out nucleic acid contamination, which could lead to RNA bridging, the combined data suggest that the interaction between ORF57 and PYM is RNA independent.
ORF57 facilitates the binding of PYM to KSHV intronless mRNA
To obtain further evidence that PYM has a function in ORF57-mediated translational enhancement, we next determined whether PYM is bound to a KSHV intronless mRNA. RNA immunoprecipitations were performed using PYM-, eIF4A3- and ORF57-specific antibodies on 293T cell lysates transfected with an intronless KSHV mRNA reporter vector (pORF47), in the absence or presence of pORF57GFP. We were only able to detect PYM bound to the intronless ORF47 mRNA in cells that co-expressed ORF57; in contrast, no interaction was observed in the absence of ORF57, suggesting that ORF57 facilitates the binding of PYM to intronless KSHV ORF47 mRNA (Figure 2Di). Importantly, no recruitment of the core EJC protein, eIF4A3 was observed either in the absence or presence of ORF57, demonstrating that ORF57 licenses the recruitment of PYM to intronless mRNAs in the absence of the core EJC protein, eIF4A3. The recruitment of ORF57GFP to the intronless ORF47 mRNA served as a positive control (Boyne et al, 2008b). Moreover, to confirm that the eIF4A3 antibody could precipitate RNA, experiments were repeated using 293T cell lysates transfected with a spliced KSHV mRNA reporter vector (pORF50), in the absence and presence of ORF57. Here, both PYM and eIF4A3 bound to the spliced mRNA independent of ORF57 (Figure 2Dii). These data, therefore, support the hypothesis that ORF57 promotes the recruitment of PYM to ORF47 mRNA and that this recruitment occurs in the absence of an EJC.
ORF57 redistributes PYM and some components of the pre-initiation complex to the nucleolus
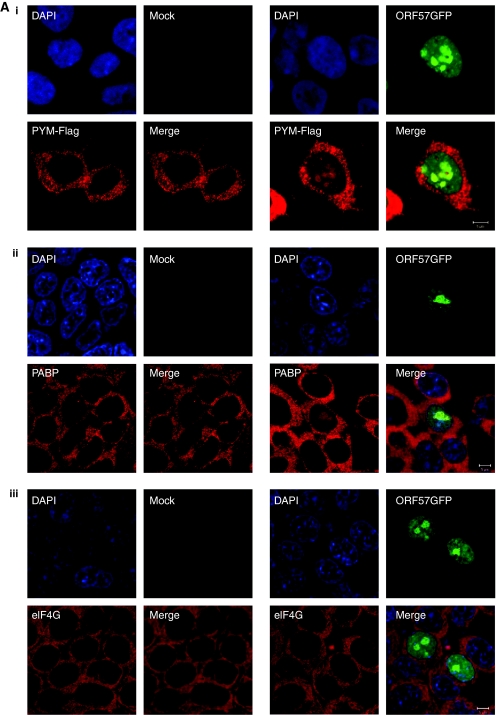
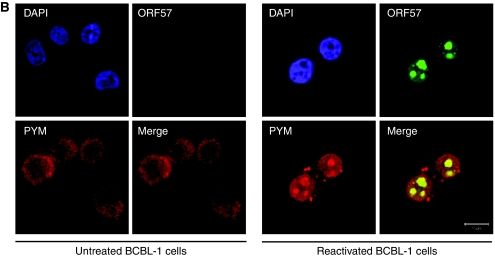
ORF57 is a nucleo-cytoplasmic protein that localizes predominately to the nucleus, specifically nuclear speckles and nucleoli (Majerciak et al, 2006; Boyne and Whitehouse, 2009), whereas PYM and components of the translation machinery are generally located in the cytoplasm (Diem et al, 2007). Considering ORF57 interacts with PYM and several translation factors, we were interested to determine whether ORF57 expression had any effect on the subcellular localization of PYM and members of the pre-initiation complex. To this end, 293T cells were cultured on poly-L lysine-coated coverslips and transfected with pORF57GFP either alone or in combination with PYM-FLAG. Indirect immunofluorescence was performed for PYM-FLAG, PABP and eIF4G and staining analysed by confocal microscopy. We were intrigued to discover that expression of ORF57GFP resulted in the redistribution of a small proportion of PYM and PABP to the nucleolus (Figure 3Ai and ii). This redistribution was evident in ∼60% of cells analysed (n=300). In contrast, and in way of an internal control for non-specific detection of GFP-fluorescence in the TRITC channel, no relocalization of eIF4G was observed in the presence of ORF57 (Figure 3Aiii). To confirm these results in KSHV-infected cells and to ensure relocalization is not an overexpression artefact, indirect immunofluorescence was performed on BCBL-1-reactivated cells using PYM- and ORF57-specific antibodies and staining analysed by confocal microscopy. We observed a clear co-localization of PYM with ORF57 in the nucleolus in reactivated BCBL-1 cells, which is absent in the unreactivated control (Figure 3B). Moreover, these experiments were performed with two previously characterized PYM mutants, termed PYMΔN19-54 and PYMΔC53 that were no longer able to interact with the EJC and the 48S pre-initiation complex, respectively (Diem et al, 2007). Results show that a small proportion of the deletion mutants are also redistributed to the nucleolus in the presence of ORF57, suggesting that ORF57 can interact with PYM in the nucleolus in the absence of the domains required for PYM's interaction with translation factors or EJC components (Supplementary Figure S4).
Figure 3a.
KSHV ORF57 redistributes PYM and some components of the pre-initiation complex to the nucleolus. (Ai) 293T cells were co-transfected with pORF57GFP and pPYM-FLAG, incubated for 24 h, fixed and immunofluorescence staining performed using a FLAG-specific antibody. 293T cells were transfected with pORF57GFP, incubated for 24 h, fixed and immunofluorescence staining performed using (Aii) a PABP-specific antibody and (Aiii) an eIF4G-specific antibody.
Figure 3b.
(B) BCBL-1 cells were either left untreated or were reactivated by treating with TPA (20 ng/ml) for 16 h, fixed and immunofluorescence staining performed using ORF57- and PYM-specific antibodies.
PYM links ORF57 to the translational machinery
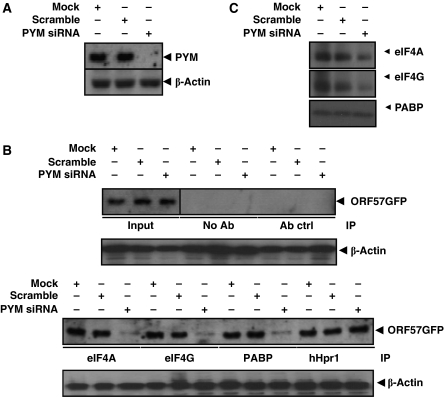
Having established that ORF57 associates with the components of the pre-initiation complex and interacts directly with PYM, we next sought to determine whether PYM functions as an adapter between ORF57 and pre-initiation complex factors. To this end, 293T cells were transfected with pORF57GFP with either mock, scramble or PYM-specific siRNAs. An almost complete knockdown of PYM levels was achieved 48 h post-transfection, when compared with β-actin loading control (Figure 4A). Control or PYM-depleted cell lysates were then immunoprecipitated using eIF4A-, eIF4G- or PABP-specific antibodies in addition to negative (no antibody and α-p53) and positive (the hTREX component, hHpr1 specific) controls. Western blot analysis revealed that in each case, the amount of ORF57GFP precipitated by components of the translation machinery (eIF4A, eIF4G and PABP) was dramatically reduced in PYM-depleted cells. In contrast, the interaction between ORF57GFP and hHpr1 was not affected by depletion of PYM, demonstrating that the reduction in ORF57GFP precipitation in PYM-depleted cells was specific for components of the translation machinery (Figure 4B). Western blot analysis was also performed to compare starting levels of eIF4A, eIF4G and PABP in mock, scrambled and PYM-depleted samples, results show that PABP levels are unaffected by PYM depletion; however, there are observable decreases in eIF4A and eIF4G, although this does not detract from the finding that association with ORF57 and the translation factors are significantly reduced in PYM-depleted cells (Figure 4C). These data suggest that PYM is required for the efficient association of ORF57 with components of the pre-initiation complex.
Figure 4.
PYM mediates the association of ORF57 and the pre-initiation complex. (A) 293T cells were transfected with pORF57GFP in addition to either mock, scramble siRNA or PYM-specific siRNA and incubated for 48 h. Total protein (100 μg) was analysed by western blotting using PYM- and β-actin-specific (loading control) antibodies to confirm depletion of PYM. (B) PYM-depleted cell lysates were generated as described in Figure 4A and immunoprecipitations were performed using either eIF4A-, eIF4G-, PABP- or hHpr1-specific antibodies, in addition to negative controls. Western blot analysis was then performed using a GFP-specific antibody to detect precipitated ORF57. Total transfected cell lysate served as a positive control for expression of ORF57 (input). Loading controls for each co-immunoprecipitation are also shown. (C) Total protein was isolated at 48 h from PYM-depleted cells and western blot analysis performed to assess eIF4A, eIF4G and PABP levels.
PYM is essential for ORF57-mediated translational enhancement of intronless KSHV mRNA
We next investigated whether PYM was required for the efficient translation of intronless viral mRNA during KSHV replication. To this end, siRNA-mediated depletion of PYM was used in the 293T BAC36 cell system, which harbours the recombinant KSHV-BAC36 genome (Gao et al, 2003; Wilson et al, 2007). However, one caveat of this system is that PYM depletion could inhibit production of the ORF57 protein itself. Therefore, to prevent inhibition of ORF57 protein production, cells were concurrently reactivated using TPA and either mock transfected or transfected with scramble or PYM-specific siRNA, which allowed sufficient ORF57 protein to be produced before optimal PYM depletion. Protein and RNA samples were then taken at t=0 h (untreated), t=24 h and t=48 h time points for analysis by semi-quantitative RT–PCR and western blotting. To confirm the knockdown of PYM, western blot analysis was performed for each time point using a PYM-specific antibody. As shown in Figure 5, we observed limited knockdown of PYM at 24 h post-transfection; however, by 48 h, almost complete knockdown of PYM was achieved. Subsequently, we assessed whether the depletion of PYM had any effect on the efficiency of translation of two intronless lytic KSHV mRNAs (ORF54 and ORF59). These were chosen due to the availability of suitable antibody reagents. Western blot analysis revealed that 293T-BAC36 cells depleted for PYM produced significantly less ORF54 and ORF59 protein compared with mock or scramble-transfected controls. To ensure that this reduction in protein levels was due to an effect on translation and not mRNA export or stability, semi-quantitative RT–PCR was performed on RNA samples taken from each time point to determine the mRNA levels for ORF54 and ORF59. We observed no difference in ORF54 or ORF59 mRNA levels, irrespective of PYM status (Figure 5). Importantly, protein levels for the spliced ORF57 gene product were comparable between control and siRNA-transfected samples at each time point, suggesting that the observed reduction of ORF54 and ORF59 protein was not due to depletion of ORF57 and as such intronless viral mRNA export. It should be noted that as predicted, due to the temporal nature of expression during reactivation, a decrease in ORF57 expression was seen at 48 h, compared with 24 h (Figure 5).
Figure 5.
Knockdown of PYM impairs intronless KSHV mRNA translation. 293T BAC36 cells were transfected with either mock, scramble siRNA or PYM-specific siRNA and concurrently reactivated using TPA (20 ng/ml). Total protein and RNA was isolated at the indicated time points and analysed by western blotting and RT–PCR, respectively, using the indicated antibodies and oligonucleotides.
Furthermore, to exclude the possibility that PYM depletion alters the nuclear–cytoplasmic distribution of viral mRNA, 293T BAC36 cells were transfected with either mock, scramble or PYM-specific siRNA and concurrently reactivated using TPA. RNA was then isolated at 48 h post-reactivation from nuclear and cytoplasmic fractions and ORF59 mRNA levels assessed using qRT–PCR. Results show no significant variation in nuclear–cytoplasmic mRNA levels in mock, scrambled and PYM-depleted samples (Supplementary Figure S5). Furthermore, to confirm the specificity of the PYM-specific siRNAs, a second control siRNA was used in similar studies. 293T BAC36 cells were transfected with either mock or a second control luciferase-specific siRNA (Griffiths and Whitehouse, 2007) and concurrently reactivated using TPA. Results show that the second control siRNA does not significantly affect mRNA nuclear–cytoplasmic levels or protein levels (Supplementary Figure S6). Therefore, together these data suggest that PYM is essential for the efficient translation of intronless KSHV mRNAs during lytic replication.
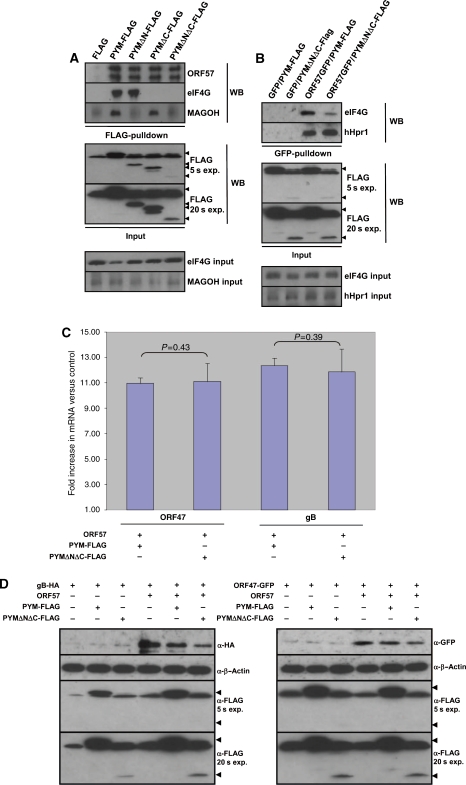
However, one caveat of using siRNA-mediated PYM depletion to assess the effect on intronless KSHV mRNA translation is that PYM depletion is likely to also alter the expression of some cellular mRNAs. Therefore, it was desirable to develop an ORF57-specific functional assay to test the function of PYM in ORF57-mediated translational enhancement, without effecting cellular translation. As mentioned earlier, Diem et al (2007) characterized two PYM mutants, termed PYMΔN19-54 and PYMΔC53 that were no longer able to interact with the EJC and the 48S pre-initiation complex, respectively. We hypothesized that should a PYM mutant lacking the N- and C-terminus retain an interaction with ORF57, then it will function in a dominant negative capacity by specifically sequestering ORF57 away from wild-type PYM, while not affecting cellular translation, due to the removal of the EJCs and pre-initiation complex-binding domains. To test this hypothesis, we generated a FLAG-tagged PYM mutant, PYMΔNΔC-FLAG, which has 54 amino acids deleted from the N-terminus and 53 amino acids deleted from the C-terminus, and assessed the binding of PYMΔNΔC-FLAG to ORF57, Magoh (EJC) or eIF4G (pre-initiation complex). 293T cells were transiently transfected with either pFLAG, pPYM-FLAG, pPYMΔN19-54-FLAG, pPYMΔC53-FLAG or pPYMΔNΔC-FLAG and Co-IPs performed using FLAG-affinity beads. As expected, PYM-FLAG precipitated ORF57, Magoh and eIF4G, whereas PYMΔN19-54-FLAG and PYMΔC53-FLAG both precipitated ORF57, but failed to interact with Magoh and eIF4G. respectively (Figure 6A), in agreement with earlier findings (Diem et al, 2007). As predicted, PYMΔNΔC-FLAG did not precipitate Magoh or eIF4G, in contrast, ORF57 was still readily precipitated (Figure 6A). It should be noted that ORF57 is sometimes observed as a doublet due to caspase 7 cleavage (Majerciak et al, 2010). Moreover, the different amounts of PYM deletion mutants detected using α-FLAG antibody reflects the fact that PYMΔNΔC is fused to a single FLAG epitope, whereas PYM, PYMΔN19-54 and PYMΔC53 are fused to three FLAG epitopes (Figure 6A, lower panel, short and long exposure). This interpretation is supported by the fact that similar amounts of ORF57 are precipitated by PYMΔNΔC-FLAG compared with each of the other PYM-FLAG fusions. Together, these data suggest that PYMΔNΔC-FLAG is an ideal construct to act as an ORF57-specific dominant negative mutant for PYM function.
Figure 6a-d.
An ORF57-specific dominant negative PYM mutant prevents ORF57 association with the 48S pre-initiation complex and perturbs ORF57-mediated intronless KSHV mRNA translation. (A) 293T cells were transfected with pORF57GFP in addition to either pFLAG, pPYM-FLAG, pPYMΔN19-54-FLAG, pPYMΔC53-FLAG and pPYMΔNΔC-FLAG, and immunoprecipitations were performed using FLAG-affinity beads. Western blot analysis was then performed using the indicated antibodies to detect the precipitated proteins. To detect the expression of each PYM-FLAG protein (indicated by the solid triangles), total cell lysate was analysed by western blotting using an FLAG-specific antibody. Two different autoradiograph exposures (exp.) are shown (5 and 20 s), as PYMΔNΔC is fused to a single FLAG epitope (as opposed three for the other fusion proteins) and therefore generates a weaker signal. Loading controls for each co-immunoprecipitation are also shown. (B) 293T cells were transfected with either pGFP or pORF57GFP in addition to either pPYM-FLAG or PYMΔNΔC-FLAG and an immunoprecipitation performed using a GFP-specific antibody. Western blot analysis was then performed using the indicated antibodies to detect precipitated proteins. Loading controls for each co-immunoprecipitation are also shown. (C) 293T cells were triple transfected with either pORF47GFP or pgB-HA, in the presence or absence of pCherry or pORF57Cherry and either pPYM-FLAG or pPYMΔNΔC-FLAG. At 24 h post-transfection, cytoplasmic RNA was isolated from each sample and 1 μg of RNA was used to generate cDNA using reverse transcriptase. A measure of 10 ng of cDNA was then used in qRT–PCR reactions. Fold increase was determined by ΔΔcT and statistical significance by a non-paired t-test. All data are representative of three independent experiments and presented as fold increase versus pCherry-transfected controls. (D) 293T cells were triple transfected with either pCherry or pORF57Cherry in addition to either pFLAG, pPYM-FLAG or pPYMΔNΔC-FLAG and either pORF47GFP or pgB-HA. At 24 h post-transfection, western blot analysis was performed using 25 μg of total protein extract and the indicated antibodies to detect protein expression levels. To detect the expression of PYM-FLAG and PYMΔNΔC-FLAG, total cell lysate was analysed by western blotting using a FLAG-specific antibody. Two different autoradiograph exposures (exp.) are shown (5 and 20 s) as PYMΔNΔC is fused to a single FLAG epitope (as opposed three for the other fusion proteins) and therefore generates a weaker signal.
To test whether PYMΔNΔC-FLAG functions as an ORF57-specific dominant negative mutant, we assessed the binding of ORF57 to the 48S-pre-initiation complex in the absence or presence of PYMΔNΔC-FLAG. We have shown earlier that PYM is required for the association of ORF57 with components of the 48S pre-initiation complex, therefore, if PYMΔNΔC-FLAG functions to titrate ORF57 away from endogenous PYM one would predict that the proportion of ORF57 associated with the 48S pre-initiation complex components, such as eIF4G, would decrease, whereas PYM-independent interactions, such as hHpr1 (hTREX protein), would be unaffected. 293T cells were co-transfected with either pGFP or pORF57GFP and pFLAG or pPYMΔNΔC-FLAG, before immunoprecipitation using an ORF57-specific antibody. Western blot analysis was then performed using eIF4G- and hHpr1-specific antibodies to assess the relative amounts of each protein precipitated by ORF57 in the absence or presence of PYMΔNΔC-FLAG. As shown in Figure 6B, expression of PYMΔNΔC-FLAG resulted in a significant reduction in the amount of eIF4G bound to ORF57, compared with cells co-expressing PYM-FLAG alone. In contrast, no decrease was observed in the amount of hHpr1 precipitated by ORF57 (Figure 6B), suggesting that PYMΔNΔC-FLAG specifically ablates PYM-dependent ORF57 interactions. These data show that PYMΔNΔC-FLAG functions as a dominant negative mutant that specifically disrupts PYM-dependent ORF57 interactions.
We next sought to determine whether the ORF57-specific loss of PYM function induced by PYMΔNΔC-FLAG resulted in a decrease in the translation of KSHV intronless mRNAs. The observation that hTREX proteins still interact with ORF57 in the presence of PYMΔNΔC-FLAG (Figure 6B) suggests that the mRNA nuclear export function of ORF57 is not effected by co-expression of PYMΔNΔC-FLAG. To confirm this, 293T cells were triple transfected with either pORF47GFP or pgB-HA, in the absence or presence of ORF57 and either pPYM-FLAG or pPYMΔNΔC-FLAG. At 24 h post-transfection, cytoplasmic RNA and total protein was isolated from each sample. Cytoplasmic levels of ORF47 and gB-HA mRNA were determined by qRT–PCR to assess whether co-expression of PYMΔNΔC-FLAG reduced the ability of ORF57 to export mRNA. As reported earlier, we observed a dramatic increase in the cytoplasmic levels of each intronless KSHV mRNA reporter in the presence of ORF57 compared with control (Nekorchuk et al, 2007). Figure 6C shows a comparative enhancement of KSHV ORF47 and gB mRNA nuclear export in cells expressing ORF57 in addition to either PYM-FLAG or PYMΔNΔC-FLAG. We observed no significant difference in the fold increase of cytoplasmic mRNA levels of ORF47 or gB (P=0.43 and 0.39, respectively) present in cells expressing ORF57 irrespective of whether these cells co-expressed PYM-FLAG or PYMΔNΔC-FLAG (Figure 6C). These data, alongside biochemical data in Figure 6B, suggest that PYMΔNΔC-FLAG does not interfere with ORF57-mediated intronless KSHV mRNA nuclear export.
To test whether expression of PYMΔNΔC-FLAG effected the translational enhancement of intronless KSHV mRNA by ORF57, western blot analysis was performed on the protein samples isolated from the transfections described above. To ensure that autoradiograph signals were not saturated, which would limit densitometry analysis, a small amount of total protein (15 μg) was analysed (Figure 6D). As shown in Figure 6E, both ORF47GFP and gB-HA were expressed at significantly higher levels in the presence of ORF57GFP or ORF57mCherry, compared with GFP or mCherry alone (14 and 11 fold, respectively). The co-expression of PYM-FLAG had no significant effect on the levels of each reporter (P=>0.05 for gB and ORF47); however, co-expression of PYMΔNΔC-FLAG resulted in a significant decrease of both gB-HA (decrease of 77.82%) and ORF47GFP (decrease of 55.84%), compared with FLAG control (P=0.04 and 0.01, respectively) (Figure 6E). Together, these data show that PYMΔNΔC-FLAG has a specific dominant negative effect on ORF57-mediated intronless mRNA translational enhancement and that this effect is independent of ORF57-mediated intronless KSHV mRNA export.
Figure 6e-f.
(E) Densitometry analysis was performed on three independent transfection experiments described in Figure 6D to determine the relative levels of ORF47GFP and gB-HA protein compared with controls and the standard error calculated (n=3). (F) 293T-BAC36 cells were transfected with the indicated vectors and concurrently reactivated using TPA. After 72 h, lytic virus replication was assayed by harvesting the supernatant. The supernatant was used to infect 293T cells, and 48 h later, the level of virus infection was scored by direct immunofluorescence (n=3).
Moreover, to determine whether expression of PYMΔNΔC-FLAG had any effect on virus replication, the KSHV-latently infected 293T-BAC36 cell line was transfected with pFLAG, pPYM-FLAG or pPYMΔNΔC-FLAG and concurrently reactivated using TPA and incubated for 72 h. The supernatants from each flask were then harvested and used to re-infect 293T cells. The level of virus replication was determined by scoring the percentage of GFP positive cells 48 h post-infection, as described earlier (Wilson et al, 2007). Similar levels of lytic replication and virus production were observed from pFLAG and pPYM-FLAG pre-transfected cells. Strikingly, virus production from pPYMΔNΔC-FLAG pre-transfected cells was reduced by approximately five-fold (Figure 6F). These data show that wild-type PYM function is required for normal levels of KSHV replication.
In summary, KSHV ORF57 appears to possess multi-functionality with regards to viral mRNA processing. We show that by interacting directly with PYM, ORF57 is able to link intronless KSHV mRNAs to the 48S pre-initiation complex and thereby enhance the translation of these viral mRNAs. The function of PYM in this process is essential as perturbation of PYM functionality ablates ORF57-mediated translational enhancement.
Discussion
The regulatory mechanisms responsible for the efficient translation of a newly exported mRNA are beginning to be elucidated. In particular, the function of splicing, and specifically the splicing-dependent deposition of the EJC, has recently come to the fore. The nuclear–cytoplasmic shuttle protein, PYM, was recently shown to link EJC-bound mRNAs to the 48S pre-initiation complex during the so-called pioneer round of translation (Ishigaki et al, 2001), thus increasing translational efficiency (Diem et al, 2007). We have reported earlier that KSHV ORF57 facilitates the export of intronless KSHV mRNAs by recruiting the hTREX complex but fails to recruit an EJC, prompting the question of how KSHV intronless mRNAs are efficiently translated. Here, we show for the first time that KSHV ORF57 interacts directly with PYM to facilitate the efficient translation of intronless KSHV transcripts. The observation that ORF57 is able to enhance translation of an intronless viral mRNA has parallels in HSV-1, CMV and EBV, where ICP27, UL69 and SM, respectively, have been shown to enhance the translation of viral genes (Fontaine-Rodriguez et al, 2004; Larralde et al, 2006; Fontaine-Rodriguez and Knipe, 2008; Ricci et al, 2009; Aoyagi et al, 2010). Moreover, these findings may have wider implications for other viruses that require the export of intronless transcripts, such as influenza. The non-structural-1 (NS1) protein of influenza A has also been reported to associate with components of the translation machinery, it would be of interest, therefore, to assess whether PYM associates with NS1 and influenza A intronless mRNA to enhance their translation. It is also plausible that the PYM-dependent increase of viral intronless mRNA translation described herein, may mirror existing cellular pathways that govern intronless mRNA translation. In support of this hypothesis, cellular SR proteins that direct the nuclear export of certain cellular intronless mRNA (e.g. Histone 2A), have also been shown to enhance the recruitment of the translational machinery (Sanford et al, 2004). It is therefore tempting to suggest that cellular SR proteins, which are functionally very similar to ORF57, may interact with and use PYM to efficiently translate intronless mRNAs that are unable to access EJC-dependent translational enhancement machinery. The observation that PYM directly enhances the translation of gM in the in vitro translation assay is, however, not unexpected, as this assay identifies general translational enhancers, a category occupied by PYM due to its ability to stabilize ribosomes (Diem et al, 2007).
Interestingly, KSHV ORF57 and homologues interact with several translation factors (Fontaine-Rodriguez et al, 2004; Aoyagi et al, 2010), suggesting that the assembly of such factors onto viral mRNA may represent a common mechanism used by herpesviruses. Importantly, the direct interaction reported here between ORF57 and PYM, suggests a compelling model for how ORF57 facilitates the assembly of the pre-initiation complex on an intronless KSHV mRNA, whereby ORF57 uses PYM as a linker protein to the 48S pre-initiation complex. Such a model is reinforced by siRNA-mediated depletion of PYM, which results in a significant decrease in the amount of ORF57 associated with translation factors. To date, no other viral protein has been reported to interact with PYM, and it will be of interest to determine whether ORF57 homologues also interact with PYM to link with cellular translation machinery. During functional studies, we observed that both the spliced ORF57 mRNA and a cellular, spliced mRNA (β-actin) remained relatively unaffected by siRNA-mediated PYM depletion. This was in direct contrast to two intronless KSHV mRNAs (ORF54 and ORF59), both of which exhibited a dramatic decrease in translation in PYM-depleted cells. One possible explanation is that spliced mRNAs have access to multiple mechanisms by which to promote translation, whereas KSHV has evolved a single, specific mechanism to enhance translation of intronless transcripts. Recent work by Ma et al (2008) shows that SKAR can recruit the mTOR-target kinase, S6K1, to newly exported mRNA by acting as a linker protein between the EJC and S6K1. Recruitment of S6K1 leads to an increase in translation through the phosphorylation of pre-initiation translation factors.
Given that the EJC appears to have multiple mechanisms for increasing translation, it seems counter intuitive that KSHV has not evolved to recruit a complete EJC to intronless viral mRNA. One possible explanation is that in addition to promoting translation, the EJC is also involved in negatively regulating protein synthesis. Specifically, during translation, a premature termination codon causes the ribosome to stall and elicits the recruitment of a complex termed SURF. Recruitment of SURF leads to a decrease in translation and eventually degradation of the transcript (Isken et al, 2008). Given the fact that an EJC is able to both positively and negatively regulate translation, it is perhaps not surprising that ORF57 fails to recruit an EJC, preferring instead to associate with direct enhancers of translation, such as PYM. Work is currently underway to determine whether ORF57 interacts with SKAR or SURF components. However, while this manuscript was in preparation, it was reported that PYM also functions to disassemble EJCs. PYM-dependent disassembly of EJCs occurs immediately after nuclear export, and it has been suggested that by coupling ribosome recruitment to EJC disassembly, PYM ensures that newly exported mRNAs are given preferential access to ribosomes. As KSHV intronless mRNAs lack EJCs (Boyne et al, 2008b), ORF57 may subvert PYM solely on the basis that it enables efficient access to ribosomes. Whether PYM remains bound to ORF57 during steady-state translation, rather than being recycled with EJC components following the pioneer round of translation will be of interest to determine.
One important question is how and when ORF57 interacts with PYM, an event that presumably then triggers the recruitment of other translation factors to the intronless KSHV mRNA. A clue may lie in the subcellular localization of ORF57 and PYM. ORF57 exhibits a nuclear speckled and nucleolar pattern of staining (Boyne and Whitehouse, 2009), whereas PYM is localized exclusively in the cytoplasm (Diem et al, 2007). Intriguingly, after treatment with the CRM1 inhibitor, leptomycin B, the localization pattern of PYM alters and rather than being located in the cytoplasm, it is instead retained in the nucleus and nucleolus. Interestingly, we observed a clear colocalization of PYM and ORF57GFP in the nucleolus of ORF57GFP-transfected cells and in KSHV-reactivated cells, suggesting that ORF57 may redistribute a proportion of PYM into the nucleolus or alternatively traffick through the nucleolus in a complex with PYM. We have reported earlier that HVS ORF57 is able to redistribute essential mRNA export proteins to the nucleolus (Boyne and Whitehouse, 2006). These data raise the possibility that the nucleolus is important for the export and subsequent translation of viral RNPs. This is a hypothesis that is not without precedence and represents an emerging theme in messenger ribonucleoprotein particle (mRNP) biogenesis. For example, the involvement of the nucleolus in mRNP biogenesis has been studied extensively in plants, where the nucleolus has key functions in nonsense-mediated decay and mRNA export (Brown and Shaw, 2008). More recently, work on Saccharomyces cerevisiae has shown that nucleolar trafficking of the mRNA-transport protein, She2p, is essential for the correct localization and translation of its target mRNA, ASH1 (Du et al, 2008). Although the functional significance of KSHV ORF57 nucleolar localization remains unclear (Nekorchuk et al, 2007; Boyne and Whitehouse, 2009), nucleolar trafficking of the HVS ORF57 protein is essential for its function in viral mRNA nuclear export (Boyne and Whitehouse, 2009). It will, therefore, be interesting to establish whether the observed redistribution of cellular mRNA processing factors to the nucleolus is functionally important with regards to KSHV replication.
In summary, our data show that KSHV ORF57 increases the translational efficiency of intronless viral mRNA through a direct interaction with PYM. Once recruited, PYM allows ORF57-bound intronless KSHV mRNAs access to the pre-initiation complex, an event that is essential for ORF57-mediated enhancement of translation.
Materials and methods
Plasmid and antibody details
Details of oligonucleotides used in RT–PCR, cloning and site-directed mutagenesis are presented in Supplementary Table S1. pORF57GFP and pORF47 are described earlier (Boyne et al, 2008a). PYM constructs and the PYM-specific mouse monoclonal antibody were gifts from Gideon Dreyfuss (University of Pennsylvania). To generate GST-PYM and GST-PYMΔC, the PYM ORF was PCR amplified from pPYM-FLAG and cloned into pGEX4T-1 (Pharmacia). To generate pPYMΔNΔC-FLAG, a region of PYM lacking the first 54 and last 53 amino acids was PCR amplified from pPYM-FLAG, incorporating a single FLAG tag and then cloned into pcDNA3.1. pORF57ΔRGG2 was generated using the QuickChange II site-directed mutagenesis kit (Stratagene). Purified bunyamwera virus nucleocapsid protein and purified His-tagged HIV Nef protein was provided by John Barr and Mark Harris, respectively (Leeds University). PABP-, eIF4A-, eIF4G- and eIF4E-specific antibodies were gifts from Simon Morley (University of Sussex) and Ian Goodfellow (Imperial College, London). ORF54-specific antibodies were provided by Friedrich Grässer (Universitätskliniken, Germany). ORF57-specific antibody was described earlier (Boyne et al, 2008a). ORF59- (Autogen Bioclear), p53- (Pharmagen Inc), GFP- (BD Biosciences) FLAG-, β-actin-, SC-35- (Sigma) antibodies were purchased from their respective suppliers. Unless stated otherwise, all antibodies were used at a dilution of 1:1000 for western blot analysis.
Cell culture, viruses and transfection
HEK-293T and 293T-BAC36 cells were cultured in Dulbecco's modified Eagle medium (DMEM, Invitrogen, Paisley, UK), KSHV-infected BCBL-1 cells were cultured in RPMI medium (Invitrogen, Paisley, UK) both media were supplemented with 10% foetal calf serum (FCS, Invitrogen), glutamine and penicillin-streptomycin. Sf9 cells were cultured in Sf-900 II SFM (Invitrogen, Paisley, UK). BCBL-1 cells were reactivated using TPA (20 ng/ml) for the designated time. Plasmid transfections were carried out using Lipofectamine 2000 (Invitrogen, Paisley, UK), as per the manufacturer's instructions. KSHV replication assays were performed as described earlier (Wilson et al, 2007).
Expression and purification of recombinant proteins
Recombinant GST-ORF57, GST-PYM and GST-PYMΔC were expressed and purified as described earlier (Malik et al, 2004). Further purification was performed using a Superdex 200 10/300 GL Column (GE Healthcare). The nucleocapsid bunyamwera virus protein was expressed and purified as described earlier (Rodgers et al, 2006). Baculovirus recombinant ORF57-6xHis was produced, expressed and purified as described by the manufacturer (Invitrogen) using the pFASTBac protocol.
In vitro transcription and translation
In vitro translation reactions were performed using the Flexi rabbit reticulocyte lysate system (Promega Inc) or human in vitro protein expression kit (Pierce) using 25 μg/ml of in vitro transcribed RNA. The in vitro transcription of gM or luciferase was performed using the MAXIscript kit (Ambion Inc). The concentration of RNA used was chosen to give a linear yield of translated product over the time course of translation (90 min). In reactions that required the addition of heterologous protein (0.5, 1 or 2.5 μg), or (0, 1, or 2.5 μg), the reactions were pre-incubated at 30°C for 15 min before the addition of RNA. After 90 min, the reactions were terminated by the addition of SDS–PAGE sample buffer, resolved on 9% polyacylamide gels and subsequently the dried gel was exposed to autoradiograph film for 16 h.
Sucrose gradients
Sucrose gradient fractionation was essentially carried out as described elsewhere (Diem et al, 2007). Briefly, BCBL-1 cells were reactivated using TPA (20 ng/ml) for 16 h, before 2 h treatment with cyclohexamide (100 μg/ml). Cytoplasmic extracts were isolated and loaded onto a 5–20% linear sucrose gradient then centrifugated at 28 800 r.p.m. in a Beckman SW41.1 rotor for 3 h at 4°C. Fractions were collected from the top of the gradient using a Biocomp fractionator (Biocomp, NB Canada) and total protein or RNA was extracted from 100 μl of each fraction by boiling with 2 × protein loading solution or extraction with Trizol (Invitrogen), respectively.
Immunoprecipitation assays and RNA analysis
GST pull downs and Co-IPs were performed as described earlier (Hall et al, 2002; Gould et al, 2009). RNA –immunoprecipitations were performed as described earlier (Boyne and Whitehouse, 2006). Quantitative RT–PCR was performed as described earlier (Boyne and Whitehouse, 2009).
Immunofluorescence
Immunofluorescence staining was carried out as described earlier (Boyne and Whitehouse, 2006). Immunofluorescence was visualized on an LSM 510 Meta confocal microscope (Zeiss) and images analysed using the manufacturer's software.
Supplementary Material
Acknowledgments
We are indebted to Gideon Dreyfuss (Howard Hughes Medical School, Pennsylvania, PA), Robin Reed (Harvard University, Boston, MA), Ian Goodfellow (Imperial College, London), Simon Morley (University of Sussex), John Barr, Mark Harris (University of Leeds) and Gary Hayward (Johns Hopkins, Baltimore, MD) for providing reagents and to Simon White (University of Leeds) for his technical assistance. This work was supported in part by Wellcome Trust (086168/Z08/Z) and BBSRC (BB/F012101/1) project grants. AW is the recipient of a BBSRC Research Development Fellowship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aoyagi M, Gaspar M, Shenk TE (2010) Human cytomegalovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4EBP1. Proc Natl Acad Sci USA 107: 2640–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Unterholzner L, Guttler T, Izaurralde E, Conti E (2004) Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep 5: 304–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Colgan KJ, Whitehouse A (2008a) Herpesvirus saimiri ORF57: a post-transcriptional regulatory protein. Front Biosci 13: 2928–2938 [DOI] [PubMed] [Google Scholar]
- Boyne JR, Colgan KJ, Whitehouse A (2008b) Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog 4: e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Whitehouse A (2006) Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc Natl Acad Sci USA 103: 15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Whitehouse A (2009) Nucleolar disruption impairs Kaposi's sarcoma-associated herpesvirus ORF57-mediated nuclear export of intronless viral mRNAs. FEBS Lett 583: 3549–3556 [DOI] [PubMed] [Google Scholar]
- Braddock M, Muckenthaler M, White MR, Thorburn AM, Sommerville J, Kingsman AJ, Kingsman SM (1994) Intron-less RNA injected into the nucleus of Xenopus oocytes accesses a regulated translation control pathway. Nucleic Acids Res 22: 5255–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Shaw PJ (2008) The role of the plant nucleolus in pre-mRNA processing. Curr Top Microbiol Immunol 326: 291–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot V (1987) Introns increase gene expression in cultured maize cells. Genes Dev 1: 1183–1200 [DOI] [PubMed] [Google Scholar]
- Chang Y, Moore PS (1996) Kaposi's sarcoma (KS)-associated herpesvirus and its role in KS. Infect Agents Dis 5: 215–222 [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF (2007) The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chen IH, Li L, Silva L, Sandri-Goldin RM (2005) ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J Virol 79: 3949–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R (2006) Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127: 1389–1400 [DOI] [PubMed] [Google Scholar]
- Colgan K, Boyne JR, Whitehouse A (2009) Uncoupling of hTREX demonstrates that UAP56 and hTHO-complex recruitment onto herpesvirus saimiri intronless transcripts is required for replication. J Gen Virol 90 (Part 6): 1455–1460 [DOI] [PubMed] [Google Scholar]
- Diem MD, Chan CC, Younis I, Dreyfuss G (2007) PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol 14: 1173–1179 [DOI] [PubMed] [Google Scholar]
- Du TG, Jellbauer S, Muller M, Schmid M, Niessing D, Jansen RP (2008) Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep 9: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine-Rodriguez EC, Knipe DM (2008) Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J Virol 82: 3538–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine-Rodriguez EC, Taylor TJ, Olesky M, Knipe DM (2004) Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology 330: 487–492 [DOI] [PubMed] [Google Scholar]
- Gao SJ, Deng JH, Zhou FC (2003) Productive lytic replication of a recombinant Kaposi's sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J Virol 77: 9738–9749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin DJ, Whitehouse A (2001) A gamma-2 herpesvirus nucleocytoplasmic shuttle protein interacts with importin alpha 1 and alpha 5. J Biol Chem 276: 19905–19912 [DOI] [PubMed] [Google Scholar]
- Gould F, Harrison SM, Hewitt EW, Whitehouse A (2009) Kaposi's sarcoma-associated herpesvirus RTA promotes degradation of the Hey1 repressor protein through the ubiquitin proteasome pathway. J Virol 83: 6727–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R, Whitehouse A (2007) Herpesvirus saimiri episomal persistence is maintained via interaction between open reading frame 73 and the cellular chromosome-associated protein MeCP2. J Virol 81: 4021–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudikote JP, Imam JS, Garcia RF, Wilkinson MF (2005) RNA splicing promotes translation and RNA surveillance. Nat Struct Mol Biol 12: 801–809 [DOI] [PubMed] [Google Scholar]
- Hall KT, Giles MS, Calderwood MA, Goodwin DJ, Matthews DA, Whitehouse A (2002) The herpesvirus saimiri open reading frame 73 gene product interacts with the cellular protein p32. J Virol 76: 11612–11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M, Coates K (1993) Identification of cellular proteins that bind to the human immunodeficiency virus type 1 nef gene product in vitro: a role for myristylation. J Gen Virol 74 (Part 8): 1581–1589 [DOI] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Walsh MJ, Snijders AP, Chang CT, Jones R, Ponting CP, Dickman MJ, Wilson SA (2009) UIF, a new mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr Biol 19: 1918–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE (2008) Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ (2003) Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12: 5–14 [DOI] [PubMed] [Google Scholar]
- Koffa MD, Clements JB, Izaurralde E, Wadd S, Wilson SA, Mattaj IW, Kuersten S (2001) Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J 20: 5769–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E (2007) Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Larralde O, Smith RW, Wilkie GS, Malik P, Gray NK, Clements JB (2006) Direct stimulation of translation by the multifunctional herpesvirus ICP27 protein. J Virol 80: 1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR (2003) Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J (2008) SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133: 303–313 [DOI] [PubMed] [Google Scholar]
- Majerciak V, Kruhlak M, Dagur PK, McCoy JP Jr, Zheng ZM (2010) Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J Biol Chem 285: 11297–11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerciak V, Yamanegi K, Nie SH, Zheng ZM (2006) Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J Biol Chem 281: 28365–28378 [DOI] [PubMed] [Google Scholar]
- Malik P, Blackbourn DJ, Clements JB (2004) The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J Biol Chem 279: 33001–33011 [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Wassarman KM, Wolffe AP (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J 17: 2107–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekorchuk M, Han Z, Hsieh TT, Swaminathan S (2007) Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J Virol 81: 9990–9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ (2004) Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev 18: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sabatini DM (2005) eIF3: a connecTOR of S6K1 to the translation preinitiation complex. Mol Cell 20: 655–657 [DOI] [PubMed] [Google Scholar]
- Reed R, Cheng H (2005) TREX, SR proteins and export of mRNA. Curr Opin Cell Biol 17: 269–273 [DOI] [PubMed] [Google Scholar]
- Ricci EP, Mure F, Gruffat H, Decimo D, Medina-Palazon C, Ohlmann T, Manet E (2009) Translation of intronless RNAs is strongly stimulated by the Epstein-Barr virus mRNA export factor EB2. Nucleic Acids Res 37: 4932–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JW, Zhou Q, Green TJ, Barr JN, Luo M (2006) Purification, crystallization and preliminary X-ray crystallographic analysis of the nucleocapsid protein of Bunyamwera virus. Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 361–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo V, Gupta AK, Swaminathan S (2001) Epstein-Barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J Virol 75: 6033–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Caceres JF (2004) A novel role for shuttling SR proteins in mRNA translation. Genes Dev 18: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant A, Gruffat H, Manet E (2008) The Epstein-Barr virus (EBV) protein EB is an mRNA export factor essential for virus production. Front Biosci 13: 3798–3813 [DOI] [PubMed] [Google Scholar]
- Swaminathan S (2005) Post-transcriptional gene regulation by gamma herpesviruses. J Cell Biochem 95: 698–711 [DOI] [PubMed] [Google Scholar]
- Williams BJ, Boyne JR, Goodwin DJ, Roaden L, Hautbergue GM, Wilson SA, Whitehouse A (2005) The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem J 387: 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SJ, Tsao EH, Webb BL, Ye H, Dalton-Griffin L, Tsantoulas C, Gale CV, Du MQ, Whitehouse A, Kellam P (2007) X box binding protein XBP-1s transactivates the Kaposi's sarcoma-associated herpesvirus (KSHV) ORF50 promoter, linking plasma cell differentiation to KSHV reactivation from latency. J Virol 81: 13578–13586 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.