Abstract
MicroRNA (miRNA) biogenesis proceeds from a primary transcript (pri-miRNA) through the pre-miRNA into the mature miRNA. Here, we identify a role of the Caenorhabditis elegans nuclear export receptor XPO-1 and the cap-binding proteins CBP-20/NCBP-2 and CBP-80/NCBP-1 in this process. The RNA-mediated interference of any of these genes causes retarded heterochronic phenotypes similar to those observed for animals with mutations in the let-7 miRNA or core miRNA machinery genes. Moreover, pre- and mature miRNAs become depleted, whereas primary miRNA transcripts accumulate. An involvement of XPO-1 in miRNA biogenesis is conserved in Drosophila, in which knockdown of Embargoed/XPO-1 or its chemical inhibition through leptomycin B causes pri-miRNA accumulation. Our findings demonstrate that XPO-1/Emb promotes the pri-miRNA-to-pre-miRNA processing and we propose that this function involves intranuclear transport and/or nuclear export of primary miRNAs.
Keywords: CBP20/NCBP-2, CBP80/NCBP-1, microRNA biogenesis, nuclear export, XPO1/CRM1/Embargoed
Introduction
According to the current model of miRNA biogenesis, miRNAs are transcribed by RNA polymerase II as capped and polyadenylated primary miRNAs (pri-miRNA) of several hundred or thousands of nucleotides in length (Bracht et al, 2004; Cai et al, 2004; Lee et al, 2004). The microprocessor complex, composed of Drosha and DGCR8 (DRSH-1 and PASH-1, respectively, in Caenorhabditis elegans), cleaves the pri-miRNAs in the nucleus to generate pre-miRNAs, characterized by their hairpin structures and size of ∼70 nt (Denli et al, 2004; Gregory et al, 2004; Han et al, 2004; Landthaler et al, 2004). Subsequently, cleavage of the pre-miRNA by the cytoplasmic RNase Dicer (DCR-1) releases the mature miRNA (Grishok et al, 2001; Hutvágner et al, 2001; Ketting et al, 2001), which is loaded into a functional miRNA-induced silencing complex (miRISC) containing an Argonaute (AGO; ALG-1 and ALG-2 in C. elegans) protein (Grishok et al, 2001; Hutvágner et al, 2004) and a GW182 protein (AIN-1 and AIN-2; Ding et al, 2005; Liu et al, 2005; Rehwinkel et al, 2005; Zhang et al, 2007) at its core.
In vertebrates and flies, Exportin-5 (Exp5) connects the two nucleolytic processing steps by exporting the nuclear pre-miRNA into the cytoplasm for further cleavage by Dicer (Yi et al, 2003; Bohnsack et al, 2004; Lund et al, 2004). However, although the miRNA biogenesis machinery is generally conserved in C. elegans, the nematode genome contains no orthologue of Exp5 (Supplementary Figure S1 and see, Bohnsack et al, 2004; Murphy et al, 2008).
The depletion of several components of the miRNA core machinery in C. elegans results in developmental phenotypes that resemble those seen upon the loss of the let-7 miRNA, such that these phenotypes provided the first indication for a function of DCR-1, ALG-1/2, and AIN-1/2 in the miRNA pathway (Grishok et al, 2001; Ketting et al, 2001; Ding et al, 2005; Zhang et al, 2007). These so-called heterochronic phenotypes are particularly apparent in a subset of skin cells, the seam cells. In wild-type animals, these cells exit the cell cycles at the larval-to-adult (L/A) transition, fuse into a syncytium, and contribute to the formation of a specific cuticular structure, the adult alae. In let-7 mutant and miRNA pathway mutant animals, cell cycle exit and/or cell differentiation fail, resulting in extra seam cell divisions, delay, or lack of formation of the seam cell syncytium and/or the alae. Moreover, on more complete loss of let-7 or general miRNA activity, animals die by vulval bursting at the L/A transition.
In this study, we show that depletion of the nuclear export receptor XPO-1 or either subunit of the nuclear cap-binding complex (CBC), NCBP-1/CBP-80 and NCBP-2/CBP-20, causes vulval bursting and heterochronic phenotypes in C. elegans. This is caused by a defect in the miRNA biogenesis at the level of primary miRNAs, and a similar function in miRNA biogenesis is also observed for the Drosophila XPO-1 orthologue Embargoed. We propose that XPO-1, possibly in conjunction with the CBC, mediates the intranuclear transport and/or nuclear export of primary miRNAs.
Results
xpo-1 is a heterochronic gene in C. elegans
Exp5 is a member of the importin β-superfamily that mediates the nuclear export of pre-miRNAs in flies and mammals. As C. elegans lacks an Exp5 orthologue (Supplementary Figure S1 and see, Bohnsack et al, 2004; Murphy et al, 2008), we were interested in testing whether other nuclear export receptors support miRNA biogenesis in C. elegans. The CSE1L/CAS orthologue, XPO-2, has previously been identified as a suppressor of the let-7(n2853) mutation (Ding et al, 2008), indicating a negative role—by genetic criteria—in miRNA function, and thus arguing against a miRNA biogenesis-promoting activity. We therefore investigated the other two C. elegans exportins, XPO-1 and XPO-3. The exportin XPO-1 is the orthologue of yeast and human CRM1/XPO1, which mediates nuclear export of the spliceosomal U snRNAs (Hutten and Kehlenbach, 2007), whereas XPO-3 is the orthologue of human Exportin-t and yeast Los1p, which mediates tRNA nuclear export (Großhans et al, 2000). xpo-1 has also previously been identified as one among >200 genes, depletion of which enhanced vulval bursting for a weak let-7 allele in an RNAi-sensitized, eri-1 mutant background, although a function in miRNA biogenesis remained elusive (Parry et al, 2007).
To obtain evidence for a possible function of either transport receptor in miRNA biogenesis or function, we exposed wild-type animals to RNAi by feeding against xpo-1, xpo-3 or a control plasmid and scored animal survival and alae defects in young adults. To avoid sterility or embryonic lethality phenotypes, we initiated RNAi on synchronized first larval (L1) stage animals. Animals treated with mock RNAi exhibited wild-type vulvae and alae (Figures 1A, D and 2A, F). By contrast, depletion of the C. elegans Argonaute, alg-1, caused both vulval bursting and alae defects (Figures 1B, D and 2B, F).
Figure 1.
RNAi against xpo-1, ncbp-1/cbp-80, or ncbp-2/cbp-20 causes animals to die by vulval bursting. Unlike (A) the healthy control animals, (B) alg-1(RNAi) and (C) xpo-1(RNAi) adults have protruding vulvae and often die by bursting through the vulva. (D) This phenotype is also penetrant on depletion of cbp-20 or cbp-80, whereas RNAi against xpo-3 or phax-1 has no effect (independent experiments n⩾2, each n⩾165 animals). ‘Control' in this and subsequent figures denotes animals that were fed bacteria carrying the insertless L4440 parental RNAi vector. Error bars=s.e.m. Scale bars are 20 μm.
Figure 2.
xpo-1(RNAi), ncbp-1/cbp-80(RNAi), and ncbp-2/cbp-20(RNAi) cause alae defects. (A) Control animals display strong and complete alae (arrows), whereas (B–F) alae in animals treated with RNAi as indicated are broken or absent altogether (brackets indicate alae breaks; for (F), independent experiments n⩾3, each n⩾22 animals for control, xpo-1(RNAi), cbp-20(RNAi) and cbp-80(RNAi); for alg-1(RNAi) one experiment with 34 animals). Residual alae in the mutant animals (arrows) are much weaker than in the control. Error bars=s.e.m. Scale bars are 20 μm.
Animals exposed to xpo-3(RNAi) appeared wild type (Figures 1D and 2F), although RT–PCR confirmed efficient mRNA depletion (data not shown). This finding suggests that under our experimental conditions sufficient XPO-3 protein might still be available to promote tRNA nuclear export. Alternatively, as in yeast and Drosophila, in which Exp-t orthologues are non-essential or not encoded in the genome, respectively (Supplementary Figure S1 and see, Großhans et al, 2000; Shibata et al, 2006), partially redundant tRNA nuclear export pathways might compensate for the loss of XPO-3 in larvae.
By contrast, xpo-1(RNAi) caused the characteristic vulval bursting and alae break phenotypes (Figures 1C, D and 2C, F), previously observed for depletion of other core components of the C. elegans miRNA pathway (Grishok et al, 2001; Ketting et al, 2001; Denli et al, 2004), including alg-1 (Figures 1B, D and 2B, F). Surviving animals were sterile for reasons that we have not investigated.
A more detailed analysis confirmed that xpo-1(RNAi) caused true heterochronic phenotypes. Thus, xpo-1(RNAi) animals displayed unfused seam cells at the young adult stage, when seam cells in wild-type animals would be fused (Supplementary Figure S2). Moreover, the number of seam cells in young adult xpo-1(RNAi) animals was increased relative to mock RNAi animals (Supplementary Figure S3), and this was due to extra seam cell division in the young adult stage and not cell-fate transformations or extra cell divisions during larval stages (Supplementary Figure S3).
In summary, we have shown that xpo-1(RNAi) phenocopies multiple aspects of the let-7 heterochronic phenotype, including lethality and defects in seam cell differentiation and proliferation control, establishing xpo-1 as a bona fide heterochronic gene.
XPO-1 is required for normal let-7 accumulation
The extensive resemblance of xpo-1(RNAi) and let-7 mutant phenotypes is consistent with a function of XPO-1 in let-7 biogenesis. In accord with this idea, we also observed that xpo-1(RNAi)-induced vulval bursting was largely suppressed by a loss-of-function mutation in the let-7 target lin-41, which also suppresses vulval bursting of let-7 mutant animals (data not shown). However, Parry et al (2007) had previously examined whether depletion of xpo-1 affected mature and/or pre-let-7 levels and failed to find any evidence to support this idea. We wished to re-examine this issue in the light of the stronger vulval bursting phenotypes that we observed in comparison to Parry et al (2007), who required xpo-1 depletion in the eri-1(mg366); let-7(mg279) background to observe significant bursting. Indeed, when we examined the abundance of mature let-7 in xpo-1(RNAi) animals, we observed a ∼50% decrease relative to control RNAi animals (Figure 3A). This finding supports a possible function of XPO-1 in let-7 biogenesis.
Figure 3.
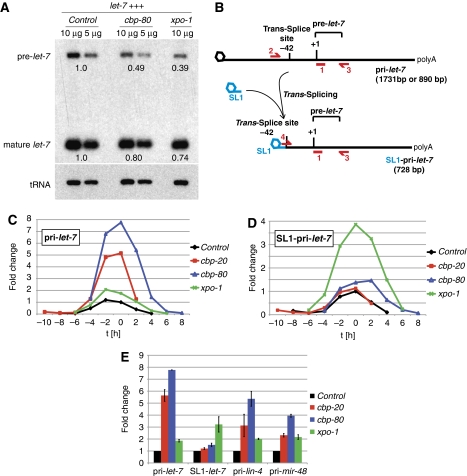
Depletion of xpo-1, cbp-20, or cbp-80 causes a widespread decrease in mature miRNA, but not in mature mirtron levels. (A–D) Northern blots using total RNA from synchronized late L4 stage animals exposed to RNAi as indicated. Oligonucleotides complementary to the indicated mature miRNAs or tRNAGly(TCC) were used. To facilitate a comparison, two different amounts of total RNA were loaded in (B) as indicated. (D) The accumulation of the mirtron mir-62 is not affected by the depletion of xpo-1 and cbp-80 (same membrane as in (C), re-probed and tRNA shown again for comparison). Numbers represent the quantification by phosphoimager, normalized to tRNAGly levels.
The cap-binding complex is a potential co-factor of XPO-1 in let-7 biogenesis
To mediate nuclear export of U snRNAs, vertebrate XPO1/CRM1 functions with three adaptor proteins—the cap-binding complex (CBC) comprising CBP20 and CBP80, and the PHAX protein (Izaurralde et al, 1995; Ohno et al, 2000). More recently, CBC was shown to be required for efficient miRNA accumulation in plants and to affect pre- and pri-miRNA levels in flies and mammals, respectively (Gregory et al, 2008; Kim et al, 2008; Laubinger et al, 2008; Gruber et al, 2009; Sabin et al, 2009). Finally, both ncbp-2/cbp-20 (F26A3.2) and ncbp-1/cpb-80 (F37E3.1) caused vulval bursting when depleted in eri-1(mg366); let-7(mg279) animals (Parry et al, 2007). Thus, to test whether PHAX and CBC also function in C. elegans miRNA biogenesis, we depleted them by RNAi.
Animals exposed to RNAi against phax-1 (Y71H2B.2) displayed neither vulval bursting nor alae defects (Figures 1D and 2F) although phax-1 mRNA was efficiently (∼70%) depleted as determined by RT–PCR (data not shown). Although it remains possible that residual PHAX-1 suffices for function, these data argue against a major role of PHAX-1 in let-7 biogenesis. In contrast, and similar to xpo-1(RNAi), depletion of cbp-20 or cbp-80 by RNAi caused penetrant alae defects and vulval bursting (Figures 1D and 2D–F); the latter phenotype being suppressed by the lin-41(ma104) mutation (data not shown). Finally, the levels of the mature let-7 miRNA were significantly reduced on CBC depletion (Figure 3B), supporting the idea that CBC, like XPO-1 might be involved in miRNA biogenesis. As a parsimonious explanation, we propose that CBP-20 and CBP-80 function together with XPO-1 in the nuclear export of let-7.
XPO-1 and the CBC are widely required for miRNA accumulation
Mutations in the core miRNA machinery cause let-7-like phenotypes even for factors generally required for miRNA function (Figures 1B, D and 2B, F; Grishok et al, 2001; Denli et al, 2004). Therefore, we tested the possibility that XPO-1 and CBC are required for the accumulation of other miRNAs. As depletion of cbp-20 caused less penetrant developmental phenotypes than RNAi against cbp-80, we focused our analysis on xpo-1 and cbp-80. We noted that these differences in cbp-20 and cbp-80 depleted animals occurred although relative depletion efficiency was comparable for both mRNAs (Supplementary Figure S4A), possibly indicating that differences in protein stability or differences in protein abundance already before depletion might render CBP-20 more refractory to efficient depletion by RNAi.
We examined the abundance of lin-4, mir-75, mir-77, and mir-237, four larvally expressed miRNAs (Lim et al, 2003), and found that their levels were decreased on xpo-1 and cbp-80 knockdown (Figure 3C), although the effect on lin-4 was modest (but see below), presumably due to its early expression in L1. We conclude that XPO-1/CRM1 and CBC are required for the biogenesis of many C. elegans miRNAs, including let-7, providing a molecular explanation for the developmental phenotypes.
XPO-1 and CBC act upstream of mature miRNA
If XPO-1 and CBC act directly and jointly in miRNA biogenesis, the involvement of CBC might suggest a function linked to a capped miRNA precursor, that is, pri-miRNA, rather than the uncapped pre- or mature miRNAs. To test this possibility, we examined the abundance of let-7 biogenesis intermediates. Although low pre-let-7 levels in wild-type animals preclude efficient detection, depletion of dcr-1 yields a readily detectable accumulation of pre-let-7 (Grishok et al, 2001; Ketting et al, 2001). Our failure to detect pre-let-7 on xpo-1 or CBC depletion thus ruled out a significant accumulation (data not shown). Indeed, when we overexpressed let-7 ∼five-fold from an integrated DNA array (Weidhaas et al, 2007 and Supplementary Figure S5A), depletion of xpo-1 or cbp-80 decreased pre-let-7 levels relative to control RNAi (Figure 4A), indicating that xpo-1 and cbp-80 function upstream of Dicer-mediated pre-let-7 processing.
Figure 4.
Depletion of XPO-1 or CBC reduces pre-let-7 levels and increases pri-miRNA accumulation. (A) Pre-let-7 levels are reduced in let-7-overexpressing animals (let-7+++) exposed to xpo-1(RNAi) or cbp-80(RNAi). Total amounts of RNA were loaded as indicated. (B) Schematic representation (not to scale) of the primary (pri-let-7) and trans-spliced SL1-pri-let-7 transcripts. The positions of oligonucleotides used for RT–qPCR and Northern blot are highlighted in red. Mature and pre-let-7 are detected by probe (1); pri-let-7 by primers (2) and (3); SL1-pri-let-7 by primers (3) and (4). (C) The levels of pri–let-7 and (D) SL1-pri-let-7 change dynamically during the L4 stage and are elevated in the xpo-1(RNAi), cbp-80(RNAi), and cbp-20(RNAi) animals. Time (x-axis) is relative to the peak of lin-42 mRNA levels in L4, which we defined as t=0 h (see Supplementary data). Pri- and SL1-pri-let-7-levels were arbitrarily set to 1 in the control RNAi strain at t=0 h. The experiment was performed in biological duplicate, a representative example is shown. (E) Pri-let-7, SL1-pri-let-7, pri-lin-4, and pri-mir-48 levels were determined in biological duplicates for the time points of peak lin-42 expression. The average fold change, compared with the RNAi control, is shown. Error bars indicate actual measurements.
We used RT–qPCR to examine the accumulation of pri-let-7 and C. elegans-specific SL1-pri-let-7, which is derived from the pri-miRNA by trans-splicing (Figure 4B; Bracht et al, 2004). Unlike for pre-let-7 and mature let-7, we observed that levels of these potential export substrates did not decline in xpo-1-, cbp-20-, or cbp-80-depleted animals but instead increased (data not shown). However, the extent of accumulation varied substantially among different experiments. As pri-let-7 expression is dynamic during the L4 stage (A Pasquinelli, personal communication), we addressed the possibility that slightly divergent staging of the animals might account for this variability among the different trials. We performed a time-course analysis using lin-42, an mRNA expression of which peaks once during each larval stage (Jeon et al, 1999), as a reference (Supplementary Figure S5B; Materials and methods section). Both pri-let-7 and SL1-pri-let-7 were dynamically expressed during the L4 stage, starting from low levels, peaking around the time of maximum lin-42 levels, and subsequently declining (Figure 4C and D). This dynamic was unchanged on xpo-1, cbp-20, or cbp-80 depletion, but the levels of both transcripts were consistently increased at all time points relative to the control animals (Figure 4C–E). Although xpo-1(RNAi) enhanced the accumulation of SL1-let-7 particularly strongly, cbp-20/-80(RNAi) preferentially affected pri-let-7 accumulation.
Next, we extended our study to the primary transcripts of lin-4, mir-237, mir-48, and mir-77, none of which has been reported to undergo trans-splicing, and for none of which we could amplify a trans-spliced product using RT–PCR with an SL1-specific primer and a pri-miRNA specific primer (data not shown). We observed that all four pri-miRNAs accumulated on xpo-1(RNAi), cbp-20(RNAi), and cbp-80(RNAi) relative to the control (Figure 4E and data not shown), suggesting that XPO-1 and the CBC act on the primary transcripts, and confirming that XPO-1 and CBC are widely required for miRNA biogenesis.
A mirtron miRNA is not affected by depletion of xpo-1 or CBC
To further test the idea that XPO-1 and CBC act on pri-miRNAs, we examined accumulation of mir-62. mir-62 belongs to the mirtron subclass of miRNAs, which reside in short introns of host mRNA genes, from which the—uncapped—pre-miRNA is released through nuclear mRNA splicing, bypassing processing by Drosha (Okamura et al, 2007; Ruby et al, 2007). We expected that reduced levels of XPO-1 and CBP-80 would not affect the accumulation of this mature miRNA if these factors acted on primary miRNAs. Indeed, depletion of xpo-1 or cbp-80 failed to decrease the levels of the mature mir-62, as predicted by our model (Figure 3D). In addition, this result indicates that splicing activity is not appreciably impaired in the xpo-1(RNAi) and cbp-80(RNAi) animals, as mature mirtron accumulation requires splicing.
Expression of miRNA pathway components is not affected by XPO-1 or CBC depletion
The fact that a mirtron miRNA accumulates normally in the presence of reduced XPO-1 or CBC levels suggests that not only splicing but also dicing and Argonaute binding are not adversely affected. However, to directly examine whether XPO-1 or CBC might affect the levels of miRNA pathway components, we examined the level of Dicer (DCR-1) using an antibody against the endogenous protein (Duchaine et al, 2006). We also examined its mRNA levels and those of Drosha (drsh-1), Pasha (pash-1), the miRNA Argonautes (alg-1 and alg-2), and the GW182 orthologues (ain-1 and ain-2) by RT–qPCR. Our experiments revealed that none of these factors was depleted by RNAi against xpo-1 or CBC (Supplementary Figure S4B and C). Notably, we saw some elevation of Dicer protein and Drosha, Argonaute, and GW182 mRNAs consistent with the suggestion of widespread autoregulation of miRNA pathway components by miRNAs (Zisoulis et al, 2010). Regardless of the cause of this effect, these results argue against an impairment of miRNA activity through depletion of core miRNA pathway genes on xpo-1 or CBC knockdown. These data thus further support the idea that XPO-1 and CBC have a direct role in supporting miRNA biogenesis at the level of pri-miRNA.
Emb, the Drosophila XPO-1 orthologue, also regulates pri-miRNA processing
The miRNA biogenesis pathway is well conserved in diverse organisms and recent data show a requirement of the CBC for efficient miRNA processing and/or activity in plants, mammals, and flies (Gregory et al, 2008; Kim et al, 2008; Laubinger et al, 2008; Gruber et al, 2009; Sabin et al, 2009) (JSY and ECL, unpublished data). In contrast, similar data are not available for XPO-1.
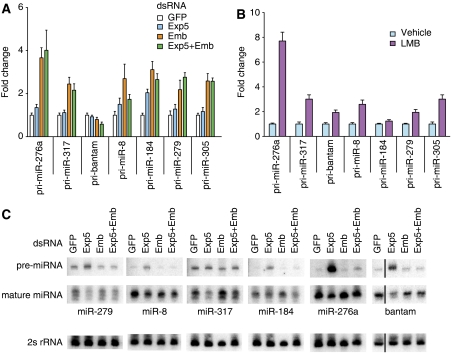
To elucidate if Embargoed (Emb), the Drosophila XPO1 orthologue, has a function in the biogenesis of miRNAs, we depleted it in S2 cells by soaking of Emb dsRNA. As in C. elegans, we observed an accumulation of several pri-miRNAs when Emb was depleted (Figure 5A, Supplementary Figure S6A). We confirmed the specificity of this effect by obtaining comparable results when blocking Emb activity with Leptomycin B (LMB; Figure 5B). Leptomycin B specifically inhibits XPO1/CRM1 by binding covalently to a conserved cysteine residue (Kudo et al, 1999), and this modification prevents substrate binding by occupying the substrate-binding site (Dong et al, 2009). Within as little as 2 h, LMB treatment caused an accumulation (at least two-fold) of several pri-miRNAs relative to the vehicle-treated control (Figure 5B). Thus, acute and chronic depletion of Emb activity causes pri-miRNA accumulation, with the rapidity of the effect arguing for a direct involvement of Emb in miRNA biogenesis.
Figure 5.
Accumulation of pri-miRNAs on depletion of Emb/Crm1 activity in Drosophila. (A) Depletion of Emb by RNAi increases the accumulation of several pri-miRNAs. Fold changes are relative to the cells soaked with control dsRNA (GFP) and normalized against pre-rp49. Knockdown efficiencies are depicted in Supplementary Figure S6. (B) Inhibition of Emb activity by LMB (25 ng/ml) confirms the increased accumulation of pri-miRNAs. Abundance of pri-miRNAs was analysed after 2-h treatment. (C) Northern blots using total RNA from cells exposed to dsRNA as indicated. The accumulation of pre-miRNAs on depletion of Exp5 is suppressed when Emb is depleted simultaneously. For bantam, non-adjacent lanes of a single autoradiograph are shown.
Although some mature miRNAs seemed to be moderately changed on Emb depletion, the effect was generally weak (Figure 5C), as previously observed on CBC depletion in flies (Sabin et al, 2009). Thus, it seems that compensatory effects downstream in miRNA biogenesis or turnover can compensate for the decreased pri-miRNA processing. Alternatively, incomplete Emb depletion might sustain sufficient export capacity in cells exposed to RNAi against Emb. Finally, Emb might only have a minor or partially redundant role in miRNA biogenesis in Drosophila.
To test whether a redundant function was performed by the pre-miRNA export receptor, Exportin-5, we co-depleted Emb and Exp5. Surprisingly, we found that the double depletion did not decrease mature miRNA levels beyond what was seen with Exp5 single depleted cells. Nonetheless more surprisingly, RNAi against Emb suppressed the accumulation of the pre-miRNA that occurs in an Exp5 single knockdown (Figure 5C). Although these experiments involve partial knockdown and not null mutations, precluding strong statements about epistasis, these findings suggest that in Drosophila Emb acts upstream of, rather than in parallel to, Exportin-5.
Discussion
We have shown in this study that C. elegans xpo-1, cbp-20, and cbp-80 are heterochronic genes that are required for proper execution of the L/A switch mediated by let-7. We have further observed that all three factors are important for the accumulation of miRNAs, including let-7, providing a molecular explanation for the developmental phenotypes. We note that a previous study failed to observe a significant decrease in let-7 on xpo-1 depletion (Parry et al, 2007). However, as xpo-1(RNAi)-induced vulval bursting in that study required the sensitized eri-1(mg366); let-7(mg279) background, less efficient xpo-1 depletion than under our experimental conditions seems a likely cause of the discrepancy (Gregory et al, 2008).
The fact that depletion of xpo-1 and CBC both decreased mature and pre-miRNA levels, but increased pri-miRNA levels, points to their function in miRNA biogenesis at a step upstream of the pre-miRNA, that is, at the level of pri-miRNAs. Formally, we cannot rule out that these functions might differ for XPO-1 and CBC. However, as XPO-1, CBP-20, and CBP-80 complexes are known in vertebrates (Ohno et al, 2000), the shared molecular and developmental phenotypes seen in the C. elegans RNAi mutants suggests that they also function as a complex in the C. elegans miRNA biogenesis pathway. As C. elegans lacks the canonical pre-miRNA export receptor, Exp5, a function in miRNA nuclear export is a strong possibility.
Pri-miRNA nuclear export would require cytoplasmic processing of the pri-miRNA (generally considered a nuclear event), and it is therefore of particular interest that CBC and Drosha have recently been shown to co-immunoprecipitate in flies and humans (Gruber et al, 2009; Sabin et al, 2009), suggesting the possibility of a large shuttling complex that contains the pri-miRNA processing activity. Processing of pri-miRNAs in C. elegans might then occur at, or during transit through, the nuclear pore. Nonetheless, as Drosha localization in C. elegans is currently unknown, and localization using various GFP-tagged Drosha transgenes has yielded inconsistent results (IB and HG, unpublished data), alternative explanations remain possible.
Previous studies on fly and human CBC reported a function in miRNA biogenesis that involved an interaction with the serrate homologous protein ARS2 (Gruber et al, 2009; Sabin et al, 2009). However, ARS2 is only present in proliferating cells, and impairs the accumulation of a specific subset of miRNAs (Gruber et al, 2009). If CBC functioned in miRNA biogenesis exclusively through its interaction with ARS2, one would predict a similarly specific function. As the effect of CBC depletion on mature miRNA levels has not been reported for humans and only for one miRNA in flies—bantam, the levels of which remained unchanged (Sabin et al, 2009)—this possibility remains to be addressed. However, the fact that depletion of E01A2.2, the C. elegans ARS2 homologue, does not result either in vulval bursting or in alae defects (IB and HG, unpublished data), and that all miRNAs that we had investigated were affected by the depletion of CBC, suggests that in C. elegans some, or possibly all, CBC functions in miRNA biogenesis are independent of ARS2. Consistent with a difference in CBC function between C. elegans and humans or flies, Gruber et al, 2009 and Sabin et al (2009) also observed a reduction of pri-miRNA levels on depletion of CBC, whereas we observed that C. elegans pri-miRNAs accumulate in this situation.
It thus seems possible that CBC has a conserved yet diverging function in miRNA biogenesis in different organisms, and this also seems to be true for XPO-1: our studies of Drosophila Emb reveal that this XPO-1 orthologue also regulates the miRNA biogenesis at the step of pri-miRNA processing, although Drosophila does harbour a miRNA export receptor, Exp5. However, the fact that Emb depletion does not enhance Exp5 phenotypes at the level of mature miRNA accumulation, but does suppress pre-miRNA accumulation, suggests that Emb functions upstream of, rather than in parallel to, Exp5. One possible function could be intranuclear transport of the pri-miRNA, as previously demonstrated for U3 snoRNA in human cells (Boulon et al, 2004).
While this paper was under preparation, CRM1/XPO-1 was reported to regulate the nuclear–cytoplasmic localization of mature miRNAs in cultured mammalian cells, suggesting that this nuclear export receptor might additionally modulate miRNA activity after miRNA biogenesis has been completed (Castanotto et al, 2009). It is unclear whether this function would be conserved in C. elegans. However, if it were, it would be insufficient to explain several of our observations, that is, the accumulation of pri-miRNA, the depletion of pre-miRNA, and the lack of an effect on the levels of the mir-62 mirtron. Nonetheless, we cannot rule out that beyond the functions in pri-miRNA biogenesis that we describe here, XPO-1 would additionally affect mature miRNA localization.
When considering the possibility of conserved, yet differing functions in miRNA biogenesis, one striking feature of the C. elegans miRNA pathway is that many or all of its canonical miRNAs are expressed from their own promoters (Supplementary data and Martinez et al, 2008), whereas a large fraction of vertebrate miRNAs are ‘intronic' such that nuclear Drosha processing releases them from their host genes (Kim and Kim, 2007). It is tempting to speculate that a varying dependence on CBC for miRNA biogenesis in C. elegans and in humans might explain these divergent gene organization patterns, with a more general requirement in C. elegans necessitating the production of capped transcripts, from ‘intergenic' miRNA loci.
Materials and methods
C. elegans strains
C. elegans strains used were: wild-type N2; MT7626: let-7(n2853) (Reinhart et al, 2000); CT19: N2;zaIs3[let-7(+) myo-3∷gfp] (Weidhaas et al, 2007); him-5;[ajm-1∷gfp, rol-6]; JR672: N2;wIs54[scm∷gfp] (Koh and Rothman, 2001); and GR1434: wIs54[scm∷gfp]V;let-7(n2853) (Hayes et al, 2006).
The C. elegans cap-binding complex and PHAX-1
Using reciprocal BLAST search, we identified F26A3.2 (ncbp-2) and F37E3.1 (ncbp-1) as the closest CBP20 and CBP80 homologues, respectively, in C. elegans (data not shown). The genes encoding these proteins are named ncbp-1 (CBP80) and ncbp-2 (CBP20), following the human nomenclature. For clarity, we used cbp-20/-80 and CBP-20/-80 throughout the text when referring to gene and protein, respectively. A PHAX homologue has already been identified previously (Ohno et al, 2000). We verified that this gene, Y71H2B.2, was indeed the closest PHAX homologue in C. elegans (data not shown) and named it phax-1.
RNAi and RNAi constructs
The RNA-mediated interference was performed by feeding, starting with synchronized L1 larvae. ‘Control' in all RNAi experiments denotes animals that were fed bacteria carrying the insertless L4440 parental RNAi vector. Appropriate developmental stages of worms were verified by vulval and gonad development using DIC optics. The RNAi constructs targeting ncbp-1/cbp-80 and ncbp-2/cbp-20 are from an RNAi library (Kamath et al, 2003). xpo-1(RNAi), xpo-3(RNAi), and phax-1(RNAi) were constructed as described in the Supplementary data.
RNA isolation and northern blot
Worms were mixed with Trizol (Invitrogen) and either ground in liquid nitrogen or freeze-thawed as described previously (Bethke et al, 2009). The RNA was extracted according to the manufacturer's instructions. Total RNA was separated on 10 or 15% PAGE–urea gels and transferred on to a membrane (Zeta-Probe GT, BioRad for UV cross-linking and Hybond-Nx, Amersham for chemical cross-linking) by wet or semidry blotting. Cross-linking was carried out either by UV irradiation plus baking or by chemical cross-linking as described previously (Pall et al, 2007). Single-stranded DNA oligonucleotides complementary to the sequence of interest were used except for let-7 and mir-62, in which an LNA-modified oligonucleotide (Exiqon) was used to facilitate detection. Probes were 5′ end-labelled with ATP-γ-[32P] and polynucleotide kinase according to standard protocols. Hybridization was carried out overnight in 4 × SSPE (0.6 M NaCl, 40 mM NaH2PO4, 4 mM EDTA), 7% SDS, 25% formamide at 37°C for the DNA oligonucleotides and in 4 × SSPE, 6% SDS, 50% formamide at 60°C (mir-62 LNA) or 65°C (let-7 LNA).
- let-7 LNA (hsa-let-7a):
5′-AACTATACAACCTACTACCTCA-3′;
- Cel-lin-4:
5′-TCACACTTGAGGTCTCAGGGA-3′;
- Cel-mir-75:
5′-AAGCCGGTTGGTAGCTTTAA-3′;
- Cel-mir-77:
5′-TGGACAGCTATGGCCTGATGAA-3′;
- Cel-mir-237:
5′-AAGCTGTTCGAGAATTCTCAGGGA-3′;
- Cel-mir-62 LNA:
5′-CTGTAAGCTAGATTACATATCA-3′;
- Cel-tRNA (tGly):
5′-GCTTGGAAGGCATCCATGCTGACCATT-3′.
For Drosophila cell culture experiments, endogenous total RNAs were isolated from dsRNA- or drug-treated S2R+ cells by Trizol (Life Technologies). Northern blot analyses were performed to analyse the pre- and mature miRNA levels: 15–20 μg total RNA per lane were separated by 12% polyacrylamide gels, transferred onto GeneScreen plus-charged nylon membranes (PerkinElmer), and probed with γ-32P-labelled LNA oligonucleotides (pre-designed by Exiqon) antisense to miR-8, miR-276a, miR-279, and miR-317 or DNA oligonucleotides (IDT) antisense to bantam (5′-AATCAGCTTTCAAAATGATCTCA-3′), miR-184 (5′-GCCCTTATCAGTTCTCCGTCCA-3′), and 2S rRNA (5′-TACAACCCTCAACCATATGTAGTCCAAGCA-3′).
RT–qPCR
The RT–qPCR analysis was performed to examine the abundance of primary miRNAs. Total RNA was diluted to 500 ng/μl and treated with DNaseI (Ambion; DNA-free) according to the manufacturer's protocol. The cDNA synthesis was performed with the ImProm-II reverse transcription system (Promega) using oligo-dT primers following the manufacturer's protocol. The resulting cDNA was used for real-time PCR with the Absolute qPCR SYBR green ROX mix (ABgene), gene-specific oligonucleotides, and an ABI Prism 7000 machine. Detailed description of normalization, time adjustments by expression of lin-42 and sequences of gene-specific oligonucleotides can be found in the Supplementary data.
The primer sets for primary transcripts of bantam, miR-8, miR-276a, miR-279, miR-305, and miR-317 were designed as previously described (Martin et al, 2009), and the primer sets for pri-miR-184, Exp5, Emb, H2B, and pre-rp49 can be found in the Supplementary data. To analyse gene expression, pri-miRNA levels were normalized to pre-rp49, and means and s.e.m. values of technical triplicates were plotted. Two additional biological replicates are shown in Supplementary Figure S6.
Knockdown of endogenous gene expression in Drosophila
To investigate the effect of knockdowns in miRNA biogenesis, we performed dsRNA soaking in S2R+ cells. The GFP dsRNA sequence was obtained from a published template (Förstemann et al, 2005). Approximately 500-bp fragments of other target genes were amplified from D. melanogaster w− genomic DNA using the primers listed below:
- Dme-Exp5-dsRNA_F_XhoI:
5′-AGAGCTCGAGCTGGAGGATCAGCTCAATCG-3′
- Dme-Exp5-dsRNA_R_XbaI:
5′-AGAGGTCTAGAGACGGAGCAGCTCGTAGAAC-3′
- Dme-emb-dsRNA_F_XhoI:
5′-CCGCTCGAGACTGGGAGACATTCATCAG-3′
- Dme-emb-dsRNA_R_XbaI:
5′-GCTCTAGAGAACCATGCTTAAACACATG-3′
The PCR-amplified fragments were cloned into the XhoI/XbaI sites of pLitmus (NEB), which contains opposing T7 promoters flanking the cloning site. The dsRNAs were synthesized from pLitmus using MEGAscript T7 Kit (Ambion).
To knock down the expression of endogenous genes, 2.5 × 106 S2R+ cells were soaked with 15 μg dsRNA in a 6-well plate for 4 days and transferred into another 6-well plate and soaked with 15 μg dsRNA for another 4 days.
Inhibition of Emb activity by leptomycin B treatment
To analyse the effect of direct inhibition of Emb protein activity on pri-miRNA level, we treated 8 × 105 S2R+ cells with 25 ng/ml LMB (Sigma) or vehicle control (70% methanol) in 12-well plates for 2 h. The treatment with a higher dosage of LMB (50 ng/ml) resulted in a similar accumulation of pri-miRNAs. In a time-course experiment, 8 × 105 S2R+ cells in 12-well plate were treated with 75 ng/ml LMB or vehicle control for 0, 1, 2, and 4 h. As pri-mir-317 level progressively increased upon treatment (data not shown), we selected the 2 h as a representative mid-level time point.
Supplementary Material
Acknowledgments
We thank Amy Pasquinelli for communicating unpublished observations and suggesting the time-course experiment. We also thank Aurora Esquela-Kerscher, Joy Alcedo, and Rafal Ciosk for critical comments on the paper; Thomas Duchaine for the anti-DCR-1 antibody; and Frank Slack and Gary Ruvkun for strains. HG is grateful to Frank Slack for support during the initial stages of this work. Research in HG's lab is supported by the SNF (Grant no. 31003A_127052) and by FMI, which is a part of the Novartis Research Foundation. The part of the study carried out by ECL's group is supported by the Sidney Kimmel Cancer Foundation, the Alfred Bressler Scholars Fund, and NIH (R01-GM083300).
Author contribution. HG conceived the project; IB and HG designed and analysed C. elegans experiments, which IB performed; JSY and ECL designed and analyzed Drosophila experiments, which JSY performed. The authors jointly wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A (2009) Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science 324: 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonné R, Bertrand E (2004) PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 16: 777–787 [DOI] [PubMed] [Google Scholar]
- Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE (2004) Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA 10: 1586–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Lingeman R, Riggs AD, Rossi JJ (2009) CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci USA 106: 21655–21659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, Han M (2005) The developmental timing regulator AIN-1 interacts with miRISCs and may target the Argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell 19: 437–447 [DOI] [PubMed] [Google Scholar]
- Ding XC, Slack FJ, Großhans H (2008) The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell Cycle 7: 3083–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Biswas A, Süel KE, Jackson LK, Martinez R, Gu H, Chook YM (2009) Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 458: 1136–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, Yates JR, Mello CC (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD (2005) Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Großhans H, Simos G, Hurt E (2000) Review: transport of tRNA out of the nucleus-direct channeling to the ribosome? J Struct Biol 129: 288–294 [DOI] [PubMed] [Google Scholar]
- Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Rawlings J, Cherry S, Ihle JN, Dreyfuss G, Thompson CB (2009) Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 138: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN (2004) The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes GD, Frand AR, Ruvkun G (2006) The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133: 4631–4641 [DOI] [PubMed] [Google Scholar]
- Hutten S, Kehlenbach RH (2007) CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol 17: 193–201 [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ, Mello CC, Zamore PD (2004) Sequence-specific inhibition of small RNA function. PLoS Biol 2: E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW (1995) A cap-binding protein complex mediating U snRNA export. Nature 376: 709–712 [DOI] [PubMed] [Google Scholar]
- Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE (1999) Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH (2008) Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol 49: 1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim VN (2007) Processing of intronic microRNAs. EMBO J 26: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Rothman JH (2001) ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128: 2867–2880 [DOI] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA 96: 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T (2004) The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D (2008) Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP (2003) The microRNAs of Caenorhabditis elegans. Genes Dev 17: 991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, Parker R, Hannon GJ (2005) A role for the P-body component GW182 in microRNA function. Nat Cell Biol 7: 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Martin R, Smibert P, Yalcin A, Tyler DM, Schaefer U, Tuschl T, Lai EC (2009) A Drosophila pasha mutant distinguishes the canonical miRNA and mirtron pathways. Mol Cell Biol 29: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, Walhout AJ (2008) Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res 18: 2005–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D, Dancis B, Brown JR (2008) The evolution of core proteins involved in microRNA biogenesis. BMC Evol Biol 8: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW (2000) PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC (2007) The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A (2007) Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res 35: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DH, Xu J, Ruvkun G (2007) A whole-genome RNAi screen for C. elegans miRNA pathway genes. Curr Biol 17: 2013–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E (2005) A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11: 1640–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S (2009) Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 138: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J, Yoneda Y (2006) Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res 34: 4711–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, Slack FJ (2007) MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res 67: 11111–11116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR, Han M (2007) Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell 28: 598–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW (2010) Comprehensive discovery of endogenous Argonaute-binding sites in Caenorhabditis elegans. Nat Struct Mol Biol 17: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.