Abstract
Integrins are αβ heterodimeric receptors that mediate divalent cation-dependent cell-cell and cell-matrix adhesion through tightly regulated interactions with ligands. We have solved the crystal structure of the extracellular portion of integrin αVβ3 at 3.1 Å resolution. Its 12 domains assemble into an ovoid “head” and two “tails.” In the crystal, αVβ3 is severely bent at a defined region in its tails, reflecting an unusual flexibility that may be linked to integrin regulation. The main inter-subunit interface lies within the head, between a seven-bladed β-propeller from αV and an A domain from β3, and bears a striking resemblance to the Gα/Gβ interface in G proteins. A metal ion–dependent adhesion site (MIDAS) in the βA domain is positioned to participate in a ligand-binding interface formed of loops from the propeller and βA domains. MIDAS lies adjacent to a calcium-binding site with a potential regulatory function.
Integrins are large heterodimeric cell surface receptors found in many animal species ranging from sponges to mammals [reviewed in (1)]. These receptors are involved in fundamental cellular processes such as attachment, migration, proliferation, differentiation, and survival. Integrins also contribute to the initiation and/or progression of many common diseases including neoplasia, tumor metastasis, immune dysfunction, ischemia-reperfusion injury, viral infections, osteoporosis, and coagulopathies [reviewed in (2, 3)]. An integrin is ~280 Å long and consists of one α (150 to 180 kD) and one β (~90 kD) subunit, both of which are type I membrane proteins. Eighteen α and eight β mammalian subunits are known, which assemble noncovalently into 24 different heterodimers. Contacts between the α and β subunits primarily involve their NH2-terminal halves [reviewed in (1)], which together form a globular head; the remaining portions form two rod-shaped tails (4–7) that span the plasma membrane.
Like other receptors, integrins transmit signals to the cell interior (so-called “outside-in” signaling), which regulate organization of the cytoskeleton, activate kinase signaling cascades, and modulate the cell cycle and gene expression [reviewed in (8)]. Unlike other receptors, however, ligand binding with integrins is not generally constitutive but is regulated to reflect the activation state of the cell. This “inside-out” regulation of integrin affinity protects the host from pathological integrin-mediated adhesion (2). Inside-out and outside-in signaling are associated with distinct conformational changes in the integrin extracellular segment. These changes vary with cell type and the state and nature of the ligand, and are modulated by divalent cations that are also required for integrinligand interaction (9–11). The structural bases of activation and regulation of integrins are unknown. Previous attempts to address this question were limited to the structure determination of the ligand-binding, ~180 –amino acid A-type domain (αA) (12) present in one-half of the integrin α subunits (13). The structural basis of ligand binding to αA-lacking integrins and its regulation are unknown. One such inte-grin is αVβ3 (CD51/CD61), a receptor important in tumor angiogenesis and metastasis, inflammation, and bone resorption [reviewed in (14)]. αVβ3 is one of the most promiscuous integrins, as it binds to multiple ligands including vitronectin, angiostatin, and osteopontin, and also serves as a receptor for several viruses such as foot-and-mouth disease virus, adenovirus, and human immunodeficiency virus [reviewed in (15)]. Here we report the crystal structure of the extracellular region of human αVβ3. The structure has both known and previously unknown domains with creative designs, and reveals unexpected quaternary arrangements that provide insights into integrin function and regulation. These studies also have major bearing on development of drugs to control angiogenesis, inflammation, viruses, and osteoporosis by targeting integrins.
Structure Determination and Refinement
Expression and purification of a soluble αVβ3 fragment, which terminates after Pro957 (in αV) and Asp692 (in β3) and therefore lacks the transmembrane and short cytoplasmic tails of both subunits, was carried out essentially as described (16). Soluble αVβ3 displayed the same epitopes, ligand-binding specificities, and divalent cation requirements as the active form of native αVβ3 [(16) and data not shown]. Protein crystals were grown by vapor diffusion using 100 mM MES, pH 6.0, 100 mM NaCl, 5 mM CaCl2, and 10% polyethylene glycol 4000 as precipitant. The structure was solved with the use of single isomorphous replacement-anomalous scattering (SIRAS) and multiwavelength anomalous diffraction (MAD) (Table 1). The final model contains all the extracellular residues with the exception of αV residues 839 – 867 and 957, and β3 residues 1–54, 435–531, and 691–692. The final model contains 58 cysteines, all of which are paired. It also contains 6 calcium ions and 11 glycans [with either one or two N-acetyl glucosamine (Glc-NAc) modeled at each site]. The final crystallographic R factors between 20 and 3.1 Å are 25.5% (working set) and 33.5% (free set). Refinement statistics are listed in Table 1. Figure 1A shows a representative electron density map. The secondary structure assignment and an alignment with integrins α5, αIIb, and β1 are given in Web fig. 1 (17).
Table 1.
Crystallographic data and refinement statistics. Methods are as follows: αVβ3 crystals belong to space group P3221, with unit cell dimensions of a = b = 130.0 Å, and c = 307.3 Å. Each asymmetric unit contains a single heterodimer and ~61% solvent. Crystals were cryoprotected in glycerol, and native and derivative data were collected at 100 K at ID-19 beamline at the Argonne National Laboratory. Data were processed with HKL (43). Heavy atom refinement and phase calculations were performed using SHARP (44), and phases were then improved using SOLOMON (45). A high degree of nonisomorphism between the Pt2+ and Lu3+ derivative data sets prevented phase combination, and so the polypeptide chain was traced using two separate electron density maps, one based on MAD phases (Lu3+) and the other calculated with SIRAS phases (Pt2+). Most of the main chain and many side chains could be recognized in these maps, with the exception of the PSI and the predicted EGF-1 and -2 domains of β3. Model building was carried out in O (46), and refinement was performed with XPLOR (47) using bulk solvent correction and simulated annealing protocols. After several rounds of refinement and model building into the resulting σA-weighted electron density maps, atomic B-factor refinement (grouping residues) was carried out. The average B factor for the whole structure is 60 Å2. The average B factors for the individual domains are as follows: propeller, 47 Å2; thigh, 78 Å2; calf-1, 75 Å2; calf-2, 60 Å2; βA domain, 56 Å2; hybrid, 68 Å2; EGF-3, 82 Å2; EGF-4, 66 Å2 and βTD, 71 Å2. Values in parentheses indicate the highest resolution shell. n/a, not available; No., number.
| Data collection | |||||
|---|---|---|---|---|---|
| Space group | P3221 | ||||
| Unit cell dimensions | 130.0 Å, 130.0 Å, 307.3 Å, 90°, 90°, 120° | ||||
| Native |
MAD(Lu3+) |
SIRAS(Pt2+) |
|||
| Remote | Edge | Peak | Peak | ||
| Wavelength (Å) | 1.0332 | 1.2398 | 1.3412 | 1.3407 | 0.9840 |
| Resolution (Å) | 50-3.1 | 50-3.1 | 50-3.6 | 50-3.2 | 50-3.5 |
| Unique reflections | 53802 | 50490 | 34310 | 46024 | 32413 |
| % completeness | 96.9 (95.0) | 91.4 (70.4) | 99.5 (99.1) | 91.6 (90.7) | 84.4 (20.2) |
| Redundancy | 6.1 | 9.0 | 10.0 | 8.9 | 8.2 |
| I/σ | 8.7 (3.7) | 5.8 (1.0) | 5.5 (2.7) | 6.6 (2.2) | 6.6 (1.0) |
| Rsym(%)* | 10.5 (55.1) | 12.1 (63.6) | 14.5 (58.2) | 14.7 (56.0) | 9.6 (48.4) |
| Phase determination | |||||
| No. of heavy atoms | 3 | 8 | |||
| Phasing power† | |||||
| Centrics | n/a¶ | 0.69 | 0.99 | 0.75 | |
| Acentrics | |||||
| Isomorphous | n/a | 0.79 | 1.20 | 0.72 | |
| Anomalous | 0.94 | 2.55 | 1.72 | 1.37 | |
| Mean figure of merit (overall) | 0.43 | 0.23 | |||
| R cullis ‡ | |||||
| Centrics | n/a | 0.84 | 0.77 | 0.96 | |
| Acentrics | |||||
| Isomorphous | n/a | 0.84 | 0.78 | 1.0 | |
| Anomalous | 0.91 | 0.70 | 0.80 | 0.83 | |
| Refinement statistics | Model statistics | ||||
| Resolution range (Å) | 20.0-3.1 | rms deviation from ideality | |||
| Rfactor§ (%) (work set) | 25.5 | Bond lengths (Å) | 0.008 | ||
| Rfactor (%) (free set) | 33.5 | Bond angles (degree) | 1.6 | ||
| Average B value (Å2) | 60 | ||||
| No. of Glc-NAc | 18 | ||||
| No. of Ca2+ | 6 | ||||
Rsym = Σ|I–〈I〉|/Σ I, where I is the observed intensity and 〈I〉 is the average intensity from multiple observations of symmetry-related reflections.
Phasing power = FH(calc)/E, where E is phase-integrated lack-of-closure. The three values for each wavelength are for centric/acentric isomorphous/acentric anomalous contribution.
Rcullis = Σhkl ||FPH + FP| – FH(calc)|/Σhkl |FPH + FP|
§Rfactor = Σhkl |Fobs(hkl) – Fcalc(hkl)|/ΣhklFobs(hkl).
Fig. 1.
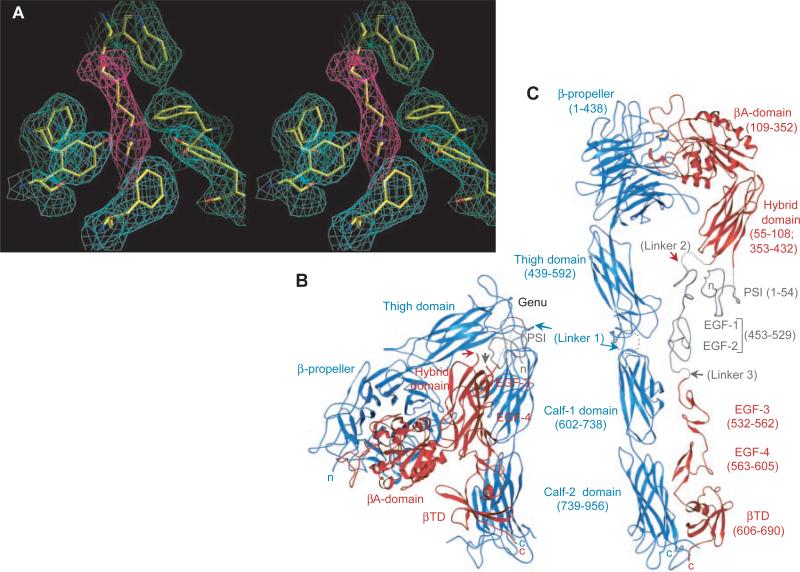
Structure of the extracellular segment of αVβ3. (A) Stereoview of a simulated-annealing omit map at 1.5 σ, in the vicinity of Arg261 (from β3) (magenta). Surrounding densities (cyan) (from α V) are from the same map. (B) Ribbon drawing (40) of crystallized αVβ3 [shown in blue (αV) and red (β3)]. (C) Model of the straightened extracellular segment of αVβ3. The two tails would extend into the plasma membrane in the native integrin. Translated and rotated EGF-3 and -4 show the approximate location of EGF-1 and -2 (gray). The PSI tracing (gray) is approximate. Connections of the untraced domains are in dotted lines. αV was straightened by extending the structure by 135° at the thigh–calf-1 interface (circled) and then rotating the calf module ~120° around its “long” axis to avoid clashes at the thigh–calf-1 interface. The same transformations were then applied to β3 (residue 445 onward). Arrows point to the position of the three longest inter-domain linkers 1, 2, and 3 in the structure. Amino acid domain boundaries are indicated in parenthesis. In this and other figures, “n” and “c” indicate NH2- and COOH-terminus, respectively.
Overall Structure of the Heterodimer
The NH2-terminal segments of the α and β subunits assemble into an ovoid “head” from which two nearly parallel “tails” emerge (Fig. 1, B and C). The head consists of a seven-bladed β-propeller from αV and a βA domain looping out from a unique immunoglobulin (Ig)-like “hybrid” domain in β3. The integrin head has dimensions of ~90 Å by 60 Å by 45 Å. The αV tail is composed of three β-sandwich domains: an Ig-like “thigh” domain and two very similar domains that form the “calf ” module. The β3 tail consists of a PSI [for plexins, semaphorins, and integrins (18)] domain, four epidermal growth factor (EGF) domains, and a β-tail domain (βTD). The α and β integrin tails fold back at a ~135° angle, forming a V-shaped structure with a “genu” between the thigh domain and the calf module of αV. This profound bending of both subunits is surprising but unambiguous. When extended, each integrin tail forms a thin cylinder, ~160 Å long and ~20 Å in diameter. These values are in agreement with those derived from rotary shadowing images (4–7).
Domain Architecture and Interdomain Interfaces αV Subunit Domains
β-propeller domain
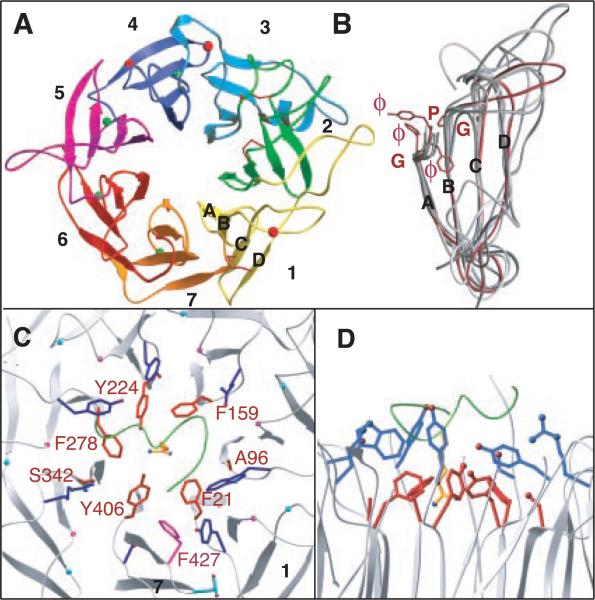
The β-propeller is formed from the NH2-terminal seven-fold ~60 residue sequence repeats of αV and consists of seven radially arranged “blades,” each formed from a four-stranded antiparallel sheet (Fig. 2A). The inner strand (strand A) of each blade lines the channel at the center of the propeller, with strands B and C of the same repeat radiating outward, and strand D of the next repeat forming the outer edge of the blade [Fig. 2A and Web fig. 2 (17)]. The β-propeller is circularized by juxtaposition of the C7 and D1 strands. The central channel is lined predominantly with amide and carbonyl oxygen groups from the seven A strands, with only a few side chains projecting into the cavity.
Fig. 2.
Structure of the integrin β-propeller. (A) Bottom view of the seven-bladed (numbered) αV propeller. Disulfides (sticks) and glycans (spheres) are in red and Ca2+ ions are in green. (B) Super-imposition of the seven blades, with blade five shown in red. The cage residues (ϕ, aromatic; G, Gly; and P, Pro) are in red. The ϕϕGϕ sequence assumes the shape of a cup. Strands A to D are labeled. Top (C) and side (D) views of the central propeller–βA domain interface. The propeller's lower (red) and upper (blue) aromatic rings surrounding Arg261 of β3 are shown. The 310-helix of β3 is in green. The lower ring residues are labeled in red. The prolines and glycines of the cage motif are shown as magenta and cyan spheres, respectively. F427 and D430 replace the proline and glycine residues in blade seven, respectively (41).
Superposition of the seven integrin blades reveals a unique consensus sequence repeat, the “cage” motif [Fig. 2, B through D, and Web fig. 2 (17)]. This motif (X17-33-{ϕϕGϕX13-20PX2-15GX5-8})7, where X is any residue and ϕ is an aromatic residue) is defined by a mostly aromatic four-residue “cup” (ϕϕGϕ) that precedes strand A, a proline immediately following strand B (Pro-B), and a glycine at the beginning of strand C (Gly-C). The first and fourth residues of the cup form the upper and lower rings of a cage-like structure that lies at the center of the contact region between αV and β3, holding Arg261 of β3 in place (Fig. 1A and Fig. 2, C and D). The second and third residues form the floor of the cup. Pro-B introduces an acute turn and is tightly sandwiched between sequential cups, providing a rigid scaffold for the upper and lower ring residues. Pro-B is replaced with Phe427 in blade 7, pushing the side chains of the lower ring residues Phe21 and Tyr406 toward Arg261 (Fig. 2C). The small side chains of the lower ring residues Ala96 and Ser342 are required to avoid clashes in this region. Gly-C is required because any side chain at this position would interfere with Pro-B.
Four solvent-exposed Ca2+-binding sites are found in the A-B β hairpin loops of blades 4-7 at the propeller's bottom (opposite the αβ interface) [see metal coordination in Web fig. 3 (17)]. The calcium-binding residues span a nine-residue segment with the consensus sequence Asp-h-Asp/Asn-x-Asp/Asn-Gly-h-x-Asp, where “h” is hydrophobic and “x” is any residue; the invariant Gly is required because of its unusual main chain torsion angles. Ca2+ is usually coordinated by oxygen atoms from side chains of residues 1, 3, 5, and 9 and the carbonyl oxygen of residue 7. Sequential β hairpin loops contact each other in a chain-like arrangement. The calcium-containing loop of blade seven makes extensive contacts with the thigh domain; the presence of calcium is likely to make this interface more rigid.
β sandwich domains
The remainder of αV consists of the thigh and calf-1 and -2 domains [Fig. 1C and Web fig. 4 (17)]. The β sheet structure of the thigh domain is similar in topology to the C2-set Ig fold (19), although the thigh domain is significantly larger. The contact area between the propeller and thigh domains buries a ~700 Å2 surface area. The elongated shape and intermediate size of this interface suggest that some inter-domain movement, likely as a rotation around the long axis of the contact area, is possible. Such movement could be regulated by calcium ions in the propeller.
Each calf domain contains two antiparallel β sheets, one with four strands (ABGD) and the other with five (EFCHI). The two domains are related by an approximate twofold screw axis symmetry. The known proteolytic cleavage site after Arg860, which generates the heavy and light chains of αV, is located in the EE′ loop of the calf-2 domain. The residues flanking the cleavage site (Gln839–Gly867) are not seen in the electron density map and are presumably disordered.
A small ~200 Å2 interface exists in the crystal at the genu between the thigh and calf-1 domains. The base of the thigh domain and the top of the calf-1 domain both contain acidic patches, and these would be expected to face each other in an extended integrin. A well-coordinated calcium ion lies at the genu; in an extended structure it may help to neutralize the negative charge at this interface. The ~500 Å2 interface between calf-1 and -2 domains is fairly extensive and largely hydrophobic, suggesting that these two domains form a rigid structural entity, the calf module.
β3 Subunit Domains
βA and hybrid domains
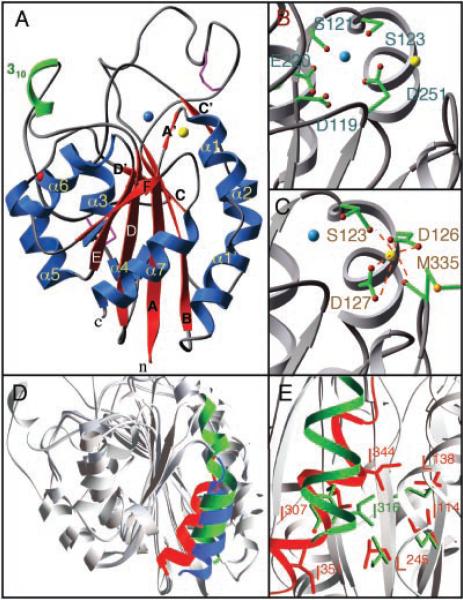
The β3 portion of the integrin head is composed of the βA and hybrid domains (Fig. 1C). The βA domain (Fig. 3A) is inserted into the B-C loop of the hybrid domain [Fig. 1C and Web fig. 5 (17)] and assumes the nucleotide-binding (or Ross-mann) fold found in Gβ and integrin αA domains (13). The βA domain consists of a central six-stranded β sheet surrounded by eight helices. A MIDAS motif occupies a crevice at the top of the central β strand, as in αA domains (13). β3 MIDAS is formed by the side chains of Asp119, Ser121, Ser123, Glu220 (equivalent to Thr209 in αA of CD11b), and Asp251 (equivalent to Asp242 in αA of CD11b) (Fig. 3B). Thus, the overall geometry of the metal-ligand coordination is similar to that of αA. However, we do not clearly see a metal ion in β3 MIDAS: although there is a density feature present, it likely represents a water molecule or a metal ion with partial occupancy. Adjacent to MIDAS lies a metal ion–binding site (ADMIDAS) (Fig. 3C). We assigned a calcium ion at ADMIDAS because calcium is present in the crystallization buffer and because the ion is octahedrally coordinated. In addition, the ion can be replaced by the calcium analog Lu3+ in the derivatized crystal. Calcium is coordinated by the carbonyl oxygens of Ser123 (from A′-α1 loop) and Met335 (from α7) and by the side chains of Asp126 and Asp127 (from α1) (Fig. 3C), thus linking α1 to the top of α7. Asp126 and Asp127 are invariant in all βA domains except that of β8, where they are both replaced with asparagines.
Fig. 3.
Structural features of the βA domain. (A) Ribbon drawing of the βA domain. Disulfides and the glycan are in purple and red, respectively. (B) Residues [single letter (41)] forming MIDAS that could participate in metal ion (blue) coordination. (C) Calcium (yellow) coordination at ADMIDAS. (D) Superimposition, based on the central β sheet, of the βA domain (minus the insertion in the B-C loop) and the “open” and “closed” forms of αA of CD11b (all gray). Shown are the α7-helices of the βA domain (red) and the “open” (blue) and “closed” (green) forms of αA of CD11b. (E) Coordination of SILEN in the βA domain (red) and in “closed” αA of CD11b (green). The βA domain SILEN residues [single letter, (41)] I114, L138, L245, and I307 (red) [equivalent to I135, L164, I236, and Y267, respectively, in αA of CD11b (green) (23)] coordinate I344 (equivalent in sequence to L312 in αA of CD11b). I351 (corresponding to I316 in αA of CD11b) is not coordinated in β3 SILEN but is partially exposed at the bottom of the structure.
αA exists in two conformations, “open” and “closed” (20, 21), corresponding to “high” and “low” affinity states, respectively (22, 23). It has been suggested that both forms are present in an equilibrium, with a switch from the “closed” to the “open” state being a necessary feature of activation in αA-integrins (22). The “closed” and “open” conformations are distinguished primarily by the length and relative position of helix α7. In the “closed” form, α7 has an extra NH2-terminal turn and is tethered to the domain body through hydrophobic contacts that include insertion of a conserved isoleucine (I316 in αA of CD11b) from α7 into the socket-for-isoleucine (SILEN) (23). Removal of this contact shifts α7 by ~10 Å, switching αA to the “open” high-affinity form.
Except for the two insertions in the B-C and D-α5 loops, the βA domain is largely superimposable on that of αA. However, α7 of the βA domain is more similar to α7 in the “open” state of αA: it lacks an extra turn at its top (Fig. 3D) and is positioned similarly relative to the body of the domain (Fig. 3E). Both features suggest that the βA domain is “open” in our αVβ3 structure, though it is not liganded. The NH2- and COOH-termini connecting the βA domain to the hybrid domain appear to be rigidly held in place through numerous contacts with the hybrid domain, suggesting that the βA domain, in contrast to αA, always exists in the “open” state and may, therefore, be regulated differently.
The hybrid domain (Fig. 1, B and C) is similar to the I-set Ig domains (19). Its core most closely resembles that of the I-set domain 1 of CD102 [see Web fig. 5 (17)]. The hybrid and βA domains make extensive contacts with each other. The βA domain–hybrid interface is circular (diameter, ~15 Å), fairly large (650 Å2), and has a mixed hydrophilic and hydrophobic nature. Its architecture and size suggest that interdomain movement across the βA domain–hybrid interface is minimal.
PSI domain
On the basis of sequence homology, the NH2-terminal 54 amino acids of β3 are predicted to fold into a PSI domain (18), located in the region connecting the hybrid domain to the expected EGF-1 domain (Fig. 1, B and C). The electron density for PSI is weak, and its Cα chain cannot be traced with certainty at the current resolution. Thus, the PSI domain is not included in the present model, although the nature of the electron density suggests that it is partially ordered.
EGF domains
Four tandem cysteine-rich repeats follow the hybrid domain in the primary sequence. Good electron density allowed us to build a model for cysteine-rich repeats 3 and 4 [Fig. 1, B and C, and Web fig. 6 (17)]. Each assumes the structure of a class I EGF fold (24) and contains the three disulfide bonds typical for EGF domains. An additional pair of cysteines links the two domains. EGF-4 contains an extra disulfide bridge that helps define the COOH-terminal boundary of the short D strand. EGF-3 and -4 form a rod-like, extended module in which the two domains are related by an approximate two-fold screw axis symmetry (Fig. 1C). A consensus site found in calcium binding (cb) EGF domains such as fibrillin (24) is lacking in the integrin EGF domains, and there is no evidence in our electron density map for calcium (or heavy atom) binding. A small (~200 Å2), but presumably rigid, interface exists between EGF-3 and -4: it includes the disulfide bridge as well as main chain hydrogen bonds, hydrophobic interactions, and a glycan.
The electron density is not sufficiently featured to trace the predicted EGF-1 and -2 domains. Most likely, these domains are partially disordered because of the severe bending of the integrin in this region. Examination of their sequence [Web fig. 6 (17)] shows, however, that EGF-1 and -2 are highly homologous to EGF-3 and -4; each is expected to contain the typical three cysteine pairs and to lack the metal ion site found in cbEGF domains. Thus, EGF-1 and -2 are also likely to form a non–calcium-binding rod-shaped module.
Fig. 6.
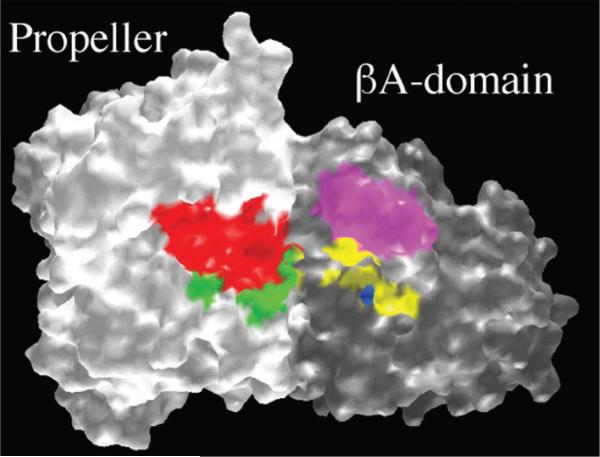
The putative ligand-binding site at the integrin αβ interface. GRASP representation of the αV propeller (white) and the α3 A-domain (gray), viewed from the top of the head (the tails lying in the back). Arg143–Phe154 (red) and Gly172–Gly181 (green) of αV correspond to the identified ligand-binding residues Arg147–Tyr166 and Gly184–Gly193, respectively, in αIIb. Ligand-binding residues in β3 (Asp179–Thr183, “ligand specificity” region (magenta); Asp119, Ser121, Ser123, Glu220 (all MIDAS residues), and Arg214 (yellow) are shown. Asp217 in β3 (also in yellow) is involved in binding of a ligand-mimetic monoclonal antibody. The expected MIDAS metal ion is in blue.
βTD. βTD consists of a four-stranded β sheet that contains antiparallel and parallel strands and faces an NH2-terminal α helix (Fig. 1C). Interactions between the α helix and the β sheet are mostly hydrophobic and involve a disulfide bond [see Web fig. 7 (17)]. The rear of the β sheet is covered with a long A-B loop. A DALI search (25) showed only a weak homology to cystatin C, an inhibitor of the papain-family cysteine pro-teases (26). However, the βTD structure differs from that of cystatin C in the direction of one of the β strands in the sheet. Hence βTD has to be classified as a new fold. Only two weak hydrophobic contacts are found between βTD and EGF-4, suggesting flexibility at this interface.
The αVβ3 Interface
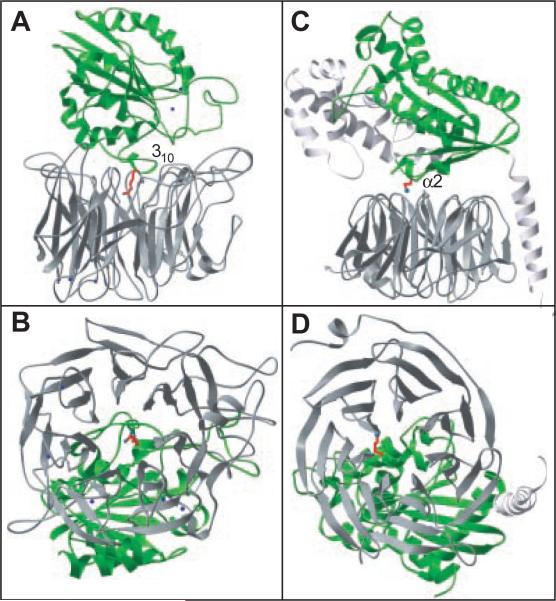
The βA domain–propeller interface bears a striking resemblance to the Gα-Gβ interface of G proteins (Fig. 4). The seven-bladed propellers of αV and Gβ have similar dimensions, and they also have almost identical orientations of the β strands with respect to the central channel (Fig. 4). Furthermore, both βA and Gα domains rest off-center with respect to the propellers, and in each case contact six of the seven blades. In both cases, the axis of the propeller's channel is aimed directly at a short helix: the 310- and α2-helices of βA and Gα domains, respectively (Fig. 4). In the case of Gα, the α2-helix marks switch region II, which undergoes a major conformational change upon activation of G proteins.
Fig. 4.
Architecture of integrin and G protein interfaces. The nucleotide-binding folds are in green, and the propellers are in gray. The integrin is shown in (A) and (B) and the G protein in (C) and (D). The helices at the core of each interface are indicated. Integrin's Arg261 and G protein's Lys210 project toward the respective propeller's central cavity. Gα (Ala30–Leu348) and Gβ (Ser2–Asn340) are shown. G protein coordinates were from (33).
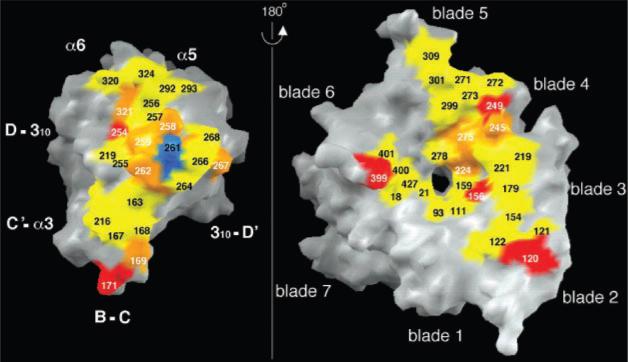
Residue Arg261 lies at the core of the βA domain–propeller interface (Fig. 1A, Fig. 2, C and D, and Fig. 5). Arg261 (Lys in all other integrin β subunits except β4) protrudes off-center into the propeller's channel, and is caged into place by two concentric rings of predominantly aromatic propeller residues (Fig. 1A and Fig. 2, C and D). Residues in the upper and lower rings lie at positions 1 and 4, respectively, of the seven cups in αV (Fig. 2, B and C). Side chains of Phe21, Phe159, Tyr224, Phe278, and Tyr406 form the lower ring and contact Arg261 directly (Fig. 2C). Some of these contacts occur through cation-π bonding (27). Residues Tyr18, Trp93, Tyr221, Tyr275, and Ser403 in the upper ring (Fig. 2D) contact side chains in the lower ring and also provide a hydrophobic interface for residues flanking Arg261 in the 310-helix. The βA domain–propeller interface has a number of additional contacts (Fig. 5) and buries a large surface area of ~1600 Å2.
Fig. 5.
Surface representation, done with GRASP (42), of the main αVβ3 interface in the head region. Contacting residues (distance cutoff 3.5 Å for hydrogen bonds and salt bridges, and 4.0 Å for van der Waals contacts) are shown. Arg261 is in blue. Hydrophobic, ionic, and mixed contacts are in yellow, red, and orange, respectively. The β3 strands and helices contributing to the interface are indicated.
Naturally occurring mutations that disrupt the αβ heterodimer in integrins have been described [reviewed in (2, 28)]. These occur most frequently in the βA domain of β2 and β3 integrin subunits, resulting in life-threatening bacterial infections and thrombasthenia (a severe bleeding disorder), respectively. Our model helps explain how such mutations cause disruption of the heterodimer. For example, Gly251 to Arg or Pro156 to Leu mutations are known to prevent formation of β2 integrin heterodimers. Gly260 in β3 (corresponding to Gly251 in β2) lies in the 310-helix next to Arg261, which makes the central contact with the propeller (Fig. 2, C and D). A Gly to Arg substitution here would result in clashes with other βA residues and would likely alter the position of the 310 helix relative to the propeller. Pro163 in β3 (equivalent to Pro156 of β2) lies in a loop adjacent to the 310-helix and contacts the propeller (Fig. 5); the longer leucine side chain would disrupt the αβ interface. In β3, a Ser162 to Leu mutation results in an unstable αIIbβ3 heterodimer and causes thrombasthenia. Ser162 lies above blade 2 of the propeller; replacing it with leucine would result in unfavorable contacts at the αβ interface.
Although the propeller–βA domain interface is the main contact between the αV and β3 subunits, in the crystal, additional contacts are seen between the propeller–EGF-3 and -4 domains, thigh–EGF-3, calf-2–EGF-4, and calf-2–βTD. All these contacts are small, not very hydrophobic, and for the most part discontinuous. Therefore, they are not expected to occur in the membrane-bound receptor.
Implications for Integrin Function and Regulation
The structure described in this communication is that of αVβ3 in its active (ligand-competent) state. Activation in this case is artificially induced by truncating the transmembrane and cytoplasmic segments that normally restrain integrins into a default low-affinity state (29, 30). Naturally occurring β3 mutants (from some patients with thrombasthenia) and mutagenesis studies of the related integrin αIIbβ3 [reviewed in (1, 31)] identified three ligand-binding regions. Two of these are located in the αIIb propeller and the third involves the βA domain MIDAS. The βA domain also contains a fourth “ligand specificity” region. Replacing this region in β1 with that of β3 switched ligand-binding specificity of integrin αVβ1 into that of αVβ3 [reviewed in (1, 31)]. The two β3 regions and the αV residues that correspond to the two regions identified in αIIb cluster at the top of the integrin head (Fig. 6). We propose that ligand binding occurs in this area. Confirmatory evidence comes from rotary-shadowed preparations where fibrinogen is found associated with the top of αIIbβ3 (6). The putative αVβ3 ligand-binding site is formed primarily by residues in loops D3-A3 (between blades 2 and 3) and B3-C3 (within blade 3) of the propeller and by residues in loop B-C and the MIDAS motif of the βA domain. The αA domain loops out from the COOH-terminal end of the D3-A3 loop in αA-integrins.
Major conformational changes throughout the β subunit and reorientation of the extracellular domains of the α and β subunits appear to coincide with integrin activation, whether induced physiologically or artificially (9, 32). Our crystal structure reveals a severely bent αVβ3 conformation. This arrangement differs dramatically from the extended conformations seen in cryoelectron microscopy images of integrins reconstituted in lipid bilayers (7) or rotary shadowing of isolated integrins (4–6). However, a minority of rotary shadowing images suggests that the tails may have collapsed on top of (or beneath) the head (5, 6). The dimensions of this oblong form (120 Å by 80 Å) agree nicely with our structure, suggesting that similar bending may occur in solution. The fact that extended and severely bent conformations of the same molecule are seen suggests that a highly flexible site, the genu, exists in the integrin. The extreme degree of bending found in the crystal structure is less likely to occur in a membrane-anchored integrin because such a conformation would likely constrain access of polymerized extracellular matrix to the ligand-binding site. Nevertheless, the high degree of flexibility at the genu provides a first glimpse of the spectrum of the quaternary changes possible during bi-directional signaling in integrins. The presence of a metal ion at the genu suggests one mechanism by which these changes could be regulated.
The structural similarities between the propeller–βA domains and the Gα-Gβ domains are intriguing (Fig. 4). In G proteins, GTP hydrolysis induces a major conformational shift in Gα, a component of which is the destabilization of the α2-helix in switch region II. This results in physical separation of the two subunits in the active state and unmasks ligand-binding sites on the propeller (33). Does the structural homology between integrins and G proteins translate into analogous models of allosteric control? A large interface between the propeller and βA domains is seen in our structure, in contrast to G proteins where in the active state, the equivalent Gβ and Gα are completely dissociated. Other observations suggest, however, that the propeller–βA domain contact area may also be a dynamic interface that can be modulated. First, integrin αIIbβ3 can be reversibly dissociated into its individual subunits by brief treatment with EDTA (34). Second, a conformational shift of the propeller relative to the βA domain has been reported (32). Third, a split of the αIIbβ3 head into two distinct knobs was observed in the presence of RGD peptides (11). In our structure, cation-π bonding between Arg261 and surrounding aromatic residues contributes to the αβ interface. Amide-aromatic interactions can be attractive or repulsive (27), unlike ion pairs. Thus, it is conceivable that small changes in side chain orientation could modulate the stability of the αβ interface, perhaps allowing for dissociation under certain conditions.
The location of the metal ion– binding sites in αVβ3 helps clarify some of the complex and unresolved regulatory effects exerted by metal ions on integrin-ligand binding. Cells expressing a mutant α4β1 (lacking one or more metal-binding sites in the propeller) detach easily from substrate under shear flow (35). The calcium ions in the propeller are not close to the proposed ligand-binding site shown in Fig. 6, but they may help to make more rigid the propeller-thigh interface and may thus regulate integrin-ligand interactions in an allosteric manner. The calcium ion at the thigh–calf-1 interface may play a similar role. Studies with β3 and α5β1 integrins have also shown that a high-affinity Ca2+ site is required for ligand-binding, and that a low-affinity Ca2+ site allosterically inhibits ligand binding (36, 37). The presence of two adjacent metal-binding sites (MIDAS and ADMIDAS) in the βA domain suggests underlying mechanisms for these effects.
The structure of αVβ3 reveals new domains, previously unpredicted domains, and creative use of the Ig scaffold. Integrin-ligand interactions are regulated not only by changes in affinity but also by altered avidity (receptor clustering) [reviewed in (38)]. Integrins also bind in cis to several membrane receptors, an interaction that modulates their signaling functions [reviewed in (39)]. The mosaic of domains revealed in the integrin structure can now serve as a foundation for future investigations into the structural basis of these interactions.
References and Notes
- 1.Humphries MJ. Biochem. Soc. Trans. 2000;28:311. [PubMed] [Google Scholar]
- 2.Arnaout MA. Immunol. Rev. 1990;114:145. doi: 10.1111/j.1600-065x.1990.tb00564.x. [DOI] [PubMed] [Google Scholar]
- 3.Hynes RO. Cell. 1992;69:11. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 4.Parise LV, Phillips DR. J. Biol. Chem. 1985;260:1750. [PubMed] [Google Scholar]
- 5.Nermut MV, Green NM, Eason P, Yamada SS, Yamada KM. EMBO J. 1988;7:4093. doi: 10.1002/j.1460-2075.1988.tb03303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisel JW, Nagaswami C, Vilaire G, Bennett JS. J Biol Chem. 1992;267:16637. [PubMed] [Google Scholar]
- 7.Erb E-M, Tangemann K, Bohrmann B, Muller B, Engel J. Biochemistry. 1997;36:7395. doi: 10.1021/bi9702187. [DOI] [PubMed] [Google Scholar]
- 8.Yamada KM, Geiger B. Curr. Opin. Cell Biol. 1997;9:76. doi: 10.1016/s0955-0674(97)80155-x. [DOI] [PubMed] [Google Scholar]
- 9.Sims PJ, Ginsberg MH, Plow EF, Shattil SJ. J. Biol. Chem. 1991;266:7345. [PubMed] [Google Scholar]
- 10.Du X, et al. J. Biol. Chem. 1993;268:23087. [PubMed] [Google Scholar]
- 11.Hantgan RR, Paumi C, Rocco M, Weisel JW. Biochemistry. 1999;38:14461. doi: 10.1021/bi9907680. [DOI] [PubMed] [Google Scholar]
- 12.Michishita M, Videm V, Arnaout MA. Cell. 1993;72:857. doi: 10.1016/0092-8674(93)90575-b. [DOI] [PubMed] [Google Scholar]
- 13.Lee J-O, Rieu P, Arnaout MA, Liddington R. Cell. 1995;80:631. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 14.Eliceiri BP, Cheresh DA. J. Clin. Invest. 1999;103:1227. doi: 10.1172/JCI6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. J. Biol. Chem. 2000;275:21785. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 16.Mehta RJ, et al. Biochem J. 1998;330:861. doi: 10.1042/bj3300861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Online at www.sciencemag.org/cgi/content/full/1064535/DC1. For additional information, see Web figs. 1 through 7, available at Science
- 18.Bork P, Doerks T, Springer TA, Snel B. Trends Biochem. Sci. 1999;24:261. doi: 10.1016/s0968-0004(99)01416-4. [DOI] [PubMed] [Google Scholar]
- 19.Chothia C, Jones EY. Annu. Rev. Biochem. 1997;66:823. doi: 10.1146/annurev.biochem.66.1.823. [DOI] [PubMed] [Google Scholar]
- 20.Lee J-O, Anne-Bankston L, Arnaout MA, Liddington RC. Structure. 1995;3:1333. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 21.Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Cell. 2000;100:47. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Rieu P, Griffith DL, Scott D, Arnaout MA. J. Cell Biol. 1998;143:1523. doi: 10.1083/jcb.143.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong JP, Li R, Essafi M, Stehle T, Arnaout MA. J. Biol. Chem. 2000;275:38762. doi: 10.1074/jbc.C000563200. [DOI] [PubMed] [Google Scholar]
- 24.Downing AK, et al. Cell. 1996;85:597. doi: 10.1016/s0092-8674(00)81259-3. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. J. Mol. Biol. 1993;223:123. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 26.Janowski R, et al. Nature Struct. Biol. 2001;8:316. doi: 10.1038/86188. [DOI] [PubMed] [Google Scholar]
- 27.Gallivan JP, Dougherty DA. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9459. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogg N, Bates PA. Matrix Biol. 2000;19:211. doi: 10.1016/s0945-053x(00)00066-4. [DOI] [PubMed] [Google Scholar]
- 29.Dana N, Fathallah DF, Arnaout MA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3106. doi: 10.1073/pnas.88.8.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes PE, et al. J. Biol. Chem. 1996;271:6571. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 31.Calvete JJ. Proc. Soc. Exp. Biol. Med. 1999;222:29. doi: 10.1111/j.1525-1373.1999.09993.x. [DOI] [PubMed] [Google Scholar]
- 32.Mould AP, Garratt AN, Puzon-McLaughlin W, Takada Y, Humphries MJ. Biochem J. 1998;331:821. doi: 10.1042/bj3310821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wall MA, et al. Cell. 1995;83:1047. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 34.Fitzgerald LA, Phillips DR. J. Biol. Chem. 1985;260:11366. [PubMed] [Google Scholar]
- 35.Pujades C, et al. Mol. Biol. Cell. 1997;8:2647. doi: 10.1091/mbc.8.12.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mould AP, Akiyama SK, Humphries MJ. J. Biol. Chem. 1995;270:26270. doi: 10.1074/jbc.270.44.26270. [DOI] [PubMed] [Google Scholar]
- 37.Hu DD, Barbas CF, Smith JW. J. Biol. Chem. 1996;271:21745. doi: 10.1074/jbc.271.36.21745. [DOI] [PubMed] [Google Scholar]
- 38.van Kooyk Y, van Vliet SJ, Figdor CG. J. Biol. Chem. 1999;274:26869. doi: 10.1074/jbc.274.38.26869. [DOI] [PubMed] [Google Scholar]
- 39.Hemler ME. Curr. Opin. Cell Biol. 1998;10:578. doi: 10.1016/s0955-0674(98)80032-x. [DOI] [PubMed] [Google Scholar]
- 40.Carson M. J. Mol. Graph. 1987;5:103. [Google Scholar]
- 41.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 42.Nicholls A, Sharp KA, Honig B. Proteins. 1991;11:281. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.de La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:47. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 45.CCP4 Acta Crystallogr. D. 1994;50:760. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr. A. 1991;47:110. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 47.Brünger AT, et al. Acta Crystallogr. D. 1998;54:905. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.We thank A. Viel, M. Frech, and D. Cheresh for valuable assistance. Supported by NIH grants DK48549, DK50305, HL54227, and AI45716 and a contract from the DOE under contract W-31-109-Eng-38. The coordinates have been deposited in the Protein Data Bank (PDB1JV2).