INTRODUCTION
Aminopeptidases are ubiquitous hydrolases that cleave the N-terminal residues of proteins and peptides for maturation, activation, or degradation, and therefore are involved in numerous biological processes.1,2 They are broadly distributed throughout all kingdoms of life and are found in subcellular organelles, cytoplasm, and in membrane-bound fractions.3 Many aminopeptidases use a set of conserved residues within a structural scaffold to form an active site capable of binding either one or two divalent metal ions that aid catalysis. Zn+2, Co+2, and Mn+2 are being the most common metals found in the active site.4–8 One of more extensively studied members of the aminopeptidase family is aminopeptidase N (APN) [alternative names: alanine aminopeptidase; aminopeptidase M; microsomal aminopeptidase; GP150; CD13; (EC 3.4.11.2)]. The APN sequence family is large and broadly distributed and includes members found in bacteria and eukaryotes, including plants and mammals. PsiBlast search identified over 1000 APN family members. Typically APNs are monomeric or homodimeric. In higher eukaryotes these enzymes are expressed in many tissues, with the highest level found in the intestinal and kidney brush border membranes, brain, lung, blood vessels, and primary cultures of fibroblasts. The sequence analysis indicates that aminopeptidase N is a member of the M1 family of the MA clan of peptidases, also termed gluzincins.5 The amino acid sequence fingerprints of the M1 family of zinc-metallopeptidases are the HEXXH(X18)E (a zinc binding motif) and GXMEN (an exopeptidase motif).9 Prominent members of this family include mammalian membrane-bound aminopeptidases [P-LAP, aminopeptidase A (APA), thyrotropin-releasing hormone degrading enzyme (TRHDE)], cytosolic proteins [puromycin-sensitive aminopeptidase (PSA) and leukotriene A4 hydrolase (LTA4H)], and secretory proteins such as [adipocyte-derived leucine aminopeptidase (A-LAP) and aminopeptidase B (APB)].5,9
The APNs catalyze liberation of N-terminal amino acids from a broad spectrum of substrates including small peptides, amide, or arylamide. The N-terminal residue is a preferably neutral or basic amino acid, although it has been reported that an intact XPro dipeptide was released when the terminal hydrophobic residue was followed by a prolyl residue.10 The diversity of function that APNs play depends on their location and source tissue. Some APNs have been used commercially, such as the APN from Lactococcus lactis, which has been used in the food industry.11 Aminopeptidases N are also present in many pathogenic bacteria and represent potential drug targets.9 In this article, we report the crystal structure of APN from N. meningitides at 2.05-Å resolution.
RESULTS AND DISSCUSION
Structure determination
The structure of APN from N. meningitides (APN-Nm) was determined using diffraction data obtained from a single crystal of selenomethionine (SeMet) labeled protein and single wavelength anomalous diffraction (SAD) method.12 The rhombohedral crystal (H3) contains one monomer in the asymmetric unit. The electron density after phasing and density modification was of high quality for all amino acids except the first three N-terminal residues, which are disordered, and a few solvent-exposed loop regions that show high temperature factors. Never-the-less the polypeptide chain could be reliably modeled for residues 4–867. The model was refined with REFMAC of the CCP4 suite13 against 2.05 Å data to a final crystallographic R factor of 16.6% and Rfree of 21.1% (Table I). Prior to deposition of the structure in the Protein Data Bank (PDB) the quality of the structure was verified with the set of validation tools in the program COOT,14 as well as PROCHECK.15 Crystal packing analysis using PISA16 showed limited contacts between symmetry-related molecules strongly suggesting that APN-Nm monomer (the asymmetric unit content) represents a biologically relevant unit. This is in agreement with the size exclusion chromatography experiment that showed APN-Nm molecular weight of ~95 kDa (data not shown).
Table I.
Aminopeptidase NData Collection and Refinement Statistics
| Data collection statistics (19-ID,SBC) | |
| Resolution (Å) | 2.05 |
| Number of observed reflections | 126,825 |
| Number of unique reflections | 67,810 |
| Rmergea (%) | 6.8 (24.8) |
| Completeness (%) | 93.5 (85.9) |
| I/σ | 10.3(2.1) |
| Refinement statistics | |
| Resolution range (Å) | 40.0–2.05 |
| Rcryst (%) | 16.6 |
| Rfree (%) | 21.1 |
| Number of nonhydrogen atoms | |
| Protein | 6916 |
| Zn | 1 |
| Solvent/buffer molecules | 823/83 |
| rmsd from target values | |
| Bond lengths (Å) | 0.016 |
| Bond angles (deg) | 1.492 |
| Average B factors (Å2) | |
| Protein main chain | 25.6 |
| Protein side chain | 27.3 |
| Solvent | 19.2 |
| Zn | 36.4 |
| Ramachandran statistics | |
| Most favorable region | 93.1 |
| Additional allowed region | 6.8 |
| Generously allowed region | 0.1 |
| Disallowed region | 0 |
Highest resolution shell in parentheses.
Rmerge = ΣΣ|Ii σ− 〈Ii〉|/Σ|Ii| where Ii is the scaled intensity of the ith measurement, and 〈Ii〉 is the mean intensity for that reflection. Rcryst = Σ||Fobs − Fcalc||/Σ|Fobs| where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively. Rfree = as for Rcryst, but for 5.1% of the total reflections chosen at random and omitted from refinement.
Structural comparison with members of peptidase M1 family
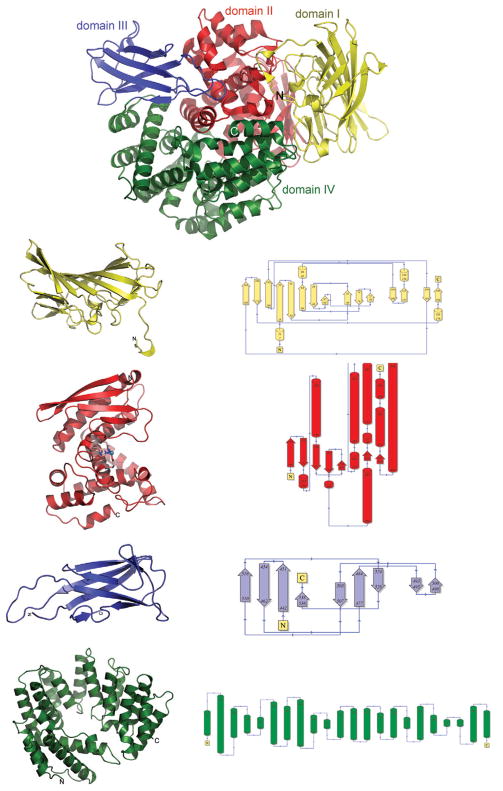
The APN-Nm structure represents multidomain α/β-fold with dimensions of 92 Å × 65Å × 68 Å × that has mostly α-helical central core and several peripheral domains predominantly made of β-strands (see Fig. 1). The overall structure of APN shows homology to the structures observed for the archeal tricorn interacting factor F3 and human leukotriene A4 hydrolase,17 as indicated by the DALI18 structural comparison searches. In spite of low sequence identity, APN-Nm structure superimposes quite well for such a large structure with protease interacting factor F3 (PDB code 1z1w, Z-score 41.9, r.m.s.d for 780 Ca atoms = 3.1 Å, sequence identity 19%), and leukotrine A4 hydrolase (PDB code 1hs6, Z-score 31.4, r.m.s.d for 610 Ca atoms = 3.6 Å, sequence identity 19%). Based on the structural homology the APN-Nm model could be further subdivided into smaller distinct structural and functional domains. As indicated by the higher structural homology and comparable size the overall APN-Nm structure closely resembles organization of four-domain tricorn interacting factor F319 and is also arranged into four distinct domains: the N-terminal domain I (residues 4–188), the catalytic domain II (residues 189–438), domain III (residues 439–539), and the C-terminal domain IV (residues 540–866).
Figure 1.
Ribbon presentation of the overall fold of aminopeptidase N from N.meningitides showing domain organization. The N-terminal domain (domain I) is colored yellow, the catalytic domain (domain II) is colored red, domain III is colored blue, the C-terminal (domain IV) is colored green. Each of the domains is also displayed side by side with the schematic representation of the domain’s topology.
The N-terminal domain is predominantly composed of β-strands with only four small, three-residue long 310 helices (see Fig. 1). The 15 β-strands are arranged into three anti-parallel β-sheets, organized into two layers. The top layer is formed entirely by a highly twisted and mixed eight-stranded β-sheet which resembles the saddle-like structure (β13β14β11β1β2β8β4β5). The second layer is composed of three structural elements: a small three-stranded sheet on one side (β3β6β7), a four-stranded β-sheet (β9β10 and β12β15) on the opposite side, and in the middle two 310 helices which are inserted between the β-sheets.
The catalytic domain has an α/β structure. The N-terminal part of the catalytic domain is almost solely made of β-strands arranged in a five-stranded β-sheet, with the exception of two α-helices (η5, α1) insertions connecting β-strands β18 and β19. Contrary to the N-terminal part of the domain, the bulk of the domain is composed mainly of α-helices, with only a small two-stranded β-sheet inclusion (β23, β24). The active site of the APN-Nm is located in the cleft at the center of this domain (Figs. 1 and 2).
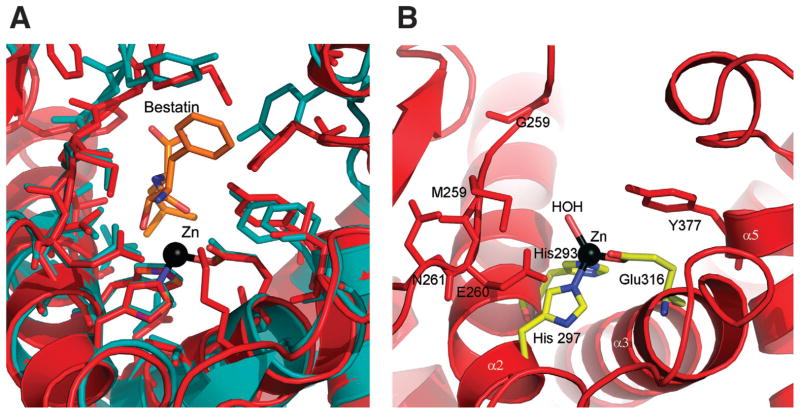
Figure 2.
(A) Close-up view showing the active site environment of APN-Nm (domain II). (B) Close-up view showing the relative positioning of the bestatin molecule(orange) in the active site of APN-Nm based on superimposition of the structure of human leukotriene A4 hydrolase (1hs6) (teal) over the structure of APN-Nm (red) using SSM server.20
The sequence alignment of the M1 family of proteases indicates the highest conservation of the sequence similarity in this domain; not surprisingly the catalytic domain also shows a high degree of structural homology among the family members (Figs. 1 and 3). The catalytic domain superimposes well with the corresponding domain of protease interacting factor F3 (PDB code 1z1w, Z-score 22.2, r.m.s.d for 216 Ca atoms = 2.4 Å, sequence identity 21%), and leukotrine A4 hydrolase (PDB code 1hs6, Z-score 19.1 r.m.s.d for 227 Ca atoms = 2.3 Å, sequence identity 22%) as well as showing some structural homology to thermolysine (PDB code 1hyt, Z-score 10.3, r.m.s.d for 195 Ca atoms = 3.8 Å, sequence identity 11%).
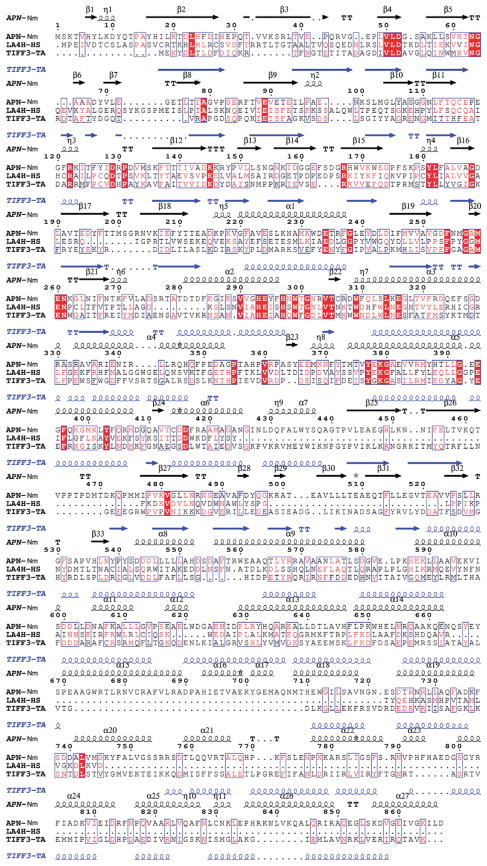
Figure 3.
Multiple sequence alignment of APN with two members of the M1 family of the MA clan of peptidases - tricorn interacting facor F3 from T. acidophilum and human leukotriene A4 hydrolase. Sequence identities are highlighted in red and similarities are shown as red letters. The corresponding secondary structures of APN from N. meningitides and protease interacting factor F3 from Thermoplasma acidophilum are shown on the top (black) and the bottom (blue). Helices (α,α-helix; η, 310 helix) appear as small squiggles, beta strands (β, β-strand) as arrows. The following abbreviations were used, with gi accession numbers indicated in parentheses: (GI:7226654) aminopeptidase N N. meningitidis MC58; (GI:67463738) tricorn interacting facor F3 from T. acidophilum; (GI:48146253) human leukotriene A4 hydrolase.
Domain III is positioned at the interface of the catalytic domain and the C-terminal domain. This domain is composed of the nine β-strands β25-β33. They are organized in two pleated anti-parallel β-sheets (β25,β26,β31 and β27,β30,β32,β33) which are packed against each other forming a small β-barrel (see Fig. 1). The β-sheets in the β-barrel are connected via a β-hairpin composed of two short β29 and β30 as well as a number of multiple cross-sheet connections. The structural comparison of domain III against the structures deposited in the PDB using DALI identified ~20 proteins with Z-scores ranging between 6 and 3. The top hits are found in the integrin, alpha v fragment, thigh domain (PDB code 1jv2, Z-score 6, r.m.s.d for 94 Ca atoms = 3.2 Å, sequence identity 9%), and tricorn protease interacting factor F3, domain III (PDB code 1z1w, Z-score 5.7, r.m.s.d for 73 Ca atoms = 3.5 Å, sequence identity 16%).
The C-terminal domain (domain IV) is composed exclusively of 19 α-helices (α8-α27) and forms a right-handed super-helical solenoid structure resembling a baseball glove. This structure could be further subdivided into two layers: fingers and a pocket. The finger layer is formed by the repeating eight units of α/α-helical hairpins. At the second repetition the finger structure is disrupted by the helix α13 which winds across the solenoid toward the C-terminal α26 leading to the formation of the pocket layer. This organization of the domain creates a cavity on the interface with catalytic domain. The structural comparison of the C-terminal domain against PDB deposits using DALI identified ~40 proteins with Z-score ranging between 10 and 3. The top hits are found in the cullin homolog 1 (cul-1) ring-box protein 1 (PDB code 1u6g, Z-score 10.5, r.m.s.d for 256 Ca atoms = 9.2Å, sequence identity 9%), and tricorn protease interacting factor F3, domain III (PDB code 1z1w, Z-score 8.7, r.m.s.d for 237 Ca atoms = 4.5 Å, sequence identity 10%).
Active site
The domains I, II, and III are arranged in a nearly linear manner forming a bottom layer whereas domain IV forms a top layer which encapsulates domains III and II leading to the formation of the large internal cavity with solvent accessible area 4268.3 Å2 and an accessible volume of 4027.1 Å3 as assessed by GPSS server.21 Three sulfate ions are found inside this cavity (two in close proximity of the active site and one on the opposite side of the cavity). The active site is located in the cleft at the center of the catalytic domain facing the cavity, just opposite the interface between domains I and III. Two long α-helices (α2, α3) contribute key conserved residues H293, H297, and E316. These three residues bind a single zinc ion found in the structure. The Zn+2 is coordinated by NE2 of His293 (2.08 Å) and NE2 of His297 (2.09 Å), OE1 of Glu316 (2.00 Å), and a water molecule (2.26 Å) [Fig. 2(A) ]. The active site is flanked from one side by the edge of β-sheet of the catalytic domain, with the strand β20 being the closest to the active site, and by the α-helix (α5) from the opposite side. Both of these regions contain highly-conserved residues among APN and the M1 protease family members. The strand β5 accommodates the G257-X-M259-E260-N261 endopeptidase fingerprint motif, while the α5 carries conserved Y377. Superimposition of the catalytic domain of human leukotriene A4 hydrolase with the inhibitor of aminopeptidases, bestatin bound in the active site with the catalytic domain of APN-Nm shows almost identical positioning of the inhibitor in the active site of APN-Nm. Few close contacts between the bestatin molecule and M256 and Y372 are found, indicating that some rearrangement of these residues would be required in order to accommodate bestatin in the active site of APN-Nm [Fig. 2(B)]. The structural comparisons suggest that bestatin could inhibit APN-Nm.
The APN-Nm appears to be in “closed” conformation and it is unclear how peptides can reach the active site. There are a number of small openings in the domain IV leading to the central cavity. These openings would allow relatively easy access of small peptide or similar molecule to inner cavity as indicated by the presence of three sulfate ions. Therefore, the diameter of the cavity “gates” put significant constraints on the size of the substrates. To process larger peptides, major conformational changes leading to formation of either a larger entrance to the inner chamber or the creation of a large opening would be required. It is possible that a small conformational rearrangement between domains III and IV could lead to the formation of an opening between domains I and IV to allow the passing of larger peptides. Small conformational changes of these domains leading to the increase/decrease of the opening have been observed in the structure of tricorn protease interacting factor F3.19
The crystal structure of aminopeptidase N from N. meningitides determined in this study provides insight into domain organization and the active site of this important enzyme from a human pathogen.
METHODS
Gene cloning and protein expression
The ORF of aminopeptidase N from N. meningitides was amplified by PCR using MC58 genomic DNA (ATCC) with KOD DNA polymerase, using conditions and reagents provided by the vendor (Novagen, Madison, WI). The gene was cloned into a pMCSG7 vector using a modified ligation-independent cloning protocol.22,23 This process generated an expression clone of a fusion protein with an N-terminal His6-tag and a TEV protease recognition site (ENLYFQ↓S). The fusion protein was over-expressed in an E. coli BL21-derivative that harbored a plasmid pMAGIC encoding three rare E. coli tRNAs (Arg [AGG/AGA] and Ile [ATA]) as described earlier.24 This construct provides an N-terminal His6-tag separated from the gene by a TEV protease recognition sequence.
A SeMet derivative of the APN-Nm was prepared as described previously.12 The transformed BL21 cells were grown at 37°C in M9 enriched medium supplemented with 0.4% glucose, 8.5 mM NaCl, 0.1 mM CaCl2, 2 mM MgSO4, and 1% thiamine.25 After the OD600 reached 1.2, 0.01% (w/v) each of leucine, isoleusine, lysine, phenylalanine, threonine, and valine was added to inhibit the metabolic pathway of methionine and encourage SeMet incorporation. About 60 mg of SeMet was added to 1 L of culture and 15 min later protein expression was induced by 1 mM isopropyl-β-d-thiogalactoside (IPTG). The cells were then incubated at 20°C overnight. The harvested cells were resuspended in lysis buffer (500 mM NaCl, 5% glycerol, 50 mM HEPES pH 8.0, 10 mM imidazole, and 10 mM 2-mercaptoethanol) and stored at −80°C.
Protein purification and crystallization
The SeMet-labeled protein was purified according to a standard protocol.23 The harvested cells were resuspended in lysis buffer and lysozyme was added at a final concentration of 1 mg/mL, as was 100 mL of protease inhibitor (Sigma, P8849) per 2 g of wet cells. This mixture was kept on ice for 20 min and then sonicated. The lysate was clarified by centrifugation at 36,000g for 1 h and filtration with a 0.44-μm membrane. The clarified lysate was applied to a 5-mL HiTrap Ni-NTA column (GE Health Systems) on an AKTAexpress system (GE Health Systems). The His6-tagged protein was released with an elution buffer (500 mM NaCl, 5% glycerol, 50 mM HEPES, pH 8.0, 250 mM imidazole, 10 mM 2-mercaptoethanol), and the fusion tag was removed by treatment with recombinant His6-tagged TEV protease (a gift from Dr. D. Waugh, NCI). A Ni-NTA affinity chromatography step was performed using a standard bench-top procedure to remove the His6-tag, uncut protein, and His6-tagged TEV protease. Proteins were concentrated using Centricon concentrators with 5000-MW cutoff (Amicon), flash frozen, and stored in liquid nitrogen.
The aminopeptidase N protein was dialyzed against crystallization buffer containing 200 mM NaCl, 20 mM HEPES pH 8.0, and 2 mM DTT and was concentrated to 40 mg/mL. Crystallization was performed by the sitting-drop vapor-diffusion method at 18°C. Three hundred commercially available conditions were used for the initial screening. One condition produced small crystals and was further optimized using the hanging-drop vapor diffusion method. Triangular prism shaped crystals were grown from 2M ammonium sulfate, 0.1M Bis TRIS pH 6.5. The crystals reached the size 0.3 mm × 0.2 mm × 0.2 mm × 0.4 mm within 5 days and belonged to the rombohedral space group H3 with unit cell parameters a = 224.2 Å, b = 224.2 Å, c = 57.7 Å, and α = β = 90°, γ = 120°. The asymmetric unit contains one molecule with the VM value of 2.93 Å3/Da (solvent content 58%). For data collection the mother liquid containing 20% glycerol was used as a cryoprotectant. The crystal was picked up with a nylon loop (Hampton Research) and flash-frozen in liquid nitrogen.
Data collection and structure determination
The SAD data set was collected on Q315 CCD detector (ADCS) at the 19ID beamline of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory, to 2.05-Å resolution from a single crystal of SeMet-labeled aminopeptidase N (Table I). The experiment set-up involved taking two test diffraction images 90° apart from which we evaluated the optimal exposure time, oscillation range, orientation of the crystal, and resolution of the data to be collected. About 120 diffraction images were recorded near the selenium white line (λ1 = 0.9795 Å). All diffraction data were collected at 100 K. All data sets were processed using the HKL300026 suite of programs. Data collection statistics are presented in Table I.
The structure of the aminopeptidase N has been determined using SAD phasing, density modification, and initial protein model building as implemented in HKL3000 software package.13,14,26–28 The density modified map outputted by SHELXE was subsequently submitted for model building with programs ARP/wARP.29 The almost completed model (90%) obtained from ARP/wARP after 600 cycles of model building was further extended manually by COOT. Cycles of manual corrections of the model were carried on in COOT and with REFMAC of the CCP4.13 The final model was refined against all reflections in the resolution ranges of 40.0–2.05 Å except for 5% randomly selected reflections, which were used for monitoring Rfree. The final round of refinement was carried out by using TLS refinement.30 The quality of the structural model was checked with PROCHECK.15 Final refinement statistics are presented in Table I. All figures were prepared using PyMOL (DeLano, W.L., the PyMOL Molecular Graphics System http://www.pymol.org).
Coordinates
The atomic coordinates for the APN have been deposited in the PDB with the accession code 2GTQ.
Acknowledgments
National Institutes of Health; Grant numbers: GM62414, GM074942; Grant sponsor: U.S. Department of Energy, Office of Biological and Environmental Research; Grant number: W-31-109-Eng-38
We thank Monica Chodkiewicz-Nocek for assistance with figure generation, Andrea Cipriani for preparing the manuscript, and all members of the Structural Biology Center at Argonne National Laboratory for their help in conducting experiments.
Footnotes
The submitted manuscript has been created by the University of Chicago as Operator of Argonne National Laboratory (“Argonne”) under Contract No. W-31-109-ENG-38 with the U.S. Department of Energy. The U.S. Government retains for itself, and others acting on its behalf, a paid-up, nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the Government.
References
- 1.Holz RC, Bzymek KP, Swierczek SI. Co-catalytic metallopeptidases as pharmaceutical targets. Curr Opin Chem Biol. 2003;7:197–206. doi: 10.1016/s1367-5931(03)00033-4. [DOI] [PubMed] [Google Scholar]
- 2.Wilcox DE. Binuclear metallohydrolases. Chem Rev. 1996;96:2435–2458. doi: 10.1021/cr950043b. [DOI] [PubMed] [Google Scholar]
- 3.Gonzales T, Robert-Baudouy J. Bacterial aminopeptidases: properties and functions. FEMS Microbiol Rev. 1996;18:319–344. doi: 10.1111/j.1574-6976.1996.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 4.Copik AJ, Nocek BP, Swierczek SI, Ruebush S, Jang SB, Meng L, D’Souza VM, Peters JW, Bennett B, Holz RC. EPR and X-ray crystallographic characterization of the product-bound form of the MnII-loaded methionyl aminopeptidase from Pyrococcus furiosus. Biochemistry. 2005;44:121–129. doi: 10.1021/bi048123+. [DOI] [PubMed] [Google Scholar]
- 5.Hooper NM. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 6.Lowther WT, Matthews BW. Metalloaminopeptidases: common functional themes in disparate structural surroundings. Chem Rev. 2002;102:4581–4608. doi: 10.1021/cr0101757. [DOI] [PubMed] [Google Scholar]
- 7.Rawlings ND, Barrett AJ. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 8.Wilce MC, Bond CS, Dixon NE, Freeman HC, Guss JM, Lilley PE, Wilce JA. Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3472–3477. doi: 10.1073/pnas.95.7.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawlings ND, Tolle DP, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noble F, Luciani N, Da Nascimento S, Lai-Kuen R, Bischoff L, Chen H, Fournie-Zaluski MC, Roques BP. Binding properties of a highly potent and selective iodinated aminopeptidase N inhibitor appropriate for radioautography. FEBS Lett. 2000;467:81–86. doi: 10.1016/S0014-5793(99)01645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan PS, van Kessel TA, van de Veerdonk FL, Zuurendonk PF, Bruins AP, Konings WN. Degradation and debittering of a tryptic digest from beta-casein by aminopeptidase N from Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1993;59:1430–1436. doi: 10.1128/aem.59.5.1430-1436.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Taking MAD to the extreme: ultrafast protein structure determination. Acta Crystallogr D Biol Crystallogr. 1999;55 (Part 6):1168–1173. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- 13.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50 (Part 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 14.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60 (Part 12, Part 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 15.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 16.Krissinel E, Henrick K. Detection of protein assemblies in crystals. Lect Notes Comput Sci. 2005;3695:163–174. [Google Scholar]
- 17.Thunnissen MM, Nordlund P, Haeggstrom JZ. Crystal structure of human leukotriene A(4) hydrolase, a bifunctional enzyme in inflammation. Nat Struct Biol. 2001;8:131–135. doi: 10.1038/84117. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 19.Kyrieleis OJ, Goettig P, Kiefersauer R, Huber R, Brandstetter H. Crystal structures of the tricorn interacting factor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations. J Mol Biol. 2005;349:787–800. doi: 10.1016/j.jmb.2005.03.070. [DOI] [PubMed] [Google Scholar]
- 20.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60 (Part 12, Part 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 21.Binkowski TA, Joachimiak A, Liang J. Protein surface analysis for function annotation in high-throughput structural genomics pipeline. Protein Sci. 2005;14:2972–2981. doi: 10.1110/ps.051759005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y, Dementieva I, Zhou M, Wu R, Lezondra L, Quartey P, Joachimiak G, Korolev O, Li H, Joachimiak A. Automation of protein purification for structural genomics. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure (Camb) 2001;9:1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stols L, Millard CS, Dementieva I, Donnelly MI. Production of selenomethionine-labeled proteins in two-liter plastic bottles for structure determination. J Struct Funct Genomics. 2004;5:95–102. doi: 10.1023/B:JSFG.0000029196.87615.6e. [DOI] [PubMed] [Google Scholar]
- 26.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62 (Part 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 27.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58(Part 10, Part 2):1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 29.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 30.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62 (Part 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]