Abstract
High-throughput structural genomics projects seek to delineate protein structure space by determining the structure of representatives of all major protein families. Generally this is accomplished by processing numerous proteins through standardized protocols, for the most part involving purification of N-terminally His-tagged proteins. Often proteins that fail this approach are abandoned, but in many cases further effort is warranted because of a protein’s intrinsic value. In addition, failure often occurs relatively far into the path to structure determination, and many failed proteins passed the first critical step, expression as a soluble protein. Salvage pathways seek to recoup the investment in this subset of failed proteins through alternative cloning, nested truncations, chemical modification, mutagenesis, screening buffers, ligands and modifying processing steps. To this end we have developed a series of ligation-independent cloning expression vectors that append various cleavable C-terminal tags instead of the conventional N-terminal tags. In an initial set of 16 proteins that failed with an N-terminal appendage, structures were obtained for C-terminally tagged derivatives of five proteins, including an example for which several alternative salvaging steps had failed. The new vectors allow appending C-terminal His6-tag and His6- and MBP-tags, and are cleavable with TEV or with both TEV and TVMV proteases.
Keywords: LIC, TEV, C-terminal, His-tag, High-throughput, Structural genomics
Introduction
The availability of genome sequence data allows more comprehensive approaches to studies of complex cellular systems. The Structural Genomics programs, such as the Protein Structure Initiative, attempted to use this available genomic information to systematically analyze genomic sequences and select proteins for structure determination based on the need of structural data. The long term Structural Genomics objective is to determine a large number of structures of novel proteins in order to expand structural and functional knowledge. This approach is based on the notion that a structure available for one member of a protein sequence family will provide structural and some functional information for the whole family [1]. Various groups have developed numerous technical approaches to accelerate protein structure determination; these are sometimes referred to as “pipelines” [2]. The high-throughput approach involves processing numerous proteins through standardized protocols that are not optimized for any given protein but work reasonably well and can be applied to many samples. For example, purification of many different proteins from complex mixtures using one protocol and aided by robotic workstations required the use of specific affinity tags. The use of a cleavable N-terminal histidine tag has proven to be the most effective thus far [3]. However, in some cases the N-terminal His-tag may not be accessible for affinity purification. Moreover, the presence of a bulky His-tag or even a few additional “tag artifact” amino acid residues on a protein’s N-terminus may be detrimental to protein folding, solubility or oligomerization. These N-terminally modified proteins will fail in the “pipeline” and may need to be abandoned. In many cases the protein’s intrinsic biological or biomedical value warrant further effort and the use of additional “salvage” pathways. Failure can occur at any stage in the process from gene to structure but many of the failed proteins pass the first critical step, expression as a soluble protein. Several methods exist to salvage proteins that fail by these standard protocols, including reductive methylation, limited proteolysis in situ and in vitro, use of orthologues, nested truncations, co-expression of interacting proteins, ligand screening, mutagenesis and surface entropy reduction [4–10]. In addition, recloning the protein or its domains with or without specific mutations or with alternative tags or fusion partners or cell free expression can sometimes yield structures where the initial efforts failed [11–14].
The protocols applied within the Midwest Center for Structural Genomics (MCSG) approximate the consensus approach of many structural genomics projects [3], and involve ligation-independent-cloning (LIC) of amplified genes into vectors that attach a cleavable N-terminal His-tag to the proteins, purification by Ni-IMAC, specific protease treatment to remove the tag, and subtractive IMAC [15]. LIC vectors are highly flexible and allow cloning of a full-length gene or virtually any fragment using simple protocols.
It has been reported that as an alternative, genes can be cloned into vectors that append C-terminal tags and these constructs can be used for protein purification and structure determination [12, 16–18]. Because no commercially available LIC vectors attach cleavable C-terminal tags, we developed a series of such vectors in an effort to salvage failed proteins. The vectors (Figs. 1 and 2) allow production of native proteins, proteins with a C-terminal His-tag either with or without a TEV protease cleavage site [19, 20], and can also append maltose-binding protein (MBP) associated with a TVMV protease cleavage site to improve protein solubility and allow in vivo removal of MBP [21, 22]. Cloning protocols for the vectors are identical to those for the family of N-terminal vectors developed earlier [23], except for the use of different primers in PCR and different dNTP’s in the LIC reaction. All proteins are expressed and purified by the same standard protocols used for N-terminally tagged proteins [15]. A set of sixteen genes encoding proteins that were soluble when produced as N-terminal His-tagged derivatives but that failed to give structures were recloned into one of the new vectors and reprocessed. Five of the proteins were successfully crystallized and their structures were determined, confirming that recloning genes into C-terminal vectors is an effective alternative to the standard N-terminal tagging approach and should be used in conjunction with other salvaging approaches.
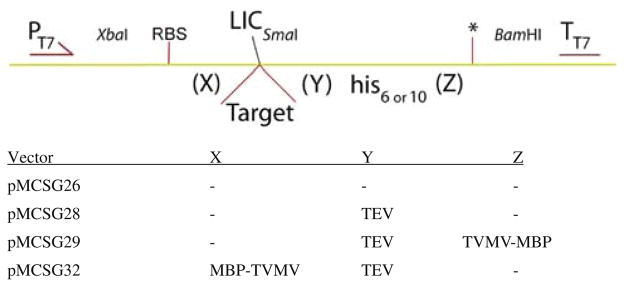
Fig. 1.
Schematic of design of C-terminal LIC vectors
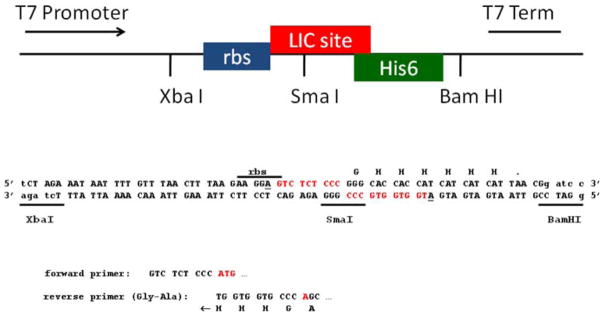
Fig. 2.
Diagrammatic representation of pMCSG26
LIC vectors appending a C-terminal His-tag were constructed by replacement of the cloning region of pMCSG7 [24] between XbaI and BamHI with hybridized nucleotides defining a ribosome binding site followed by a SmaI site and nucleotides encoding a His6-tag followed by a stop codon (*), giving pMCSG26. LIC inserts the target gene into the SmaI site and expression generates a C-terminally His-tagged protein. In derivatives pMCSG28, pMCSG29 and pMCSG32, additional sequences add the TEV protease recognition sequence to the target protein at position Y to allow removal of the His-tag, and either the TVMV protease recognition sequence followed by MBP at position Z or MBP followed by the TVMV site at position X to allow production of MBP fusion proteins and in vivo removal of MBP [21, 22]. Details of primers for the different vectors are given in Table 1.
Table 1.
C-terminal LIC vectors
| Vector | Appended to protein | Primers (N, C)a |
|---|---|---|
| pMCSG26 | C-His6 (or C-TEVb-His6) | 1, 2 |
| pMCSG28 | C-TEV- His6 | 1, 3 |
| pMCSG29c | C-TEV-His10-TVMV-MBP | 1, 3 |
| pMCSG32 | N-MBP-TVMV, C-TEV-His6 | 4d, 5 |
Primers are: 1, GTCTCTCCCATG; 2, TGGTGGTGCCCAGCb; 3, GGTTCTCCCCAGC; 4, GTCTCTCCCAGATGd; 5, GGTTCTCCC CAGC followed by the sequence complementary to the target gene. Inclusion of the gene’s stop codon in reverse primers 2 or 3 will result in production of the native protein without any additional amino acids appended
A TEV recognition site can be introduced by appending its coding sequence to the primer 2 to give TGGTGGTGCCCAGCGGATTGGAAGTACAGGTTCTC
Vector pMCSG29 was derived from pMCSG27, and validation of its elements confirms those of pMCSG27 as well
The final ATG of this primer is complementary to the target gene, and can be eliminated if production of a protein without the encoded methionine is preferred
Cleavage of pMCSG26 with SmaI and treatment with T4 polymerase in the presence of dATP creates 9- and 11-base LIC overhangs, indicated in red. The underscored A codons limit the overhangs. Amplification of target genes with the indicated primers and subsequent treatment with T4 polymerase in the presence of dTTP generate the complementary overhangs and the start codon, indicated in red. The listed reverse primer appends an ala-gly spacer between the protein and the His-tag, generating a protein with 8 additional amino acids at the carboxy terminus: AGHHHHHH. For vectors pMCSG28 and 29, which include a TEV site in the vector, the reverse primer is GGTTCTCCCCAGC.
Materials and methods
Construction of pMCSG26
The cloning region of pMCSG7 was excised by digestion with XbaI and BamHI, and the vector was dephosphorylated and purified by agarose gel electrophoresis. The synthetic oligonucleotides CTAGAAATAATTTTGTTTAACTT-TAAGAAGGAGTCTCTCCCGGGCACCACCATCATCATCATTAACG and GATCCGTTAATGATGATGATGGTGGTGCCCGGGAGAGACTCCTTCTTAAAGTTAAACAAAATTATTT, defining a ribosome binding site, a SmaI restriction site and encoding the His6-tag and stop codon, were hybridized, phosphorylated and ligated into the linearized pMCSG7. The identity of the final vector, pMCSG26, was confirmed by sequencing with the T7 promoter and T7 terminator primers. The sequence of the pMCSG26 LIC region is shown in Fig. 2.
Construction of pMCSG28
The TEV protease recognition sequence was inserted into pMCSG26 to create the vector pMCSG28. The synthetic oligonucleotides CTAGAAATAATTTTGTTTAACTTTAAGAAGGAGTCTCTCCCGGGGAGAA-CCTGTACTTCCAATCCG and CCGGCGGATTGGAAGTACAGGTTCTCCCCG-GGAGAGACTCCTTCTTAAAGTTAAACAAAATTATTT encoding the TEV protease recognition sequence were annealed, phosphorylated and ligated into pMCSG26 that had been digested with XmaI and XbaI and dephosphorylated. The resulting plasmid, pMCSG28, was verified by sequencing the modified region.
Construction of pMCSG29
A sequence encoding the E. coli MBP preceded by a TVMV protease recognition sequence was added to pMCSG26 to make the vector pMCSG27. MBP was amplified by PCR with Platinum Pfx Polymerase (Invitrogen, Carlsbad, CA) using the 81-mer forward primer CACCCCGGGCACCACCATCATCATCATCACCA-CCATCACGAAACCGTGCGTTTCCAGTCTAAAATCGAAGAAGGTAAACTG, which defined the TVMV recognition sequence, and the 36-mer reverse primer GTGGGATCCTTACGAATTAGTCTGCGCGTCTTTCAG. The reaction conditions were 1× Pfx polymerase buffer, 1.5 mM MgSO4, 0.3 mM dNTPs, 1 mM each primer, approximately 250 ng E. coli genomic DNA and 2 units of Platinum Pfx polymerase in a final volume of 100 μl. The reaction was denatured at 94°C for 5 min followed by 35 cycles at 94°C for 30 s, 56°C for 45 s and 68°C for 60 s, followed by a single cycle at 68°C for 10 min. The resulting product was purified by agarose gel electrophoresis, extracted with the Qiaex II Gel Extraction Kit (Qiagen, Valencia, CA), digested with BamHI and XmaI, and ligated into the plasmid pMCSG26 that had been digested with the same enzymes and dephosphorylated. The TEV protease recognition sequence was added to the resulting plasmid, pMCSG27, as described above for construction of pMCSG28, giving vector pMCSG29. The modified regions were verified by sequencing.
Construction of pMCSG32
A variant vector that will produce a cleavable N-terminal MBP and cleavable C-terminal His-tag was made by inserting DNA encoding MBP and a TVMV protease recognition sequence in front of the LIC site of pMCSG28. The DNA fragment was amplified from vector pMCSG19 [21] by PCR with the primers CCCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGG and GAGACCCGGGAGAGACTCTGGAAACGCACGGTTT. The forward primer included the XbaI restriction site from pMCSG19 and the reverse primer included an XmaI restriction site as part of the LIC region. The PCR product was purified, digested with XbaI and XmaI, and ligated into pMCSG28 that had been digested with the same enzymes and dephosphorylated. The identity of the final vector, pMCSG32, was confirmed by sequencing with the T7 primers and primers specific for MBP.
Cloning genes into the vectors
All cloning protocols were identical to those described for the pMCSG series of N-terminal vectors [23] with the following exceptions. Use of SmaI as the LIC site requires that different dNTPs be used in LIC, dATP for the vectors and dTTP for the PCR products. In addition, different PCR primers are required. For vectors pMCSG26-29, the forward primer is GTCTCTCCCATG followed by the sequence of the target gene. For vector pMCSG32 the forward primer requires the addition of 2 bases to put the cloned gene in the same reading frame as MBP, giving GTCTCTCCCAGATG, in which the added AG result in a glutamine preceding the methionine encoded by the final ATG. The final ATG is complementary to the target gene, and can be eliminated if desired. Reverse primers, followed by the complement of the gene, begin: TGGTGGTGCCCAGC for pMCSG26 and pMCSG27 and GGTTCTCCCCAGC for pMCSG28, pMCSG29 and pMCSG32. A TEV recognition site prior to the final His-tag can be introduced into proteins cloned into vectors pMCSG26 and pMCSG27 if TGGTGGTGCCCAGAGGATTGGAAGTACAGGTTCTC is used as the reverse primer. Details of cloning the AsbF gene are given below as an example.
Cloning of B. anthracis asbF gene
The petrobactin biosysnthesis gene asbF of B. anthracis was amplified from a previous clone of the gene inserted into pMCSG7 by standard protocols [23]. PCR was performed using Platinum Pfx with primers GTCTCTCCCATGAA-ATATTCACTATGT and TGGTGGTGCCCAGCAGAAGTTA-CTACTTCTAAATT. Samples were denatured at 94°C for 3 min followed by 35 cycles at 94°C for 30 s, 52°C for 45 s and 68°C for one minute, with a final incubation at 68°C for 10 min in a Robocycler Gradient 96 thermocycler (Stratagene, La Jolla, CA). The PCR product was purified as described above. A second version of the protein with a TEV protease recognition sequence before the His-tag was created by PCR using the same forward primer and the synthetic oligonucleotide TGGTGGTGCCCAGCGGATTGGAAGTACAGGTTCTCAGAAGTTACTACTTCTA-AATT as reverse primer. When expressed in pMCSG26, the protein encoded by this clone includes the TEV protease recognition sequence in addition to the His-tag (in total, -ENLYFQSAGHHHHHH) at the C-terminus. Subsequent cleavage with TEV protease will leave the first six residues of the TEV site (ENLYFQ).
Standard LIC protocols [24] were modified slightly to accommodate the different LIC site. PCR products were treated with T4 DNA polymerase (Novagen, Madison, WI) and dTTP. The pMCSG26 vector was linearized by digestion with SmaI, purified, then treated with T4 DNA polymerase and dATP. The PCR product was annealed to vector DNA by mixing 3 μl T4-treated pMCSG26 with 2 μl T4-treated PCR product in a 14 ml polypropylene tube and incubating on ice for 30 min. Transformation into E. coli was accomplished by the addition of 50 ml Library Efficiency DH5α cells (Invitrogen, Carlsbad, CA) followed by the standard protocol. Clones containing the asbF gene were identified by restriction enzyme digestion and confirmed by sequencing.
Expression and purification of C-terminally tagged proteins
After LIC, vectors were transformed into BL21(DE3) MAGIC cells containing a plasmid encoding rare tRNAs [25]. For small scale analyses, cells were grown in LB and induced with IPTG as described previously [26]. Functionality of the vectors was evaluated by analysis of proteins after purification on a Maxwell 16 Instrument (Promega, Madison, WI) [27], followed by treatment with TEV protease. For vectors pMCSG28 and pMCSG32, proteins were coexpressed with TVMV protease to evaluate in vivo cleavage of MBP [21, 22]. For large scale purifications, proteins were produced as their selenomethionyl (SeMet) derivatives in 2L pop bottles [28], and purified using the AKTA Express System (GE Healthcare) by IMAC, buffer-exchange column, followed by TEV cleavage, and subtractive IMAC [3, 15].
Results
Construction and validation of C-terminal vectors
All four vectors are derivatives of vector pMCSG7 [24] in which that vector’s entire cloning region is replaced by a synthetic DNA sequence or PCR product that defines the new LIC region. For the basal vector, pMCSG26, this region defines a ribosome-binding site followed by a SmaI site, to be used for LIC, and nucleotides encoding a His6-tag followed by a stop codon. Additional sequences in the other three vectors encode the TEV protease recognition site and, for pMCSG29 and pMCSG32, MBP and the TVMV protease site. Sequencing of the cloning region of this vector confirmed the expected nucleotide sequence. All four vectors generated successful clones of PCR products when annealing reactions were performed on ice in spite of their shorter than conventional, 9–11 base pair overhangs [29]. Introduction of a gene encoding a well-behaved, highly soluble protein demonstrated the expected functionality: in all cases, His-tagged targets were generated, TEV treatment removed the tag, and, in the case of pMCSG29 and pMCSG32, coproduction of TVMV protease resulted in in vivo cleavage of MBP. For these two vectors the product after in vivo cleavage consists of a target protein with cleaveable His-tag and resembles the product of pMCSG28.
Salvaging failed proteins as C-terminal fusion derivatives
Sixteen proteins that had failed at various stages (Table 2) as N-terminal derivatives were recloned and reprocessed as C-terminal derivative using pMCSG26 or pMCSG28. All proteins were expressed as their SeMet derivatives and processed by standard MCSG high-throughput protocols. Results obtained with N-terminal and C-terminal derivatives are compared in Table 2. The target proteins were selected based on production of a soluble protein that purified successfully as an N-terminal derivative, so the high degree of success in these two steps with the C-terminal derivatives was expected. Most proteins were produced in similar amounts and show comparable solubilities. However, a few targets faired significantly worse as C-terminal derivatives; targets AsbE, RimM and PmbA showed poor production of soluble protein. The latter two, though cleaved efficiently by TEV protease, did not generate sufficient material for crystallization. On the other hand, other targets performed better in production of suitable crystals as their C-terminal derivatives. AsbB, which generated crystals that diffracted poorly when processed as an N-terminal derivative, produced good-quality crystals as a C-terminal derivative allowing structure determination. TEV protease treatment of N-terminally tagged AsbF failed completely to remove the His-tag and crystals produced from the uncut protein did not diffract X-rays. The C-terminal derivative, in contrast, was readily cleaved, generated excellent crystals, and the structure was solved. Similar success occurred with the regulatory protein TetR. Purification of the N-terminally tagged derivative generated a doublet of two His-tagged proteins, which can be attributed to either partial proteolysis or premature termination of translation for monomeric proteins. The resulting crystal diffracted X-rays, but resolution was poor (3.5 Å). The C-terminally tagged protein gave a single band (as only full-length protein would result in a His-tagged construct) and produced sufficiently ordered crystals to allow the structure to be solved. Interestingly, in the latter case TEV protease failed to cleave the His-tag, whereas it had effectively removed the N-terminal tag, but the protein still produced better crystals even with the His6-tag attached to its C-terminus. The CD3330 and DapE produced soluble protein with N-terminal His6-tag but TEV protease failed to cleave the tag off and these proteins did not crystallize. With a C-terminal tag, both proteins behaved well and produced well diffracting crystals leading to structure. For some of the other proteins, C-terminal derivatives gave somewhat improved results in crystallization. For example AsbB with a C-terminal tag produced diffraction quality crystals allowing structure solution. Proteins AsbC, CaiA and Crl produced crystals as C-terminal derivatives whereas none were obtained from the N-terminal tagged proteins though their structures were not solved, thus far. The remaining proteins gave comparable results with both N- and C-terminal derivitization. In summary, from a set of 16 proteins, 7 with N-terminal tags produced crystals, but no structure could be determined. In contrast, 10 proteins with C-terminal tags crystallized and 5 structures have been determined: TetR (PDB id 3f0c), AsbB (Kim et al., manuscript in preparation), AsbF (PDB id 3dx5, [30]), DapE (Nocek et al. manuscript in preparation, PDB id 3ic1) and CD3330 (Tan et al. manuscript in preparation, PDB id 3ivp).
Table 2.
Comparison of processing of N-terminally and C-terminally tagged proteins
| Protein | N-terminally taggeda |
C-terminally tagged |
||||||
|---|---|---|---|---|---|---|---|---|
| Solubilityb | Cleavedc | Crystals, diffraction limit | Solvedd | Solubilityb | Cleavedc (%) | Crystals, dif fraction limit | Solvedd | |
| AsbA | 0.5 | + | Microcrystals | – | 0.5 | 10–20e | – | – |
| AsbB | 3 | 100% | 4.5 Å | – | 3 | 80 | 2 Å | In progress |
| AsbC | 2 | 50% | – | – | 3 | 80 | + no diffraction | – |
| AsbD | 3 | 90% | – | – | 2 | 100 | – | – |
| AsbE | 3 | 100% | + no diffraction | – | 0 | – | – | – |
| AsbF | 3 | 0% | + no diffraction | – | 3 | 100 | 2 12 Å | 3dx5 |
| RimM | 3 | 40% | Microcrystals | – | 1 | 100 | – | – |
| CaiA | 3 | 0% | – | – | 3 | 100 | 3.1 Å | – |
| DapE | 3 | 0% | – | – | 3 | 100 | 2.5 Å | 3ic1 |
| Crl | 3 | 100% | 7.5 Å | – | 2 | 100 | + no diffraction | – |
| RecF | 2 | 100% | 10 Å | – | 1 | 0 | – | – |
| PmbA | 3 | 50% | 7.5 Å | – | 0.5 | n/a | – | – |
| YwiE | 2f | – | – | – | 2 | 0 | 7Å | – |
| BF1701 | 3f | – | – | – | 3 | 50 | 3.5 Å | – |
| CD3330 | 2f | – | – | – | 3 | 100 | 2.02 Å | 3ivp |
| TetR | 3 doubletg | 100% doubletg | 3.5 Å | – | 3 | 0 | 2.96 Å | 3f0c |
Expression from vector pMCSG7 or pMCSG19 gave N-terminal tags with His6-TEV site or MBP-TVMV site—His6-TEV site, respectively
Solubility scores were based on yields of soluble protein on gels, where 0 indicates no soluble protein, and 0.5 means very low; 1—low; 2—moderate; and 3—very good production of soluble protein
Cleavage with TEV protease is given in approximate percentage (cleavage estimated by visual evaluation of gels)
Protein Data Bank ID numbers are given for deposited structures
Since cleavage was very poor, only uncut protein was used in crystallization experiments
Poor or no Ni-binding of the protein at the purification stage
Purification yielded a doublet of proteins of similar molecular weight, both of which cleaved with TEV protease
Discussion
As structural genomics pipelines are being applied to a broader range of proteins, it becomes clear that the current standard protocols need to be amended to include approaches better suitable for specific classes of proteins. For example, it would be unwise to purify oxidation sensitive proteins under oxidative conditions. Instead, effective high-throughput methods must be developed to handle such proteins. In the current structural genomics pipelines, the bioinformatics approaches help to select proteins for a particular set of protocols. A significant fraction of proteins still fail to yield structures using these standard protocols. Analysis of large sets of data may reveal which alternative strategies may be more effective in expression, purification and crystallization of these proteins. The use of surface entropy reduction or reductive methylation to alter proteins’ properties, the use of detergents to improve solubility or of ligands to improve protein stability or reduce conformational flexibility can serve as examples. Salvaging approaches can have very different costs associated and success rates. For example, in situ proteolysis or reductive methylation [9, 10], although having not very high success rates, can take advantage of existing preparations of purified proteins, require little effort and are very cost effective. Similarly, re-cloning of the gene with specific mutations, such as those that reduce surface entropy, or to produce nested truncations can also rescue failed proteins [8, 14], although at a higher cost. However, these approaches still cannot rescue all proteins and additional high-throughput approaches are needed. Expression of proteins with C-terminal tags has been reported previously [12, 16–18] and was used extensively with GFP reporters for evaluation of protein expression and solubility [31]. One of limitation of C-terminal fusions is difficulty in generating protein targets with no or small sequence “artifacts” on the C-terminus.
Here we exploited an alternative to the N-terminal location of affinity and solubility tags to expand the number of salvaging approaches for addressing recalcitrant proteins. We developed a series of LIC vectors that allow, using simple protocols, appending different C-terminal tags to proteins that had failed as N-terminal derivatives. At this same time we allow these fusion proteins to be processed by the standard structure determination pipeline protocols. The procedure requires re-amplification and re-cloning of the gene, but because of the design of the vectors, established automated cloning protocols can be used. In addition, because only targets that expressed as soluble proteins are processed, the success rate of obtaining soluble proteins is much higher (81%) as compared with untested targets (44%; www.mcsg.anl.gov-statistics tab, progress, soluble/expressed). The vectors performed as designed and allow purification of His-tagged proteins by standard methods, removal of the tag with TEV protease, and attachment of MBP, which can be removed by in vivo or in vitro proteolysis by TVMV protease. In the examples presented here, sixteen proteins that had failed with N-terminal tags were reprocessed in one of these vectors to give C-terminal derivatives, yielding five structures, TetR—a trancription factor, AsbB and AsbF—a siderophore condensing enzyme and 3-dehydroshikimate dehydratase, CD3330—transposon-related DNA-binding protein, and DapE—N-succinyl-L,L-diaminopimelic acid desuccinylase, respectively, involved in petrobactin biosynthesis.
The rate of success, based on a small set of 16 proteins, is quite high—10 out of 16 tested crystallized (62.5%) and 5 out of 16 (31.3%) yielded high-quality structures. Though based on a small set, this number is notably higher than that obtained routinely for new clones. However, these proteins represent a subset that was in effect prescreened for solubility and purification as N-terminal derivatives, thus eliminating highly insoluble proteins. Compared to the normal rate of success from purification to structure, the rate of success is higher than that achieved for untested targets. When compared to the rate of success with other salvage methods, the success rate is also high. In the protocols used within the MCSG, reductive methylation generates a successful solution of structures for approximately 7% of the proteins passed through the process. Proteolysis yields approximately 12% success from failed proteins. Importantly, however, both of these latter procedures involve the use of purified protein and do not require backing up in the process to reclone and purify the protein, so lower success rates are acceptable.
C-terminal tags have been shown previously to be preferable to N-terminal tags for producing some proteins [12], but are generally thought less attractive for structural studies because of the lack of a specific protease that removes the tag. For N-terminal tags, the TEV protease and related enzymes offer very high-specificity and cutting at the C-terminal end, leaving only a single amino acid of the recognition sequence attached to the protein. No similar set of proteases is yet available that cuts with such high specificity near the N-terminal end of the recognition sequence. The use of the TEV protease cleavage site at the C-terminus of the proteins results in 6 residues being added, and for this reason LIC vectors appending N-terminal tags are preferred for the initial cloning of target genes. However, previous results [18] as well as the results described here show that these extra residues often do not preclude solution of the proteins’ structure. One of these, TetR, was solved in spite of the fact that TEV failed to cleave the C-terminally tagged derivative, leaving the final His-tag attached as well. These results indicate the inclusion of these extra amino acids is not fundamentally detrimental and should not rule out this approach for salvaging failed proteins. Use of the vectors as a salvage rather than a parallel pathway saves cost, and the inclusion of the TEV protease site before the terminal His-tag also allows use of the common structural genomics step of subtractive IMAC to remove E. coli proteins that bind Ni-IMAC resins [15].
So one can ask the question, why do C-terminal tags make a difference? Analysis of our five structures showed that for AsbF, the N-terminus is partially buried; the first helix of the protein is beneath the N-terminal amino acid itself and seems to turn down into the protein. Most likely, with the N-terminal tag, the cleavage-site may not be accessible to a TEV protease to cut the N-terminal tag off. Consequently, the N-terminal tag could interfere with proper folding or ability to assume the final conformation needed for favorable packing for well-ordered crystals. In contrast, as the C-terminus is open completely to the solvent, the C-terminal tag has little effect on structure or packing. Similarly in DapE N-terminal Met is partially buried and is involved in hydrophobic interactions on the protein surface, but C-terminus is open to solvent. In the case of AsbB, both the C- and the N-termini are on the surface; however, the N-terminus is packed between protein chains. Although an N-terminal tag could be cleaved, it leaves three additional residues, which may disturb interaction between the N-terminus and the protein body as well as crystal packing. But the C-terminus is more open to the surface partially disordered, suggesting the structure there is less critical to protein folding or crystallization. In the case of CD3330 the N-terminus is involved in dimer formation and also in crystal packing but the C-terminus is open to solvent. The case of TetR is more ambiguous because the His-tag is not visible in electron density and both N- and C-termini are accessible.
C-terminal tags offer certain advantages that may contribute to their apparent effectiveness. Because the His-tag is at the end of the protein, only full-length proteins will be tagged and proteins truncated by premature termination of translation will not be purified by the Ni-IMAC column. Such premature termination occurs with some mammalian proteins expressed in prokaryotes, resulting in multiple bands of overexpressd proteins on gels, and can in many cases be attributed to frame shifts or release of message from the ribosome at rare codons [32–34]. The use of strains that produce elevated amounts of rare tRNAs largely obviates this problem, but does not eliminate it completely. Trace amounts of truncated proteins not readily detected on gels may in some cases interfere with crystallization or result in poorly ordered crystals or mosaicity. One of the solved proteins in this study, TetR, produced two bands as an N-terminally tagged derivative, possibly due to such truncation. However, the C-terminally modified protein yielded a single band even before purification, so proteolysis could not be ruled out. Another possible advantage to C-terminal tags is that overexpression produces the native N-terminus, avoiding possible disruption of the normal folding process by the appended leader sequence. In addition, there are many examples in which the N-terminus of a protein is involved in its function, making the ability to produce C-terminally tagged proteins a valuable capability.
The new set of vectors affords another salvage pathway for important proteins, providing not just C-terminal His-tags, but also flexibility in cloning and processing. MBP can be attached either C-terminally (pMCSG29) or N-terminally (pMCSG32), and in both cases can be removed separately from the His-tag by the use of TVMV protease, either in vivo during expression or after lysis or purification if it is beneficial to either folding or processing of the protein. The primary vectors, pMCSG28 and pMCSG29, encode the TEV recognition site and accept the same PCR products, which in turn require only short primers, reducing the chance of synthesis errors. Vector pMCSG26 lacks the encoded TEV site. Introduction of the site into cloned genes requires the use of longer primers, but this approach also allows introduction of different protease sites if desired or, through the inclusion of a stop codon in the reverse primer, production of the native protein. This flexibility could be very advantageous if alternative proteases are discovered with desirable properties, or if native proteins are preferred, for example in co-expression with a tagged interacting protein to minimize possible interference with protein interaction or eliminate the need of full cleavage of two tags in the interacting pair. If one of the tags is not a His-tag, partial cleavage, while not interfering with purification, could interfere with crystallization by generating heterogeneity in the complex.
Conclusions
We report here development of a series of LIC vectors that allow production of native proteins with a C-terminal His-tag either with or without a TEV protease cleavage site [19, 20], or with a C-terminal His-tag plus maltose-binding protein (MBP) associated with a TVMV protease cleavage site to improve protein solubility and allow in vivo removal of MBP [21, 22]. Cloning protocols for the vectors are identical to those for the family of N-terminal vectors developed earlier [23], except for the use of different primers in PCR and different dNTP’s in the LIC reaction. All proteins are expressed and purified by the same standard protocols used for N-terminally tagged proteins [15]. A set of sixteen genes encoding proteins that were soluble when produced as N-terminal His-tagged derivatives but that failed to give structures were recloned into one of the new vectors and reprocessed. The proteins included ten arbitrarily chosen structural genomics targets and all six genes of the B. anthracis petrobactin biosynthesis pathway. Collectively these latter proteins synthesize petrobactin, a virulence factor for B. anthracis and are potential drug targets [35–37]. Five of the proteins: TetR (a transcriptional factor of the TetR family from C. hutchinsonii, PDB id 3f0c), DapE (a N-succinyl-L,L-diaminopimelic acid desuccinylase from Haemophilus influenzae, PDB id 3ic1), CD3330 (a transposon-related DNA-binding protein from Clostridium difficile, PDB id 3ivp), AsbB (a siderophore condensing enzyme of B. anthracis petrobactin biosysthesis), and AsbF (the dehydroshikimate dehydratase of petrobactin biosynthesis, PDB id 3dx5) were successfully crystallized and their structures were determined. The current rate of successful structure determination, five structures determined from among sixteen target proteins evaluated, confirms that recloning genes into C-terminal vectors is an effective alternative to the standard N-terminal tagging approach, and should be used in conjunction with other salvaging approaches. The C-terminal vectors also provide an attractive platform for the initial cloning and expression of target genes, in addition to serving in salvaging functions.
Acknowledgments
We thank David Sherman for template DNA for recloning the B. anthracis asb genes. This work was supported by the National Institutes of Health Grant GM074942, the Contract No. HHSN272200700058C, and by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Footnotes
The submitted manuscript has been created by the University of Chicago as operator of Argonne National Laboratory under Contract No. W-31-109-ENG-38 with the U.S. Department of Energy. The U.S. government retains for itself, and others acting on its behalf, a paid-up, nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the government.
Contributor Information
William H. Eschenfeldt, Midwest Center for Structural Genomics, Biosciences Division, Argonne National Laboratory, Bldg. 202/Rm. BE111, 9700 South Cass Avenue, Argonne, IL 60439, USA
Natalia Maltseva, Center for Structural Genomics of Infectious Diseases, Computational Institute, University of Chicago, Chicago, IL 60667, USA.
Lucy Stols, Midwest Center for Structural Genomics, Biosciences Division, Argonne National Laboratory, Bldg. 202/Rm. BE111, 9700 South Cass Avenue, Argonne, IL 60439, USA.
Mark I. Donnelly, Midwest Center for Structural Genomics, Biosciences Division, Argonne National Laboratory, Bldg. 202/Rm. BE111, 9700 South Cass Avenue, Argonne, IL 60439, USA
Minyi Gu, Midwest Center for Structural Genomics, Biosciences Division, Argonne National Laboratory, Bldg. 202/Rm. BE111, 9700 South Cass Avenue, Argonne, IL 60439, USA; Center for Structural Genomics of Infectious Diseases, Computational Institute, University of Chicago, Chicago, IL 60667, USA.
Boguslaw Nocek, Center for Structural Genomics of Infectious Diseases, Computational Institute, University of Chicago, Chicago, IL 60667, USA.
Kemin Tan, Midwest Center for Structural Genomics, Biosciences Division, Argonne National Laboratory, Bldg. 202/Rm. BE111, 9700 South Cass Avenue, Argonne, IL 60439, USA.
Youngchang Kim, Midwest Center for Structural Genomics, Biosciences Division, Argonne National Laboratory, Bldg. 202/Rm. BE111, 9700 South Cass Avenue, Argonne, IL 60439, USA; Center for Structural Genomics of Infectious Diseases, Computational Institute, University of Chicago, Chicago, IL 60667, USA.
Andrzej Joachimiak, Email: andrzejj@anl.gov, Midwest Center for Structural Genomics, Biosciences Division, Argonne National Laboratory, Bldg. 202/Rm. BE111, 9700 South Cass Avenue, Argonne, IL 60439, USA; Center for Structural Genomics of Infectious Diseases, Computational Institute, University of Chicago, Chicago, IL 60667, USA.
References
- 1.Marsden RL, Orengo CA. Methods Mol Biol. 2008;426:3–25. doi: 10.1007/978-1-60327-058-8_1. [DOI] [PubMed] [Google Scholar]
- 2.Fox BG, Goulding C, Malkowski MG, Stewart L, Deacon A. Nat Methods. 2008;5:129–132. doi: 10.1038/nmeth0208-129. [DOI] [PubMed] [Google Scholar]
- 3.Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC. Nat Methods. 2008;5:135–146. [Google Scholar]
- 4.Cooper DR, Boczek T, Grelewska K, Pinkowska M, Sikorska M, Zawadzki M, Derewenda Z. Acta Crystallogr D Biol Crystallogr. 2007;63:636–645. doi: 10.1107/S0907444907010931. [DOI] [PubMed] [Google Scholar]
- 5.Kim Y, Quartey P, Li H, Volkart L, Hatzos C, Chang C, Nocek B, Cuff M, Osipiuk J, Tan K, Fan Y, Bigelow L, Maltseva N, Wu R, Borovilos M, Duggan E, Zhou M, Binkowski TA, Zhang RG, Joachimiak A. Nat Methods. 2008;5:853–854. doi: 10.1038/nmeth1008-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wernimont A, Edwards A. PLoS One. 2009;4:e5094. doi: 10.1371/journal.pone.0005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao X, Bain K, Bonanno JB, Buchanan M, Henderson D, Lorimer D, Marsh C, Reynes JA, Sauder JM, Schwinn K, Thai C, Burley SK. J Struct Funct Genomics. 2005;6:129–134. doi: 10.1007/s10969-005-1918-5. [DOI] [PubMed] [Google Scholar]
- 8.Derewenda ZS. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Dong A, Xu X, Edwards AM, Chang C, Chruszcz M, Cuff M, Cymborowski M, Di Leo R, Egorova O, Evdokimova E, Filippova E, Gu J, Guthrie J, Ignatchenko A, Joachimiak A, Klostermann N, Kim Y, Korniyenko Y, Minor W, Que Q, Savchenko A, Skarina T, Tan K, Yakunin A, Yee A, Yim V, Zhang R, Zheng H, Akutsu M, Arrowsmith C, Avvakumov GV, Bochkarev A, Dahlgren LG, Dhe-Paganon S, Dimov S, Dombrovski L, Finerty P, Jr, Flodin S, Flores A, Graslund S, Hammerstrom M, Herman MD, Hong BS, Hui R, Johansson I, Liu Y, Nilsson M, Nedyalkova L, Nordlund P, Nyman T, Min J, Ouyang H, Park HW, Qi C, Rabeh W, Shen L, Shen Y, Sukumard D, Tempel W, Tong Y, Tresagues L, Vedadi M, Walker JR, Weigelt J, Welin M, Wu H, Xiao T, Zeng H, Zhu H. Nat Methods. 2007;4:1019–1021. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayment I. Methods Enzymol. 1997;276:171–179. [PubMed] [Google Scholar]
- 11.Vinarov DA, Newman CL, Tyler EM, Markley JL, Shahan MN. Curr Protoc Protein Sci. 2006;Chapter 5(Unit 5):18. doi: 10.1002/0471140864.ps0518s44. [DOI] [PubMed] [Google Scholar]
- 12.Dyson MR, Shadbolt SP, Vincent KJ, Perera RL, McCafferty J. BMC Biotechnol. 2004;4:32. doi: 10.1186/1472-6750-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox JD, Waugh DS. Methods Mol Biol. 2003;205:99–117. doi: 10.1385/1-59259-301-1:99. [DOI] [PubMed] [Google Scholar]
- 14.Klock HE, Koesema EJ, Knuth MW, Lesley SA. Proteins. 2008;71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y, Dementieva I, Zhou M, Wu R, Lezondra L, Quartey P, Joachimiak G, Korolev O, Li H, Joachimiak A. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biedendieck R, Yang Y, Deckwer WD, Malten M, Jahn D. Biotechnol Bioeng. 2007;96:525–537. doi: 10.1002/bit.21145. [DOI] [PubMed] [Google Scholar]
- 17.Lee J, Kim SH. Protein Expr Purif. 2009;63:58–61. doi: 10.1016/j.pep.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisedchaisri G, Chou CJ, Wu M, Roach C, Rice AE, Holmes RK, Beeson C, Hol WG. Biochemistry. 2007;46:436–447. doi: 10.1021/bi0609826. [DOI] [PubMed] [Google Scholar]
- 19.Dougherty WG, Carrington JC, Cary SM, Parks TD. EMBO J. 1988;7:1281–1287. doi: 10.1002/j.1460-2075.1988.tb02942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 21.Donnelly MI, Zhou M, Millard CS, Clancy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. Protein Expr Purif. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nallamsetty S, Kapust RB, Tozser J, Cherry S, Tropea JE, Copeland TD, Waugh DS. Protein Expr Purif. 2004;38:108–115. doi: 10.1016/j.pep.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Eschenfeldt WH, Stols L, Millard CS, Joachimiak A, Donnelly MI. Methods Mol Biol. 2009;498:105–115. doi: 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 25.Dieckman L, Gu M, Stols L, Donnelly MI, Collart FR. Protein Expr Purif. 2002;25:1–7. doi: 10.1006/prep.2001.1602. [DOI] [PubMed] [Google Scholar]
- 26.Stols L, Zhou M, Eschenfeldt WH, Millard CS, Abdullah J, Collart FR, Kim Y, Donnelly MI. Protein Expr Purif. 2007;53:396–403. doi: 10.1016/j.pep.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Frederick RO, Bergeman L, Blommel PG, Bailey LJ, McCoy JG, Song J, Meske L, Bingman CA, Riters M, Dillon NA, Kunert J, Yoon JW, Lim A, Cassidy M, Bunge J, Aceti DJ, Primm JG, Markley JL, Phillips GN, Jr, Fox BG. J Struct Funct Genomics. 2007;8:153–166. doi: 10.1007/s10969-007-9032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stols L, Millard CS, Dementieva I, Donnelly MI. J Struct Funct Genomics. 2004;5:95–102. doi: 10.1023/B:JSFG.0000029196.87615.6e. [DOI] [PubMed] [Google Scholar]
- 29.Aslanidis C, de Jong PJ, Schmitz G. PCR Methods Appl. 1994;4:172–177. doi: 10.1101/gr.4.3.172. [DOI] [PubMed] [Google Scholar]
- 30.Pfleger BF, Kim Y, Nusca TD, Maltseva N, Lee JY, Rath CM, Scaglione JB, Janes BK, Anderson EC, Bergman NH, Hanna PC, Joachimiak A, Sherman DH. Proc Natl Acad Sci U S A. 2008;105:17133–17138. doi: 10.1073/pnas.0808118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabantous S, Rogers Y, Terwilliger TC, Waldo GS. PLoS One. 2008;3:e2387. doi: 10.1371/journal.pone.0002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao W, Tyagi S, Kramer FR, Goldman E. Mol Microbiol. 1997;25:707–716. doi: 10.1046/j.1365-2958.1997.5081871.x. [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Lee SB. Protein Expr Purif. 2006;50:49–57. doi: 10.1016/j.pep.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Spanjaard RA, Chen K, Walker JR, van Duin J. Nucleic Acids Res. 1990;18:5031–5036. doi: 10.1093/nar/18.17.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abergel RJ, Wilson MK, Arceneaux JE, Hoette TM, Strong RK, Byers BR, Raymond KN. Proc Natl Acad Sci U S A. 2006;103:18499–18503. doi: 10.1073/pnas.0607055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cendrowski S, MacArthur W, Hanna P. Mol Microbiol. 2004;51:407–417. doi: 10.1046/j.1365-2958.2003.03861.x. [DOI] [PubMed] [Google Scholar]
- 37.Miethke M, Marahiel MA. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]