Abstract
Pax6, a mammalian homolog of the Drosophila paired box gene family member expressed in stem and progenitor cells, resides at the top of the genetic hierarchy in controlling cell fates and morphogenesis. While Pax6 activation can lead to mitotic arrest, premature neurogenesis, and apoptosis, the underlying molecular mechanisms have not been resolved. Here we report that either Pax6(+5a) or Pax6(−5a) was sufficient to promote, whereas their knockdown reduced the expression of δ-catenin (CTNND2), a neural specific member of the armadillo/β-catenin superfamily. Pax6(+5a) elicited stronger effects on δ-catenin than Pax6(−5a). Inducible Pax6(+5a) expression demonstrated a biphasic and dose-dependent regulation of δ-catenin expression and cell fates. A moderate upregulation of Pax6(+5a) promoted δ-catenin expression and induced neurite-like cellular protrusions, but increasing expression of Pax6(+5a) reversed these processes. Furthermore, sustained high expression of Pax6(+5a) triggered apoptosis as determined by the reduction of phospho-Bad, Bcl-2, survivin and procaspases, as well as the increases in Bax and cleaved poly(ADP-ribose) polymerase. Importantly, re-introducing δ-catenin by ectopic expression elicited a feedback suppression on Pax6(+5a) expression and reduced Pax6(+5a) induced apoptosis. Therefore, δ-catenin expression is not only controlled by Pax6, but it also provides a feedback suppression mechanism for their functional interactions with important implications in cellular morphogenesis, apoptosis, and cancer.
Keywords: Paired box gene, δ-catenin, cell fates, apoptosis, feedback regulation
Introduction
Transcription factor Pax6 (Paired box 6) was first identified as a Pax family member and cloned based on its homology to the Drosophila gene, paired [1]. Pax6 resides at the top of the genetic hierarchy in the development of several ectoderm-derived organs and controls a series of critical steps in early eye development and neuronal differentiation [2–4]. Pax6 is a highly conserved member of a family of genes encoding transcription factors that contain two DNA-binding domains—the paired domain (PD) and the homeodomain (HD)—and a transcriptional activation domain. Pax6 gene produces two major isoforms by alternative splicing, namely, Pax6(+5a) and Pax6(−5a). Pax6(−5a) differs from Pax6(+5a) by the absence of an exon 5a-encoded 14 amino acid insertion in its paired-type DNA-binding domain (paired domain or PD). Pax6(+5a) and Pax6(−5a) show distinct DNA-binding properties to the promoters of their downstream target proteins [1, 5]. Pax6 activates many downstream target genes to control cell fate and morphogenesis although the underlying molecular mechanisms are not well understood [6–8]. Pax6(+5a) inhibits cell proliferation in a dose-dependent manner but not cell fate, and exogenously expressed Pax6 can activated the downstream target in dosage-dependent manner [9]; while Pax6(−5a) affects both proliferation and cell fate [10]. Full-length Pax6 has also been suggested to induce caspase-3 independent apoptosis in a small percentage of cells [11].
δ-Catenin (CTNND2), or neural plakophilin-related arm protein (NPRAP)/Neurojungin, is a brain specific synaptic junction protein of armadillo/β-catenin superfamily implicated in brain and eye development [12–14], learning and cognitive functions [15], and cancer formation [16, 17]. It was mapped to human chromosome 5p15, and was initially identified as interacting with Alzheimer’s disease protein presenilin [13, 18, 19]. Deletion of chromosome 5p15.2 where δ-catenin gene is located appears to link to the severe mental retardation phenotypes of Cri-du- Chat syndrome [20]. Pax6 (Chromosome 11p13), with its expression in the brain, eye, and pancreas during embryogenesis and postnatal development, shows remarkably similar distribution to that of δ-catenin [13, 21]. Interestingly, both Pax6 and δ-catenin can induce neurite-like extensions in non-neuronal cells associated with cell shape change [22, 23]. They also show profound effects on cell cycle and cell survival gene profiles [11, 17].
Recent studies showed that δ-catenin expression in the eye and brain was severely reduced when Pax6 was mutated in mice, suggesting that Pax6 is essential for the expression of δ-catenin [24]. Furthermore, examination of human EST data bank revealed δ-catenin mRNA sequences in prostate, kidney, ovarian, brain, breast, and esophageal tumors. Altered expression of δ-catenin was associated with cancer formation [16, 25], and Pax6 enhanced δ-catenin expression in prostate cancer cells [26]. In this study, we demonstrate, for the first time, that Pax6(+5a) and Pax6(−5a) regulate δ-catenin expression in an isoform- and dose-sensitive manner, but δ-catenin also exerts a feedback suppression on Pax6 with important implications in cellular morphogenesis, apoptosis, and cancer.
Materials and Methods
Cell lines
Y79 (Human retinoblastoma line), ARPE-19 (Human retinal pigment epithelial cell line), CWR22Rv-1 (Human prostate cancer derived cell line), HeLa (Human cervical cancer cells taken from Henrietta Lacks), NIH3T3 (mouse, NIH Swiss, embryo) were obtained from the American Type Culture Collection (ATCC, Rockville, MD). Y79 cells were suspension cultured in RPMI 1640 supplement with 15% fetal bovine serum (FBS), penicillin (100 units/ml), and streptomycin (100 units/ml) (Gibco BRL, Rockville, MA). ARPE-19 cells were grown in Dulbecco's modified Eagle's medium/nutrient F12 (DMEM-F12) supplemented with 10% fetal bovine serum and 25 mg/ml gentamycin. HeLa and NIH3T3 were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 units/ml) (Gibco BRL, Rockville, MA). A stable tetracycline repressor HeLa cell line (HeLa Tat-TetR-Pax6) in which expression of Pax6(+5a) was under the control of a tetracycline inducible promoter was cultured in DMEM supplemented with 10% fetal bovine serum and), penicillin (100 units/ml), and streptomycin (100 units/ml) (Gibco BRL, Rockville, MA) with G418 in the medium. This cell line expresses both the bacterial tetracycline repressor (TetR or TR) from the CMV promoter, as well as Pax6 under the control of a tetracycline inducible promoter called CMVTetO2 (Invitrogen). Pax6(+5a) expression was induced by the addition of 0.4 µg/ml Doxycycline (derivative of tetracycline) into medium, a dose that did not induce cell death. All cultures were maintained at 37°C with 5% CO2 atmosphere.
Plasmids constructions of Pax6(+5a), Pax6(−5a), δ-catenin and shRNAs for Pax6(+5a) and Pax6(−5a)
To subclone Pax6(+5a) and Pax6(−5a) into pcDNA3.1 vector, we amplified Pax6(−5a) CDS from pCMV-Pax6(−5a) vector [23] with primers: forward sequence, 5’-CCGAATTCTGCAGACCCATGCAGA-3’; reverse sequence, 5’-CATTTGGCCCTTCGATTAGA-3’. An exon 5a-encoded 42 base pair in addition to linker sequence was amplified with primers: forward sequence, 5’-CCGAATTCTGCAGACCCATGCAGA-3’; reverse sequence, 5’-CATTTGGCCCTTCGATTAGA-3’. Pax6(+5a) and Pax6(−5a) were subcloned into pcDNA3.1 by ligation of the HindIII-BamHI fragment. After plasmid construction, the sequences of all vectors were confirmed by DNA sequencing and found to be in frame. Full-length δ-catenin in pEGFP-C1 was constructed as described [22].
Specific shRNAs directed against human Pax6(+5a) or Pax6(−5a) nucleotide sequences were designed using the criteria established by Tuschl [27] and generated by Origene Technologies (USA). The target oligonucleotide sequences were as follows: Pax6(+5a) 5'-ATGCAGATGCAAAAGTCCAAGTGCTGGACA-3' and Pax6(−5a) 5'-ACACTTGAGCCATCACCAATCAGCATAGG-3'. A shRNA pRS plasmid was used as a vector control.
Transfection of cultured Y79 cells
Y79 cells were grown in RPMI 1640 as described [28]. After reaching 85% confluence, cells were respectively transiently transfected with 2.0 µg Pax6(+5a) or Pax6(−5a) plasmids + 20 µl Lipofectamine™ 2000 (Invitrogen, 1 mg/mL) per plate according to the manufacturer’s instructions. Cells transfected with pcDNA3.1 (Clontech) were used as a vector control. The shRNA for Pax6(+5a) or Pax6(−5a) were transfected into Y79 cells using Lipofectamine 2000 (Invitrogen) as follows: 1.0 µg Pax6(+5a) or Pax6(−5a) shRNA + 20 µl Lipofectamine™ 2000 (Invitrogen, 1 mg/mL) per plate, and the empty shRNA pRS were used as a vector control.
HeLa cell line with Pax6 transfection and HeLa Tat-TetR-Pax6 cell line with δ-catenin transfection
HeLa cells were grown in DMEM as described [29]. After reaching 85% confluence, HeLa cells were transiently transfected with 2.0 µg Pax6(+5a) plasmids + 20 µl Lipofectamine™ 2000 (Invitrogen, 1 mg/mL) per plate, according to the manufacturer’s instructions. Cells transfected with pcDNA3.1 (Clontech) were used as a vector control.
HeLa Tat-TetR-Pax6 cells were grown in DMEM as described [23]. To generate HeLa Tat-TetR-Pax6 cells, TetR and Pax6(+5a) were first subcloned into pDONR221 (Invitrogen). Subsequently, TetR was recombined with the CMV promoter in the 2K7bsd construct and Pax6 was recombined in with the CMVTetO2 promoter (Invitrogen) in the 2K7bsd construct, as described [30]. Lentivectors were produced, and 2ml of supernatant was used to transduce HeLa-Tat cells. HeLa-Tat cells transfetced with TetR were designated as HeLa Tat-TetR-Pax6 cells and induced to express Pax6 under the control of the inducible CMVTetO2 promoter. After reaching 85% confluence, HeLa Tat-TetR-Pax6 cells were transiently transfected with 2.0 µg δ-catenin plasmids + 20 µl Lipofectamine™ 2000 (Invitrogen, 1 mg/mL) per plate according to the manufacturer’s instructions. Cells transfected with pEGFP (Clontech) were used as a vector control.
Total RNA isolation and RT-PCR
Total RNA was extracted from each culture sample using the RNeasy Mini kit (Qiagen Science, MD) following the manufacturer’s instructions. The concentration of RNA was measured by spectrophotometry. Total RNA was reverse-transcribed to cDNA with reverse transcriptase (RT) reagents (Ambion) according to the manufacturer’s protocol. Briefly, the RT reaction was carried out in a final volume of 20 ul that contained RT buffer, dNTP, RNase inhibitor, Oligo (dT) primers, MMLV-RT retroviridase, water free from enzyme, and total RNA. The mixture was incubated at 85°C for 3 min and 44°C for 60 min, and RT was inactivated by heating at 92°C for 10 min, followed by incubation at 4°C for 5 min.
Semi-quantitative RT-PCR analysis was used to determine the gene expression of δ-catenin and Pax6(+5a) or Pax6(−5a) at mRNA level. The PCR primers used for detection of δ-catenin mRNA expression level and sequencing for coding region were listed as follows: forward sequence: 5’-ATGTTTGCGAGGAAGCCGC-3’; reverse sequence: 5’- GTCTGGTTGCTATGGTAGCTGGC-3’; product length 510bp, annealing temperature 59°C. For comparing the expression of either Pax6(+5a) or Pax6(−5a) in each group, the pairs of primers for Pax6(+5a) or Pax6(−5a) were designed according to their coding regions: Pax6(+5a) forward sequence: 5’-CCCATGCAGATGCAAAAGTC −3’; reverse sequence: 5’-CATTTGGCCCTTCGATTAGA −3’; product length 694bp, annealing temperature 56°C. Pax6(−5a) forward sequence: 5’-ACACACTTGAGCCATCACCA-3’; reverse sequence: 5’-CTAGCCAGGTTGCGAAGAAC-3’; product length 741bp, annealing temperature 62°C. GAPDH was used as a control in which forward sequence was 5'-GTGGATATTGTTGCCATCAATGACC-3', and reverse sequence was 5'-GCCCCAGCCTTCATGGTGGT-3', with the product length of 248bp and annealing temperature set at 56°C.
Western blot analysis
Cultured cells were lysed in 10 mM HEPES, pH 7.3, 150 mM NaCl, 2mM EDTA, 1% Triton X-100, 0.5% deoxycholate, 0.2% SDS with protease inhibitor cocktails, and then centrifuged at 14,000 × g at 4°C. The extracted protein sample was mixed with the same volume of sample buffer and subjected to denaturation at 95°C for 5 min. The samples were electrophoresed and separated by SDS-PAGE, and transferred onto supported nitrocellulose membrane (Whatman, Dassel, Germany). The membrane was blocked Tris-Tween buffer with 5% non-fat dry milk (Blocking buffer) at room temperature, followed by incubation with the primary antibody. The dilutions for primary antibodies in blocking buffer were as follows: mouse anti-δ-catenin monoclonal antibody (1/300) (Transduction Laboratory, BD Biosciences); rabbit anti-Pax6(−5a) polyclonal antibody (1/300) (Santa Cruz Biotechnology); goat anti-Pax6(+5a) polyclonal antibody (1/1000) (Santa Cruz Biotechnology); rabbit anti-Pax6(−5a) and Pax6(+5a) polyclonal antibody (1/500) (Covance). After being washed with blocking buffer, the corresponding secondary antibody anti-HRP (1:2500 dilution) (Promega) was added and incubated with the membrane. Protein of interests were revealed using ECL solution A and B that contained a chemiluminescence generator. GAPDH or actin protein expression was used as loading control.
Immunofluorescent light microscopy
At different days following transfection, cells seeded on the coverslips were fixed with 4% paraformaldehyde (w/v). After washes in PBS, cells were permeabilized in 0.5% (v/v) Triton X-100. The fixed cells were then treated with PBS containing 10% BSA (blocking buffer) and incubated with primary antibodies. The dilutions for the primary antibodies in blocking buffer were as follows: anti-δ-catenin monoclonal antibody (1:50 dilution) (Transduction Laboratory, BD Biosciences); anti-Pax6(+5a) goat polyclonal antibody (1:300 dilution) (Santa Cruz Biotechnology); anti-Pax6(−5a) rabbit polyclonal antibody (1:100 dilution) (Santa Cruz Biotechnology). Following 3 washes in washing buffer (1% BSA), cells were incubated with the corresponding secondary antibodies conjugated to Cy3 or FITC (1/400) (Molecular Probes). Cell nuclei were stained with 1 ug/ml Hoechst. Cell staining was studied and photographed under the Zeiss LSM 510 laser confocal scanning light microscope (Carl Zeiss, Thornwood, NY). Morphometric analyses were performed using a MetaMorph 4.6 imaging software system (Universal Imaging Corp., West Chester, PA).
Analyses of inducible Pax6(+5a) expression and proteins involved in apoptosis
HeLa Tat-TetR-Pax6 cells were grown in DMEM with G418. Under the culture conditions without doxycycline, there was low basal Pax6(+5a) expression. To induce Pax6 (+5a) overexpression, doxycycline was added to a final concentration of 0.4 µg/ml and refreshed everyday [11]. Some cells were either treated with caspase inhibitor (Z-VAD-FMK) or transfected with δ-catenin. RNA, protein or coverslip were collected as described before to determine the expression of Pax6(+5a) and δ-catenin in HeLa Tat-TetR-Pax6 cell line at different time points. For Western blots, proteins involved in apoptosis were analyzed as well. The dilutions for primary antibody in blocking buffer were as follows: rabbit anti-PARP polyclonal antibody (1/500) (Cell Signaling Technology); rabbit anti-cleaved-PARP polyclonal antibody (1/500) (Cell Signaling Technology); rabbit anti-caspase-3 polyclonal antibody (1/500) (Cell Signaling Technology); rabbit anti-caspase-9 polyclonal antibody (1/500) (Cell Signaling Technology); rabbit anti-survivin polyclonal antibody (1/500) (Cell Signaling Technology); anti-Bax rabbit polyclonal antibody (1:500 dilution) (Santa Cruz Biotechnology); anti-phospho-Bad polyclonal antibody (1/500) (Cell Signaling Technology); anti-Bcl-2 mouse monoclonal antibody (1:500 dilution) (Santa Cruz Biotechnology). The corresponding secondary antibodies anti-HRP (1:2500 dilution) (Promega) were incubated with the membranes. GAPDH expression was used as control and the results were analyzed semi-quantitatively using Quantity One (BioRad). Statistical analysis was performed using SigmaPlot (SPSS Science).
Flow cytometry analysis
In order to analyze cell cycle progression and the associated cell death events induced by Pax6(+5a) overexpression and ectopic δ-catenin expression in addition to the Western blot profiles, HeLa Tat-TetR-Pax6 cells treated with doxycycline (0.4ug/ml) were incubated with caspase inhibitor (Z-VAD-FMK) or transfected with δ-catenin. At day 3, cells in each group were collected and treated with propidium iodide (PI) to stain DNA. The cells were then analyzed for PI levels with flow cytometry (Becton Dickinson FACScan), using Modfit LT software V3.1 to generate a profile of the cell populations indicative of their cell cycle stages and apoptotic states. All data were presented as means ± SEM and t tested by SAS12.0. The p-values were assigned with confidence level set at 0.05.
Results
We first analyzed 3145 base-pair (bp) DNA sequences upstream of human δ-catenin transcription start site (promoter region) using Genomatix software system (http://www.genomatix.de/) to simulate the predicted Pax6 binding domain. We identified that human δ-catenin promoter region contains multiple potential binding sites for Pax6 as described before [26]. We constructed both Pax6(+5a) and Pax6(−5a) plasmids and overexpressed the cDNAs into several cell lines to determine their effects on δ-catenin expression.
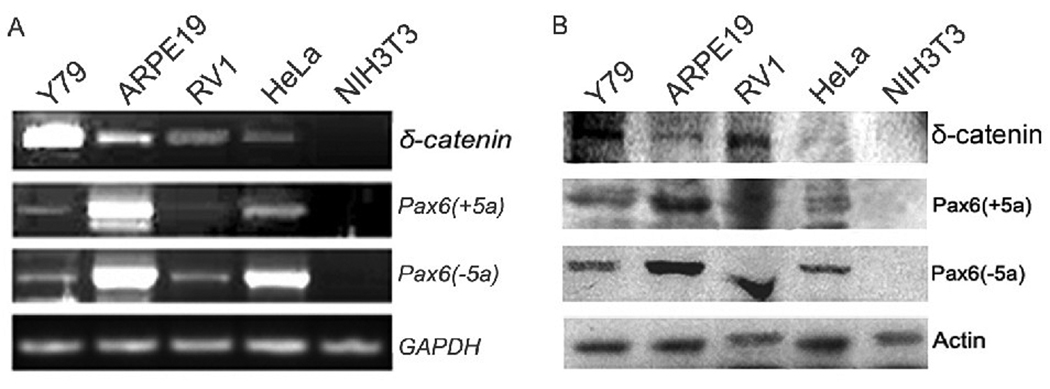
RT-PCR and Western blot analyses demonstrated the expression of Pax6(+5a), Pax6(−5a), and δ-catenin in Y79 (human retinoblastoma cell line), ARPE-19 (human retinal pigment epithelial cell line), CWR22Rv-1 (human prostate cancer derived cell line), and HeLa (human cervical cancer cells taken from Henrietta Lacks) cells, but not in NIH3T3 (mouse fibroblast cell line) cells (Fig 1). Y79 cells showed highest expression for δ-catenin, therefore was selected to be the model to explore initially how pax6 regulates δ-catenin expression.
Fig. 1. Pax6(+5a), Pax6(−5a) and δ-catenin expression in cell lines.
A: RT-PCR showing Pax6(+5a), Pax6(−5a), and δ-catenin mRNA expression in Y79, ARPE-19, CWR22Rv-1 (Rv-1), HeLa and NIH3T3 cells. GAPDH is used as internal mRNA control. B: Western blots showing Pax6(+5a), Pax6(−5a), and δ-catenin protein expression in Y79, ARPE-19, CWR22Rv-1 (Rv-1), HeLa and NIH3T3 cells. Actin is used as protein loading control.
Pax6 (+5a) elicits the stronger effects on δ-catenin expression
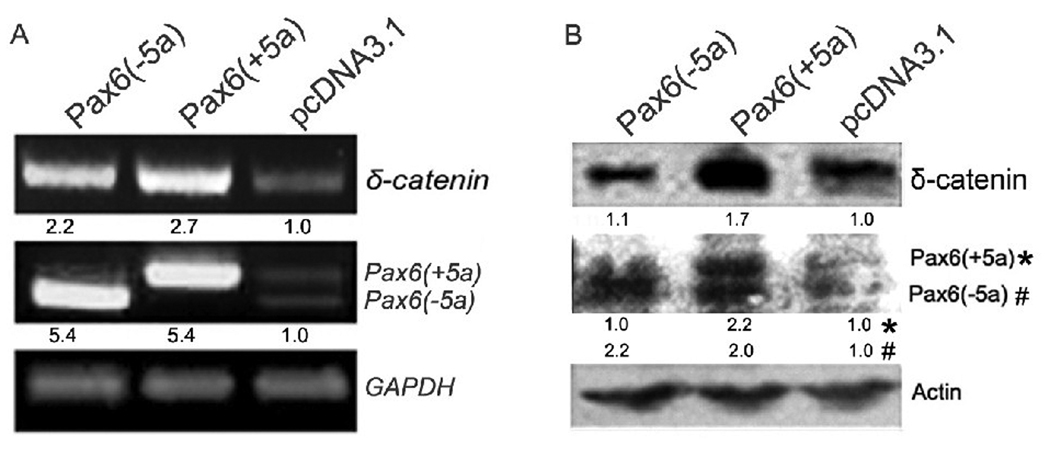
The best characterized Pax6 alternative splicing event involves the insertion of a 42 bp exon (exon 5a) into the PAI subdomain of the PD [31]. This event results in two major Pax6 isoforms, Pax6(+5a) and Pax6(−5a), with different DNA-binding properties. We transiently transfected either Pax6(+5a) or Pax6(−5a) into Y79 cell line and directly assessed their effects on δ-catenin expression through RT-PCR (Fig 2A) and Western blot (Fig 2B). Both gel images and semi-quantitative measurements underneath each lane showed that overexpression of either Pax6(+5a) or Pax6(−5a) was sufficient to promote δ-catenin expression. In addition, Pax6(+5a) expression induced higher expression level of δ-catenin (Fig 2A and B).
Fig. 2. Overexpression of either Pax6(+5a) or Pax6(−5a) is sufficient to promote δ-catenin expression.
A. RT-PCR showing Pax6(+5a), Pax6(−5a), and δ-catenin mRNA expression in Y79 cells transiently transfected with Pax6(+5a), Pax6(−5a), or pcDNA3.1 as vector control. GAPDH is used as internal mRNA control. B. Western blots showing Pax6(+5a), Pax6(−5a), and δ-catenin protein expression in Y79 cells transiently transfected with Pax6(+5a), Pax6(−5a), or pcDNA3.1. Note: semi-quantitative measurements of the RT-PCR and Western blot are shown underneath each lane. Actin is used as protein loading control.
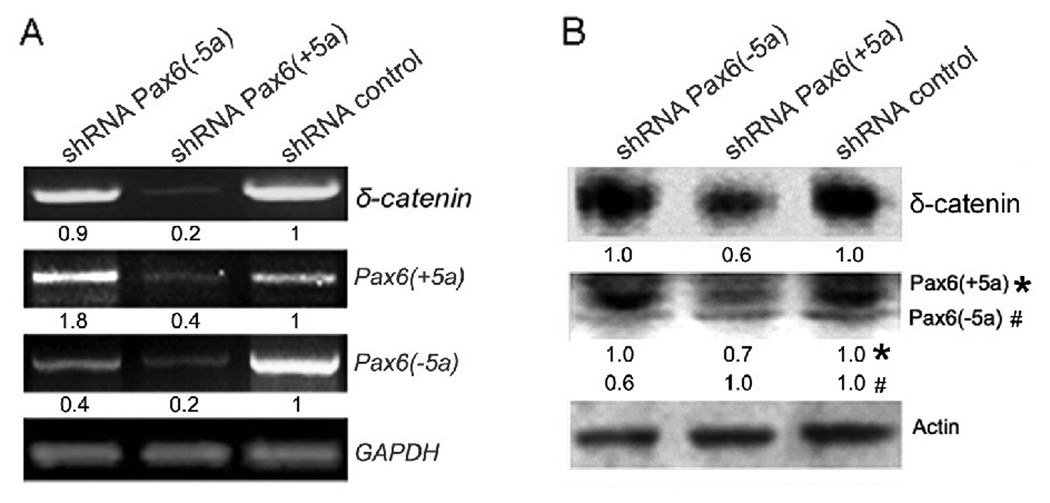
We then introduced Pax6 isoform specific shRNA to knock down Pax6(+5a) and Pax6(− 5a), respectively in Y79 cells. The gel images as well as semi-quantitative measurements underneath each lane showed that knockdown of either Pax6(+5a) or Pax6(−5a) reduced δ-catenin expression (Fig 3A, RT-PCR; Fig 3B Western-blot). When Pax6(−5a) was suppressed by more than 50%, Pax6(+5a) expression level was not affected, and δ-catenin expression was decreased moderately [Fig 3A and B, shRNA Pax6(−5a)]. However, when Pax6(+5a) was suppressed by more than 50%, Pax6(−5a) was also reduced. In this case, δ-catenin expression was barely detected [Fig 3A and B, shRNA Pax6(+5a)]. Therefore, both the overexpression (Fig 2A and B) and knockdown experiments (Fig 3A and B) supported the notion that Pax6(+5a) elicits the stronger effects on δ-catenin expression.
Fig. 3. Knockdown of Pax6(+5a) or Pax6(−5a) suppresses δ-catenin expression.
Pax6 isoform specific shRNA is designed to knockdown Pax6(+5a) or Pax6(−5a) respectively in Y79 cells to explore either Pax6 isoform’s function on δ-catenin expression. A: RT-PCR showing Pax6(+5a), Pax6(−5a), and δ-catenin mRNA expression in Y79 cells after knockdown of Pax6(+5a) or Pax6(−5a) respectively. GAPDH is used as internal mRNA control. B: Western blots showing Pax6(+5a), Pax6(−5a), and δ-catenin protein expression in Y79 cells after knockdown of Pax6(+5a) or Pax6(−5a) respectively. Note: semi-quantitative measurements of the RT-PCR and Western blot are shown underneath each lane. The results showed that knockdown of Pax6(+5a) elicits the stronger effects on δ-catenin while the effects of Pax6(−5a) is more moderate. Actin is used as protein loading control.
Pax6 regulation of δ-catenin expression is isoform- and dose-dependent
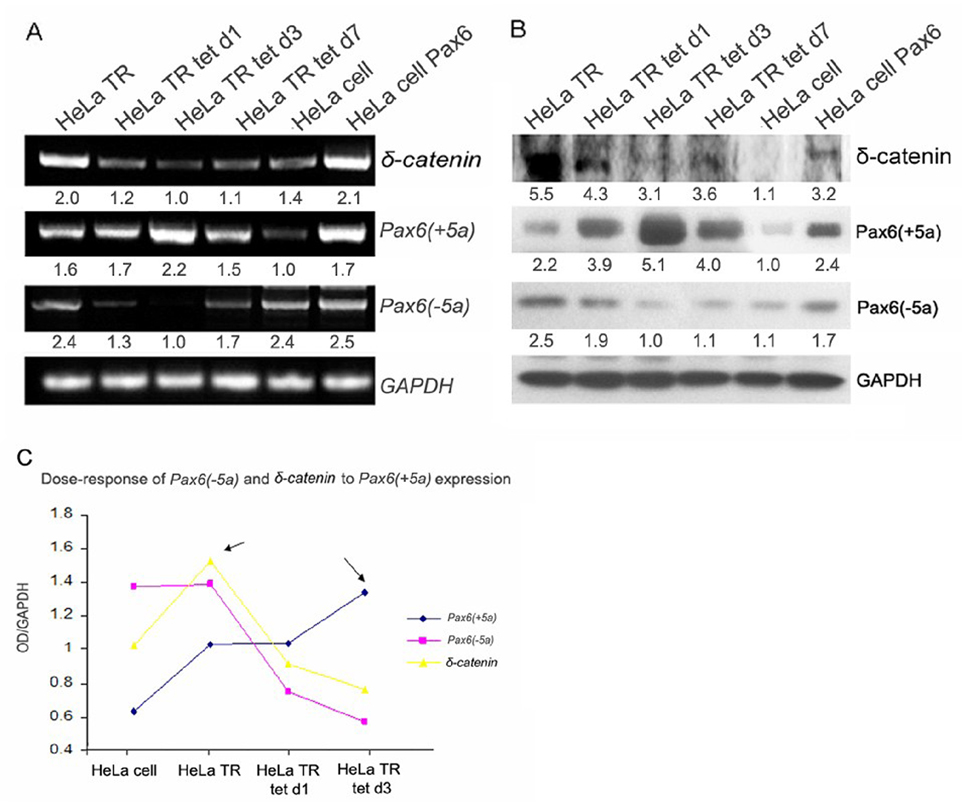
To investigate further the effects of Pax6 on δ-catenin expression, we analyzed a stable tetracycline repressor HeLa cell line (HeLa Tat-TetR-Pax6) in which the expression of Pax6(+5a) is under the control of a tetracycline inducible promoter element. Both RT-PCR and Western blot analyses were employed with the semi-quantitative measurements performed and shown underneath each lane of gels (Fig 4A and B). As demonstrated in Fig 4A (RT-PCR) and Fig 4B (Western blots), both Pax6(+5a) mRNA and protein levels were moderately higher in HeLa Tat-TetR-Pax6 cell line (HeLa TR) without doxycycline induction than its parental line (HeLa cell). To induce Pax6(+5a) overexpression, doxycycline was added to the culture medium at a final concentration of 0.4µg/ml (Fig 4A and B). Pax6(+5a) expression increased very quickly to peak at about 48–64 hours post-induction (HeLa TR d3). Under the condition of moderate Pax6(+5a) overexpression in non-induced HeLa Tat-TetR-Pax6 (HeLa TR) cells, δ-catenin expression was increased compared to the parental HeLa cells [Fig 4A, RT-PCR; Fig 4B, Western blot and Fig 4C, compare δ-catenin mRNA with Pax6(+5a) expression level]. However, with the continued increases of high Pax6(+5a) expression from day 1 to day 3 post-induction, δ-catenin expression reduced gradually, while Pax6(−5a) expression showed significant reduction as well (Fig 4A, B and C). When doxycycline induction was stopped and the HeLa Tat-TetR-Pax6 cells were allowed to grow to day 7, Pax6(+5a) level was reduced as expected, whereas the expression of Pax6(−5a) as well as δ-catenin resumed the upward trend (Fig 4A and B, HeLa TR tet d7). These data indicated that a moderate upregulation of Pax6(+5a) promoted δ-catenin expression whereas a sustained high expression of Pax6(+5a) actually inhibited δ-catenin expression. Furthermore, while Pax6(−5a) expression did not change significantly between the parental HeLa and HeLa Tat-TetR-Pax6 (HeLa TR) cell lines, its expression level decreased along with that of δ-catenin following the induction of high Pax6(+5a) expression (Fig 4C). These data showed that the regulation of δ-catenin expression by Pax6 is isoform- and dose-dependent.
Fig. 4. Dose responses of Pax6(−5a) and δ-catenin expression to induced overexpression of Pax6(+5a).
A. RT-PCR showing Pax6(+5a), Pax6(−5a), and δ-catenin expression in time course in HeLa Tat-TetR-Pax6 cells with (HeLa tet) or without (HeLa TR) induction of Pax6(+5a) overexpression as well as parental HeLa cells (HeLa) and HeLa cells transiently transfected with Pax6(+5a)(HeLa Pax6). Note: a moderate upregulation of Pax6(+5a) promotes δ-catenin expression, whereas a sustained high expression of Pax6(+5a) inhibits δ-catenin expression. GAPDH is used as internal mRNA control. B: Western blots showing Pax6(+5a), Pax6(−5a), and δ-catenin protein expression in HeLa cell and HeLa Tat-TetR-Pax6 cells with or without induction of Pax6(+5a) overexpression. Note: semi-quantitative measurements of the RT-PCR and Western blot are shown underneath each lane in A and B. Actin is used as protein loading control. C. Dose-response curve of Pax6(−5a) and δ-catenin to Pax6(+5a) expression shows that the regulation of δ-catenin expression by Pax6 is isoform- and dose-dependent. With the increasing Pax6(+5a) expression, Pax6(−5a) expression decreases gradually. δ-Catenin expression shows a parabolic curve, which reaches a peak when Pax6(+5a) is moderately upregulated, and then downregulates with the increasing Pax6(+5a) expression. Note: HeLa TR: HeLa Tat-TetR-Pax6 cells without doxycycline induction of Pax6(+5a) overexpression; HeLa Tat tet d1 and d3 indicates day1 and day 3 after HeLa Tat-TetR-Pax6 cells are induced to overexpress Pax6(+5a) by doxycycline treatment. HeLa Tat tet d7 indicates that HeLa Tat-TetR-Pax6 cells are induced to overexpress Pax6(+5a) by doxycycline treatment for 3 days, then treatment was stopped and the cells were grown to day 7 before analysis. HeLa: parental HeLa cells without any treatment or transfection. HeLa cell Pax6: HeLa cells transiently transfected with Pax6(+5a).
Induced overexpression of Pax6(+5a) is accompanied by the reduction of Pax6(−5a)
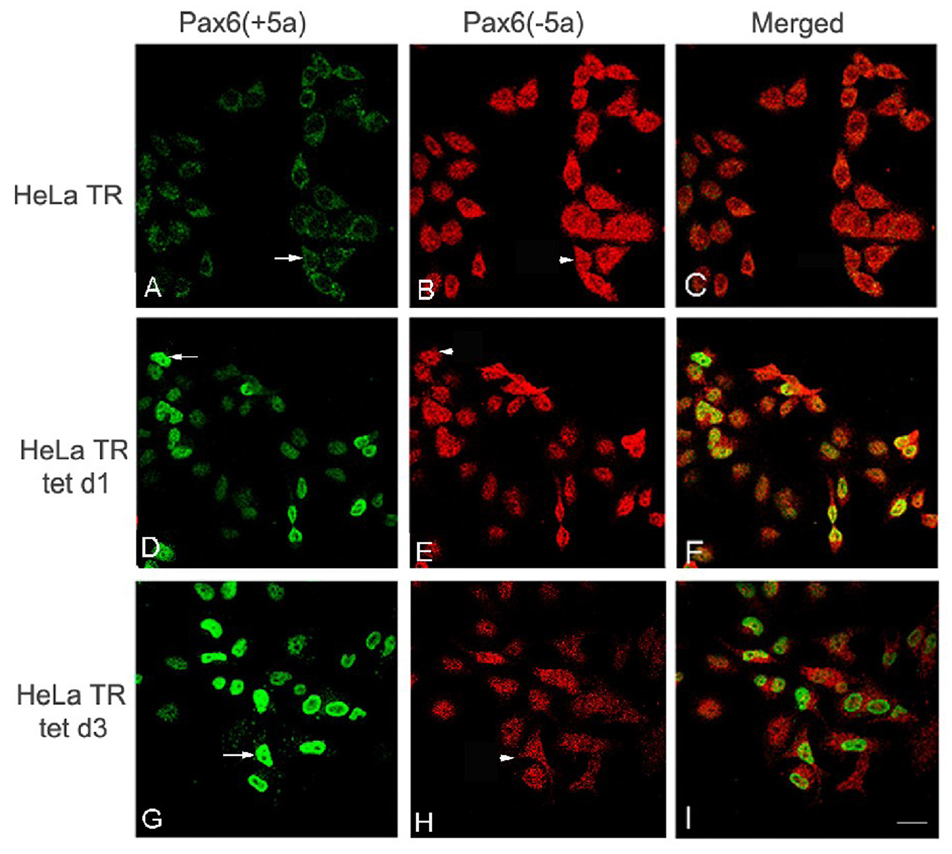
To explore further the relationship between the expression of Pax6(+5a) and Pax6(−5a), we performed an indirect immunofluorescence staining of Pax6. Laser confocal fluorescence light microscopy showed that HeLa Tat-TetR-Pax6 cells without doxycycline induction (HeLa TR) had a moderate expression of Pax6(+5a) (Fig 5B), as compared to a strong staining of Pax6(−5a) (Fig 5A). At day1 and day3 post-induction with doxycycline, Pax6(+5a) showed stronger staining (Fig 5E and H) than that without doxycycline (Fig 5B), whereas Pax6(−5a) expression (Fig 5D and G) was weaker than that without doxycycline (Fig 5A). As expected for the inducible expression, Pax6(+5a) expression reached the maximal peak at day 3 post-induction (Fig 5H), while Pax6(−5a) expression reduced (Fig 5G). These results were consistent with our RT-PCR and Western blot data in which we showed that the reductions of Pax6(−5a) did not result in significant decreases in δ-catenin expression [see Fig 3, which showed that knockdown of Pax6(−5a) can only elicit slight downregulation of δ-catenin expression]. These data indicated that it was Pax6(+5a) overexpression that reduced δ-catenin expression which was accompanied by the reduction of Pax6(−5a) expression.
Fig. 5. Induced overexpression of Pax6(+5a) is accompanied by the reduction of Pax6(−5a).
Immunofluorescent laser confocal light microscopy shows the time course of Pax6(+5a) and Pax6(−5a) expression in HeLa Tat-TetR-Pax6 cells double labeled using anti-Pax6(+5a) and anti-Pax6(−5a). A–C: Pax6(+5a) and Pax6(−5a) expression in HeLa Tat-TetR-Pax6 cells without (HeLa TR) induction of Pax6(+5a) overexpression. D–F: HeLa Tat-TetR-Pax6 cells at day 1 (HeLa tet d1) with induction of Pax6(+5a) overexpression. G–I: HeLa Tat-TetR-Pax6 cells at day 3 (HeLa tet d3) with induction of Pax6(+5a) overexpression. Note: compared with A, many cells in D show moderately increased Pax6(+5a) expression and show dramatically increased expression in G (arrows). On the other hand, Pax6(−5a) shows slightly decreased expression in cells in D but more pronounced decrease in expression in H when compared to A (arrowheads). A, D, and G: Pax6(+5a); B, E, and H: Pax6(−5a); C, F, and I: Merged Images. Bar: 25 µm.
Induced overexpression of Pax6 (+5a) alters cell cycle progression and triggers apoptosis
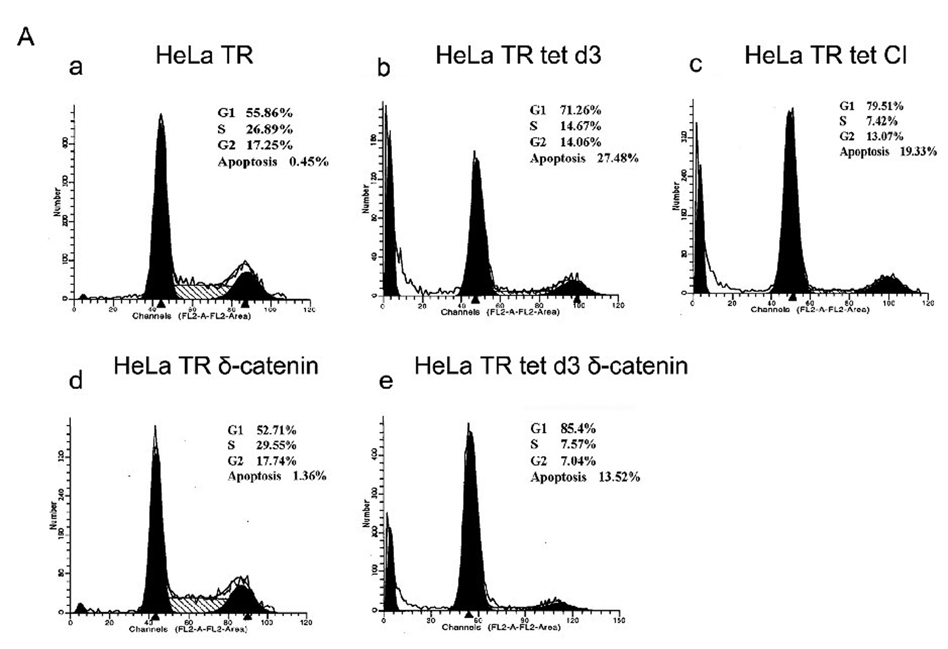
It has been reported that overexpression of Pax6(+5a) inhibited cell proliferation [11]. The reduction in cell proliferation could be due to slower progression of individual cells through cell cycle or to increased cell death. HeLa Tat-TetR-Pax6 cells in the presence or absence of doxycycline were either treated with caspase inhibitor (Z-VAD-FMK) or were transfected with δ-catenin. At day 3 following the treatment, cells were collected and analyzed by flow cytometry with propidium iodide (PI) staining. We found that while cells in G1 phase increased from 55.86% (Fig 6A–a, HeLa TR) to 71.26% (Fig 6A–b, HeLa TR tet d3), cells in S and G2 phases reduced from 26.89% and 17.25% (Fig 6A–a, HeLa TR) to 14.67% and 14.06% (Fig 6A–b, HeLa TR tet d3), respectively. This result supports the literature that Pax6 overexpression retards cell cycle [11].
Fig. 6. Mitotic and apoptotic changes in HeLa Tat-TetR-Pax6 cell line.
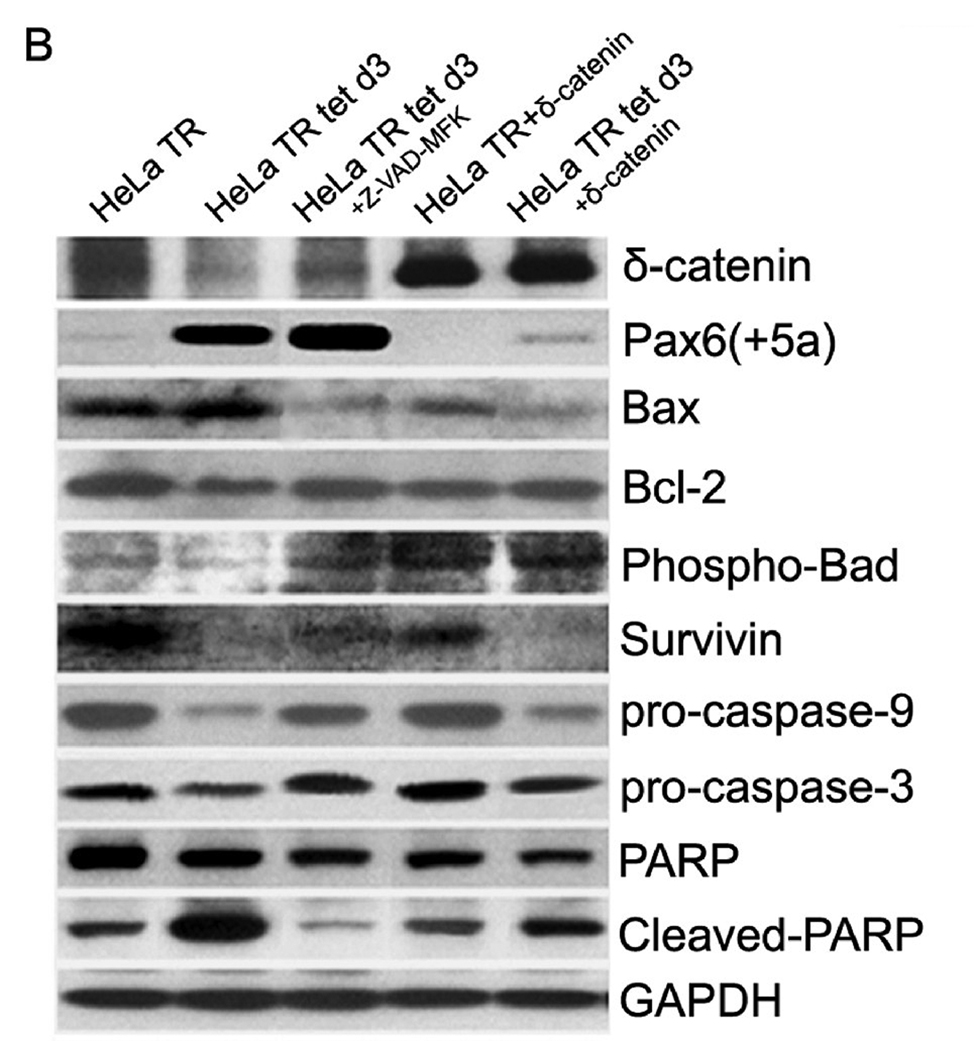
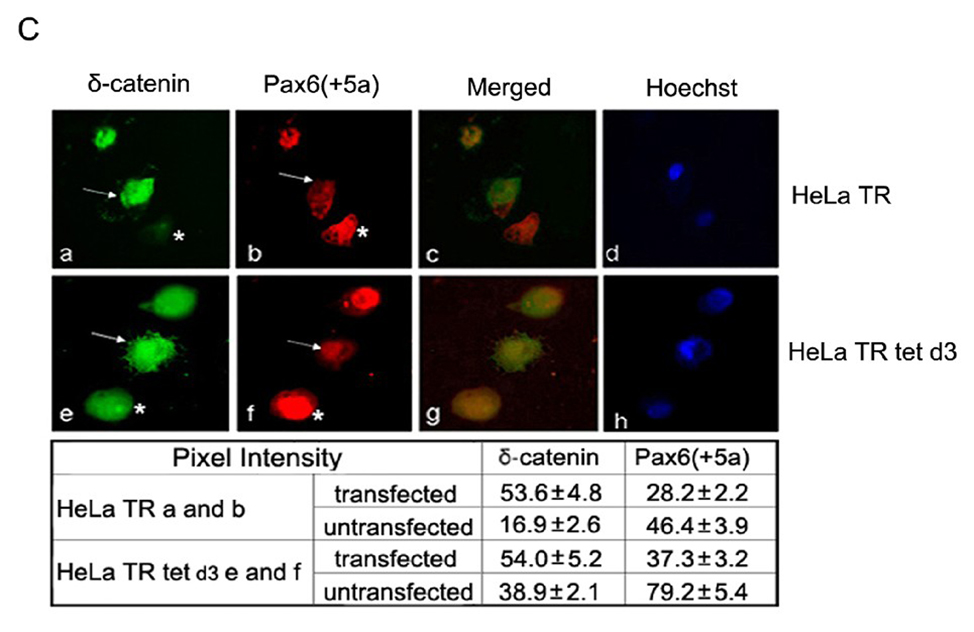
A. Cell cycle kinetics analysis. HeLa Tat-TetR-Pax6 cells after doxycycline induction is treated respectively with caspase inhibitor (Z-VAD-FMK) or transfected with δ-catenin. At day3, cells in each group are collected and analyzed by flow cytometry with propidium iodide (PI) staining. a: HeLa Tat-TetR-Pax6 cells without doxycycline induction (HeLa TR); b: HeLa Tat-TetR-Pax6 cell 3 days after doxycycline induction (HeLa TR tet d3); c: HeLa Tat-TetR-Pax6 cell with Pax6(+5a) overexpression in the presence of Z-VAD-FMK (HeLa TR tet CI); d: Overexpression of δ-catenin in HeLa Tat-TetR-Pax6 cells without Pax6(+5a) overexpression (HeLa TR δ-catenin); e: Overexpression of δ-catenin in HeLa Tat-TetR-Pax6 cells with Pax6(+5a) overexpression (HeLa TR tet δ-catenin). B. The effects of Pax6 and δ-catenin expression on apoptotic protein expression. Western blots show protein expression profiles of Pax6(+5a), δ-catenin, PARP, cleaved-PARP, procaspase-9, procaspase-3, Bax, Bcl-2, Phospho-Bad and survivin. Note that δ-catenin expression in HeLa TR tet d3 cells is reduced with high Pax6(+5a) expression when compared to that in HeLa TR cells. Blocking caspase activation using caspase inhibitor (HeLa TR tet d3+Z-VAD-FMK) reduces the inhibitory effects of high Pax6(+5a) expression on δ-catenin expression. Pax6(+5a) and δ-catenin expression is both increased when compared to that of no-caspase inhibitor treatment. On the other hand, overexpression of δ-catenin (HeLa TR tet d3+δ-catenin) also reduces Pax6(+5a) expression and the apoptotic protein expression induced by Pax6(+5a) overexpression. The results show that overexpression of δ-catenin reduces Pax6(+5a) expression in HeLa Tat-TetR-Pax6 cells, which provides a negative feedback between Pax6 and δ-catenin expression. C. Double immunofluorescent light microscopy showing the effects of δ-catenin overexpression on Pax6(+5a) expression. a–d: Pax6(+5a) (red) and δ-catenin (green) expression in HeLa TR cells; c: Merged image of a and b; d: Nuclei staining. e–h: Pax6(+5a) (red) and δ-catenin (green) expression in HeLa tet d3 cells. g: Merged image of e and f; h: Nuclei staining. Note: arrows indicate δ-catenin transfected cells; asterisks indicate untransfected cells. The arrows show that the expression of Pax6(+5a) (red) is reduced in cells transfected with δ-catenin (green). The table in C shows the morphometric density of pixel values in cells stained by immunofluorescence light microscopy. Bar: 20 µm.
Furthermore, compared with 0.45% of sub-G1 populations in HeLa TR cells with moderate Pax6(+5a) expression (Fig 6A–a), the percentage of cells in sub-G1 was much higher at 27.48% in HeLa Tat-TetR-Pax6 cells in the presence of doxycycline for 3 days therefore with highest Pax6(+5a) overexpression (Fig 6A–b). The sub-G1 populations represent cells containing less than 1 N chromosomal copy, including apoptotic cells undergoing chromosomal degradation. The addition of the caspase inhibitor (CI) Z-VAD-FMK reduced the percentage of sub-G1 cells from 27.48% (Fig 6A–b) to 19.33% (Fig 6A–c). Compared with 0.45% of sub-G1 populations in HeLa TR ells with moderate Pax6(+5a) expression, δ-catenin transfection showed slightly increased sub-G1 cells to 1.36% (Fig 6A–d, HeLa TR δ-catenin). But when Pax6(+5a) overexpression was induced by doxycycline, transfection with δ-catenin reduced the percentage of sub-G1 cells from 27.48% (Fig 6A–b) to 13.52% (Fig 6A–d, HeLa tet δ-catenin). These data showed that when there was Pax6(+5a) expression at the basal level, δ-catenin overexpression slightly promoted apoptosis. When there was high Pax6(+5a) expression, δ-catenin overexpression suppressed apoptosis.
Sustained high expression of Pax6 (+5a) induces changes in intracellular regulators of apoptosis
To confirm that a sustained high expression of Pax6(+5a) induced apoptosis we performed Western blot analyses (Fig 6B). We treated HeLa Tat-TetR-Pax6 cells after 3 days of doxycycline induction (HeLa TR tet d3) with caspase inhibitor Z-VAD-FMK. We also investigated whether ectopic expression of δ-catenin had inhibitory effects to feedback regulate Pax6(+5a) expression and apoptosis. We determined the expression levels of Pax6(+5a), δ-catenin and several intracellular regulators of apoptosis. Compared with HeLa TR cells with moderate Pax6(+5a) expression, high Pax6(+5a) expression (HeLa TR tet d3) resulted in the reduction of δ-catenin, total poly(ADP-ribose) polymerase (PARP), Phospho-Bad, Bcl-2, survivin, procaspase 9, and procaspase 3 (Fig 6B). This was accompanied by the increased expression of Bax and cleaved PARP, consistent with the literature that Pax6(+5a) overexpression led to apoptotic changes [11]. We found that blocking caspase activation by Z-VAD-FMK reduced the inhibitory effects of the high Pax6(+5a) expression on δ-catenin expression. After treatment with Z-VAD-FMK, Bax and cleaved PARP were reduced, while procaspase-3, procaspase-9 and δ-catenin were increased (Fig 6B).
In addition, ectopic overexpression of δ-catenin suppressed basal Pax6(+5a) expression in HeLa TR cells without doxycycline (Fig 6B and 6C), although it did not alter the expression of most of other genes involved in apoptosis (Fig 6B). As an exception, total PARP was slightly reduced but the cleaved PARP showed no significant changes (Fig 6B). However, the ectopic overexpression of δ-catenin repressed Pax6(+5a) expression in HeLa Tat-TetR-Pax6 cells after 3 days of doxycycline induction, suggesting a possible feedback inhibition impinged on Pax6(+5a) when Pax6(+5a) expression was too high (Fig 6B and 6C). Furthermore, the expression of Bax and cleaved PARP were both reduced significantly when compared to HeLa TR tet d3 cells in the presence of doxycycline but without ectopic δ-catenin overexpression (Fig 6B). As exceptions, there were no changes in the expression level of survivin, indicating that δ-catenin overexpression only attenuated the apoptosis mediated through caspase and Bcl-2 family pathways, such as Bcl-2 and Bax, but not through the Inhibitor of Apoptosis Protein (IAP), such as survivin.
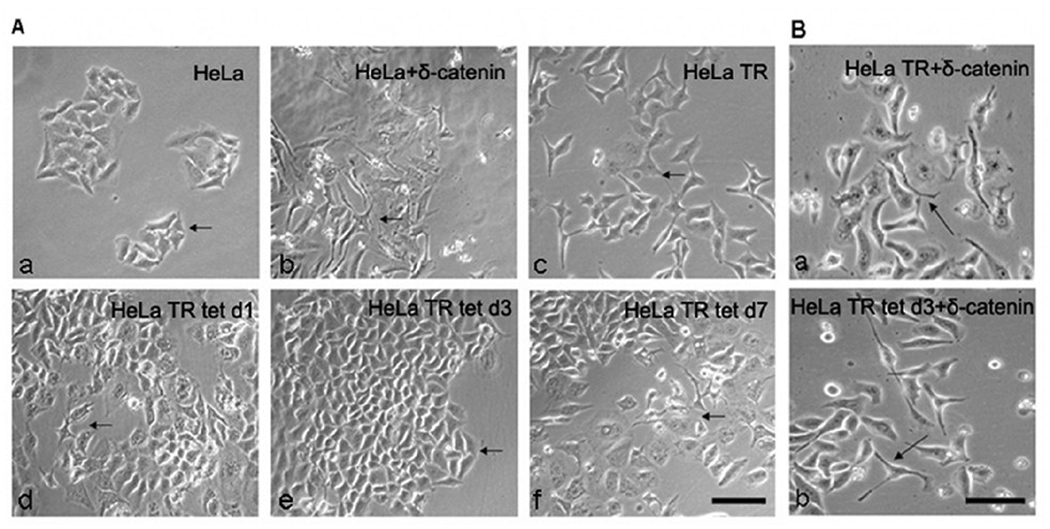
Pax6(+5a) and δ-catenin expression modulates morphological changes in HeLa cells
We further investigated the effects of Pax6 and δ-catenin on HeLa cell morphology since overexpression of either of these two proteins have been shown to extend cellular protrusions and processes [22, 23]. Parental HeLa cells showed typical epithelial cell clusters (Fig 7A–a, arrow) with low expression of Pax(+5a) and δ-catenin (Fig 4). They extended cellular processes when they were transiently transfected with Pax6(+5a)(Fig 7A–b, arrow), which was accompanied by δ-catenin upregulation (Fig 4A). HeLa Tat-TetR-Pax6 cells without doxycycline induction (HeLa TR) showed the dispersion of cell clusters with cellular processes (Fig 7A–c, arrow), in which a moderate upregulation of Pax6(+5a) was observed (Fig 4A). After doxycycline induction, Pax6(+5a) showed progressively increased expression from day 1 to day 3 (Fig 4A), but the cellular processes was reduced at day 1 and suppressed at day 3 (Fig 7A–d and e, arrow), which was accompanied by the downregulation of δ-catenin (Fig 4A). When doxycycline was stopped, and HeLa Tat-TetR-Pax6 cells were allowed to grow to day 7, Pax6(+5a) expression levels began to decline (Fig 4A). Under such condition, cellular processes remerged (Fig 7A–f, arrow).
Fig. 7. Morphological changes of HeLa and HeLa Tat-TetR-Pax6 cells influenced by Pax6 and δ-catenin expression.
A. A moderate upregulation of Pax6(+5a) promotes, whereas its sustained high expression suppresses the extension of cellular protrusions and processes. a: Parental HeLa cells without any treatments as negative control. Arrows: epithelial cell clusters. b: HeLa cells transiently transfected with Pax6(+5a) as positive control. Arrows: cells extending processes. c: HeLa TR cells without doxycycline induction [with moderate level of Pax6(+5a) expression]. Arrows: cells extending processes. d: HeLa TR tet d1 cells at day 1 after doxycycline induction [with increasing Pax6(+5a) expression]. Arrow: cells still showing processes; Arrowheads: cells no longer extending processes. e: HeLa TR tet d3 cells at day 3 after doxycycline induction [with increasingly high Pax6(+5a) expression]. Arrowheads: cells no longer extending processes. f: HeLa TR tet d7 cells with doxycycline induction [with the highest Pax6(+5a) expression] for 3 days and followed by in the absence of induction until day 7. Arrows: cells extending processes. Bar: 50 µm. B. δ-Catenin overexpression promotes the extension of cellular protrusions and processes regardless whether Pax6 is overexpressed. a: HeLa TR cells without doxycycline induction transfected with δ-catenin; b: HeLa TR tet d3 cells with a 3-day doxycycline induction transfected with δ-catenin. Arrows indicate the extension of cellular processes. Bar: 30 µm.
Finally, HeLa Tat-TetR-Pax6 cells showed cellular processes when they were transiently transfected with δ-catenin, regardless whether they were incubated in the absence of doxycycline therefore expressed a moderate level of Pax6(+5a) (Fig 7B–a) or induced in the presence of doxycycline to express high levels of Pax6(+5a) (Fig 7B–b). These results showed that transient overexpression with ectopic δ-catenin can compensate the reduction of δ-catenin expression and the inhibition of cellular processes due to sustained high expression of Pax6(+5a).
Discussion
Pax6 is an evolutionarily conserved transcription factor which controls critical steps in the morphogenesis of several ectoderm derived organs including the development of vertebrate eye and brain [1, 32]. Pax6 shows remarkably similar distribution to that of δ-catenin [13, 21]. Recent studies showed that Pax6 is essential for the expression of δ-catenin, which is severely reduced when Pax6 is mutated in the mouse model [24], and they also adopted EMSA to show that Pax6 can bind to δ-catenin promoter. Using DNA microarray hybridization, δ-catenin was identified as a target of Pax6 in the CNS. Pax6 could effectively and specifically bind a DNA sequence in δ-catenin promoter that is conserved between mouse and human [24]. Furthermore, altered expression of δ-catenin is associated with prostate cancer [16, 25], and Pax6 enhances δ- catenin expression in prostate cancer cells [26]. However, these studies did not reveal how Pax6 regulates δ-catenin expression and functions.
In this study, we demonstrated, for the first time, that regulation of δ-catenin expression by Pax6 is not only isoform- and dose-dependent, but also provides important feedback mechanisms of their functional interactions in cellular morphogenesis, apoptosis, and cancer. The overexpression of either Pax6 isoforms, Pax6(+5a) or Pax6(−5a), is sufficient to upregulate δ-catenin expression whereas knockdown of either isoforms reduces δ-catenin expression. Furthermore, Pax6(+5a) showed a stronger effect than Pax6(−5a). These studies demonstrate that the PD domain of Pax6(+5a) have greater binding affinity for δ-catenin promoter sequence, consequently triggering a stronger upregulation of δ-catenin expression. We have applied δ-catenin promoter-driven luciferase reporter assay to detect either Pax6 isoforms. Consistent with earlier report using EMSA method [21], we observed that both Pax6(+5a) and Pax6(−5a) have functional binding in δ-catenin promoter, but Pax6(+5a) elicited stronger effects on promoting δ-catenin luciferase activity (data not shown).
δ-Catenin expresses in neuroepithelial cells and can alter epithelial cell morphology, and induce dramatic extension of dendrite-like processes [22]. Interestingly, HeLa cells showed significant process extensions under the condition of moderate but not high expression of Pax6(+5a). This points to the possibility that it is the moderate level of Pax6(+5a) that induces δ-catenin expression which is involved in the modulation of cellular process formation.
We have also observed that a moderately increased expression of Pax6(+5a) upregulates the expression of p120ctn, a close family member of δ-catenin, which supports the study of Duncan et al [33] in lens fiber cells. Interestingly, we found a dose-dependent regulation of p120ctn expression much like what is to δ-catenin, indicating that p120ctn expression is also tightly controlled in response to Pax6 gene dosage (data not shown).
Feedback mechanism of Pax6 regulation on δ-catenin expression and apoptosis
It is reported that overexpression of Pax6(+5a) inhibits cell proliferation in a dose-dependent manner but not cell fate, while Pax6(−5a) affects both proliferation and cell fate [10]. Thus, the reduction in the rate of cell proliferation could be due to slower progression of individual cells through the cell cycle or to increased cell death. Indeed, overexpression of Pax6 in neuroblastoma cells suppressed cell growth by increasing the number of cells in G1 and decreasing the number of cells in the S-phase [34]. Full-length Pax6 induced caspase-3 independent apoptosis in a small percentage of cells [11]. Our current findings are consistent with the literature by showing the alteration of cell cycle progression when Pax6(+5a) is overexpressed.
Based on our study, caspase-3 pathway is involved in the cell death induced by Pax6(+5a) in cancer cells such as HeLa cell lines. We found that when caspase dependent apoptotic pathway is inhibited by Z-VAD-FMK, procaspase-9, procaspase-3 is increased. Consequently, caspase substrate PARP is accumulated and cleaved PARP is reduced. Furthermore, when there is higher Pax6(+5a) expression, Bcl-2, Phospho-Bad and survivin are reduced but Bax is increased, which may increase the percentage of cells in apoptosis. After Z-VAD-FMK treatment, survivin, Phospho-Bad and Bcl-2 are increased but Bax is reduced, resulting in the decreases of cells in apoptosis as shown by flow cytometry. These studies strongly indicate that the high Pax6(+5a) expression induced apoptosis involves the activation of caspase cascade and survivin.
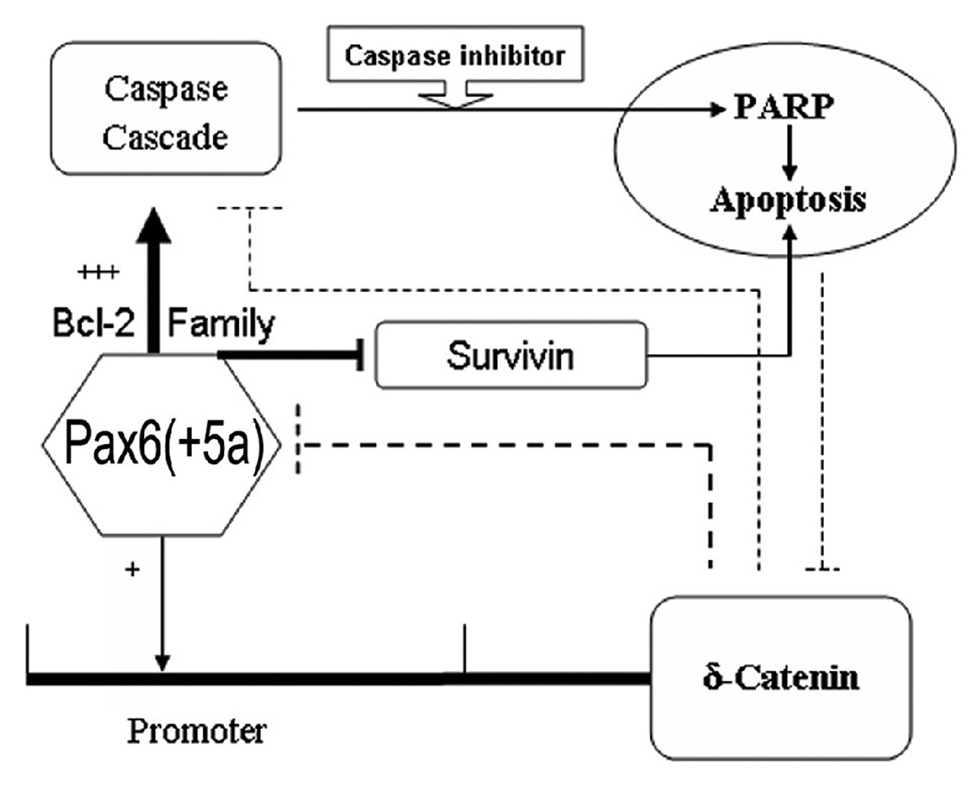
Therefore, we propose a model of feedback mechanism of Pax6 regulation on δ-catenin expression and apoptosis (Fig 8). In the presence of high Pax6(+5a) expression levels, Bax translocates from the cytoplasm to the mitochondria and forms pores in the mitochondrial membrane; Bad phosphorylation is decreased, which inhibits the anti-apoptotic functions of Bcl-2. These changes lead to membrane depolarization and consequently the release of cytochrome c which facilitates the interaction between APAF-1 and caspase-9, resulting in the activation of the caspase cascade [35, 36]. Caspase-3 is required for Bcl-2 cleavage, and the Bcl-2 cleavage product localizes to mitochondria further promoting caspase activation to ensure the execution of apoptotic cell. On the other hand, members of the inhibitor of apoptosis (IAP), such as survivin, are decreased, which could also add onto the apoptotic process.
Fig. 8. Schematic illustration of the model highlighting putative Pax6 and δ-catenin feedback regulation and their roles in apoptosis.
Note: +: indicates moderate expression of Pax6(+5a); +++: indicates higher Pax6(+5a) overexpression; →: indicates promote or upregulate □□ : indicates inhibit or downregulate
While a moderate expression of Pax6(+5a) and its interaction with the promoter of δ-catenin is required for δ-catenin expression, a sustained high expression of Pax6(+5a) triggers apoptosis that involves caspase cascade components including Bax, Bad, Bcl-2, and PARP as well as survivin, which could invoke a feedback regulation to reduce δ-catenin expression (Fig 8). When caspase activation was blocked by Z-VAD-FMK and the apoptosis was reduced, δ-catenin expression can maintain at the same level as that of being upregulated by the moderate Pax6(+5a) expression. As another evidence for the feedback regulation of δ-catenin on Pax6, δ-catenin overexpression reduces Pax6(+5a) expression and suppresses the apoptosis induced by Pax6(+5a) overexpression (Fig 8). δ-Catenin has potential binding sites in Pax6 promoter such as Kaiso box [37]. Therefore, under certain conditions, δ-catenin may act upon Pax6 directly or indirectly as a negative feedback control element.
Previous studies showed that Bax expression is controlled by p53 [38], but we didn’t find p53 protein expression in this HeLa Tat-TetR-Pax6 cell line (data not shown). HeLa cells were derived from a cervical carcinoma and have human papilloma virus. The E6 protein targets p53 protein for degradation [39]. So in HeLa cells, any p53 protein that is synthesized is rapidly degraded. This indicates that Pax6(+5a) induced a p53 independent and caspase-3 dependent apoptosis in HeLa Tat-TetR-Pax6 cells (Fig 8).
Pax6 and cancer
Pax6 regulation in cancer is complex. Although Pax6 is normally expressed in stem cells during development of eye, brain and pancreas, its expression is correlated with pancreatic adenocarcinomas and the differentiation causes its downregulation in the pancreas [40]. On the other hand, Pax6 expression suppresses glioblastoma cell invasiveness [41]. While Pax6 is required for some endocrine cell types in the gastrointestinal tract, it is also found in tumors of the intestine [42–44]. Interestingly, δ-catenin is also normally expressed in brain but becomes upregulated in cancers of peripheral tissues [16, 25]. We showed previously that transcriptional upregulation plays important roles in δ-catenin overexpression in at least some prostate cancer cases. Recently, we found that ectopic expression of transcription factors such as Pax6 and E2F-1 upregulates δ-catenin expression in prostate cancer cells in culture [26]. Therefore, it is possible that Pax6 and δ-catenin functionally interact in prostate cancer development. Finally, the expression of Pax6 in tumor cells are not neuroendocrine in origin but are of common embryonic ancestry to Pax6 expressing cells [40]. This may be explained in part by the tumor arising from multipotent stem cells. In the future, it will be important to determine how Pax6 is involved in the regulation of δ-catenin overexpression and the control of cell growth-death decision in carcinogenesis.
Acknowledgement
We thank Melissa Clark and Christi Boykin for excellent technical assistance, and Lu laboratory members for many helpful discussions. This study was supported in part by NIH/NCI (CA111891) and the Department of Defense (PC040569) grants (Q.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare that they have no conflict of interest.
Reference
- 1.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 2.Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- 3.Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- 4.Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 5.Tang HK, Singh S, Saunders GF. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene PAX6. J Biol Chem. 1998;273:7210–7221. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- 6.Estivill-Torrus G, Pearson H, van Heyningen V, Price DJ, Rashbass P. Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development. 2002;129:455–466. doi: 10.1242/dev.129.2.455. [DOI] [PubMed] [Google Scholar]
- 7.Chapalamadugu KC, Robison BD, Drew RE, Powell MS, Hill RA, Amberg JJ, Rodnick KJ, Hardy RW, Hill ML, Murdoch GK. Dietary carbohydrate level affects transcription factor expression that regulates skeletal muscle myogenesis in rainbow trout. Comp Biochem Physiol B Biochem Mol Biol. 2009 doi: 10.1016/j.cbpb.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuoc TC, Stoykova A. Er81 is a downstream target of Pax6 in cortical progenitors. BMC Dev Biol. 2008;8:23. doi: 10.1186/1471-213X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haubst N, Berger J, Radjendirane V, Graw J, Favor J, Saunders GF, Stoykova A, Gotz M. Molecular dissection of Pax6 function: the specific roles of the paired domain and homeodomain in brain development. Development. 2004;131:6131–6140. doi: 10.1242/dev.01524. [DOI] [PubMed] [Google Scholar]
- 11.Ouyang J, Shen YC, Yeh LK, Li W, Coyle BM, Liu CY, Fini ME. Pax6 overexpression suppresses cell proliferation and retards the cell cycle in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2006;47:2397–2407. doi: 10.1167/iovs.05-1083. [DOI] [PubMed] [Google Scholar]
- 12.Paffenholz R, Franke WW. Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation. 1997;61:293–304. doi: 10.1046/j.1432-0436.1997.6150293.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou J, Liyanage U, Medina M, Ho C, Simmons AD, Lovett M, Kosik KS. Presenilin 1 interaction in the brain with a novel member of the Armadillo family. Neuroreport. 1997;8:2085–2090. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]
- 14.Paffenholz R, Kuhn C, Grund C, Stehr S, Franke WW. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250:452–464. doi: 10.1006/excr.1999.4534. [DOI] [PubMed] [Google Scholar]
- 15.Israely I, Costa RM, Xie CW, Silva AJ, Kosik KS, Liu X. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr Biol. 2004;14:1657–1663. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- 16.Lu Q, Dobbs LJ, Gregory CW, Lanford GW, Revelo MP, Shappell S, Chen YH. Increased expression of delta-catenin/neural plakophilin-related armadillo protein is associated with the down-regulation and redistribution of E-cadherin and p120ctn in human prostate cancer. Hum Pathol. 2005;36:1037–1048. doi: 10.1016/j.humpath.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Zeng Y, Abdallah A, Lu JP, Wang T, Chen YH, Terrian DM, Kim K, Lu Q. delta-Catenin promotes prostate cancer cell growth and progression by altering cell cycle and survival gene profiles. Mol Cancer. 2009;8:19. doi: 10.1186/1476-4598-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanahashi H, Tabira T. Isolation of human delta-catenin and its binding specificity with presenilin 1. Neuroreport. 1999;10:563–568. doi: 10.1097/00001756-199902250-00022. [DOI] [PubMed] [Google Scholar]
- 19.Levesque G, Yu G, Nishimura M, Zhang DM, Levesque L, Yu H, Xu D, Liang Y, Rogaeva E, Ikeda M, Duthie M, Murgolo N, Wang L, VanderVere P, Bayne ML, Strader CD, Rommens JM, Fraser PE, St George-Hyslop P. Presenilins interact with armadillo proteins including neural-specific plakophilin-related protein and beta-catenin. J Neurochem. 1999;72:999–1008. doi: 10.1046/j.1471-4159.1999.0720999.x. [DOI] [PubMed] [Google Scholar]
- 20.Medina M, Marinescu RC, Overhauser J, Kosik KS. Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics. 2000;63:157–164. doi: 10.1006/geno.1999.6090. [DOI] [PubMed] [Google Scholar]
- 21.Davis J, Duncan MK, Robison WG, Jr, Piatigorsky J. Requirement for Pax6 in corneal morphogenesis: a role in adhesion. J Cell Sci. 2003;116:2157–2167. doi: 10.1242/jcs.00441. [DOI] [PubMed] [Google Scholar]
- 22.Kim K, Sirota A, Chen Yh YH, Jones SB, Dudek R, Lanford GW, Thakore C, Lu Q. Dendrite-like process formation and cytoskeletal remodeling regulated by delta-catenin expression. Exp Cell Res. 2002;275:171–184. doi: 10.1006/excr.2002.5503. [DOI] [PubMed] [Google Scholar]
- 23.Cartier L, Laforge T, Feki A, Arnaudeau S, Dubois-Dauphin M, Krause KH. Pax6-induced alteration of cell fate: shape changes, expression of neuronal alpha tubulin, postmitotic phenotype, and cell migration. J Neurobiol. 2006;66:421–436. doi: 10.1002/neu.20225. [DOI] [PubMed] [Google Scholar]
- 24.Duparc RH, Boutemmine D, Champagne MP, Tetreault N, Bernier G. Pax6 is required for delta-catenin/neurojugin expression during retinal, cerebellar and cortical development in mice. Dev Biol. 2006;300:647–655. doi: 10.1016/j.ydbio.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 25.Burger MJ, Tebay MA, Keith PA, Samaratunga HM, Clements J, Lavin MF, Gardiner RA. Expression analysis of delta-catenin and prostate-specific membrane antigen: their potential as diagnostic markers for prostate cancer. Int J Cancer. 2002;100:228–237. doi: 10.1002/ijc.10468. [DOI] [PubMed] [Google Scholar]
- 26.Kim K, Oh M, Ki H, Wang T, Bareiss S, Fini ME, Li D, Lu Q. Identification of E2F1 as a positive transcriptional regulator for delta-catenin. Biochem Biophys Res Commun. 2008;369:414–420. doi: 10.1016/j.bbrc.2008.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuschl T. RNA interference and small interfering RNAs. Chembiochem. 2001;2:239–245. doi: 10.1002/1439-7633(20010401)2:4<239::AID-CBIC239>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 28.Murakami T, Yano O, Takahashi H, Akiya S, Higashi K. [Effects of cadmium on the gene expression of retinoblastoma (Y79) cells in culture] Nippon Ganka Gakkai Zasshi. 1992;96:737–741. [PubMed] [Google Scholar]
- 29.Hayashi T, Huang J, Deeb SS. RINX(VSX1), a novel homeobox gene expressed in the inner nuclear layer of the adult retina. Genomics. 2000;67:128–139. doi: 10.1006/geno.2000.6248. [DOI] [PubMed] [Google Scholar]
- 30.Suter DM, Tirefort D, Julien S, Krause KH. A Sox1 to Pax6 switch drives neuroectoderm to radial glia progression during differentiation of mouse embryonic stem cells. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0319. [DOI] [PubMed] [Google Scholar]
- 31.Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci U S A. 1998;95:13720–13725. doi: 10.1073/pnas.95.23.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- 33.Duncan MK, Kozmik Z, Cveklova K, Piatigorsky J, Cvekl A. Overexpression of PAX6(5a) in lens fiber cells results in cataract and upregulation of (alpha)5(beta)1 integrin expression. J Cell Sci. 2000;113(Pt 18):3173–3185. doi: 10.1242/jcs.113.18.3173. [DOI] [PubMed] [Google Scholar]
- 34.Zhou YH, Wu X, Tan F, Shi YX, Glass T, Liu TJ, Wathen K, Hess KR, Gumin J, Lang F, Yung WK. PAX6 suppresses growth of human glioblastoma cells. J Neurooncol. 2005;71:223–229. doi: 10.1007/s11060-004-1720-4. [DOI] [PubMed] [Google Scholar]
- 35.Wong WW, Puthalakath H. Bcl-2 family proteins: the sentinels of the mitochondrial apoptosis pathway. IUBMB Life. 2008;60:390–397. doi: 10.1002/iub.51. [DOI] [PubMed] [Google Scholar]
- 36.Brunelle JK, Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodova M, Kelly KF, VanSaun M, Daniel JM, Werle MJ. Regulation of the rapsyn promoter by kaiso and delta-catenin. Mol Cell Biol. 2004;24:7188–7196. doi: 10.1128/MCB.24.16.7188-7196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cregan SP, MacLaurin JG, Craig CG, Robertson GS, Nicholson DW, Park DS, Slack RS. Bax-dependent caspase-3 activation is a key determinant in p53-induced apoptosis in neurons. J Neurosci. 1999;19:7860–7869. doi: 10.1523/JNEUROSCI.19-18-07860.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 40.Lang D, Mascarenhas JB, Powell SK, Halegoua J, Nelson M, Ruggeri BA. PAX6 is expressed in pancreatic adenocarcinoma and is downregulated during induction of terminal differentiation. Mol Carcinog. 2008;47:148–156. doi: 10.1002/mc.20375. [DOI] [PubMed] [Google Scholar]
- 41.Mayes DA, Hu Y, Teng Y, Siegel E, Wu X, Panda K, Tan F, Yung WK, Zhou YH. PAX6 suppresses the invasiveness of glioblastoma cells and the expression of the matrix metalloproteinase-2 gene. Cancer Res. 2006;66:9809–9817. doi: 10.1158/0008-5472.CAN-05-3877. [DOI] [PubMed] [Google Scholar]
- 42.Yamaoka T, Yano M, Yamada T, Matsushita T, Moritani M, Ii S, Yoshimoto K, Hata J, Itakura M. Diabetes and pancreatic tumours in transgenic mice expressing Pa x 6. Diabetologia. 2000;43:332–339. doi: 10.1007/s001250050051. [DOI] [PubMed] [Google Scholar]
- 43.Hill ME, Asa SL, Drucker DJ. Essential requirement for Pax6 in control of enteroendocrine proglucagon gene transcription. Mol Endocrinol. 1999;13:1474–1486. doi: 10.1210/mend.13.9.0340. [DOI] [PubMed] [Google Scholar]
- 44.Larsson LI, St-Onge L, Hougaard DM, Sosa-Pineda B, Gruss P. Pax 4 and 6 regulate gastrointestinal endocrine cell development. Mech Dev. 1998;79:153–159. doi: 10.1016/s0925-4773(98)00182-8. [DOI] [PubMed] [Google Scholar]